Abstract
Part of the reason for rejecting aquatic environments as possible vectors for the transmission of Helicobacter pylori has been the preference of this microorganism to inhabit the human stomach and hence use a direct oral-oral route for transmission. On the other hand, most enteric bacterial pathogens are well known for being able to use water as an environmental reservoir. In this work, we have exposed 13 strains of seven different Helicobacter spp. (both gastric and enterohepatic) to water and tracked their survival by standard plating methods and membrane integrity assessment. The influence of different plating media and temperatures and the presence of light on recovery was also assessed. There was good correlation between cultivability and membrane integrity results (Pearson's correlation coefficient = 0.916), confirming that the culture method could reliably estimate differences in survival among different Helicobacter spp. The species that survived the longest in water was H. pylori (>96 h in the dark at 25°C), whereas H. felis appeared to be the most sensitive to water (<6 h). A hierarchical cluster analysis demonstrated that there was no relationship between the enterohepatic nature of Helicobacter spp. and an increased time of survival in water. This work assesses for the first time the survival of multiple Helicobacter spp., such has H. mustelae, H. muridarum, H. felis, H. canadensis, H. pullorum, and H. canis, in water under several conditions and concludes that the roles of water in transmission between hosts are likely to be similar for all these species, whether enterohepatic or not.
Helicobacter pylori is a gram-negative, flagellated bacterium, closely related to Campylobacter spp. and capable of colonizing the gastrointestinal tract and causing disease in the human population (6). While H. pylori research in the medical area has evolved at a fast pace during the last 20 years, there are surprisingly few studies on the ecological aspects of the bacterium outside the human host. As a consequence, the scientific community is still struggling to understand how the bacterium is transmitted, leading to delays in the design of prophylactic measures to counteract the large prevalence found in some human populations worldwide, in which up to 90% of individuals can be infected (3).
The main view up to now has been that, as humans are the only known reservoir for H. pylori, the transmission of infection must be fecal-oral, gastric-oral, or oral-oral (3, 19). Research on direct person-to-person transmission is, however, failing to provide enough evidence to account for all infected individuals. Therefore, external environmental reservoirs such as food, water, and domestic animals have been suggested to harbor the bacterium. To support this argument, there is a growing amount of data reporting the identification of the bacterium in these environments, with particular incidence in water, by molecular techniques (e.g., references 8, 10, 24, 26, 31, and 32). Nevertheless, these routes of infection have been repeatedly dismissed on the basis that H. pylori loses its viability status very rapidly by transforming itself into a nonviable coccoid cell (9, 20). Reflecting the lack of sufficient information about the bacterium's physiology in drinking water, the U.S. Environmental Protection Agency decided to maintain H. pylori on the second candidate contaminant list, which has recently been released (30).
H. pylori is merely one of many related species that are known to colonize not only the gastrointestinal but also the hepatobiliary tracts of humans and other animals (28). These Helicobacter species have been divided into two groups, the gastric and the enterohepatic, depending on the location of the body that each organism typically colonizes (28). Like that of H. pylori, the preferred routes of transmission of these species between different hosts remain unidentified. Most other enteric bacterial pathogens, such as Salmonella spp., Shigella spp., and Escherichia coli, are recognized waterborne agents (21, 25). Accordingly, all have longer durations of cultivability in water than H. pylori (25).
In this work, the following hypothesis was devised: if only enterohepatic Helicobacter spp. are waterborne agents, then these bacteria should demonstrate longer durations of cultivability and viability in water than gastric Helicobacter spp. We have therefore exposed 13 strains of seven different Helicobacter spp. (both gastric and enterohepatic) and also two strains of Campylobacter jejuni to water and tracked their survival by standard plating methods and membrane integrity assessment. To our knowledge, this is the first work in which the durations of cultivability and viability of non-pylori Helicobacter spp. in water have been assessed and compared.
MATERIALS AND METHODS
Culture maintenance.
All H. pylori strains were maintained on Columbia agar (Biomérieux, Marcy l'Etoile, France) supplemented with 5% (vol/vol) defibrinated horse blood (Probiológica, Sintra, Portugal). Plates were incubated at 37°C in a CO2 incubator (HERAcell 150; Thermo Electron Corporation, Waltham, MA) set to 10% CO2 and 5% O2, and bacteria were streaked onto fresh plates every 2 or 3 days; the only difference in terms of culture maintenance between H. pylori and the other Helicobacter and Campylobacter species tested was that the latter were also grown on Campylobacter selective agar (CA; Sigma) supplemented with 5% (vol/vol) defibrinated horse blood. A list of all species used can be found in Table 1.
TABLE 1.
