Abstract
Free-living protozoa are thought to be of fundamental importance in aquatic ecosystems, but there is limited understanding of their diversity and ecological role, particularly in surface-associated communities such as biofilms. Existing eukaryote-specific PCR primers were used to survey 18S rRNA gene sequence diversity in stream biofilms but poorly revealed protozoan diversity, demonstrating a need for protozoan-targeted primers. Group-specific PCR primers targeting 18S rRNA genes of the protozoan phylum Ciliophora were therefore designed and tested using DNA extracted from cultured protozoan isolates. The two most reliable primer combinations were applied to stream biofilm DNA, followed by cloning and sequencing analysis. Of 44 clones derived from primer set 384F/1147R, 86% were of probable ciliate origin, as were 25% of 44 clones detected by primer set 121F/1147R. A further 29% of 121F/1147R-detected clones matched sequences from the closely related phylum Apicomplexa. The highly ciliate-specific primer set 384F/1147R was subsequently used in PCRs on biofilm DNA from four streams exhibiting different levels of human impact, revealing differences in ciliate sequence diversity in samples from each site. Of a total of 240 clones, 73% were of probable ciliate origin; 54 different putative ciliate sequences were detected from throughout seven taxonomic ciliate classes. Sequences from Oligohymenophorea were most commonly detected in all samples, followed by either Spirotrichea or Phyllopharyngea. Restriction fragment length polymorphism profile-based analysis of clones suggested a potentially higher level of diversity than did sequencing. Nevertheless, newly designed PCR primers 384F/1147R were considered to provide an effective molecular basis for characterization of ciliate diversity in stream biofilms.
Protozoa can be defined as motile unicellular eukaryotes possessing a capacity for phagotrophy and comprise an extremely diverse assemblage of organisms commonly grouped into amoebae, ciliates, flagellates, and intracellular parasitic apicomplexans (10, 17). It is clear that protozoa, thought to occur virtually everywhere that liquid water exists, are integral components of aquatic ecosystems, regulating the abundance of bacteria and phytoplankton and population dynamics of other protozoa through predation and providing a source of nutrition for invertebrate zooplankton and probably macroinvertebrates and fish larvae (6, 24, 31, 41-43, 51). Protozoa have been identified as a major microbial pathway for the transfer of carbon and phosphorus to higher trophic levels within pelagic systems such as an oligotrophic lake (20) and, more recently, running waters such as streams (28). In general, however, an understanding of the role of protozoa in microbial food webs is limited to planktonic communities, and little is known of the diversity and ecological role of protozoa in benthic communities. Bacterivory by ciliates and flagellates has been shown to enhance the decomposition of leaf detritus in streams through a presumed increased turnover of bacterial populations (37). This suggests that protozoan predation is likely to play an important role in stream biofilms, where the majority of microbes as well as microbial energy and nutrient cycling in stream ecosystems may occur (4, 11, 39).
Detection and identification of protozoa have commonly been achieved through microscopic examination of morphological features. It remains difficult and time-consuming to reliably detect or identify many protozoan species by these methods, as protozoa may be fragile and inconspicuous and as it may be difficult to determine whether a given morphological feature can be regarded as distinct or not (7, 19, 38, 46). Molecular techniques based upon analysis of small-subunit RNA gene sequences, while not foolproof (18), offer the potential for more-accurate and -efficient methods for detecting and identifying protozoan organisms and characterizing protozoan communities, including unculturable components (3, 7). Recent studies have used eukaryote-specific PCR primers to reveal previously unsuspected protistan diversity, including stramenopiles, rhizarians, and alveolates and sequences of uncertain affinity, in deep-sea and anoxic aquatic environments (26, 30, 47-50). However, little work has been specifically directed at understanding the diversity of the heterotrophic protist (protozoan) components of ecosystems.
Combined biochemical, molecular, and morphological evidence has recently been used to resolve outstanding questions of eukaryotic taxonomy, confirming the polyphyletic evolutionary history of protozoa (1, 23). This suggests that eukaryote-specific primers are likely to be of limited utility for studying only the protozoan component of eukaryote diversity. Rather, it is necessary to target monophyletic protozoan groups, and the existence of recent molecular sequence data provides an opportunity for development of group-specific PCR primers and associated molecular biological methods for this purpose. Group-specific PCR primers have previously been designed for several monophyletic protozoan groups (3, 5, 16, 21, 25), but such tools remain undeveloped for detection of most protozoan phyla, including the alveolate phylum Ciliophora. The ciliates are considered to be a very diverse and important protozoan group. Although they have been relatively well studied, their accurate identification is nevertheless challenging because it is dependent upon considerable taxonomic expertise and often involves complex fixing and staining protocols. Many ciliates are very fast moving, making observation of live cells difficult. Fortunately, ciliates are an appropriate target for molecular diversity analysis, since there exists a reasonable body of publicly available sequence data and because, in contrast to amoebae and flagellates, ciliates do constitute a monophyletic group. The main objective of this research was therefore to develop specific PCR primers with which to characterize the diversity of ciliated protozoa within stream biofilms.
MATERIALS AND METHODS
Biofilm sample collection.
Biofilm samples were collected from four streams in Auckland, New Zealand. Three streams had stony substrates: Cascade Stream (an undeveloped native forest site; map reference NZMS 260-Q11 & Pt. R11 458778), Stoney Creek (a partially developed native forest site close to houses and roads; map reference NZMS 260-Q11 & Pt. R11 503762), and Opanuku Stream (a rural site in proximity to agricultural development; map reference NZMS 260-Q11 & Pt. R11 525770), while Pakuranga Stream (a highly developed urban site; map reference NZMS 260-Q11 802765) consists of a concrete channel at the sampling location. Biofilm material was collected from an approximately 130-cm2 area of the surface of streambed boulders, or concrete channel in the case of Pakuranga Stream, by scrubbing with Speci-sponges (Nasco Ltd.) to absorb dislodged biofilm material. The sponges were then placed into sterile Whirl-Pak bags (Nasco Ltd.) with about 50 ml stream water at ambient temperature. In the laboratory, sponges were gently squeezed to release biofilm material into petri dishes containing stream water (as appropriate from each site) for culture isolation. Alternatively, sponges were subjected to repetitive compression in a Seward stomacher laboratory blender (normal speed for 120 s) and the resulting biofilm material was transferred into 50-ml centrifuge tubes and pelleted by centrifugation (3,500 × g, 10 min) for subsequent DNA extraction.
Protozoan cultures.
