Abstract
In anaerobic coastal sediments, hydrolytic and/or fermentative bacteria degrade polymeric material and produce labile intermediates, which are used by terminal metabolizers to complete the conversion of organic material to CO2. We used molecular approaches to evaluate the response of two bacterial terminal metabolizer groups from a coastal tidal creek sediments, sulfate reducers and methanogens, to controlled changes in carbon resource supply. Tidal creek sediment bioreactors were established in April and August 2004. For each date, intact sediment sections were continuously supplied with flowthrough seawater that was either unamended or amended with the high-molecular-weight polysaccharide dextran. Biogeochemical data indicate that the activity of fermenting bacteria and the terminal metabolizers was limited by organic carbon supply during both experiments, with a significant increase in net volatile fatty acid (VFA) production and rates of sulfate reduction and methanogenesis following dextran addition. Community composition (measured by using terminal restriction fragment length polymorphism analysis, and functional gene [dsrA, mcrA] clone libraries) changed from April to August. However, community composition was not different between amended and unamended cores within each month, despite the change in resource level. Moreover, there was no relationship between community richness and evenness with resource level. This lack of variation in community composition with C addition could be attributed to the dynamic environment these sediment communities experience in situ. Fluctuations in VFA concentrations are most likely very high, so that the dominant bacterial species must be able to outcompete other species at both high and low resource levels.
Coastal sediments are an important repository for terrestrially derived organic matter and may moderate carbon flux from continents to the ocean and atmosphere (9, 27, 37, 41). High rates of prokaryotic activity in coastal sediments rapidly consume oxygen, making anaerobic pathways of carbon consumption dominant (1, 25, 32, 34). The complex pathways for anaerobic carbon processing in marine sediments are best viewed as a network of interactive prokaryotic functional groups. Hydrolytic/fermentative bacteria degrade particulate-organic-matter and high-molecular-weight (HMW) polymeric material into (in part) labile, low-molecular-weight (LMW) dissolved organic intermediates such as acetate and other volatile fatty acids (VFAs). Anaerobic terminal metabolizers (sulfate reducers, denitrifiers, and methanogens) subsequently mineralize these intermediates, completing the conversion of organic material to CO2 and CH4 and releasing inorganic nutrients back into the ecosystem (3, 4, 42).
Recent advances in nucleic acid sequencing and the widespread availability of molecular techniques (12, 19, 23, 40) provides a means to dissect the component prokaryotes and gene inventories represented within a given functional group. Information on the diversity of taxa that carry out a key anaerobic metabolism, as well as the types and abundance of genes mediating that function, are now accessible by PCR amplification and sequencing of informative genes. For the first time, the question of how concepts such as species composition, species diversity, and interspecific competition relate to bulk measurements of a key prokaryotic function can be addressed.
This study focused on using molecular approaches to evaluate the response of two prokaryotic terminal metabolizer groups from coastal tidal creek sediments, i.e., sulfate reducers and methanogens, to controlled changes in the carbon resource supply. A previous study of this coastal system had shown that the activities of hydrolytic and/or fermentative (hydrolytic/fermentative) bacteria and terminal metabolizers are not always tightly coupled (52). At lower temperatures (<25°C), hydrolytic/fermentative bacteria degraded HMW organic carbon faster than terminal metabolizers could consume it, resulting in the accumulation of VFA intermediates. At higher temperatures (>25°C), the activity of the terminal metabolizers rapidly consumed all available labile intermediates, reducing the concentration of labile dissolved organic carbon (DOC) (e.g., VFAs) to very low concentrations. That previous study provided an excellent model system for investigating the response of sediment functional groups, i.e., the sulfate reducer and methanogen terminal metabolizers, to changes in resource availability under controlled conditions in which both resource levels and prokaryotic activity were thoroughly quantified. The reported differences in terminal metabolizer activity during defined manipulations of resource availability could be examined in concert with inventories of taxonomic and functional genes.
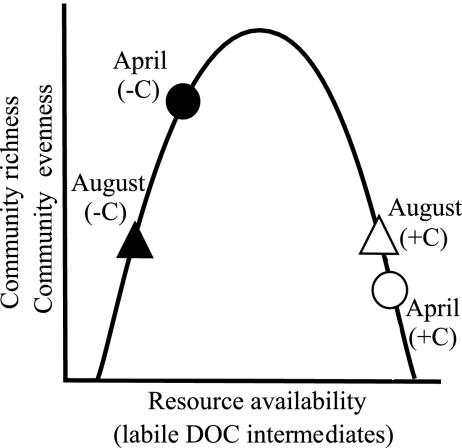
Changes in prokaryotic community composition along both experimental and naturally occurring gradients in resource supply (C, N, and P) have been studied previously, although the results are not always consistent (10, 11, 30, 44). The response of seawater prokaryotic communities to nutrient enrichment (N and P), for example, varied from year to year (24), while lake mesocosm communities showed no response to varying nutrient (N and P) levels (26). Community composition includes characteristics such as species richness and evenness, which when evaluated separately are predicted to follow a unimodal curve with respect to resource supply in plant and animal communities (39) (Fig. 1). However, published work to date addressing this relationship in prokaryotic assemblages documented a positive relationship between richness and resource availability (6, 18, 51), a negative relationship (49), or a mixture of the two (44). Deviation from the traditional unimodal model may be due to differences in the response of the phylogenetic or functional group being considered (28, 29, 51). These contradictions emphasize the need for more experimental data examining how prokaryotic communities respond to shifts in resource variability and how these changes relate to function. Experimental approaches that include both a taxonomy-centered approach (e.g., community composition and diversity) and a gene-centered approach (e.g., functional gene pool composition and diversity) are particularly valuable.
FIG. 1.
Conceptual diagram predicting the pattern in microbial community richness and evenness in response to varying resource levels in a carbon amended (carbon amended, open symbols; unamended, closed symbols) experiment conducted in coastal marine sediments on two dates (April, circles; August, triangles).
In the present study, replicate coastal sediment bioreactors were established in two seasons in 2004: spring (April) and summer (August). For each date, intact sediment sections were continuously supplied with flowthrough seawater that was either unamended or amended with a HMW polysaccharide (10-kDa dextran). Hydrolytic/fermentative bacteria produced labile intermediates for the terminal metabolizers at rates dependent on availability of substrate and on temperature, and changes in the taxonomy and functional gene inventory for the two major groups of terminal metabolizers (sulfate reducers and methanogens) were monitored. Thus, differences in resource availability to the terminal metabolizers were the result of both natural seasonal differences in carbon availability (April versus August) and experimental manipulations of carbon availability (amended versus unamended).
MATERIALS AND METHODS
Experimental manipulations.
The data assembled for the present study were generated from a manipulative laboratory experiment. Some biogeochemical results from the experiment, focused on the shallow sediments (2 to 4 cm), have been published previously (52). Deeper sections (16- to 18-cm sediment depth) of the same sediment cores were manipulated at the same time as the shallow sections. We generated molecular data from the deeper samples, because the biogeochemical data indicated that the prokaryotic community contained actively respiring methanogens despite the presence of sulfate and significant sulfate reduction rates. The presence of both microbial communities allowed us to monitor the interaction between two very distinct subgroups in response to increased carbon resources.