Species of Helicobacter and Campylobacter used in this study
| Organism | Classification |
|---|---|
| H. pylori NCTC 11367T | Gastric |
| H. pylori J99T | Gastric |
| H. pylori 26695T | Gastric |
| H. pylori 968a | Gastric |
| H. pylori 1320a | Gastric |
| H. pylori 1330a | Gastric |
| H. mustelae 2G1b | Gastric |
| H. mustelae 2H1b | Gastric |
| H. felis 2I4b | Gastric |
| H. muridarum 2A5b | Enterohepatic |
| H. canadensis CCUG 47163Tc | Enterohepatic |
| H. pullorum CCUG 33837Tc | Enterohepatic |
| H. canis CIP 104753c | Enterohepatic |
| C. jejuni 1.1d | Enteric |
| C. jejuni 1.2d | Enteric |
Clinical isolate kindly provided by Maria Lurdes Monteiro.
Isolate kindly provided by Jay Solnick.
Isolate kindly provided by Francis Mégraud.
Environmental isolate.
Suspension preparation, inoculation, and cultivable cell counts.
For all experiments, cells from 2-day-old cultures were harvested from Columbia blood agar (CBA) or CA plates, suspended in approximately 20 ml of autoclaved distilled water, and subjected to a vortex for 1 min for homogenization. The cell concentration was then assessed by determining the optical density (OD) at 640 nm, and the necessary volume of this initial inoculum was then used in order to obtain a final concentration of approximately 108 total cells/ml in 200 ml of autoclaved distilled water. The relationship between OD and total cell counts was previously established by performing microscopic cell counts and OD readings at several cell dilutions. Subsequently, 10-ml aliquots of the suspended cells were dispensed into each well of six-well tissue culture plates (Orange Scientific, Braine-l'Alleud, Belgium). The tissue culture plates were then placed in the dark inside an incubator (Shell Lab, OR) at various temperatures (15, 25, and 37°C). The experiments at 25°C were also carried out in the presence of light, provided by a 15-W cool white lamp installed inside the incubator.
Species cultivability in the wells was determined by plating the appropriate dilutions (1:10 dilutions in distilled water) onto CBA in triplicate. Plates were incubated at 37°C for 3 to 7 days (depending on the species) under the same microaerophilic conditions used for culture maintenance. For the experiments assessing the influence of culture medium, cells were also harvested and plated onto CA. Each individual experiment was carried out three times.
Membrane integrity assessment.
To assess the membrane integrity of the bacteria, the LIVE/DEAD BacLight kit (Molecular Probes, OR) was used (7). The two reagents (i.e., Syto9 and propidium iodide) were prepared according to the manufacturer's instructions and mixed in equal proportions. After exposure for 6 h in water, a sample of 100 to 200 μl of the appropriate Helicobacter or Campylobacter sp. was filtered through a 25-mm black Nuclepore polycarbonate membrane with a pore size of 0.2 μm (Whatman, Kent, United Kingdom). After filtration, cells on the membrane were stained with the mixture described above (300 μl per sample) and the membrane was incubated for 15 min in the dark. Cells were visualized under an epifluorescence microscope (BX51; Olympus Portugal, Lisbon) equipped with a charge-coupled device camera (DP71; Olympus Portugal) and a filter block that simultaneously detected the two components of the mixture. A total of 15 fields with an area of 0.0144 mm2 were subjected to counting using image analysis software, and the average result was used to calculate the total number of cells per milliliter of sample. The experiment was performed in triplicate.
Statistics.
In cultivability assessments, the datasets for most strains did not follow a normal distribution, even after the application of different types of transformations to the original data. Therefore, the symmetry of the results, calculated as the skewness/standard-error ratio, was assessed. According to Glass and Hopkins (14), if the above-mentioned ratio lies within the interval (−2, 2), parametric tests may still be applicable. As this was not the case, the nonparametric Kruskal-Wallis test was used, followed by a Tukey-based posthoc analysis, in which rank sums are used instead of means to calculate a q value, as described by Zar (33). Differences in the results were considered statistically significant if the q value was higher than the critical value of the q distribution for a significance level of 95%.
For the hierarchical cluster analysis (HCA), the area under the curve for each separate experiment was calculated by the trapezium rule, meaning that three areas were obtained for each strain and condition. The average area was then used as a single variable to perform the HCA by applying the nearest-neighbor method as a clustering method and the squared Euclidean distance as a similarity measure. Values were transformed by Z scores for standardization.
All calculations were performed by using either the statistical package SPSS (SPSS Inc., Chicago, IL) or Microsoft Office Excel (Microsoft Corporation, Redmond, CA).
Confirmative procedures.
