Abstract
Probiotic bacteria encounter various stresses after ingestion by the host, including exposure to the low pH in the stomach and bile in the small intestine. The probiotic microorganism Lactobacillus reuteri ATCC 55730 has previously been shown to survive in the human small intestine. To address how L. reuteri can resist bile stress, we performed microarray experiments to determine gene expression changes that occur when the organism is exposed to physiological concentrations of bile. A wide variety of genes that displayed differential expression in the presence of bile indicated that the cells were dealing with several types of stress, including cell envelope stress, protein denaturation, and DNA damage. Mutations in three genes were found to decrease the strain's ability to survive bile exposure: lr1864, a Clp chaperone; lr0085, a gene of unknown function; and lr1516, a putative esterase. Mutations in two genes that form an operon, lr1584 (a multidrug resistance transporter in the major facilitator superfamily) and lr1582 (unknown function), were found to impair the strain's ability to restart growth in the presence of bile. This study provides insight into the possible mechanisms that L. reuteri ATCC 55730 may use to survive and grow in the presence of bile in the small intestine.
The idea that bacteria could benefit human health was postulated almost 100 years ago by Elie Metchnikoff (23). Recently the use of probiotics, which are defined as “live microorganisms which when administered in adequate amounts confer a health benefit on the host” (10), has become increasingly popular. Probiotic microorganisms are currently being investigated for many possible health benefits in many different ailments, including inflammatory bowel disease, diarrhea, and hypercholesterolemia (9, 33, 40, 46). The mechanisms through which probiotics confer their beneficial effects are mostly unknown; examples of current theories include immunomodulation of the host by the synthesis of immunomodulatory compounds, the production of antimicrobial compounds that inhibit pathogen growth, and large-scale alterations of the microbiota (28, 31).
Probiotic bacteria encounter a variety of stresses that need to be overcome to remain viable. For example, many bacteria are packaged into food products such as yogurt and fermented milk, which exposes them to temperature and osmotic stress. After ingestion, probiotics must be able to survive the extreme acidic conditions in the stomach and the detergent properties of bile acids in the small intestine. Bile acids are amphipathic molecules that are synthesized from cholesterol and play an important role in the digestion of fats and absorption of fat-soluble vitamins. The concentration of bile acids ranges from 0.2 to 2% in the human small intestine and fluctuates based on the amount of fat intake in the diet (14). Bile acids have potent antimicrobial activity against many microbes and are known to cause damage to cells that are considered to be bile resistant, most likely via disruption of the membrane and cell wall. The resistance mechanisms of gram-negative bacteria are fairly well characterized; these mechanisms include protection from the hydrophobic outer membrane and utilization of efflux pumps to expel bile salts that do enter the cell (13). The resistance mechanisms of gram-positive organisms, which in general are less bile resistant than gram-negative bacteria, are less well understood.
Lactic acid bacteria, particularly lactobacilli, are the genus most commonly used as probiotics, in part because of their safe usage in food production. Potential new probiotic strains should include several important characteristics; they should be of human origin, nonpathogenic, able to remain viable in the gastrointestinal (GI) tract for at least short periods of time, and be resistant to various stresses (9). In the GI tract, the main sources of antimicrobial stress are the low pH encountered in the stomach and the detergent-like properties of bile acids found in the small intestine. Although the precise mechanisms by which bile acids cause cell death are not understood, their chemical nature indicates they will be able to solubilize membranes and cause significant membrane damage. This is supported by genetic and genomic studies in a variety of different species that show that the main response of gram-positive organisms to bile exposure appears to be alteration of the cellular envelope. Isolation of bile-sensitive mutants of Enterococcus faecalis and Listeria monocytogenes identified genes mainly involved in maintenance and synthesis of the cell membrane and wall, as well as genes involved in general stress responses (2, 16). Microarray analysis of Lactobacillus plantarum and Lactobacillus acidophilus identified expression changes in genes whose product is found in the cell envelope (6, 25). In addition, there is also microscopic evidence supporting the role of bile in alteration of the cellular envelope. Bron et al. demonstrated that cultures of L. plantarum cells exposed to bile contained some shrunken cells and cells that tended to clump together and had rough surfaces (5), while bile exposure also caused the appearance of shrunken and empty cells in cultures of Propionibacterium freudenreichii (17).