Individual protozoan cells and associated bacteria were isolated from biofilm samples using a De Fonbrune pneumatic micromanipulator system, transferred to petri dishes containing modified Neff's amoeba saline (34), and incubated at ambient room temperature under natural light/dark regimens. Fresh cultures were initiated every 7 to 14 days by inoculation of between 50 and 200 μl of existing cultures (depending upon density) into 20 ml of fresh medium. Ten cultures representing a range of protozoa were thus maintained: five ciliates (tentatively identified as a Chilodonella sp., a Colpidium sp., a Cyclidium sp., a Glaucoma sp., and a Tetrahymena sp. according to a protozoan identification key (35), four flagellates (a chrysophyte Spumella sp., a cryptomonad Chroomonas sp., a euglenid Entosiphon sp., and a kinetoplastid Bodo sp.), and a lobosean amoeba (an Acanthamoeba sp.). These cultures provided material for testing the reliability of molecular methods and the specificity of PCR primers for target organisms. Identification of ciliate cultures was confirmed using PCR primers developed in this study and sequencing.
DNA extraction.
DNA was released from protozoan pure cultures by boiling 50-μl samples for 10 min. DNA was extracted from pelleted biofilm samples using a phosphate-sodium dodecyl sulfate (SDS)-chloroform bead-beating method adapted from a previous study (29), as follows. Each pelleted sample was resuspended in 270 μl of 100 mM sodium phosphate buffer (pH 8.0), to which 300 μl of SDS lysis buffer (10% SDS, 500 mM Tris [pH 8.0], 1.0 M NaCl) was added, and mixed gently. The resulting mixture was transferred to a 2-ml vial containing 0.5 g each of 0.1-mm and 3.0-mm silica-zirconium beads (Biospec Products, Inc.), to which 300 μl of chloroform was then added. The mixture was shaken in a Bio101 Savant Fastprep FP120 at 4 m s−1 for 40 s, cooled in an ice bath for 1 min, shaken a second time, and then centrifuged to pellet debris (19,400 × g, 5 min). The supernatant was added to 360 μl of 7 M ammonium acetate, briefly mixed by hand, and centrifuged (19,400 × g, 5 min), resulting in two separate phases. The clear upper phase was removed, and DNA was precipitated from this solution by addition of 315 μl of ice-cold isopropanol, followed by incubation at room temperature for 15 min. Nucleic acids were pelleted by centrifugation (19,400 × g, 5 min), washed with 1 ml of 70% ethanol, centrifuged again (19,400 × g, 5 min), air dried, and resuspended in 40 μl of sterile water. The efficacy of DNA extraction was assessed by electrophoresis on a 1% agarose gel stained with ethidium bromide.
PCR primers.
Eukaryote-specific PCR primers derived from the literature and newly designed PCR primers designed to target the 18S rRNA genes of the protozoan phylum Ciliophora were both used in this study (Table 1). To design ciliate-specific primers, representative 18S rRNA gene sequences from 61 ciliates from throughout all ciliate classes and 24 nonciliate organisms common in stream biofilms were retrieved from GenBank, aligned using Geneious 2.0.1 (Biomatters Ltd., NZ; developed by A. J. Drummond, M. Kearse, J. Heled, R. Moir, T. Thierer, B. Ashton, and A. Wilson), and scrutinized for sites conserved among ciliates but not among nonciliates. Potential primer sequences were tested for ciliate specificity by conducting nucleotide-nucleotide BLAST searches within GenBank. Primer sequences showing a high degree of specificity were synthesized by Invitrogen Ltd. (New Zealand) and tested in PCRs with DNA template material extracted from ciliate and nonciliate cultures and stream biofilm samples. The reliability of different primers was determined from comparative PCR success rates. Products of PCRs with biofilm-derived template DNA were digested with HaeIII (Invitrogen). Resulting restriction fragment length polymorphism (RFLP) profiles showing comparatively more bands were presumed to indicate the ability of newly designed primers to detect greater sequence diversity.
TABLE 1.
Primers used for PCRs in this study to detect and identify ciliated protozoa in pure cultures and stream biofilm samples
| Target and primer | Sequence (5′→3′) | Reference(s) or source |
|---|---|---|
| Eukaryotic 18S rRNA genes | ||
| 82FEa | GAA DTC GYG AAY GGC TC | 13, 26, 47 |
| EK-1520Rb | CYG CAG GTT CAC CTA C | 26, 47 |
| E528F | CGG TAA TTC CAG CTC C | 50 |
| 1391REc | GGG CGG TGT GTA CAA RGR G | 13, 50 |
| Ciliophora 18S rRNA genese | ||
| 121F | CTG CGA ATG GCT CAT TAM AA | This study |
| 384F | YTB GAT GGT AGT GTA TTG GA | This study |
| 1147R | GAC GGT ATC TRA TCG TCT TT | This study |
| 1147Fd | GAA CGA AAG WTA RGG GAT CA | This study |
| 2755Rd | CGT TSA WGA TCY ANA ATT NCA AAG | This study |
| 2824Rd | CAG GGA CKT ART CAR TGC AA | This study |
Referred to as 18S-82F, with sequence GAA ACT GCG AAY GGC TC, in reference 47 and EK-82F, with sequence GAA ACT GCG AAT GGC TC, in reference 26.
Referred to as 18S-1520R in reference 47.
Referred to as Univ1391RE in reference 50.
Designed and tested but excluded from further application in this study.
Primers were newly designed.
PCR amplification and purification.
PCR amplifications were carried out using standard 50-μl reaction mixtures (1× PCR buffer, 2 mM MgCl2, and 1 U AmpliTaq DNA polymerase [all Applied Biosystems]; 100 μM deoxynucleoside triphosphates [Invitrogen]; 0.2 μM forward and reverse primers; and 0.4% bovine serum albumin [Invitrogen]), with 2 μl of template DNA extracted from either protozoan cultures or stream biofilm samples. The eukaryote-specific primer combinations tested were 82FE/EK-1520R and E528F/1391RE, expected to give amplified product sizes of about 1,600 and 1,000 bp, respectively. Ciliate-specific primers 121F and 384F were each used with primer 1147R and expected to give product sizes of about 1,000 and 750 bp, respectively. The following temperature regimen was used: initial incubation for 5 minutes at 94°C and then 30 cycles of 45 s at 94°C (denaturing), 60 s at 55°C (annealing), and 90 s at 72°C (extension), followed by a final extension step of 7 min at 72°C. The success of PCRs was determined by the presence or absence of correctly sized products upon electrophoresis on 1% agarose gels stained with ethidium bromide. PCR products were purified using an Invitrogen Purelink PCR purification kit according to the manufacturer's instructions.