To confirm the identities of H. pylori strains, hybridizations with a specific peptide nucleic acid probe for 16S rRNA were performed as described by Guimarães et al. (15). The identity of the other Helicobacter and Campylobacter spp. was confirmed by 16S rRNA sequencing.
RESULTS
Comparison of durations of cultivability of different Helicobacter and Campylobacter spp.
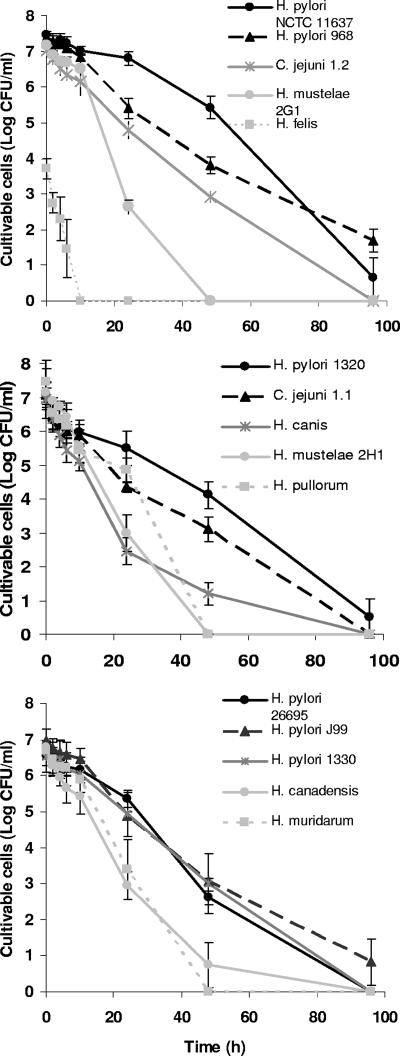
The cultivable cell counts obtained for all species can be observed in Fig. 1. H. felis was the species by far the most sensitive to water, loosing its cultivability between 6 and 10 h after the start of exposure. The sensitivity of H. felis to water was noticeable even at time zero. In fact, only 0.01% of H. felis cells were cultivable at this point, which contrasts with the approximately 10% proportion of cultivable cells obtained for all other species. To ensure that this low degree of initial cultivability was not related to the physiological state of H. felis when it was scraped from the plates, the bacterium was also scraped after 1 and 3 days, with similar results being obtained. All other species were able to remain cultivable for more than 24 h. Of these, the two strains of H. mustelae and also H. pullorum lost cultivability between 24 and 48 h, whereas only H. pylori was able to remain cultivable until the end of the experiment. Surprisingly, C. jejuni strains in this study were slightly more sensitive than H. pylori. Apart from the results for the less resistant H. felis and two less susceptible strains of H. pylori (NCTC 11637 and 968), which were statistically significant compared to those for any of the other strains, there were no statistically different results. Interestingly, H. pylori NCTC 11637 was acquired from a culture collection, whereas H. pylori 968 is a fresh clinical isolate, and the other H. pylori strains tested included two strains from culture collections and two fresh clinical isolates. It appears that, in this experiment, continued culture in artificial environments had little or no impact on times of survival in water. A summary of the statistical analysis of these data is shown in the supplemental material (see Table SA1).
FIG. 1.
Cultivability of 13 Helicobacter spp. and 2 C. jejuni strains in water on CBA at 25°C in the absence of light over time. Each experiment was performed in triplicate, and error bars represent standard deviations.
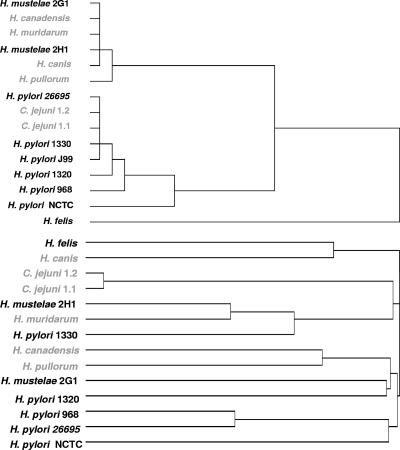
HCA and the phylogenetic tree.
From the dendrogram obtained after HCA, it can be observed that H. felis was placed alone in a separate group (Fig. 2, top panel). Among the remaining bacteria, two distinct groups could be identified: all H. pylori strains belonged to the same group, together with the two strains of C. jejuni, whereas H. mustelae, H. canadensis, H. muridarum, H. canis, and H. pullorum were placed in the other group. For the construction of the phylogenetic tree (Fig. 2, bottom panel), 16S rRNA sequences from all species were aligned using ClustalW (12). By analyzing the phylogenetic tree, it can be observed that not all enterohepatic Helicobacter species were clustered together, but the grouping of species is still markedly different from the one obtained by HCA. Both H. pylori and H. mustelae had strains clustered in the two different major groups.