Lactobacillus reuteri is a species with a broad host range, with isolates originating from many different species, including humans, pigs, chickens, dogs, mice, and hamsters (7). L. reuteri is also considered to be indigenous to the human GI tract (27). L. reuteri ATCC 55730, a strain currently marketed for probiotic usage, has been demonstrated in clinical trials to be effective against diarrhea in children, as well as to alleviate colic in infants (26, 30, 45). In addition, consumption of L. reuteri ATCC 55730 reduced the number of sick days taken by workers in a large trial in Sweden (38). How these benefits are achieved at the molecular level is still unknown. L. reuteri ATCC 55730 is known to produce a broad-spectrum antimicrobial compound, reuterin, by metabolism of glycerol under anaerobic conditions (36). Based on the observation that this strain is able to survive in the human duodenum and ileum (41), it is an appropriate organism to use in the investigation of bile resistance mechanisms.
This research investigated the gene expression response of L. reuteri ATCC 55730 to bile exposure and has begun to uncover the mechanisms this strain uses to survive and grow in the presence of bile. Microarray experiments were conducted to determine the gene expression profiles of cells upon initial bile exposure (bile shock) and cells that had resumed growth in the presence of bile (bile adaptation). Based on the microarray results, nine genes were chosen for mutational analysis. These results indicate some of the mechanisms important in the bile shock and adaptation responses of L. reuteri ATCC 55730 in vitro and may provide a further understanding of characteristics important for survival in the GI tract.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. All liquid cultures of lactobacilli were grown under microaerobic conditions (2% O2, 5% CO2, balanced with N2) in MRS broth (BD Difco) at 37°C, unless otherwise specified. All plate cultures of lactobacilli were grown under anaerobic conditions using the GasPack EZ anaerobe container system (BD Difco) at 37°C, unless otherwise specified. Lactobacillus reuteri strains containing pVE6007 were grown at 35°C. All Escherichia coli cells were grown under aerobic conditions at 37°C in LB broth (BD Difco). When specified, drugs were added to the following concentrations: 10 μg/ml (L. reuteri) or 400 μg/ml (E. coli) erythromycin, 10 μg/ml chloramphenicol, and 40 μg/ml kanamycin. L. reuteri mutants (containing the pORI28 disruption) were always grown in the presence of 10 μg/ml erythromycin. Dehydrated bovine bile/oxgall (Sigma) was resuspended in MRS broth to make a 50% weight/volume solution. This mixture was sterilized by autoclaving and stored at 37°C for up to 4 weeks.
TABLE 1.
Bacterial strains and vectors used for this study
| Strain or vector | Description | Source or reference |
|---|---|---|
| Strains | ||
| EC1000 | E. coli strain containing a chromosomal copy of the pWV01 repA gene; Kanr | 15 |
| ATCC 55730 | L. reuteri strain isolated from human breast milk | Biogaia, AB, Sweden |
| PRB190 | L. reuteri ATCC 55730 clpL (lr1864) mutant; Emr | 42 |
| PRB188 | L. reuteri ATCC 55730 putative esterase (lr1516) mutant; Emr | 42 |
| PRB167 | lr0085::pKW01 in an ATCC 55730 background; Emr | This study |
| PRB186 | L. reuteri ATCC 55730 clpE (lr0004) mutant; Emr | Stefan Roos |
| PRB126 | lr1265::pKW02 in an ATCC 55730 background; Emr | This study |
| PRB130 | lr1584::pKW03 in an ATCC 55730 background; Emr | This study |
| PRB163 | lr1291::pKW04 in an ATCC 55730 background; Emr | This study |
| PRB125 | lr1351::pKW05 in an ATCC 55730 background; Emr | This study |
| PRB114 | lr1706::pKW06 in an ATCC 55730 background; Emr | This study |
| Vectors | ||
| pVE6007 | CmrrepA-positive temperature-sensitive derivative of pWV01 | 19 |
| pORI28 | EmrrepA-negative derivative of pWV01 | 15 |
| pKW01 | pORI28 + 203-bp insert from lr0085 | This study |
| pKW02 | pORI28 + 303-bp insert from lr1265 | This study |
| pKW03 | pORI28 + 331-bp insert from lr1584 | This study |
| pKW04 | pORI28 + 310-bp insert from lr1291 | This study |
| pKW05 | pORI28 + 152-bp insert from lr1351 | This study |
| pKW06 | pORI28 + 240-bp insert from lr1706 | This study |
RNA isolation.