Intact sediment cores (8.8-cm inner diameter) were collected from the unvegetated, intertidal creekbank in Umbrella Creek, a tidal creek in the Satilla River Estuary system, on 20 April and 28 August 2004. Sediment cores were transferred to the laboratory and sectioned the following day. The 16- to 18-cm depth interval from four sediment cores was used in flowthrough bioreactor experiments. Reactor experiments were conducted at 20°C in April and 29°C in August, reflecting measured in situ sediment temperatures. Sediment flowthrough reactors (8, 43, 52) were constructed of polycarbonate plastic and consisted of sediment in a 2-cm section of polycarbonate core, capped on either end with 0.6 cm of a 15-μm frit, and held in place with an upper and lower housing. The polycarbonate housings had an inlet and outlet in the center and spiral grooves on the inner surface to promote diffuse flow through the sediment. A peristaltic pump was used to pump anaerobic artificial water (292 mM NaCl, 12.6 mM MgCl2, 0.9 mM CaCl2, 5 mM Na2SO4, 4 mM NH4Cl, 1.2 mM KH2PO4, 5.7 mM KCl, and 9.5 mM NaHCO3) from reservoirs through the reactors at a slow flow rate (∼10 ml h−1). The entire assembly (reservoir, pump, and reactors) was placed in a nitrogen-flushed glove bag in an incubator. A mixture of 2.5% CO2 in N2 (to maintain a constant dissolved inorganic carbon [DIC] concentration in the reservoir) was used to purge the reservoirs for 4 h initially and then for 0.5 h per day for the duration of the experiment to maintain anaerobic conditions. The oxygen content of the reservoirs was checked periodically by microelectrode (Unisense) and was always below detection (1 μM) after the initial purge.
Duplicate reactors were used for two treatments: a control and a HMW carbon addition. Artificial water was pumped through all reactors for an initial 4-day period to establish flow and flush the water occupying the space between sediment particles (porewater). KBr was then added to the inflow reservoirs to a final concentration of 1 mM, and HMW DOC was added to the reservoir for the carbon treatment to a final concentration of 2 mM carbon as dextran (Sigma, nominal molecular weight of 10,000). Water exiting the flowthrough reactors was sampled at least once daily for 9 days following the start of carbon addition. Samples for geochemical analysis were obtained from the outflow of the sediment reactors by placing the outflow tubing in an 8-ml glass vial for 2 h (allowing the vial to overflow for at least 1 volume). A portion of the 8-ml sample was analyzed immediately for DIC by using a Shimadzu TOC 5000 infrared gas analyzer, while another portion was immediately acidified with 4.4 N phosphoric acid in an He-purged headspace vial for methane (CH4) quantification using a Shimadzu GC 14A flame ionization detector. The remaining water sample was filtered with a 0.2 micron filter, and then a portion was acidified with 0.2 N nitric acid and stored at 4°C until the DOC concentration was measured by using a Shimadzu TOC 5000 apparatus. Sulfate (SO42−) and Cl− concentrations were also measured on nitric acid-preserved filtered water by using a Dionex ion chromatograph. An additional portion of the filtered sample was frozen and later run for determination of VFA content using modified 2-nitrophenylhydrazide derivatization and high-performance liquid chromatography (2). A final portion of filtered water was preserved with 0.1 M zinc acetate for Br colorimetric analysis using a Lachat Quickchem 8000 autoanalyzer. Inflow reservoirs were also sampled several times during the experiment. Flow rates and pH of the outflow water was also monitored throughout the experiment. At the termination of the experiment, flowthrough reactors were dismantled, and the sediment was sampled for porosity, bulk density, and prokaryotic phospholipid fatty acids (PLFAs) using GC analysis after phospholipid extractions (7). At the end of the experiment, a portion of the sediment from each reactor was immediately frozen (−80°C) for prokaryotic community analyses. This created four samples for each experiment; two unamended (control) replicates at the end of the experiment and two carbon-amended samples at the end of the experiment.
Chemical analyses and metabolic calculations.
Net production or consumption rates (R) of DIC, SO42−, CH4, and VFAs in the sediment bioreactors were determined from the changes in concentration from the inflow (Cin) to the outflow (Cout) water and the flow rate (Q) through the bioreactors: R = Q(Cout − Cin).
Rates of terminal metabolism (TM) and hydrolysis/fermentation (HF) were calculated on a per-mol-of-carbon basis as described previously by Weston and Joye (53): TM = RDIC + RCH4 and HF = TM + RVFA + RDOC, where RDIC and RCH4 are the production rates of DIC and CH4, respectively. HF is then the rate of TM (assuming that all TM is fueled by fermentation end products), as well as the net rate of VFA production (RVFA) and the production of other unidentified DOC intermediates (RDOC) that have been produced via hydrolysis and fermentation but not used by the terminal metabolizers. The sulfate reduction rate (RSO4) was calculated from the consumption of SO42− during flow through the bioreactors. The rate of methanogenesis (RCH4) was calculated from the production of CH4 over time.
DNA extraction.
Genomic DNA was extracted from 1 to 2 g of frozen sediment using MoBio soil DNA extraction kits (MoBio, Solana Beach, CA). The nucleic acids were precipitated and resuspended in 30 μl of sterile water. The DNA was then diluted 1:50 to reduce the concentration of inhibitory compounds and stored at −20°C.
16S rRNA gene T-RFLPs.
Bacterial 16S rRNA genes were amplified with the general bacterial primers 27F (5′-AGAGTTTGATCMTGGCTCAG-3′) and 1522R (5′-AAGGAGGTGATCCANCCRCA-3′) (20). 27F primers were fluorescently labeled on the 5′ end with FAM (carboxyfluorescein). All PCR amplifications were carried out with Ready-To-Go PCR beads (Amersham Pharmacia, Piscataway, NJ). PCRs contained 0.2 μM concentrations of each primer and 50 to 100 ng of DNA, with a total reaction volume of 25 μl. Thermal cycling conditions for 16S rRNA gene amplification began with an initial 3 min at 95°C, followed by 30 cycles of 1 min at 95°C, 1 min at 55°C, and 1.5 min at 72°C. A final step of 10 min at 72°C was included to complete any partial polymerizations. For terminal restriction fragment length polymorphism (T-RFLP) analysis (35), restriction enzyme digestion of the PCR product was carried out in a 20-μl volume containing 100 ng of purified PCR product and 10 U of CfoI at 37°C for 6 h in the manufacturer's recommended reaction buffers. Digested DNA was precipitated in ethanol and suspended in 12.5 μl of deionized formamide with 1 μl of DNA fragment length standard Gene-Scan-2500 TAMRA (tetramethylrhodamine; Applied Biosystems). The terminal restriction fragment lengths were separated by electrophoresis on an ABI Prism 310 genetic analyzer using GENESCAN analysis software (Applied Biosystems). Only one restriction enzyme was used, because the goal was to generate a comparative analysis for considering changes in community composition, and we assumed the microbial diversity was too high to resolve through the use of multiple enzymes.