FIG. 2.
HCA, based on squared Euclidean distances, of the average area under the cultivability curve (top) and phylogenetic analysis of the strains used in this study (bottom). Names in gray represent enterohepatic or enteric species.
Influence of culture media on the survival of Helicobacter spp.
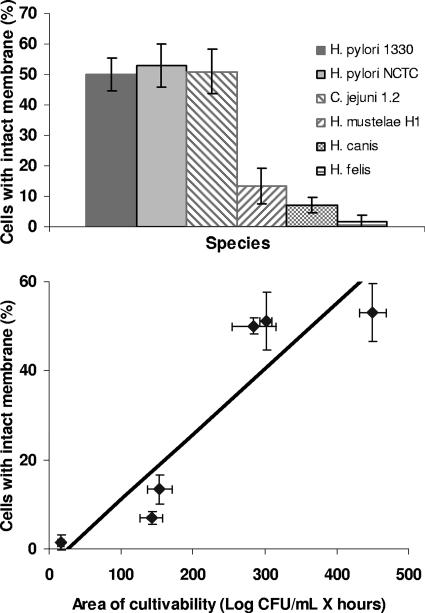
One obvious concern in this work was how to compare the lengths of survival in water among different species of bacteria. Consequently, a comparison between recovery from CBA and that from CA was performed for all Helicobacter and Campylobacter spp. apart from H. pylori (Table 2). Overall, it appeared that the influence of the medium was limited, as statistical analysis was unable to disclose significant differences between results for the two media (see Table SA2 in the supplemental material). The suitability of CBA for comparing viability data among different Helicobacter spp. was further confirmed by membrane integrity assessment (Fig. 3, top panel). Of the six strains tested, the H. felis strain exhibited the highest percentage of cells with damaged membranes, with only 1.5% of the H. felis cells presenting an intact membrane after 6 h. In contrast, propidium iodide was able to penetrate only approximately 50% of the cells of C. jejuni and both strains of H. pylori. These data were correlated with the values obtained by the cultivability assessment, and a Pearson's correlation coefficient of 0.916 was obtained, showing that both methods are reasonably in agreement when evaluating the survival of Helicobacter (and also Campylobacter) spp. in water (Fig. 3, bottom panel).
TABLE 2.
Comparison between cultivability on CBA and that on CA for different species of Helicobacter and two C. jejuni strainsa
| Medium | Mean area (log CFU·h/ml) ± SD for:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| C. jejuni 1.1 | C. jejuni 1.2 | H. mustelae 2H1 | H. mustelae 2G1 | H. muridarum | H. felis | H. canis | H. pullorum | H. canadensis | |
| CBA | 291.7 ± 21.1 | 301.4 ± 8.7 | 153.9 ± 17.3 | 161.1 ± 7.5 | 156.6 ± 19.2 | 16.2 ± 4.5 | 142.2 ± 15.5 | 187.3 ± 12.7 | 159.0 ± 19.9 |
| CA | 278.3 ± 38.1 | 282.0 ± 31.7 | 156.2 ± 16.8 | 212.4 ± 36.2 | 151.6 ± 18.3 | 14.4 ± 5.8 | 91.1 ± 13.8 | 124.8 ± 15.3 | 105.0 ± 34.2 |
Data represent the values of the area under the curve of cultivability over time as calculated by the trapezium rule. The cultivability, over time, of the strains diluted in water and plated onto CA is shown in Fig. SA1 in the supplemental material.
FIG. 3.
Membrane integrity results obtained for six different strains, including one C. jejuni strain. Error bars correspond to standard deviations of results from three replicate experiments (top) and the correlation between cultivability and membrane integrity for the same six species (bottom).
Influence of light and temperature on the cultivability of Helicobacter spp.