For each of five biological replicate experiments, a culture of L. reuteri ATCC 55730 was grown in MRS broth to an optical density at 600 nm (OD600) approximately equal to 0.5. Upon reaching this stage in growth, 0.5% oxgall was added to the culture. At the correct time points, 5-ml samples were collected from the culture and immediately mixed with an equal part of ice-cold methanol. Cell pellets were collected by centrifugation, washed with STE buffer (6.7% sucrose, 50 mM Tris-Cl [pH 8.0], 1 mM EDTA), and resuspended in STE buffer containing 0.25 U/μl mutanolysin (Sigma). Cell pellets were then incubated at 37°C for 20 min. RNA was then isolated using the Qiagen RNeasy kit according to the manufacturer's instructions.
Microarray experiments.
Long oligonucleotides (60-mers) were designed and synthesized for 1,864 open reading frames from a draft genome sequence of L. reuteri ATCC 55730 (1) and 15 open reading frames encoding known extracellular proteins from L. reuteri DSM 20016 (43) using OligoArray 1.0 software. Six control 60-mer oligonucleotides were also included. These controls are identical to DNA sequences from E. coli genes (yacF, ybaS, yciC, yfiF, ygjU, and yjcG) and have no sequence similarity to the L. reuteri genome. Once synthesized, the oligonucleotide concentrations were normalized to a concentration of 25 μM and spotted onto Corning UltraGAPS-II slides using an OmniGrid robot (GeneMachines). Each gene was represented once on the microarray. All six of the control spots were represented eight times on the array, once in each subgrid. Oligonucleotide design, synthesis, and array construction were performed at the Research Technology Support Facility at Michigan State University, East Lansing, MI. RNA isolation, labeling, and hybridization were carried out essentially as previously described (39, 42). Information regarding the microarray platform can be found at the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) under GEO platform no. GPL6366.
Five biological replicates were performed for each of the two sets of microarray experiments. In addition, technical replication was achieved by switching the dyes used for labeling each biological replicate. Therefore, each RNA sample was subjected to two hybridizations and values used for subsequent data analysis were averages of the dye swap values. The first set of experiments, referred to as the bile shock experiments, compared the gene expression profiles of cells before exposure to 0.5% bile to those that had been exposed for 15 min. The second set of experiments, referred to as the bile adaptation experiments, compared the gene expression profiles of cells before exposure to 0.5% bile to those that had begun growing again after exposure (Fig. 1). Microarray data were analyzed using iterative outlier analysis with three iterations as previously described (4, 39). Briefly, iterative outlier analysis calculates the geometric mean and standard deviation of the entire data set. Differentially expressed genes (outliers) were selected as being more than 2.5 standard deviations away from the mean of the population. To identify additional differentially expressed genes in the data set, the outliers were removed, the geometric mean and standard deviations were recalculated, and any genes that were more than 2.5 standard deviations from the mean were identified as differentially expressed.
FIG. 1.
Representative growth curve of L. reuteri ATCC 55730 used for microarray experiments. Arrows represent time points where samples were taken for RNA isolation. Open arrows represent samples for bile shock experiments; filled arrows represent samples for bile adaptation experiments. Oxgall (0.5%) was added at 250 min.
Mutant construction.
Mutants were created using the system developed by Russell and Klaenhammer (29) and modified for use in L. reuteri by Walter et al. (44). Briefly, 200 to 300 bp from the gene of interest was PCR amplified from L. reuteri ATCC 55730 and cloned into pORI28. The plasmid with the insertion was then transformed into E. coli EC1000, a carrier strain that contains the RepA protein needed for pORI28 to replicate; the transformed cells were grown in the presence of erythromycin and kanamycin (EC1000). pORI28 with the insertion was then extracted and transformed into L. reuteri ATCC 55730 cells containing pVE6007. pVE6007 is a helper plasmid that provides RepA (also allowing pORI28 to replicate). L. reuteri cells containing both plasmids were grown aerobically without shaking at the permissive temperature of 35°C in the presence of chloramphenicol and erythromycin for 18 h. This culture was then diluted 1:200 and grown aerobically in the presence of erythromycin without shaking at 45°C for 8 to 24 h. The 45°C culture was then plated onto plates containing MRS plus erythromycin and incubated at 45°C for 24 h. Isolated colonies were then obtained by streaking onto fresh MRS-erythromycin plates and incubated at 45°C for another 24 h to ensure loss of pVE6007. Individual colonies were then screened for the loss of pVE6007 by selecting for erythromycin resistance and chloramphenicol sensitivity at 37°C. Colonies that had lost pVE6007 were then screened for the correct insertion by PCR amplification of both flanking regions (one primer annealing to the chromosome outside of the region cloned into pORI28 and one primer annealing to pORI28) and confirmation of the absence of the correctly sized wild-type gene (also through PCR). PCR primers are available by request.