Cloning, RFLP analysis, and sequencing.
PCR products obtained from biofilm-extracted DNA using eukaryote-specific and ciliate-specific primers were used to construct clone libraries using an Invitrogen TOPO TA cloning kit according to the manufacturer's instructions. Transformed Escherichia coli colonies were picked at random into 150-μl aliquots of Luria-Bertani broth and incubated overnight at 37°C. In the case of clones derived from Opanuku Stream using eukaryote-specific primer sets and ciliate-specific primer sets 121F/1147R and 384F/1147R, 48 colonies were picked; in all other cases, 96 colonies were picked. Samples were subsequently incubated at 94°C for 20 min for use as template material in PCRs with the primers M13F and M13R from the TOPO kit to recover cloned inserts. Aliquots (10 μl) of all successfully recovered cloned sequences were each digested with 1 U of the restriction endonuclease HaeIII at 37°C for 4 h and visualized by electrophoresis on 1.5% agarose gels. Each different restriction profile was assumed to indicate a different sequence (and species). RFLP profiles from Opanuku Stream clones were used to construct species accumulation curves by plotting the number of different RFLP profiles detected per number of clone RFLP profiles analyzed. At least one clone corresponding to each different RFLP profile was purified and sequenced by Macrogen Inc. (Seoul, South Korea). Sequence chromatograms were visually inspected to assess sequence quality and checked for chimeric molecules using Chimera Check at the Ribosomal Database Project (9) and Bellerophon (22). Results of BLAST searches for matching nucleotide sequences in GenBank were used to determine probable identities of cloned sequences. Cloned sequences found to match the same GenBank sequence were compared by alignment in Geneious 2.0.1, and those sharing less than 97% nucleotide identity were considered to represent different taxonomic units.
Nucleotide sequence accession numbers.
The different sequences obtained in this study have been deposited in the GenBank database under accession numbers EF586072 to EF586193 and EU280168 to EU280178.
RESULTS
Eukaryote-specific primers.
PCRs using eukaryote-specific primer combinations 82FE/EK-1520R and E528F/1391RE with protozoan pure culture material as the template had a very low success rate. An investigation into the cause of these PCR failures was undertaken, in which template sample cell density, template DNA extraction method, template DNA concentration, and PCR conditions were varied singly and in combination. However, a reason for frequent reaction failures could not be conclusively identified. Comparison of primer sequences with primer binding site sequences for cultured organisms resulted in identification of several mismatches, but these were in positions considered unlikely to affect reaction outcome and did not correlate with the pattern of reaction failures. PCRs (and subsequent cloning and sequencing) with eukaryote-specific primers on Opanuku Stream biofilm DNA extracts were notably more successful, but very few protozoan sequences were detected. From 48 randomly selected clones derived from primer set 82FE/EK-1520R, only 22 cloned sequences were successfully recovered using PCR. Fifteen different RFLP profiles and matches to 14 different GenBank sequences were detected among these, matching a variety of eukaryote taxa, none of which were protozoan. Forty-three clones derived from primer set E528F/1391RE were recovered using PCR and found to contain 27 different RFLP profiles and 22 different sequences, 1 of which (representing a single clone) most closely matched a Cercozoa protozoan sequence (86% query coverage, 92% maximum identity). Among the clone libraries derived from both primer combinations, matches with diatom sequences (Bacillariophyta) far exceeded matches with sequences from all other taxa, followed by matches with sequences from various metazoan organisms, chlorophytes, and fungi (Table 2; for sequence match data, see Tables S1 and S2 in the supplemental material).
TABLE 2.
Summary of identity of 18S rRNA gene clones derived from biofilm samples from rural Opanuku Stream using eukaryote-specific primer setsa
| Sequence source | Primers 82FE/EK-1520R
|
Primers E528F/1391RE
|
||
|---|---|---|---|---|
| No. of clones | No. of different sequences | No. of clones | No. of different sequences | |
| Annelida | 1 | 1 | ||
| Arthropoda | 3 | 3 | ||
| Ascomycota | 1 | 1 | ||
| Bacillariophyta | 15 | 7 | 25 | 7 |
| Basidiomycota | 1 | 1 | ||
| Cercozoa | 1 | 1 | ||
| Chlorophyta | 1 | 1 | 1 | 1 |
| Chytridiomycota | 1 | 1 | ||
| Mollusca | 1 | 1 | 3 | 3 |
| Nematoda | 2 | 2 | 1 | 1 |
| Rotifera | 1 | 1 | ||
| Tardigrada | 1 | 1 | 5 | 2 |
| Uncultured fungus | 1 | 1 | ||
| Total | 22 | 14 | 43 | 22 |
Clones were identified according to RFLP profiles (data not shown) and BLAST matches.
Newly designed ciliate-specific primers.
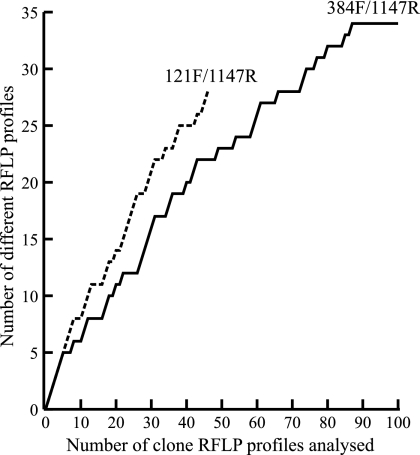
Twenty-one potential primer sequences were assessed for ciliate specificity through BLAST searches. Of these, six sequences had a high level of ciliate specificity and were synthesized as primers (Table 1). All primer combinations were used in successful PCRs with cultured ciliate material and Opanuku Stream biofilm DNA extract as the template, although primer 2755R was less reliable than the others. No primer combination resulted in a product when used in reactions with flagellate and amoeba protozoan culture material as the template, indicating that ciliate specificity was high. HaeIII digestion of aliquots of PCR products from reactions with Opanuku Stream biofilm DNA extract as the template suggested that products of primer combinations 121F/1147R and 384F/1147R contained relatively greater sequence diversity than products of other primer combinations, and these were subsequently cloned and sequenced. Species accumulation curves based on RFLP profiles derived from these clones do not approach an asymptote, indicating that the full extent of ciliate diversity associated with Opanuku Stream biofilm is not reflected in the clones analyzed (Fig. 1).
FIG. 1.
Species accumulation curves for 18S rRNA gene sequences derived from Opanuku Stream biofilm using ciliate-targeted primer combinations 121F/1147R and 384F/1147R, based on the number of different RFLP profiles detected per number of clones analyzed.