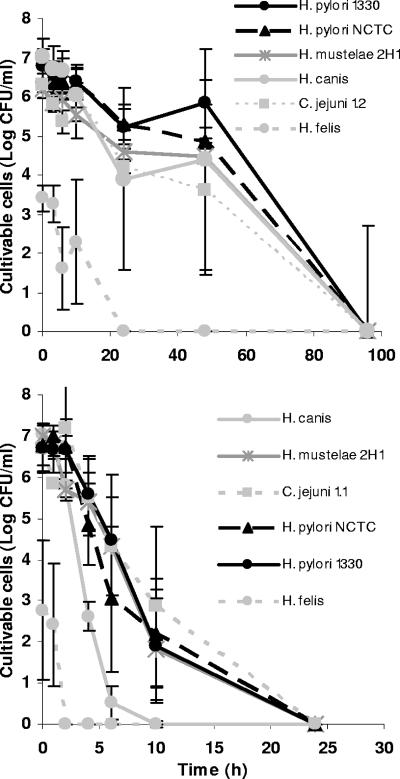
In the experiments performed at 25°C but under light exposure, all Helicobacter and Campylobacter spp. tested lost their cultivability within 24 h of water exposure (see Fig. SA2 in the supplemental material). H. felis was once more the most sensitive bacterium, but all species appeared to be more susceptible to these conditions than to those in which no light was present (Table 3). Nevertheless, statistically significant results were obtained only for H. pylori NCTC 11637.
TABLE 3.
Effect of light on the cultivability of different Helicobacter and Campylobacter spp. at 25°Ca
| Condition | Mean area (log CFU·h/ml) ± SD for:
|
|||||
|---|---|---|---|---|---|---|
| H. pylori NCTC | H. pylori 1330 | C. jejuni 1.2 | H. mustelae 2H1 | H. canis | H. felis | |
| Absence of light | 459.1 ± 33.3 | 284.8 ± 30.2 | 301.4 ± 8.7 | 153.9 ± 17.3 | 142.2 ± 15.5 | 16.2 ± 4.5 |
| Presence of light | 83.1 ± 17.1 | 88.3 ± 2.6 | 61.5 ± 9.5 | 84.2 ± 20.3 | 81.6 ± 5.4 | 10.9 ± 5.0 |
Data represent the values of the area under the curve of cultivability over time as calculated by the trapezium rule. The cultivability of the strains in water when exposed to light over time is shown in Fig. SA2 in the supplemental material.
Results obtained for growth at different temperatures are shown in Fig. 4. As can be observed by comparing Fig. 4 and 2, differences in growth at 15 and 25°C were not very large, and this lack of major differences contrasts with the noticeably shorter durations of cultivability obtained at 37°C. The statistical analysis confirmed this pattern, as results obtained at 37°C for all strains, except H. felis, were significantly different from those obtained at 15 and 25°C.
FIG. 4.
Cultivability of Helicobacter and Campylobacter spp. in water on CBA at 15°C (top) and 37°C (bottom) in the absence of light over time. Error bars correspond to standard deviations of results from three replicate experiments.
DISCUSSION
When comparing the times of survival in water among different species, it is essential to ensure that the values obtained are not greatly affected by external factors, such as different initial physiological states of the cells and different performances of the recovery media in relation to different species. As nearly all Helicobacter and Campylobacter spp. populations included cultivable cells at a level of approximately 10% in relation to the total cells at time zero, the physiological states of these species at the beginning of the experiment were considered to be similar. The only exception was H. felis, with only 0.01% of the total cells being cultivable. For this species, the experiment with inoculums of different ages showed that no matter the age of the inoculum, the initial degree of cultivability of the species remained low. Because this low-level cultivability may still be due to the inadequacy of the plating medium for the recovery of this bacterium, an alternative medium was also tested. In fact, whereas H. pylori is usually grown on CBA, many of the other Helicobacter and Campylobacter species use alternative culture media such as CA (28). Consequently, the experiment was extended to all non-H. pylori species. Differences were not statistically significant for any species, which implies that CBA could be used as an alternative plating medium to recover Helicobacter spp. not only for this work but also for other applications such as clinical isolation.
Despite the similar performances of CBA and CA, an independent method that could assess if the medium was equally suited to recover all bacteria was needed. Despite a few differences in morphology among Helicobacter species (28), we have postulated that membrane structures of the different species would be similarly permeable to propidium iodide. In fact, less phylogenetically similar species such as E. coli, Salmonella enterica, and Shigella flexneri previously showed the same sequence pattern of staining properties with this dye (5). The good correlation coefficient obtained for cultivability and membrane integrity approaches validates both methods to assess Helicobacter and Campylobacter spp. survival in water and confirms that our initial assumptions were correct.
A few studies have already determined the durations of cultivability of H. pylori in water at different temperatures (1, 4, 27). These other studies have reported a loss of cultivability at around 25°C within 48 h, which contrasts markedly with the durations of cultivability of more than 196 h reported for some of the H. pylori strains in this study. By using a range of fresh clinical isolates, we have ensured that this effect was not due to the adaptation of type strains to laboratory conditions (23). In terms of water exposure of H. pylori, the main differences between this study and another performed earlier by our group (4) were that this time the exposure occurred in the absence of daylight and agitation and that we started with a 1-log-higher inoculum concentration. Hamblin et al. have showed that H. pylori is sensitive to visible light and have even suggested that phototherapy may be used to treat infection in the human stomach caused by this pathogen (16). When we repeated the above-mentioned experiment but this time with a 15-W white lamp turned on, cultivability times for H. pylori decreased back to 10 to 24 h. All other species of Helicobacter appeared to be equally affected by light, suggesting that this response is uniform among the members of the genus.