To avoid detection of primers, terminal fragments smaller than 50 bp were excluded from the analysis. The reproducibility of patterns was confirmed by repeated T-RFLP analysis of 16S rRNA genes using the same DNA extract. The T-RFLP data were analyzed using a Visual Basic program that reconciles minor shifts in fragment sizes between successive chromatograms (47). Peaks comprising <1% of total chromatogram area were excluded from the analysis. The relative abundance of terminal restriction fragments within the samples was determined by calculating the area under the each peak as a percentage of the total area under all peaks within one sample. Communities were characterized by the number of peaks and the area under the peaks. Since individual peaks in this analysis could represent multiple taxa, the number of peaks is not considered an indication of species richness but rather used for comparative purposes. Given the poor characterization of bacterial 16S rRNA genes from sediment microbial communities, we did not try to resolve species composition in the sediments using multiple restriction enzymes but rather used the T-RFLP analysis to monitor any major shifts in bacterial community composition with resource amendment.
Functional gene clone library construction.
Genomic DNA was used to amplify two functional genes. The first was a prokaryotic gene coding for the enzyme dissimilatory sulfite reductase (dsrAB), which catalyzes the final step in biotically mediated sulfate reduction. The primers dsr1F (5′- AC[C/G]CACTGGAAGCACG-3′) and dsr4R (5′-GTGTAGCAGTTACCGCA-3′) were used to complete 30 PCR cycles, with each cycle consisting of 30 s at 94°C, 30 s at 55°C, and 90 s at 72°C, with a final step of 10 min at 72°C (50). The second functional gene coded for methyl coenzyme M reductase (mcrA), which catalyzes the terminal step in biogenic methane production. The primers ME1 (5′-GCMATGCARATHGGWATGTC-3′) and ME2 (5′-TCATKGCRTAGTTDGGRTAGT-3′) were used to complete 30 PCR cycles with each cycle consisting of 30 s at 94°C, 30 s at 50°C, and 90 s at 72°C, with a final step of 10 min at 72°C (21). The single PCR product (dsrAB = 1,900 bp, mcrA = 700 bp) was recovered from a 1% agarose gel with a QiaSpin gel extraction kit (Qiagen, Valencia, CA). The amplicons were ligated into plasmid pCR2.1 and cloned using the TOPO-TA cloning system (Invitrogen, Carlsbad, CA). Individual clones were selected and grown in freezing medium in 96-well deep plates and then subsampled and stored at −80°C. Plasmid extraction and sequencing was done commercially (SeqWright; DNA Technology Services) using the forward primer for each gene.
Phylogenetic analyses.
Cloned sequences were aligned with sequences of cultured organisms and environmental clones obtained from GenBank using a CLUSTAL W algorithm (48) in the BioEdit software package (v7.0.4.1) (22). The alignment was translated into amino acid sequences, and then similarity matrices were generated by using unambiguously aligned positions (i.e., >50% of sequences had identical amino acids). The program Protdist from the PHYLIP package (version 3.5c) (17) was used to perform Dayhoff PAM matrix-based distance matrix calculations (31). Operational taxonomic units (OTUs) were assigned using a distance of 0.10 (90% similarity) for both functional gene libraries in the DOTUR software package (45). OTU assignments over a range of distances from 0.01 to 0.30 did not change results of sample comparisons using ANOSIM (described below). Therefore, we chose the 0.10 distance based on level of coverage as indicated by rarefaction curves (not shown). Bootstrapped, neighbor-joining trees were created by using a nonredundant data set of sequences in the software package MEGA version 3.1 (33). Sequences were checked for chimeric characteristics by performing BLAST searches (5) with full-length and partial sequences.
Statistical analysis.
Chao1 (estimate of species richness) and Shannon indices (diversity index) were generated by DOTUR for functional gene libraries. Evenness was calculated as Shannon index/ln(S). For every plot of richness and evenness, the x-axis values were rates of fermentation over the 9-day experiment, expressed as micromoles of carbon per cubic centimeter of sediment, representing the production of DOC intermediates by hydrolytic/fermentative bacteria and therefore the resource supply to the terminal metabolizers. We used a generalized linear model and quadratic regression models (GLM regression, SYSTAT 10.0) to examine the relationship between resource level and richness and evenness. We used the test of Mitchell-Olds and Shaw to determine whether a curvilinear relationship reaches a maximum or minimum (38). Curvilinear relationships that have a maximum or minimum are considered unimodal or U-shaped, respectively. For all statistical analyses we used a P value of ≤0.05 for assigning statistical significance.
We used three independent methods to assess differences in community composition. One method evaluated whether differences between gene libraries were significant using webLIBSHUFF (http://libshuff.mib.uga.edu) (46; J. R. Henriksen, webLIBSHUFF [http://libshuff.mib.uga.edu]). As a second, independent method, we calculated the nonparametric chi-square value for specific OTUs to determine deviations from an expected value using the equation (O − E)2/E, where O is the observed abundance of sequences in an OTU and E is the expected abundance assuming equal distribution in all treatments. The third method involved a nonparametric analysis of similarity (ANOSIM) and had the advantage of using the replicate data when calculating significance. For the ANOSIM, we generated separate Bray-Curtis similarity matrices (16, 36) by using the fragment composition of the bacterial community, dsrA OTU distributions, and mcrA OTU distributions. The data were fourth-root transformed in order to deemphasize dominance by any one OTU. Differences in community composition among the groups were visualized by using nonmetric multidimensional scaling of the similarity matrix to produce a two-dimensional ordination figure (13). A nonparametric ANOSIM was used to test whether there were significant differences in prokaryotic community composition with treatment or between seasons (14). For samples found to be significantly different using ANOSIM, the subroutine SIMPER was used to determine which OTUs might be contributing to the differences between samples. The program calculates the average dissimilarity between all pairings of each replicate within a treatment to the replicates in another treatment and tabulates the average percent contribution of each OTU to the dissimilarity observed between the treatments (10). SIMPER does not assign significance to differences in OTU abundance but is rather an exploratory analysis (15).
The variation in six biogeochemical variables (TM, F, VFA production, PLFAs, carbon mineralization through sulfate reduction, and carbon mineralization through methanogenesis) was assessed for correlations with differences in community composition with the statistical subroutine RELATE (15). RELATE compares a similarity matrix resulting from environmental data with a Bray-Curtis similarity matrix generated using community composition data. Normalized Euclidean distances are used to generate a similarity matrix of environmental data. The two similarity matrices are compared and a Spearman rank correlation coefficient (ρ) is generated. To determine statistical significance, a simple permutation test is used, with the null hypothesis that there is no relationship whatsoever between the two similarity matrices and ρ is approximately zero. The null distribution can be obtained by permuting sample labels at random 999 times and recalculating ρ to build up a frequency histogram with which the true value of ρ can be compared. All data analyses were performed with PRIMER 5 for Windows software (Plymouth Marine Laboratory, Plymouth, United Kingdom).
Nucleotide sequence accession numbers.
Sequence data have been deposited with GenBank and assigned the accession numbers EU301850 to EU302122 (mcrA genes) and EU192370 to EU192651 (dsrA genes).