Sequencing of clones derived from newly designed primers showed that ciliate specificity varied between different primer sets. In each case 44 of 48 clones picked for analysis were successfully recovered by PCR. Twenty-five percent of 44 clones derived from primers 121F/1147R most closely matched sequences from the phylum Ciliophora, a further 29% of clones matched sequences from the protozoan phylum Apicomplexa, and the remaining clones matched various other eukaryote sequences (see Table S3 in the supplemental material). In contrast, primer combination 384F/1147R was found to be much more ciliate specific: 86% of 44 clones matched Ciliophora sequences, with the remaining clones matching sequences from the protozoan phylum Cercozoa, fungi, and a chlorophyte (see Table S6 in the supplemental material). Primer combination 384F/1147R was therefore applied to biofilm samples from further streams, resulting in construction of DNA clone libraries from a total of four different streams.
Analysis of the clone libraries revealed differences between biofilm samples from the four streams. Sixty-nine of 96 Cascade Stream clones selected for analysis were successfully recovered with PCR, among which 43 different RFLP profiles were detected. Fifty-one different RFLP profiles were detected among 88 clones recovered from Stoney Creek and 31 RFLP profiles among 77 clones from Pakuranga Stream, in addition to 27 different RFLP profiles among 44 clones from Opanuku Stream. Sequences corresponding to 13 Cascade Stream RFLP profiles (17 clones), 11 Stoney Creek RFLP profiles (18 clones), 3 Pakuranga Stream RFLP profiles (3 clones), and a single Opanuku Stream clone were of poor quality and excluded from further analysis, as was a single chimeric Opanuku Stream sequence. In all clone libraries, the number of RFLP profiles detected exceeded the number of different BLAST sequence matches.
Sequence matches, combined with RFLP profiles, resulted in identification of a total of 240 clones among the four stream samples. Of these, 176 (73%) were determined to be of probable ciliate origin. Within these putative ciliate clones, 54 different taxonomic units, representing seven ciliate classes, were identified (Table 3; for sequence match data, see Tables S4 to S7 in the supplemental material). Of the 54, only 1 was found in samples from three streams and 3 in samples from two streams. The remaining 50 were each detected in samples from only one of the four streams. Clones most frequently matched oligohymenophorean sequences, followed by phyllopharyngean, spirotrichean, colpodean, and litostomatean sequences. Least frequently matched were prostomatean and nassophorean sequences. The mean coverage of GenBank matches with ciliate sequences was 96%, while the mean maximum identity of matches with ciliate sequences was 93%.
TABLE 3.
Summary of identity of 18S rRNA gene clones derived from biofilm DNA extracts from four streams using ciliate-specific primer combination 384F/1147Ra
| Sequence source (class or phylumb) | Cascade Stream (undeveloped native forest)
|
Stoney Creek (developed native forest)
|
Opanuku Stream (rural)
|
Pakuranga Stream (urban)
|
Total
|
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| No. of clones | No. of sequences | No. of clones | No. of sequences | No. of clones | No. of sequences | No. of clones | No. of sequences | No. of clones | No. of sequences | |
| Ciliates | ||||||||||
| Colpodea | 2 | 1 | 10 | 3 | 12 | 4 | ||||
| Litostomatea | 2 | 1 | 1 | 1 | 4 | 4 | 7 | 6 | ||
| Nassophorea | 4 | 2 | 4 | 2 | ||||||
| Oligohymenophorea | 20 | 6 | 14 | 5 | 26 | 8 | 41 | 6 | 101 | 23 |
| Phyllopharyngea | 14 | 4 | 10 | 2 | 24 | 6 | ||||
| Prostomatea | 2 | 1 | 2 | 1 | 1 | 1 | 5 | 3 | ||
| Spirotrichea | 5 | 2 | 4 | 2 | 11 | 6 | 2 | 2 | 22 | 9 |
| Uncultured ciliate | 1 | 1 | 1 | 1 | ||||||
| Total ciliates | 31 (60) | 11 (73) | 49 (70) | 18 (90) | 38 (86) | 15 (79) | 58 (78) | 15 (83) | 176 (73) | 54 (87) |
| Nonciliates | ||||||||||
| Amoebozoa | 9 | 1 | 2 | 1 | 1 | 1 | 12 | 1 | ||
| Apicomplexa | 9 | 1 | 9 | 1 | ||||||
| Basidiomycota | 14 | 1 | 14 | 1 | ||||||
| Cercozoa | 2 | 1 | 2 | 1 | ||||||
| Chlorophyta | 1 | 1 | 1 | 1 | ||||||
| Chytridiomycota | 3 | 2 | 19 | 1 | 3 | 2 | 1 | 1 | 26 | 3 |
| Total nonciliates | 21 | 4 | 21 | 2 | 6 | 4 | 16 | 3 | 64 | 8 |
Clones were identified according to RFLP profiles (data not shown) and BLAST matches. Values in parentheses are percentages of ciliates.
Class for ciliates and phylum for nonciliates.
The diversity of sequences detected varied between the samples from different streams (Table 3). The proportion of clones matching ciliate sequences ranged from 60% for Cascade Stream biofilm to 86% for Opanuku Stream biofilm. The number of different putative ciliate taxonomic units detected ranged from 11 in Cascade Stream biofilm to 18 in Stoney Creek biofilm. Clones matching oligohymenophorean and spirotrichean sequences were detected in biofilm samples from all four streams. Clones matching phyllopharyngean, colpodean, litostomatean, and prostomatean sequences were detected in biofilm samples from two or three streams, but clones matching nassophorean sequences were detected only in Stoney Creek biofilm. Clones matching sequences from seven ciliate classes were detected in Stoney Creek biofilm, whereas clones matching sequences from only two ciliate classes, Oligohymenophorea and Spirotrichea, were detected in Opanuku Stream biofilm.
In terms of species, a Zoothamnium sp. (Oligohymenophorea) sequence was the most common ciliate sequence to match Cascade Stream clones (see Table S4 in the supplemental material) and was also a common match to Stoney Creek clones (see Table S5 in the supplemental material), while two Opanuku Stream clones matched a different Zoothamnium sequence (see Table S6 in the supplemental material). The other most common Stoney Creek ciliate sequence matches (Platyophrya sp., Colpodea, and Dysteria spp., Phyllopharyngea) were not detected in samples from the other streams, nor were the most common ciliate sequence matches in samples from Opanuku Stream (a Vorticella sp., Oligohymenophorea) or Pakuranga Stream (a Mesanophrys sp. and an Entorhipidium sp., both Oligohymenophorea; see Table S7 in the supplemental material). Interestingly, clones matching sequences from sessile peritrichs (Epistylis spp., a Vorticella sp., and Zoothamnium spp.) were common in biofilm samples from all three stony-bottom streams but were not detected in channelized Pakuranga Stream biofilm.