Another surprising result was the similarity of the observed cultivability characteristics of H. pylori and C. jejuni. Of the enteric waterborne pathogens, C. jejuni is perhaps one of the most sensitive to water, with a duration of cultivability of about 120 h at 25°C (e.g., reference 29), which even so is approximately double the standard duration of cultivability observed for H. pylori in water. In the previous study of C. jejuni cultivability, the medium used to cultivate C. jejuni was Karmali agar, which was not tested in the present work. Because membrane integrity assessment results appear to show that CBA reflects what is truly happening in terms of cell viability for all species, a possible explanation for this discrepancy is that the stage of development of culture media for C. jejuni is simply more advanced than that for H. pylori. This possibility makes all the more sense if we bear in mind that research on culture medium for H. pylori is a relatively new area compared to that for C. jejuni.
To gain clarity and to facilitate the comparison of the results obtained, the data on the influence of light and culture medium were presented as cultivability areas instead of the original graphics of cultivability over time. We are, however, aware that some information is lost, such has how long the bacterium remains cultivable and whether cultivability decreases throughout the experiment or if the decrease happens mostly at the end. Therefore, if this strategy is to be used, the original graphics should always appear as supplementary information.
This study also demonstrated that an increase in temperature has a negative effect on all species tested. Shahamat et al. long ago observed this effect on H. pylori (27), a result that was subsequently confirmed by Adams et al. (1). A similar result was also obtained previously for C. jejuni (29). In our case, differences between 15 and 25°C were not significant, but a clear decrease in survival time at 37°C could be observed.
The lack of correspondence between the HCA results and the phylogenetic tree based on 16S rRNA sequences appears to indicate that there is no correlation between the duration of cultivability in water and phylogeny. However, recent reports have suggested that 16S rRNA gene sequence data analyses do not faithfully reflect phylogenetic relationships for the genus Helicobacter (e.g., reference 13). A phylogenetic analysis of Helicobacter species based on partial gyrB gene sequences has therefore been performed previously and has identified two distinct clusters, one formed by gastric species and the other mostly by enterohepatic species (17). Interestingly, the only exception was the gastric species H. mustelae, which clustered with the enterohepatic species, a result similar to that of our HCA. Still, the importance of this coincidence should not be overestimated. The dendrogram obtained contains both enterohepatic H. canadensis and H. pullorum clustered together with H. pylori, and whereas H. pylori exhibits one of the longest durations of cultivability in water, H. felis, another gastric Helicobacter species, clearly exhibits the lowest. Overall, it appears sensible to state that cultivability is not related to the enterohepatic or gastric nature of Helicobacter spp. However, indications provided by the fact that different strains of H. pylori, C. jejuni, and H. mustelae had similar durations of cultivability seem to demonstrate that cultivability is species dependent. The expansion of this work to more strains of the other species may allow us to further sustain this conclusion.
While most of the scientific community remains skeptical about a role for water in H. pylori transmission, the closely related Arcobacter and Campylobacter bacteria have long been shown to survive under such conditions (18, 22). The ultimate proof for these two microorganisms to be considered waterborne was based on their consistent recovery from real water microenvironments, a situation that remains elusive for H. pylori. The lack of recovery of H. pylori from the environment has been associated with the transformation of this microorganism into a coccoid morphology when exposed to water. For many years, researchers considered that this shape was a manifestation of cell death; however, recent reports demonstrated that coccoid morphology in fact corresponds to an active, biologically led process, switched on by the bacterium as a protection mechanism (2, 11).
Conclusions.
As enteric bacteria are usually associated with waterborne outbreaks, we reasoned that enterohepatic Helicobacter spp. might exhibit longer survival in water than H. pylori. By showing that the survival of members of the Helicobacter genus in water is unrelated to the enterohepatic or gastric nature of Helicobacter spp., this work establishes that the roles of water in transmission are likely to be similar for all species and adds to the view that water may have a part in H. pylori transmission. Future work on the transmission of Helicobacter spp. in water may now focus on one bacterial species (whether enteric or gastric) and more confidently extend the conclusions to the remaining species.
Supplementary Material
Acknowledgments
We thank Maria Lurdes Monteiro, Francis Mégraud, and Jay Solnick for providing the clinical isolates and Helicobacter spp. used in this study.