RESULTS
Biogeochemical patterns.
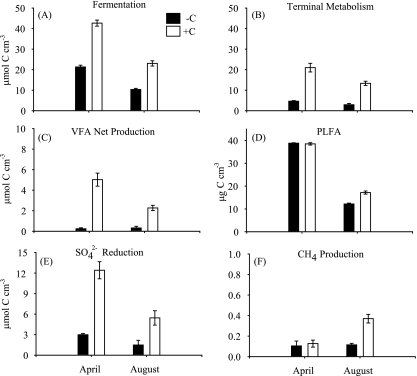
Rates of HF and TM were lower in the August unamended (control) cores than in the April unamended cores, despite higher ambient temperatures (Fig. 2). The total prokaryotic biomass (measured as PLFAs) was also lower in August than in April (Fig. 2). During both April and August, the sediment prokaryotic communities responded to the carbon amendment by increasing fermentation rates (Fig. 2A), resulting in higher VFA production in amended versus unamended cores and thus increasing the C available to the terminal metabolizers. However, net VFA production in the amended cores was lower in August than in April (Fig. 2C). The increase in fermentation rates and the resulting increase in LMW organic matter intermediates stimulated rates of TM in amended cores above the rates measured in unamended cores in both months (Fig. 2B). Carbon amendment also generated an increase in prokaryotic biomass in August as shown by an increase in prokaryotic PLFAs (Fig. 2D). Sulfate reduction rates were stimulated in both months by dextran additions, but methanogenesis only increased above control levels in August (Fig. 2E and F). As expected given the chemistry of inflow water to the cores, TM was dominated by sulfate reduction in both months, and methanogenesis accounted for a maximum of ca. 3% of the TM (August amended cores).
FIG. 2.
Rates of microbial activity and bacterial fatty acid content in flowthrough sediment bioreactor experiments in April and August of 2004. Black bars represent values from unamended cores, white bars the carbon amended treatment. The x-axis and y-axis units are the same for all graphs except where indicated. Values were summed for the entire 9-day period of the carbon amendment except for PFLAs which denote sediment content at the termination of the experiment. Error bars are ±1 standard deviation (n = 2).
Community composition.
We generated three datasets to describe the prokaryotic community composition in these experiments: the relative abundance of T-RFLP peaks, dsrA gene sequences, and mcrA gene sequences. Twelve of the thirteen T-RFLP peaks found in April were shared by both amended and unamended samples, and ten of the thirteen T-RFLP peaks in August were found in both treatments. Of the 48 and 41 OTUs (April and August, respectively) generated from dsrA gene sequences, 42% were found in both amended and unamended cores in April, whereas 49% were common between treatments in August. Similarly, of the OTUs of mcrA genes, 57% of the 18 were found in both amended and unamended cores in April, and 56% of the 14 were found in August.
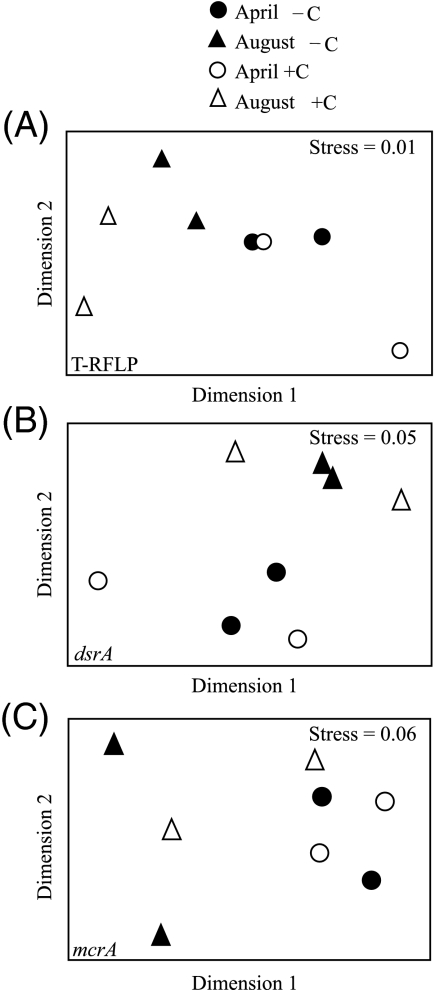
We examined differences in community composition as a function of carbon amendment (plus or minus dextran) or between the 2 months (April versus August). For dsrA and mcrA functional genes, webLIBSHUFF and chi-square analyses found no differences in amended versus unamended samples for either month (AprilmcrA, pxy = 0.837 and pyx = 0.453; AprildsrA, pxy = 0.681 and pyx = 0.918, AugustmcrA: pxy = 0.146 and pyx = 0.248, and AugustdsrA: pxy = 0.102 and pyx = 0.095). However, P values indicate a statistically significant difference between April and August communities when all sequences from each date were pooled (August versus April for mcrA pxy = 0.005 and pyx = 0.001; August versus April for dsrA pxy = 0.032 and pyx = 0.001). As a third statistical method we performed an ANOSIM using each data set, which generates an R value of >0.5 and a P value of <0.05 when significant differences between two communities exist. ANOSIM results likewise indicated that community composition did not differ between amended and unamended samples in either month. However, April and August communities were statistically different for T-RFLP peaks, mcrA, and dsrA functional gene sequences measured on day 9 (Fig. 3).
FIG. 3.
Multidimensional scaling of the Bray-Curtis similarity matrix generated using the relative abundance of T-RFLP peaks generated from 16S rRNA genes (A), OTUs assigned using dsrA genes (B), and OTUs assigned using mcrA genes (C). Carbon-amended (open symbols) and unamended (closed symbols) data are graphed for both April (circles) and August (triangles). Temporal (April versus August) differences were determined by using ANOSIM and were statistically significant for all datasets. For T-RFLP data, R = 0.573 and P = 0.029 (A); for dsrA sequences, R = 0.654 and P = 0.029 (B); and for mcrA sequences, R = 0.573 and P = 0.029 (C).
The SIMPER subroutine identified several OTUs that were possibly responsible for the differences in mcrA and dsrA composition between the 2 months as indicated by the ANOSIM analysis. Genes (dsrA) similar to those originating from the species Desulfacinum infernum and Desulfobacterium aniline (dsrA) appeared to be more abundant in August versus April sulfate-reducing communities (Table 1). Relatives of Methanoculleus thermophilus, Methanopyrus kandleri, and Methanomethylovorans thermophila (mcrA) were found in greater abundance in August methane-producing communities compared to April (Table 2). There was a larger abundance of prokaryotic genes similar to Desulfovibrio cuneatus in April compared to August in the sulfate-reducing communities (Table 2), and the dominant methanogen in April was most closely aligned with a dimethyl sulfide-degrading, obligately anaerobic archaeon, Methanomethylovorans hollandica (Table 2). However, the SIMPER subroutine is only exploratory (no significance assigned to identified differences) and, coupled with sequence errors due to inherent PCR bias, the differences indicated here should be considered only suggestive.
TABLE 1.