A small number of nonciliate sequence matches were detected (Table 3). Nine Cascade Stream clones matched a single apicomplexan sequence (a Theileria sp., see Table S4 in the supplemental material), and a further nine Cascade Stream clones matched an amoebozoan sequence (a Stemonitis sp.), which was also matched at lesser frequency by clones from Stoney Creek and Pakuranga Stream biofilm (see Tables S5 and S7, respectively, in the supplemental material). Clones matching sequences from the chytrid genus Monoblepharis were present in samples from all four streams but were by far most common in Stoney Creek biofilm (19 clones). In Pakuranga Stream biofilm 14 clones matched a basidiomycete sequence (a Schizopora sp.). In addition, two clones matching a cercozoan sequence and one clone matching a chlorophyte sequence were detected in Opanuku Stream biofilm (see Table S6 in the supplemental material). All GenBank matches with amoebozoan, apicomplexan, and chytrid sequences (47 clones) were fragmented and of relatively poor quality, with a mean match coverage of 87% and mean maximum identity of 86%. Among all four streams, only eight different nonciliate taxonomic units were detected, but these accounted for a total of 64 clones out of 240.
DISCUSSION
Eukaryote-specific primers.
The eukaryote-specific PCR primers 82FE, EK-1520R, E528F, and 1391RE have all amplified protozoan sequences from environmental samples in prior studies (13, 26, 47, 50). In the present study, these primers proved largely ineffective at amplifying 18S rRNA gene sequences from cultured protozoan DNA. When applied to environmental extracts, these primers amplified sequences which matched those from a variety of eukaryotes, as expected, but the number of sequences matching those from protozoa among cloned PCR products was low (0 of 22 clones and 1 of 43 clones from 82FE/EK-1520R and E528F/1391RE, respectively). In contrast, high proportions of diatom and metazoan sequences were detected, along with several sequences of presumed fungal and chlorophyte origin, reflecting the broad specificity of these primers. These results suggest a restricted ability on the part of these primers to detect protozoa and indicate that eukaryote-specific primers are of limited value for this application in complex stream communities due to the presence of large quantities of competing sequences from algal, fungal, plant, and metazoan organisms. This is in contrast to the suboxic (47), anoxic (13, 50), and aphotic (26) environments to which these primers have previously been applied, in which it is presumed that the relative abundances of protozoan and nonprotozoan organisms may be rather different. Indeed, a comparative survey of freshwater ponds using eukaryote-specific PCR primers has shown protozoan sequences to be much more frequent in a suboxic habitat than in an oxygenated habitat (47). Therefore, PCR primers specific to monophyletic groups of interest are required if the protozoan subset of microeukaryotic diversity in stream biofilms is to be analyzed.
Design and application of new primers to detection of ciliate diversity.
Ciliate-specific PCR primers designed in this study revealed very different DNA sequences and were more selective than eukaryote-specific PCR primers, as sequences from most nontarget organisms, particularly diatoms and metazoans, were not detected. Of primer combinations 121F/1147R and 384F/1147R, the latter combination detected the most ciliate-matching sequences and was therefore considered the most ciliate specific: at least 54 different ciliate sequences were detected, representing 7 of 11 known ciliate classes as defined by Lynn (27): Colpodea, Litostomatea, Nassophorea, Oligohymenophorea, Phyllopharyngea, Prostomatea, and Spirotrichea. Although the detected sequences are not entirely reflective of the full range of known ciliate diversity, several factors suggest that these primers should be capable of detecting a representative range of sequences from most or all ciliate classes. First, the primer design process was based on DNA fragments conserved throughout an alignment of 18S rRNA sequences from all 11 ciliate classes. Occasional unconserved nucleotides were present at primer binding sites, but these weren't found in particular classes. For example, if it is assumed that the three terminal 3′ positions are most important for correct priming (14) and that mismatched nucleotides in any of these positions will hence cause failure of the primer to bind to its intended site, then primer 384F may not produce the expected PCR product from 2 of 18 oligohymenophorean sequences and 1 of 5 litostomatean sequences investigated (of course, these mismatches may be due to sequencing errors in the deposited sequences). Since the aligned sequences reflect evolutionary relationships, it is likely that the distribution of unconserved nucleotides in the aligned sequences is representative of that of other ciliate sequences and that the primers will find appropriate annealing sites in all classes as frequently as in the aligned sequences. In any case, BLAST nucleotide searches for matches to primer sequences indicated that primers 384F and 1147R should bind to sequences from the ciliate classes that are unrepresented in these results. In addition, the type of ciliate sequences detected during cloning is broadly similar to those observed during visual inspection of samples from the same environment. For example, microscopic inspection of samples from Opanuku Stream resulted in visual identification of several oligohymenophorean and spirotrichean ciliates and occasional phyllopharyngean, prostomatean, and nassophorean ciliates, an assemblage of organisms similar to that detected by molecular analysis. Furthermore, Oligohymenophorea, Spirotrichea, Phyllopharyngea, Litostomatea, and Colpodea are the largest ciliate classes in terms of numbers of described species, considered as having some 600, 400, 340, 300, and 100 known species, respectively, compared with a range of between 70 and 8 species in Heterotrichea, Nassophorea, Prostomatea, Armophorea, Karyorelictea and Plagiopylea, in order of decreasing numbers of species, according to D. H. Lynn (Classification of the phylum Ciliophora [http://www.uoguelph.ca/∼ciliates/classification/genera.html]). The number of sequences from different classes detected in this study shows a similar distribution: oligohymenophorean sequences were most common, followed by phyllopharyngean and spirotrichean sequences, while only two nassophorean sequences and a single prostomatean sequence were detected. Figure 1 suggests that the clones analyzed in this study represent only a fraction of the total ciliate diversity present in biofilm samples, and it seems likely that rare or nonabundant ciliate species from the smaller, unrepresented classes may have been overlooked for this reason.
Sixty-four of 240 clones derived from primers 384F/1147R matched nonciliate sequences, showing that the specificity of these primers is not perfect. The range of nonciliate sequence matches was very limited, however, and most sequences were either fragmented or of poor quality, suggesting that some ambiguity surrounds these sequence matches and that interpretation of these nonciliate sequence matches remains limited.