This work was supported by the Portuguese institute Fundação para a Ciência e Tecnologia (postdoctoral grant SFRH/BPD/20484/204) and by the European Commission research project SAFER (contract no. EVK1-CT-2002-00108).
We are solely responsible for this work; the work presented does not represent the opinion of the European Community, and the Community is not responsible for the use that may be made of the data appearing herein.
Footnotes
Published ahead of print on 1 February 2008.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Adams, B. L., T. C. Bates, and J. D. Oliver. 2003. Survival of Helicobacter pylori in a natural freshwater environment. Appl. Environ. Microbiol. 69:7462-7466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Azevedo, N. F., C. Almeida, L. Cerqueira, S. Dias, C. W. Keevil, and M. J. Vieira. 2007. Coccoid form of Helicobacter pylori as a morphological manifestation of cell adaptation to the environment. Appl. Environ. Microbiol. 73:3423-3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Azevedo, N. F., N. Guimarães, C. Figueiredo, C. W. Keevil, and M. J. Vieira. 2007. A new model for the transmission of Helicobacter pylori: role of environmental reservoirs as gene pools to increase strain diversity. Crit. Rev. Microbiol. 33:1-13. [DOI] [PubMed] [Google Scholar]
- 4.Azevedo, N. F., A. P. Pacheco, M. J. Vieira, and C. W. Keevil. 2004. Nutrient shock and incubation atmosphere influence recovery of culturable Helicobacter pylori from water. Appl. Environ. Microbiol. 70:490-493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berney, M., F. Hammes, F. Bosshard, H. U. Weilenmann, and T. Egli. 2007. Assessment and interpretation of bacterial viability by using the LIVE/DEAD BacLight kit in combination with flow cytometry. Appl. Environ. Microbiol. 73:3283-3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blaser, M. J., and J. C. Atherton. 2004. Helicobacter pylori persistence: biology and disease. J. Clin. Investig. 113:321-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boulos, L., M. Prevost, B. Barbeau, J. Coallier, and R. Desjardins. 1999. LIVE/DEAD BacLight: application of a new rapid staining method for direct enumeration of viable and total bacteria in drinking water. J. Microbiol. Methods 37:77-86. [DOI] [PubMed] [Google Scholar]
- 8.Bragança, S. M., N. F. Azevedo, L. C. Simoes, C. W. Keevil, and M. J. Vieira. 2007. Use of fluorescent in situ hybridisation for the visualisation of Helicobacter pylori in real drinking water biofilms. Water Sci. Technol. 55:387-393. [DOI] [PubMed] [Google Scholar]
- 9.Bumann, D., H. Habibi, B. Kan, M. Schmid, C. Goosmann, V. Brinkmann, T. F. Meyer, and P. R. Jungblut. 2004. Lack of stage-specific proteins in coccoid Helicobacter pylori cells. Infect. Immun. 72:6738-6742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cellini, L., A. Del Vecchio, M. Di Candia, E. Di Campli, M. Favaro, and G. Donelli. 2004. Detection of free and plankton-associated Helicobacter pylori in seawater. J. Appl. Microbiol. 97:285-292. [DOI] [PubMed] [Google Scholar]
- 11.Chaput, C., C. Ecobichon, N. Cayet, S. E. Girardin, C. Werts, S. Guadagnini, M. C. Prevost, D. Mengin-Lecreulx, A. Labigne, and I. G. Boneca. 2006. Role of AmiA in the morphological transition of Helicobacter pylori and in immune escape. PLoS Pathog. 2:e97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chenna, R., H. Sugawara, T. Koike, R. Lopez, T. J. Gibson, D. G. Higgins, and J. D. Thompson. 2003. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 31:3497-3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dewhirst, F. E., Z. Shen, M. S. Scimeca, L. N. Stokes, T. Boumenna, T. Chen, B. J. Paster, and J. G. Fox. 2005. Discordant 16S and 23S rRNA gene phylogenies for the genus Helicobacter: implications for phylogenetic inference and systematics. J. Bacteriol. 187:6106-6118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Glass, G. V., and K. D. Hopkins. 1996. Statistical methods in education and psychology, 3rd ed. Allyn and Bacon, Boston, MA.