SIMPER analysis identifying the OTUs that contributed most to the differences in dsrA sequence composition between April and Augusta
| OTU | Closest match in NCBI database (% similarity) | Relative abundance (%) in:
|
% Contribution to temporal differences | |
|---|---|---|---|---|
| April | August | |||
| 21 | Desulfacinum infernum (85) | 0 | 1.75 | 3.77 |
| 35 | Desulfovibrio cuneatus (69) | 1.5 | 0 | 3.68 |
| 40 | Desulfacinum infernum (72) | 0 | 1 | 2.75 |
| 24 | Desulfacinum infernum (75) | 0.25 | 1.5 | 2.61 |
| 16 | Desulfobacterium anilini (83) | 0 | 1 | 2.61 |
| 26 | Desulfofustis glycolicus (87) | 0.75 | 2.75 | 2.53 |
| 8 | Desulfobacterium cetonicum (79) | 0.25 | 1 | 2.37 |
| 14 | Desulfobacterium anilini (92) | 0.75 | 3 | 2.37 |
| 18 | Desulfobacterium anilini (85) | 0.75 | 2 | 2.31 |
| 32 | Desulfotomaculum acetoxidans (65) | 1.5 | 0 | 2.15 |
| 39 | Desulfacinum infernum (71) | 0 | 1.25 | 2.12 |
| 58 | Desulfobacca acetoxidans (82) | 0.75 | 1.25 | 2.08 |
Data for the amended and unamended samples were combined within a month, for a total of 126 and 153 sequences (April and August, respectively) in the libraries.
TABLE 2.
SIMPER analysis identifying the OTUs that contributed most to the differences in mcrA sequence composition between April and Augusta
| OTU | Closest match in NCBI database (% similarity) | Relative abundance (%) in:
|
% Contribution to temporal differences | |
|---|---|---|---|---|
| April | August | |||
| 19 | Methanoculleus thermophilus (83) | 0 | 1.25 | 9.3 |
| 9 | Methanospirillum hungatei (63) | 2.75 | 0.25 | 8.92 |
| 5 | Methanopyrus kandleri (43) | 0.5 | 4.75 | 7.76 |
| 1 | Methanomethylovorans hollandica (52) | 14.75 | 4 | 7.53 |
| 6 | Methanomethylovorans thermophila (52) | 1 | 3.5 | 5.97 |
| 3 | Methanomethylovorans hollandica (61) | 6.25 | 1.5 | 5.74 |
Data for the amended and unamended samples were combined within a month, for a total of 128 and 145 sequences (April and August, respectively) in the libraries.
In this controlled experiment, we used correlation analyses (RELATE subroutine of Primer 5) to infer causality when relating changes in the community to changes in biogeochemical cycling. Confounding biogeochemical variables were removed by eliminating one of a pair of variables that were highly correlated (r2 > 0.95). Values for organic carbon consumption through sulfate reduction were highly correlated with carbon consumption through TM, and therefore sulfate reduction was removed from the analysis. Similarly, net VFA production was highly correlated with organic carbon consumption through fermentation (F) and therefore removed, leaving four variables in the regression model. For T-RFLP data, a Spearman rank correlation coefficient (ρ) of 0.515 was generated with P = 0.014, using the biogeochemical variables CH4 production, TM, F, and PLFA. Correlations with functional gene datasets included the same four biogeochemical variables. For dsrA gene sequences the values were ρ = 0.558 with P = 0.001, and for mcrA gene sequences the values were ρ = 0.311 with P = 0.10.
Functional gene richness, evenness, and taxonomy.
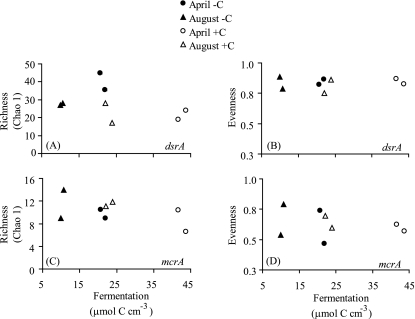
The dsrA and mcrA gene libraries included 281 and 274 sequences, respectively. At a sequence distance of 0.10, 66 unique OTUs were identified for dsrA and 21 for mcrA. The number of OTUs per sample ranged from 17 to 27 for dsrA across the eight samples and 5 to 12 for mcrA (Table 3). Neither a quadratic nor a linear regression equation was significant for correlations of dsrA or mcrA richness and evenness versus the amount of organic carbon consumed through fermentation during the experiment (a proxy for substrate supply to the terminal metabolizers) (Fig. 4).
TABLE 3.
Summary of dsrA and mcrA clone libraries in unamended and amended sediment samples in both April (spring) and August (summer)a
| Parameter |
dsrA
|
mcrA
|
||||||
|---|---|---|---|---|---|---|---|---|
| April 2004 (20°C)
|
August 2004 (29°C)
|
April 2004 (20°C)
|
August 2004 (29°C)
|
|||||
| No dextran | With dextran | No dextran | With dextran | No dextran | With dextran | No dextran | With dextran | |
| No. of sequences | 36, 39 | 34, 17 | 40, 42 | 37, 34 | 42, 40 | 17, 29 | 27, 55 | 38, 25 |
| No. of OTUs | 23, 27 | 19, 17 | 22, 26 | 20, 15 | 8, 11 | 5, 7 | 8, 12 | 10, 8 |
| Coverage estimate (%) | 32, 26 | 47, 0 | 45, 27 | 40, 60 | 93, 85 | 94, 97 | 85, 91 | 89, 84 |
| Chao 1 richness estimate | 36, 45 | 19, 24 | 27, 28 | 28, 17 | 9, 11 | 3, 6 | 9, 14 | 10, 12 |
| No. of different sequences | 34, 38 | 27, 17 | 34, 38 | 33, 29 | 21, 28 | 15, 22 | 19, 42 | 21, 19 |
For each treatment, n = 2. Data for each of two replicates are separated by a comma. For all data, the time period was 9 days.
FIG. 4.
Community richness (left panels) and evenness (right panels) in sediment microbial communities calculated using dsrA sequences (A and B) and mcrA sequences (C and D) collected on day 9. x-axis values are the amount of carbon consumed during fermentation throughout the entire experiment, which should be equivalent to the amount of organic acids produced and therefore available to the terminal metabolizers. Carbon-amended (open symbols) and unamended (closed symbols) data are graphed for both April (circles) and August (triangles). Quadratic or linear regressions are shown when statistically significant (P < 0.10).
The functional gene library was more diverse for dsrA sequences than mcrA sequences (Fig. 5 and 6). A total of 17% of the sequences in the dsrA library had their closest match to a cultured organism in the NCBI database, and the remaining 83% matched dsrA genes in uncultured bacteria. Of the sequences with best matches to cultured organisms in the dsrA library (48 sequences), all but 3 were deltaproteobacteria from four families: Desulfobacteraceae, Syntrophaceae, Desulfovibrionaceae, and Syntrophobacteraceae. The remaining three sequences had best matches to two species of the genus Desulfotomaculum, class Bacillacea. The amino acid similarity between our sequences and those of cultured organisms ranged from 74 to 93% for dsrA, with an average similarity of 85.6%. Similarity between our sequences and those of uncultured organisms ranged from 59 to 97% for dsrA, with an average similarity of 87.6%.