Representativeness of RFLP and sequence diversity.
RFLP-based surveys of molecular diversity have been found to underestimate total sequence diversity, due to conservation of restriction sites among different species (15). In this study, RFLP diversity of clones derived from ciliate-specific primers suggests a considerably higher level of diversity than sequencing. The availability of ciliate sequence data is presently limited; for example, only about one-sixth of the total recognized Phyllopharyngea ciliate species are represented by 18S sequence data in GenBank. This suggests that the ability to identify ciliates on the basis of matches to database sequence information is limited. Sequence matches in this study were frequently imperfect, and in a number of cases, two or more cloned sequences sharing pairwise similarity of less than 97% were found to match the same GenBank sequence, presumably due to the unavailability of more-representative sequences. Use of a similarity threshold that is higher than 97% when assessing different taxonomic units would, it is presumed, result in the determination of a number of different sequences closer to the number of different RFLP profiles detected. Given the difficulties inherent in morphological characterization of protozoan species, another possibility may be raised, namely, that ciliate species which have been differentiated on the basis of morphology may not actually be genetically distinct, although this would not explain the higher level of diversity suggested by RFLP profiling of clones. Further studies combining traditional taxonomic methods alongside molecular investigations of diversity using primers such as those developed in this study are necessary to investigate these points.
Species accumulation curves derived from Opanuku Stream RFLP profiles point to the existence of a high level of RFLP diversity associated with stream biofilms (Fig. 1). The curve associated with primers 121F/1147R is steeper than that associated with 384F/1147R, consistent with the lesser specificity of 121F/1147R demonstrated by cloning/sequencing results (see Tables S3 and S6 in the supplemental material). Neither curve appears to approach an asymptote, indicating that further RFLP profiles await discovery, even after analysis of 100 clones (384F/1147R), and that insufficient clones were analyzed in this study to reveal the full extent of diversity present in stream biofilms. These curves are similar to those based upon sequence data derived from other 18S rRNA-based surveys of environmental cercozoan and protistan diversity (3, 12) and suggest the existence of diverse communities of a subset of eukaryotes (121F/1147R) and ciliates (384F/1147R) in Opanuku Stream.
These results are subject to caveats regarding variability in ribosomal gene copy number among different eukaryotes, which may confound conclusions about the relative abundance of different organisms based upon sequences detected (36). Additionally, it is likely that encysted protozoa in a particular environment will be underrepresented to some extent, depending upon the DNA extraction method applied (17). It has been shown that use of multiple different “universal” primers results in detection of a higher proportion of different sequences (50), and it is presumed that this may also be true, to some extent, of primers targeted at specific phyla. This implies that the diversity detected in this study may be only a subset of the total and that application of multiple ciliate-specific primers, in addition to greater cloning and sequencing efforts, may increase the range of ciliate sequence diversity detected.
Primer coverage of protozoan diversity.
The ciliates comprise only one of several major protozoan groups, and molecular assessment of ciliate diversity does not therefore constitute a complete evaluation of protozoan diversity. Group-specific primers for the protozoan phylum Cercozoa (3), the Foraminifera (21), the Dinophyceae (25), and the Chrysophyceae (5) have also been published. It is presently unclear whether the excavates—a loose grouping of assorted flagellates—form a monophyletic group (8, 44, 45), and development of PCR primers targeting all excavates may therefore not be feasible, instead requiring individual targeting of excavate subgroups. Amoebozoa spp. are a well-supported monophyletic protozoan group (2, 33) and an appropriate target for group-specific primer design, as are Choanozoa and Cryptophyceae (unrelated flagellate groups) (23), Acantharea and Polycystinea (formerly included in Radiolaria) (32, 52), Actinophryida and Centrohelida (formerly included in Heliozoa) (32, 40), and Apicomplexa, sister taxa to the Ciliophora and Dinophyceae. BLAST searches for sequence matches to primer 121F suggested that it would detect sequences from all three alveolate groups (ciliates, apicomplexans, and dinoflagellates). In this study several apicomplexan sequences were detected by primer combination 121F/1147R, suggesting that primer 121F or a modification thereof, in combination with a suitable reverse primer, may provide a basis for development of Apicomplexa- or Alveolata-specific primers.
This is the first study of Ciliophora 18S rRNA gene sequence diversity in environmental samples and the first molecular investigation of microeukaryotes in stream biofilms. This study has demonstrated that molecular methods can be used to characterize microeukaryote diversity in stream environments, and more specifically ciliate diversity in stream biofilms, through design and application of primers targeted to this monophyletic protozoan taxon. This is possible due to recent advances in understanding protozoan phylogeny and the associated availability of sequence data. Comparing results of molecular analyses using eukaryote-specific primers and ciliate-specific primers shows that phylum-specific PCR primers can provide more-detailed data regarding specific taxa of interest. Primers 384F and 1147R were effective for the detection of a broad range of ciliate diversity and revealed the existence of diverse microeukaryote communities associated with stream biofilms. Group-specific primers remain to be developed for many other protozoan groups. This study has defined a feasible route to achieving a comprehensive measure of molecular protozoan diversity, and the process which led to the method applied in this study can be applied in the same way to other protozoan groups. Application of the methodology developed in this study has the potential to contribute to improved understanding of the role of protozoa in aquatic ecosystems.
Supplementary Material
Acknowledgments
Funding for this research was provided by New Zealand Foundation for Research, Science and Technology Public Good Science Fund UOA306 and a University of Auckland Masters Scholarship.