- 15.Guimarães, N., N. F. Azevedo, C. Figueiredo, C. W. Keevil, and M. J. Vieira. 2007. Development and application of a novel peptide nucleic acid probe for the specific detection of Helicobacter pylori in gastric biopsy specimens. J. Clin. Microbiol. 45:3089-3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hamblin, M. R., J. Viveiros, C. M. Yang, A. Ahmadi, R. A. Ganz, and M. J. Tolkoff. 2005. Helicobacter pylori accumulates photoactive porphyrins and is killed by visible light. Antimicrob. Agents Chemother. 49:2822-2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hannula, M., and M. L. Hanninen. 2007. Phylogenetic analysis of Helicobacter species based on partial gyrB gene sequences. Int. J. Syst. Evol. Microbiol. 57:444-449. [DOI] [PubMed] [Google Scholar]
- 18.Ho, H. T. K., L. J. A. Lipman, and W. Gaastra. 2006. Arcobacter, what is known and unknown about a potential foodborne zoonotic agent! Vet. Microbiol. 115:1-13. [DOI] [PubMed] [Google Scholar]
- 19.Kivi, M., and Y. Tindberg. 2006. Helicobacter pylori occurrence and transmission: a family affair? Scand. J. Infect. Dis. 38:407-417. [DOI] [PubMed] [Google Scholar]
- 20.Kusters, J. G., M. M. Gerrits, J. A. G. Van Strijp, and C. M. J. E. Vandenbroucke-Grauls. 1997. Coccoid forms of Helicobacter pylori are the morphologic manifestation of cell death. Infect. Immun. 65:3672-3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leclerc, H., L. Schwartzbrod, and E. Dei-Cas. 2002. Microbial agents associated with waterborne diseases. Crit. Rev. Microbiol. 28:371-409. [DOI] [PubMed] [Google Scholar]
- 22.Miller, W. G., and R. E. Mandrell. 2005. Prevalence of Campylobacter in the food and water supply: incidence, outbreaks, isolation and detection, p. 101-163. In J. M. Ketley and M. E. Konkel (ed.), Campylobacter: molecular and cellular biology. Horizon Bioscience, Norwich, United Kingdom.
- 23.Owen, R. J., S. A. Chisholm, G. Brick, J. V. Lee, S. Surman-Lee, S. Lai, B. Said, and G. Nichols. 2006. Culture of Helicobacter pylori from domestic water samples: the impact of strain variation on growth on solid and in liquid media. Water Sci. Technol. 54:147-152. [DOI] [PubMed] [Google Scholar]
- 24.Park, S. R., W. G. Mackay, and D. C. Reid. 2001. Helicobacter sp. recovered from drinking water biofilm sampled from a water distribution system. Water Res. 35:1624-1626. [DOI] [PubMed] [Google Scholar]
- 25.Percival, S., R. Chalmers, M. Embrey, P. Hunter, J. Sellwood, and P. Wyn-Jones. 2004. Microbiology of waterborne diseases. Elsevier Academic Press, London, United Kingdom.
- 26.Piqueres, P., Y. Moreno, J. L. Alonso, and M. A. Ferrus. 2006. A combination of direct viable count and fluorescent in situ hybridization for estimating Helicobacter pylori cell viability. Res. Microbiol. 157:345-349. [DOI] [PubMed] [Google Scholar]
- 27.Shahamat, M., U. Mai, C. Paszkokolva, M. Kessel, and R. R. Colwell. 1993. Use of autoradiography to assess viability of Helicobacter pylori in water. Appl. Environ. Microbiol. 59:1231-1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Solnick, J. V., and D. B. Schauer. 2001. Emergence of diverse Helicobacter species in the pathogenesis of gastric and enterohepatic diseases. Clin. Microbiol. Rev. 14:59-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tatchou-Nyamsi-Konig, J. A., A. Moreau, M. Federighi, and J. C. Block. 2007. Behaviour of Campylobacter jejuni in experimentally contaminated bottled natural mineral water. J. Appl. Microbiol. 103:280-288. [DOI] [PubMed] [Google Scholar]
- 30.U.S. Environmental Protection Agency. 2005. Announcement of the drinking water contaminant candidate list 2. Fed. Regist. 70:9071-9077. [Google Scholar]
- 31.Voytek, M. A., J. B. Ashen, L. R. Fogarty, J. D. Kirshtein, and E. R. Landa. 2005. Detection of Helicobacter pylori and fecal indicator bacteria in five North American rivers. J. Water Health 3:405-422. [DOI] [PubMed] [Google Scholar]
- 32.Watson, C. L., R. J. Owen, B. Said, S. Lai, J. V. Lee, S. Surman-Lee, and G. Nichols. 2004. Detection of Helicobacter pylori by PCR but not culture in water and biofilm samples from drinking water distribution systems in England. J. Appl. Microbiol. 97:690-698. [DOI] [PubMed] [Google Scholar]
- 33.Zar, J. 1984. Biostatistical analysis, 2nd ed. Prentice-Hall International, Inc., Englewood Cliffs, NJ.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.