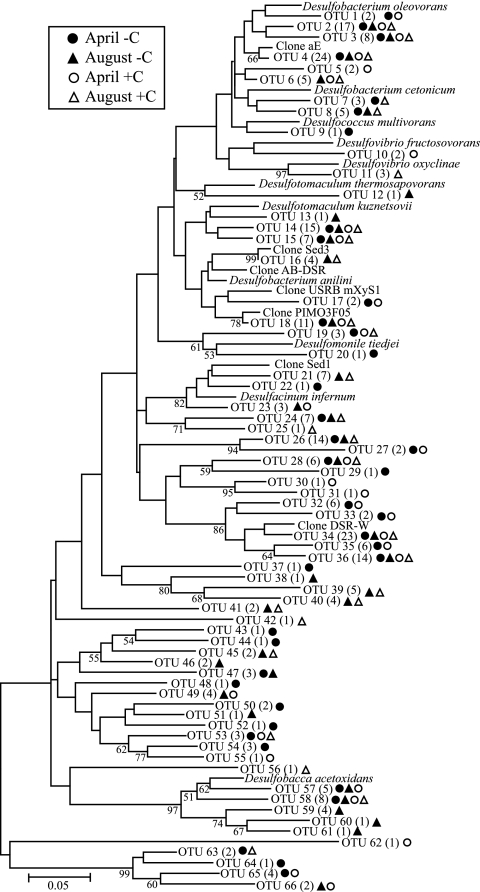
FIG. 5.
Neighbor-joining tree showing taxonomic positions of dsrA gene sequences from coastal marine sediments. Numbers in parentheses indicate the number of clones obtained for each sequence. Bootstrap values (percentage of 200 trials) for branch points are indicated when >50. Bar, 0.05 substitutions per sequence position. The presence of a particular OTU in carbon-amended (open symbols) and unamended (closed symbols) treatments are indicated for both April (circles) and August (triangles) directly adjacent to the OTU name.
FIG. 6.
Neighbor-joining tree showing taxonomic positions of mcrA gene sequences cloned from coastal marine sediments. Numbers in parentheses indicate the number of clones obtained for each sequence. Bootstrap values (percentage of 200 trials) for branch points are indicated when >50. Bar, 0.05 substitutions per sequence position. The presence of a particular OTU in carbon-amended (open symbols) and unamended (closed symbols) treatments are indicated for both April (circles) and August (triangles) directly adjacent to the OTU name.
A total of 34% of the sequences in the mcrA library had a closest match to a cultured organism in the NCBI database. The remaining 66% matched mcrA genes in uncultured bacteria. Sequences with best matches to cultured organisms in the mcrA library represented five families: Methanobacteriaceae, Methanomicrobiaceae, Methanopyraceae, Methanosarcinaceae, and Methanospirillaceae. The amino acid similarity between our sequences and those from cultured organisms ranged from 65 to 99% for mcrA, with an average similarity of 85%. The similarity between our sequences and those from uncultured organisms in the mcrA library ranged from 77 to 98%, with an average of 86% similarity.
DISCUSSION
As our ability to characterize prokaryotic communities has improved it has become possible to evaluate the relevance of classical ecological principles to patterns in prokaryotic abundance and diversity. This study contributes to a growing body of work exploring one such principle, the relationship between community species diversity and resource supply (often expressed as productivity). There is a marginal consensus in the literature that a unimodal relationship between community species diversity and productivity is the most likely model (39). Equally debated are the mechanisms that create the unimodal relationship, whether considering plants, animals, or bacteria. Testing the response of prokaryotic community diversity to changes in system productivity and/or resource supply has come from both manipulative experiments (that change one variable at a time) and studies using naturally occurring gradients in productivity (that include the effects of multiple trophic levels). Our work has elements of both approaches. We studied a naturally occurring prokaryotic community in a relatively intact environment, but manipulated one geochemical variable: the quantity of the labile organic compounds available to microorganisms.
Frequent sampling of the outflow from the bioreactors provided rates of biogeochemical cycling that could then be directly linked to community dynamics at two different times of the year (April and August). To verify that there was sufficient time for measurable changes in community composition to occur, we estimated the number of cell doublings for sulfate-reducing bacteria and methanogens. A minimum of nine doublings was calculated for either functional group using a conservative bacterial growth efficiency (30%) and a high estimate of initial community size (107 cell cm−3). The initial community size was the most important factor influencing the estimation of the number of doublings (higher starting values resulted in a lower number of doublings), whereas the estimated bacterial growth efficiency value had a small effect on calculated doublings (change of only one doubling for bacterial growth efficiency values ranging from 30 to 70%). These conservative calculations indicate that there was ample time for a new community structure to develop during the 9 days of C amendment in our experiments.
Using the estimated doublings calculated above, we sought to determine whether rare species in the samples would have sufficient time to become a measurable portion (at least 10%) of the community if VFA additions preferentially stimulated their growth. Our calculations suggest that a rare species initially comprising 0.0001 to 0.01% of the community and doubling three times faster than every other species would become at least 10% of the community within 10 to 20 doublings of the entire community (smaller starting population sizes required more doublings of the entire community before the rare species became 10% of community). These calculations provide further evidence that the design of the laboratory experiments was sufficient for allowing shifts in community composition to occur, including the establishment of initially rare species responding to VFA amendment.
The fact that no significant community shifts, and therefore no relationship between richness and evenness with resource level, were found for either functional gene (Fig. 4) shows that there is little support for resource-driven patterns in diversity in the terminal metabolizer communities of this coastal sediment. Perhaps a more interesting result is that strong shifts in resource supply that quadrupled in situ rates of substrate supply to the TM community had little apparent affect on the community structure. T-RFLP data (Fig. 3) show that community composition has a temporal (April to August) signal but not a resource supply signal. Likewise, dsrA and mcrA sequences segregated based on month (April versus August) but not resource supply.
Seasonal patterns in carbon mineralization in these sediments have been linked to the differential response of hydrolytic/fermentative bacteria and terminal metabolizers to variation in temperature (52). As temperatures increased from April to August, sulfate reduction rates likely exceeded the supply of labile intermediates produced by the hydrolytic/fermentative bacterial group. Decoupling between the two groups could have created severe carbon limitation of terminal metabolizers, lowering their activity and bacterial numbers and alleviating to some degree competition between sulfate reducers and methanogens when labile carbon was supplied in the experimental manipulation. As evidence of this change in biogeochemical cycling, H2S in situ porewater concentrations at a depth of 16 to 18 cm were fourfold higher in August (data not shown), the net VFA production was lower in August (Fig. 2C), and methane production was higher (Fig. 2F). The Spearman rank correlation coefficients generated by the RELATE analysis support the contention that shifts in dsrA and 16S rRNA gene community composition between the two time points were due to temporal changes in prokaryotic biomass and rates of fermentation and TM.