Footnotes
Published ahead of print on 25 January 2008.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Adl, S. M., A. G. B. Simpson, M. A. Farmer, R. A. Andersen, O. R. Anderson, J. R. Barta, S. S. Bowser, G. Brugerolle, R. A. Fensome, S. Fredericq, T. Y. James, S. Karpov, P. Kugrens, J. Krug, C. E. Lane, L. A. Lewis, J. Lodge, D. H. Lynn, D. G. Mann, R. M. McCourt, L. Mendoza, Ø. Moestrup, S. E. Mozley-Standridge, T. A. Nerad, C. A. Shearer, A. V. Smirnov, F. W. Spiegel, and M. F. J. R. Taylor. 2005. The new higher level classification of eukaryotes with emphasis on the taxonomy of protists. J. Eukaryot. Microbiol. 52:399-451. [DOI] [PubMed] [Google Scholar]
- 2.Bapteste, E., H. Brinkmann, J. A. Lee, D. V. Moore, C. W. Sensen, P. Gordon, L. Durufle, T. Gaasterland, P. Lopez, M. Muller, and H. Philippe. 2002. The analysis of 100 genes supports the grouping of three highly divergent amoebae: Dictyostelium, Entamoeba, and Mastigamoeba. Proc. Natl. Acad. Sci. USA 99:1414-1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bass, D., and T. Cavalier-Smith. 2004. Phylum-specific environmental DNA analysis reveals remarkably high global biodiversity of Cercozoa (Protozoa). Int. J. Syst. Evol. Microbiol. 54:2393-2404. [DOI] [PubMed] [Google Scholar]
- 4.Battin, T. J., L. A. Kaplan, J. D. Newbold, and C. M. E. Hansen. 2003. Contributions of microbial biofilms to ecosystem processes in stream mesocosms. Nature 426:439-442. [DOI] [PubMed] [Google Scholar]
- 5.Berglund, J., K. Jurgens, I. Bruchmuller, M. Wedin, and A. Andersson. 2005. Use of group-specific PCR primers for identification of chrysophytes by denaturing gradient gel electrophoresis. Aquat. Microb. Ecol. 39:171-182. [Google Scholar]
- 6.Berninger, U. G., B. J. Finlay, and P. Kuuppo-Leinikki. 1991. Protozoan control of bacterial abundances in freshwater. Limnol. Oceanogr. 36:139-147. [Google Scholar]
- 7.Caron, D. A., P. D. Countway, and M. V. Brown. 2004. The growing contribution of molecular biology and immunology to protistan ecology: molecular signatures as ecological tools. J. Eukaryot. Microbiol. 51:38-48. [DOI] [PubMed] [Google Scholar]
- 8.Cavalier-Smith, T. 2002. The phagotrophic origin of eukaryotes and phylogenetic classification on protozoa. Int. J. Syst. Evol. Microbiol. 52:297-354. [DOI] [PubMed] [Google Scholar]
- 9.Cole, J. R., B. Chai, R. J. Farris, Q. Wang, A. S. Kulam-Syed-Mohideen, D. M. McGarrell, A. M. Bandela, E. Cardenas, G. M. Garrity, and J. M. Tiedje. 2007. The Ribosomal Database Project (RDP-II): introducing myRDP space and quality controlled public data. Nucleic Acids Res. 35:D169-D172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Corliss, J. O. 2002. Biodiversity and biocomplexity of the protists and an overview of their roles in the maintenance of our biosphere. Acta Protozool. 41:199-219. [Google Scholar]
- 11.Costerton, J. W., Z. Lewandowski, D. E. Caldwell, D. R. Korber, and H. M. Lappin-Scott. 1995. Microbial biofilms. Annu. Rev. Microbiol. 49:711-745. [DOI] [PubMed] [Google Scholar]
- 12.Countway, P. D., R. J. Gast, P. Savai, and D. A. Caron. 2005. Protistan diversity estimates based on 18S rDNA from seawater incubations in the western North Atlantic. J. Eukaryot. Microbiol. 52:95-106. [DOI] [PubMed] [Google Scholar]
- 13.Dawson, S. C., and N. R. Pace. 2002. Novel kingdom-level eukaryotic diversity in anoxic environments. Proc. Natl. Acad. Sci. USA 99:8324-8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dieffenbach, C. W., T. M. J. Lowe, and G. S. Dveksler. 1995. General concepts for PCR primer design, p. 133-142. In C. W. Dieffenbach and G. S. Dveksler (ed.), PCR primer: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 15.Diez, B., C. Pedros-Alio, and R. Massana. 2001. Study of genetic diversity of eukaryotic picoplankton in different oceanic regions by small-subunit rRNA gene cloning and sequencing. Appl. Environ. Microbiol. 67:2932-2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Doherty, M., B. A. Costas, G. B. McManus, and L. A. Katz. 2007. Culture-independent assessment of planktonic ciliate diversity in coastal northwest Atlantic waters. Aquat. Microb. Ecol. 48:141-154. [Google Scholar]
- 17.Finlay, B. J., and G. F. Esteban. 1998. Freshwater protozoa: biodiversity and ecological function. Biodivers. and Conserv. 7:1163-1186. [Google Scholar]
- 18.Forney, L. J., X. Zhou, and C. J. Brown. 2004. Molecular microbial ecology: land of the one-eyed king. Curr. Opin. Microbiol. 7:210-220. [DOI] [PubMed] [Google Scholar]
- 19.Fried, J., R. Psenner, W. Ludwig, and K. H. Schleifer. 2002. Improvement of ciliate identification and quantification: a new protocol for fluorescence in situ hybridization (FISH) in combination with silver stain techniques. Syst. Appl. Microbiol. 25:555-571. [DOI] [PubMed] [Google Scholar]
- 20.Heath, R. T., S. J. Hwang, and M. Munawar. 2003. A hypothesis for the assessment of the importance of microbial food web linkages in nearshore and offshore habitats of the Laurentian Great Lakes. Aquat. Ecosyst. Health Management 6:231-239. [Google Scholar]
- 21.Holzmann, M., J. Pawlowski, A. Habura, H. Giles, and S. S. Bowser. 2003. Freshwater foraminiferans revealed by analysis of environmental DNA samples. J. Eukaryot. Microbiol. 50:135-139. [DOI] [PubMed] [Google Scholar]
- 22.Huber, T., G. Faulkner, and P. Hugenholtz. 2004. Bellerophon: a program to detect chimeric sequences in multiple sequence alignments. Bioinformatics 20:2317-2319. [DOI] [PubMed] [Google Scholar]
- 23.Keeling, P. J., G. Burger, D. G. Durnford, B. F. Lang, R. W. Lee, R. E. Pearlman, A. J. Roger, and M. W. Gray. 2005. The tree of eukaryotes. Trends Ecol. Evol. 20:670-676. [DOI] [PubMed] [Google Scholar]
- 24.Kneitel, J. M., and J. M. Chase. 2004. Disturbance, predator, and resource interactions alter container community composition. Ecology 85:2088-2093. [Google Scholar]