Bile stress assays.
To determine levels of bile resistance for the mutants, the percentages of survival of wild-type and mutant cultures were determined after 30 min of exposure to 0.3% bile. In short, cultures were grown under microaerobic conditions in MRS at 37°C to an OD600 of 0.5. Samples were taken, and colony counts were determined by dilution plating. Oxgall (0.3%) was then added to each culture, and after 30 min, colony counts were again determined. The before-bile and after-bile colony counts were used to determine the percentage of survival for each strain. All growth curves and viability plating were performed three times for each strain, with the exception of the wild-type experiments, which were repeated eight times.
RESULTS
Lactobacillus reuteri ATCC 55730 is able to grow in physiologically relevant concentrations of bile.
One important characteristic for probiotic bacterial strains is the ability to remain viable during passage through the GI tract, including the ability to overcome exposure to bile stress in the small intestine. Growth experiments were conducted to determine the response of L. reuteri ATCC 55730 to physiological concentrations of bovine bile. In general, when 0.05 to 0.1% bile was added to an early or mid-log culture (OD600 of 0.2 or 0.5), the culture continued growing, although at a slightly reduced rate. The doubling time of the culture would slow from 38 min before the addition of bile to 50 min after the addition of bile. When concentrations of bile ranging from 0.3% to 5% were added, we observed a period of growth arrest followed by a resumption of growth. However the doubling time in the presence of these higher bile concentrations was three to four times slower than prior to treatment with bile.
The growth phase of L. reuteri cells also influenced the ability of bile to affect cell growth and viability. Early-log-phase cells (OD600 of 0.2) were the most resistant to the effects of bile treatment. Addition of bile at later stages of log-phase growth and early stationary phase indicated that cells become more susceptible to bile as the culture density increases (as measured by a decrease in the OD and cell viability of the culture after the addition of bile). Active growth appears to be required for this effect as late-stationary-phase cultures were completely resistant to bile stress, even at concentrations of 5%.
Microarray analysis of genes involved in bile shock and adaptation.
We used DNA microarrays to characterize both the bile shock response and bile adaptation response of L. reuteri. When cells encounter stress, they often respond by altering their gene expression program to effectively counteract stress-induced damage. Because L. reuteri exhibits a biphasic response to bile exposure, we measured the global RNA profiles of cells that were paused for growth (which we denote as bile shock) and cells that had resumed growth in the presence of bile (bile adaptation).
(i) Bile shock.
In order to determine the genes involved in the bile shock response, microarray experiments were carried out to compare the gene expression profiles of mid-log cells that had not been exposed to bile to those of cells that had been exposed to 0.5% bile for 15 min. Eighty-eight genes were found to have significant expression changes, with 45 genes overexpressed and 43 genes underexpressed after 15 min of bile exposure. The majority of underexpressed genes are classified as being involved in substrate transport and metabolism, which is expected due to the lack of growth observed upon exposure to bile (Table 2). The overexpressed genes are found in a wide variety of classes, and several have known roles in adaptation to other types of stresses (see below and Discussion).
TABLE 2.
Classes of genes differentially expressed during the first 15 min of exposure to 0.5% bilea
| Gene classification | No. of genes during bile shock:
|
|
|---|---|---|
| Overexpressed | Underexpressed | |
| Energy production and conversion | 0 | 4 |
| Cell division and envelope biogenesisb | 1 | 3 |
| Substrate transport and metabolismc | 4 | 21 |
| Translation, ribosomal structure, and biogenesis | 2 | 2 |
| Transcription | 8 | 1 |
| Replication, recombination, and repair | 3 | 3 |
| Posttranslational modification, protein turnover, chaperones | 6 | 2 |
| Defense mechanisms | 3 | 0 |
| Unknown functions | 18 | 7 |
| Total | 45 | 43 |
Genes were classified based on COG domains found in the protein sequence through a search of the JGI Integrated Microbial Genomes database.