Although several environmental variables changed between April and August sampling periods, an increase in sediment temperature may have created the shift in community gene composition that we measured. The legacy of these changes may be reflected in differences in OTU abundance between the 2 months. Four dsrA OTUs that were more abundant in the hotter month (August) had as their closest cultured match the thermophilic Desulfacinum infernum. Similarly, three mcrA OTUs that were most abundant in August most closely matched three thermophilic species (Methanoculleus thermophilus, Methanopyrus kandleri, and Methanomethylovorans thermophila). A sequence similar to Desulfovibrio cuneatus, a low-temperature-adapted species, was more abundant in the cooler month, April. These differences in OTU abundance suggest that temperature may be one factor influencing shifts in prokaryotic community composition from April to August.
Although there was a temporal shift in community composition, the most interesting finding is that the sediment terminal metabolizer communities are inherently capable of dealing with varying resource levels. This might be indicative of a dynamic environment experienced by these communities in situ, so that the dominant prokaryotic species are able to compete well at both high and low resource levels. The data collected at our sampling location found that DOC porewater concentrations varied considerably (53). Several factors may influence DOC concentrations, including variation in runoff discharge, variation in tidal cycles, fluctuations in primary productivity, and pulses of anthropogenic inputs from septic tank usage in the watershed. The heterotrophic prokaryotic communities in these sediments appear to be well suited for exploiting increases in labile DOC, without a concomitant change in diversity. It appears that the phenotypic plasticity demonstrated by the heterotrophic prokaryotic community with respect to C utilization decoupled changes in rates of mineralization from changes in the community's genetic composition.
Acknowledgments
This study was funded by an NSF Microbial Biology postdoctoral fellowship (to J.W.E.), by a grant from the Gordon and Betty Moore Foundation (to M.A.M.), and by the Georgia Sea Grant Program (award numbers NA06RG0029-R/WQ11 and R/WQ12A to S.B.J.).
We thank D. Albert, J. Dai, H. Ding, M. Erickson, J. Menken, W. Porubsky, T. Roberts, V. Samarkin, and M. Sun for laboratory assistance. Technical support was also provided by E. J. Biers, C. Lasher, J. Lyons, X. Mou, R. Poretsky, and W. Ye.
Footnotes
Published ahead of print on 11 January 2008.
REFERENCES
- 1.Addy, K., A. Gold, B. Nowicki, J. McKenna, M. Stolt, and P. Groffman. 2005. Denitrification capacity in a subterranean estuary below a Rhode Island fringing salt marsh. Estuaries 28:896-908. [Google Scholar]
- 2.Albert, D. B., and C. S. Martens. 1997. Determination of low-molecular-weight organic acid concentrations in seawater and pore-water samples via HPLC. Mar. Chem. 56:27-37. [Google Scholar]
- 3.Albert, D. B., C. Taylor, and C. S. Martens. 1995. Sulfate reduction rates and low-molecular-weight fatty-acid concentrations in the water column and surficial sediments of the Black Sea. Deep-Sea Res. 42(Pt. I):1239-1260. [Google Scholar]
- 4.Alperin, M. J., D. B. Albert, and C. S. Martens. 1994. Seasonal-variations in production and consumption rates of dissolved organic carbon in an organic-rich coastal sediment. Geochim. Cosmochim. Acta 58:4909-4930. [Google Scholar]
- 5.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 6.Benlloch, S., F. Rodriguez Valera, and A. J. Martinez Murcia. 1995. Bacterial diversity in two coastal lagoons deduced from 16S rDNA PCR amplification and partial sequencing. FEMS Microbiol. Ecol. 18:267-279. [Google Scholar]
- 7.Boschker, H. T. S., J. F. C. de Brouwer, and T. E. Cappenberg. 1999. The contribution of macrophyte-derived organic matter to microbial biomass in salt-marsh sediments: stable carbon isotope analysis of microbial biomarkers. Limnol. Oceanogr. 44:309-319. [Google Scholar]
- 8.Brüchert, V., and C. Arnosti. 2003. Anaerobic carbon transformation: experimental studies with flow-through cells. Mar. Chem. 80:171-183. [Google Scholar]
- 9.Burdige, D. J. 2005. Burial of terrestrial organic matter in marine sediments: a re-assessment. Global Biogeochem. Cycles 19:GB4011. [Google Scholar]
- 10.Carney, K. M., P. A. Matson, and B. J. M. Bohannan. 2004. Diversity and composition of tropical soil nitrifiers across a plant diversity gradient and among land-use types. Ecol. Lett. 7:684-694. [Google Scholar]
- 11.Carrero-Colon, M., C. H. Nakatsu, and A. Konopka. 2006. Microbial community dynamics in nutrient-pulsed chemostats. FEMS Microbiol. Ecol. 57:1-8. [DOI] [PubMed] [Google Scholar]
- 12.Casamayor, E. O., C. Pedros-Alio, G. Muyzer, and R. Amann. 2002. Microheterogeneity in 16S ribosomal DNA-defined bacterial populations from a stratified planktonic environment is related to temporal changes and to ecological adaptations. Appl. Environ. Microbiol. 68:1706-1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clarke, K. 1993. Non-parametric multivariate analyses of changes in community structure. Aust. J. Ecol. 18:117-143. [Google Scholar]
- 14.Clarke, K., and R. Green. 1988. Statistical design and analysis for a “biological effects” study. Mar. Ecol. Prog. Ser. 46:213-226. [Google Scholar]
- 15.Clarke, K., and R. Warwick. 2001. Change in marine communities: an approach to statistical analysis and interpretation, 2nd ed. Plymouth Marine Laboratory, Plymouth, United Kingdom.
- 16.Everitt, B. 1980. Cluster analysis, 2nd ed. Heinemann, London, England.
- 17.Felsenstein, J. 1989. PHYLIP: phylogeny inference package (version 3.2). Cladistics 5:164-166. [Google Scholar]
- 18.Fierer, N., J. P. Schimel, and P. A. Holden. 2003. Variations in microbial community composition through two soil depth profiles. Soil Biol. Biochem. 35:167-176. [Google Scholar]
- 19.Fuhrman, J. A., and L. Campbell. 1998. Marine ecology: microbial microdiversity. Nature 393:410-411. [Google Scholar]
- 20.Giovannoni, S. J. 1991. The polymerase chain reaction, p. 177-201. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. Wiley, New York, NY.