- 25.Lin, S., H. Zhang, Y. Hou, L. Miranda, and D. Bhattacharya. 2006. Development of a dinoflagellate-oriented PCR primer set leads to detection of picoplanktonic dinoflagellates from Long Island Sound. Appl. Environ. Microbiol. 72:5626-5630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lopez-Garcia, P., F. Rodriguez-Valera, C. Pedros-Alio, and D. Moreira. 2001. Unexpected diversity of small eukaryotes in deep-sea Antarctic plankton. Nature 409:603-607. [DOI] [PubMed] [Google Scholar]
- 27.Lynn, D. H. 2003. Morphology or molecules: how do we identify the major lineages of ciliates (Phylum Ciliophora)? Eur. J. Protistol. 39:356-364. [Google Scholar]
- 28.Marxsen, J. 2006. Bacterial production in the carbon flow of a central European stream, the Breitenbach. Freshw. Biol. 51:1838-1861. [Google Scholar]
- 29.Miller, D. N., J. E. Bryant, E. L. Madsen, and W. C. Ghiorse. 1999. Evaluation and optimization of DNA extraction and purification procedures for soil and sediment samples. Appl. Environ. Microbiol. 65:4715-4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moon-van der Staay, S. Y., R. De Wachter, and D. Vaulot. 2001. Oceanic 18S rDNA sequences from picoplankton reveal unsuspected eukaryotic diversity. Nature 409:607-610. [DOI] [PubMed] [Google Scholar]
- 31.Muylaert, K., R. Van Mieghem, K. Sabbe, M. Tackx, and W. Vyverman. 2000. Dynamics and trophic roles of heterotrophic protists in the plankton of a freshwater tidal estuary. Hydrobiologia 432:25-36. [Google Scholar]
- 32.Nikolaev, S. I., C. Berney, J. F. Fahrni, I. Bolivar, S. Polet, A. P. Mylnikov, V. V. Aleshin, N. B. Petrov, and J. Pawlowski. 2004. The twilight of Heliozoa and rise of Rhizaria, an emerging supergroup of amoeboid eukaryotes. Proc. Natl. Acad. Sci. USA 101:8066-8071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nikolaev, S. I., C. Berney, N. B. Petrov, A. P. Mylnikov, J. F. Fahrni, and J. Pawlowski. 2006. Phylogenetic position of Multicilia marina and the evolution of Amoebozoa. Int. J. Syst. Evol. Microbiol. 56:1449-1458. [DOI] [PubMed] [Google Scholar]
- 34.Page, F. C. 1988. A new key to freshwater and soil Gymnamoebae. Freshwater Biological Association, Ambleside, England.
- 35.Patterson, D. J. 1996. Free-living freshwater Protozoa: a colour guide. University of New South Wales Press, Sydney, Australia.
- 36.Prokopowich, C. D., T. R. Gregory, and T. J. Crease. 2003. The correlation between rDNA copy number and genome size in eukaryotes. Genome 46:48-50. [DOI] [PubMed] [Google Scholar]
- 37.Ribblett, S. G., M. A. Palmer, and D. W. Coats. 2005. The importance of bacterivorous protists in the decomposition of stream leaf litter. Freshw. Biol. 50:516-526. [Google Scholar]
- 38.Rogerson, A., and C. Gwaltney. 2000. High numbers of naked amoebae in the planktonic waters of a mangrove stand in southern Florida, USA. J. Eukaryot. Microbiol. 47:235-241. [DOI] [PubMed] [Google Scholar]
- 39.Romani, A. M., H. Guasch, I. Munoz, J. Ruana, E. Vilalta, T. Schwartz, F. Emtiazi, and S. Sabater. 2004. Biofilm structure and function and possible implications for riverine DOC dynamics. Microb. Ecol. 47:316-328. [DOI] [PubMed] [Google Scholar]
- 40.Sakaguchi, M., T. Nakayama, T. Hashimoto, and I. Inouye. 2005. Phylogeny of the Centrohelida inferred from SSU rRNA, tubulins, and actin genes. J. Mol. Evol. 61:765-775. [DOI] [PubMed] [Google Scholar]
- 41.Sanders, R. W., D. A. Caron, and U. G. Berninger. 1992. Relationships between bacteria and heterotrophic nanoplankton in marine and fresh waters: an inter-ecosystem comparison. Mar. Ecol. Progr. Ser. 86:1-14. [Google Scholar]
- 42.Sherr, E. B., and B. F. Sherr. 1994. Bacterivory and herbivory: key roles of phagotrophic protists in pelagic food webs. Microb. Ecol. 28:223-235. [DOI] [PubMed] [Google Scholar]
- 43.Sherr, E. B., and B. F. Sherr. 2002. Significance of predation by protists in aquatic microbial food webs. Antonie van Leeuwenhoek 81:293-308. [DOI] [PubMed] [Google Scholar]
- 44.Simpson, A. G. B. 2003. Cytoskeletal organization, phylogenetic affinities and systematics in the contentious taxon Excavata (Eukaryota). Int. J. Syst. Evol. Microbiol. 53:1759-1777. [DOI] [PubMed] [Google Scholar]
- 45.Simpson, A. G. B., Y. Inagaki, and A. J. Roger. 2006. Comprehensive multigene phylogenies of excavate protists reveal the evolutionary positions of “primitive” eukaryotes. Mol. Biol. Evol. 23:615-625. [DOI] [PubMed] [Google Scholar]
- 46.Sims, G. P., R. Aitken, and A. Rogerson. 2002. Identification and phylogenetic analysis of morphologically similar naked amoebae using small subunit ribosomal RNA. J. Eukaryot. Microbiol. 49:478-484. [DOI] [PubMed] [Google Scholar]
- 47.Slapeta, J., D. Moreira, and P. Lopez-Garcia. 2005. The extent of protist diversity: insights from molecular ecology of freshwater eukaryotes. Proc. R. Soc. Biol. Sci. Ser. B 272:2073-2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stoeck, T., and S. Epstein. 2003. Novel eukaryotic lineages inferred from small-subunit rRNA analyses of oxygen-depleted marine environments. Appl. Environ. Microbiol. 69:2657-2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stoeck, T., S. S. Epstein, and G. T. Taylor. 2003. Novel eukaryotes from the permanently anoxic Cariaco Basin (Caribbean Sea). Appl. Environ. Microbiol. 69:5656-5663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stoeck, T., B. Hayward, S. S. Epstein, G. T. Taylor, and R. Varela. 2006. A multiple PCR-primer approach to access the microeukaryotic diversity in environmental samples. Protist 157:31-43. [DOI] [PubMed] [Google Scholar]
- 51.Stoecker, D. K., and J. McDowell Capuzzo. 1990. Predation on protozoa: its importance to zooplankton. J. Plankton Res. 12:891-908. [Google Scholar]
- 52.Yuasa, T., O. Takahashi, J. K. Dolven, S. Mayama, A. Matsuoka, D. Honda, and K. R. Bjørklund. 2006. Phylogenetic position of the small solitary phaeodarians (Radiolaria) based on 18S rDNA sequences by single cell PCR analysis. Mar. Micropaleontol. 59:104-114. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.