- 21.Hales, B. A., C. Edwards, D. A. Ritchie, G. Hall, R. W. Pickup, and J. R. Saunders. 1996. Isolation and identification of methanogen-specific DNA from blanket bog feat by PCR amplification and sequence analysis. Appl. Environ. Microbiol. 62:668-675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hall, T. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95-98. [Google Scholar]
- 23.Hamamura, N., S. H. Olson, D. M. Ward, and W. P. Inskeep. 2005. Diversity and functional analysis of bacterial communities associated with natural hydrocarbon seeps in acidic soils at Rainbow Springs, Yellowstone National Park. Appl. Environ. Microbiol. 71:5943-5950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haukka, K., E. Kolmonen, R. Hyder, J. Hietala, K. Vakkilainen, T. Kairesalo, H. Haari, and K. Sivonen. 2006. Effect of nutrient loading on bacterioplankton community composition in lake mesocosms. Microb. Ecol. 51:137-146. [DOI] [PubMed] [Google Scholar]
- 25.Henrichs, S. M., and W. S. Reeburgh. 1987. Anaerobic mineralization of marine sediment organic-matter: rates and the role of anaerobic processes in the oceanic carbon economy. Geomicrobiol. J. 5:191-237. [Google Scholar]
- 26.Hewson, I., and J. A. Fuhrman. 2004. Richness and diversity of bacterioplankton species along an estuarine gradient in Moreton Bay, Australia. Appl. Environ. Microbiol. 70:3425-3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hopkinson, C. S., I. Buffam, J. Hobbie, J. Vallino, M. Perdue, B. Eversmeyer, F. Prahl, J. Covert, R. Hodson, M. A. Moran, E. Smith, J. Baross, B. Crump, S. Findlay, and K. Foreman. 1998. Terrestrial inputs of organic matter to coastal ecosystems: an intercomparison of chemical characteristics and bioavailability. Biogeochemistry 43:211-234. [Google Scholar]
- 28.Horner-Devine, M. C., M. A. Leibold, V. H. Smith, and B. J. M. Bohannan. 2003. Bacterial diversity patterns along a gradient of primary productivity. Ecol. Lett. 6:613-622. [Google Scholar]
- 29.Jackson, C. R., P. F. Churchill, and E. E. Roden. 2001. Successional changes in bacterial assemblage structure during epilithic biofilm development. Ecology 82:555-566. [Google Scholar]
- 30.Jardillier, L., D. Boucher, S. Personnic, S. Jacquet, A. Thenot, D. Sargos, C. Amblard, and D. Debroas. 2005. Relative importance of nutrients and mortality factors on prokaryotic community composition in two lakes of different trophic status: microcosm experiments. FEMS Microbiol. Ecol. 53:429-443. [DOI] [PubMed] [Google Scholar]
- 31.Jones, D. T., W. R. Taylor, and J. M. Thornton. 1992. The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 8:275-282. [DOI] [PubMed] [Google Scholar]
- 32.King, G. M. 1988. Patterns of sulfate reduction and the sulfur cycle in a South Carolina salt marsh. Limnol. Oceanogr. 33:376-390. [Google Scholar]
- 33.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5:150-163. [DOI] [PubMed] [Google Scholar]
- 34.Laverman, A. M., P. Van Cappellen, D. van Rotterdam-Los, C. Pallud, and J. Abell. 2006. Potential rates and pathways of microbial nitrate reduction in coastal sediments. FEMS Microbiol. Ecol. 58:179-192. [DOI] [PubMed] [Google Scholar]
- 35.Liu, W. T., T. L. Marsh, H. Cheng, and L. J. Forney. 1997. Characterization of microbial diversity by determining terminal restriction fragment length polymorphisms of genes encoding 16S rRNA. Appl. Environ. Microbiol. 63:4516-4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Magurran, A. 1988. Ecological diversity and its measurement. Princeton University Press, Princeton, NJ.
- 37.Middelburg, J. J., G. Klaver, J. Nieuwenhuize, A. Wielemaker, W. deHaas, T. Vlug, and J. vanderNat. 1996. Organic matter mineralization in intertidal sediments along an estuarine gradient. Mar. Ecol. Prog. Ser. 132:157-168. [Google Scholar]
- 38.Mitchellolds, T., and R. G. Shaw. 1987. Regression analysis of natural selection: statistical inference and biological interpretation. Evolution 41:1149-1161. [DOI] [PubMed] [Google Scholar]
- 39.Mittelbach, G. G., C. F. Steiner, S. M. Scheiner, K. L. Gross, H. L. Reynolds, R. B. Waide, M. R. Willig, S. I. Dodson, and L. Gough. 2001. What is the observed relationship between species richness and productivity? Ecology 82:2381-2396. [Google Scholar]
- 40.Oremland, R. S., D. G. Capone, J. F. Stolz, and J. Fuhrman. 2005. Whither or wither geomicrobiology in the era of “community metagenomics.” Nat. Rev. Microbiol. 3:572-578. [DOI] [PubMed] [Google Scholar]
- 41.Roden, E. E., J. H. Tuttle, W. R. Boynton, and W. M. Kemp. 1995. Carbon cycling in mesohaline Chesapeake Bay sediments. 1. POC deposition rates and mineralization pathways. J. Mar. Res. 53:799-819. [Google Scholar]
- 42.Rossello-Mora, R., B. Thamdrup, H. Schafer, R. Weller, and R. Amann. 1999. The response of the microbial community of marine sediments to organic carbon input under anaerobic conditions. Syst. Appl. Microbiol. 22:237-248. [DOI] [PubMed] [Google Scholar]
- 43.Roychoudhury, A. N., E. Viollier, and P. Van Cappellen. 1998. A plug flow-through reactor for studying biogeochemical reactions in undisturbed aquatic sediments. Appl. Geochem. 13:269-280. [Google Scholar]
- 44.Schafer, H., L. Bernard, C. Courties, P. Lebaron, P. Servais, R. Pukall, E. Stackebrandt, M. Troussellier, T. Guindulain, J. Vives-Rego, and G. Muyzer. 2001. Microbial community dynamics in Mediterranean nutrient-enriched seawater mesocosms: changes in the genetic diversity of bacterial populations. FEMS Microbiol. Ecol. 34:243-253. [DOI] [PubMed] [Google Scholar]
- 45.Schloss, P. D., and J. Handelsman. 2005. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl. Environ. Microbiol. 71:1501-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Singleton, D. R., M. A. Furlong, S. L. Rathbun, and W. B. Whitman. 2001. Quantitative comparisons of 16S rRNA gene sequence libraries from environmental samples. Appl. Environ. Microbiol. 67:4374-4376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stepanauskas, R., M. A. Moran, B. A. Bergamaschi, and J. T. Hollibaugh. 2003. Covariance of bacterioplankton composition and environmental variables in a temperate delta system. Aquat. Microb. Ecol. 31:85-98. [Google Scholar]
- 48.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Torsvik, V., F. L. Daae, R. A. Sandaa, and L. Ovreas. 1998. Novel techniques for analysing microbial diversity in natural and perturbed environments. J. Biotechnol. 64:53-62. [DOI] [PubMed] [Google Scholar]
- 50.Wagner, M., A. J. Roger, J. L. Flax, G. A. Brusseau, and D. A. Stahl. 1998. Phylogeny of dissimilatory sulfite reductases supports an early origin of sulfate respiration. J. Bacteriol. 180:2975-2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Warren, P. H., and A. J. Weatherby. 2006. Energy input and species diversity patterns in microcosms. Oikos 113:314-324. [Google Scholar]
- 52.Weston, N. B., and S. B. Joye. 2005. Temperature-driven decoupling of key phases of organic matter degradation in marine sediments. Proc. Natl. Acad. Sci. USA 102:17036-17040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Weston, N. B., W. P. Porubsky, V. A. Samarkin, M. Erickson, S. E. Macavoy, and S. B. Joye. 2006. Porewater stoichiometry of terminal metabolic products, sulfate, and dissolved organic carbon and nitrogen in estuarine intertidal creek-bank sediments. Biogeochemistry 77:375-408. [Google Scholar]