This class represents two COG categories: cell cycle control, cell division, and chromosome partitioning and cell wall/membrane/envelope biogenesis.
This class represents multiple COG categories that include transport and metabolism of carbohydrates, amino acids, nucleotides, coenzymes, lipids, inorganic ions, and secondary metabolites.
(ii) Bile adaptation.
The expression profiles of mid-log cells that had not been exposed to bile were also compared to the profiles of cells that had resumed growth in the presence of 0.5% bile. After analysis of this set of array experiments, 84 genes were found to have significant expression changes, with 17 being overexpressed during the adaptation stage and 67 being underexpressed. Again, the majority of underexpressed genes during growth in bile are classified as being involved in substrate transport and metabolism; in addition, genes involved in energy production, translation, and ribosome structure and biogenesis and genes of unknown function are also underexpressed. Most of these changes are likely due to the dramatic reduction in growth rate of L. reuteri cells grown in the presence of bile. There were 17 genes that were significantly overexpressed during growth in bile; over half of these genes are annotated as having unknown functions (Table 3). The expression patterns of a subset of genes that were differentially expressed were found to overlap between the two sets of microarray experiments, with 8 genes being overexpressed and 12 genes being underexpressed during both bile shock and adaptation. A full description of the differentially expressed genes discovered in all of the microarray experiments can be found in Tables S1 to S4 in the supplemental material. A complete data set of microarray data can be found at NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) under GEO series accession no. GSE10155.
TABLE 3.
Classes of genes differentially expressed during growth in the presence of 0.5% bilea
| Gene classification | No. of genes during bile adaptation:
|
|
|---|---|---|
| Overexpressed | Underexpressed | |
| Energy production and conversion | 0 | 11 |
| Cell division and envelope biogenesisb | 0 | 4 |
| Substrate transport and metabolismc | 5 | 23 |
| Translation, ribosomal structure, and biogenesis | 0 | 11 |
| Transcription | 2 | 1 |
| Replication, recombination, and repair | 0 | 4 |
| Intracellular trafficking, secretion, and vesicular transport | 0 | 2 |
| Signal transduction mechanisms | 0 | 2 |
| Defense mechanisms | 1 | 0 |
| Unknown functions | 9 | 9 |
| Total | 17 | 67 |
Genes were classified based on COG domains found in the protein sequence through a search of the JGI Integrated Microbial Genomes database.
This class represents two COG categories: cell cycle control, cell division, and chromosome partitioning and cell wall/membrane/envelope biogenesis.
This class represents multiple COG categories that include transport and metabolism of carbohydrates, amino acids, nucleotides, coenzymes, lipids, inorganic ions, and secondary metabolites.
Mutations in three genes, lr0085, lr1516, and lr1864, decrease the ability of cells to survive bile shock.
Bile salts have been proposed to have a wide range of cellular effects, including cell wall or membrane damage, DNA damage, protein denaturation, oxidative stress, and low intracellular pH (3). Several genes were chosen for mutation based on their proposed functions in adapting to a variety of stresses. Disruptions were created using the pVE6007/pORI28 system in nine genes that were found to be significantly overexpressed during the bile exposure of L. reuteri ATCC 55730 (Table 4). Two Clp chaperones (lr0004 [clpE]) and lr1864 [clpL]) were disrupted; Clp chaperones have been implicated in the heat shock response of Bacillus subtilis, as well as other gram-positive organisms (8, 11). The dps gene (lr1706) and a putative esterase (lr1516) were also disrupted to investigate the proposed oxidative stress and cell wall damage effects of bile. The putative esterase (lr1516) belongs to a cluster of orthologous genes (COG) that includes β-lactamase class C and other various penicillin-binding proteins (20). Three other genes of unknown function were also chosen for disruption: lr1291, a putative metalloproteinase; lr1351, a conserved membrane protein; and lr0085, a gene of unknown function that appears to be specific to the species L. reuteri. Finally, two multidrug resistance transporters were disrupted (lr1265 and lr1584). E. coli has the ability to actively pump bile salts out of the cell, and efflux pumps, similar to drug resistance transporters, have been found to have a major role in this activity (37).
TABLE 4.
Gene expression changes in the presence of bile for genes chosen for disruption
| Gene | GenBank accession no. | Annotation | Change (fold) duringa:
|
|
|---|---|---|---|---|
| Bile shock | Bile adaptation | |||
| lr0004 | EF421856 | Clp chaperone (ClpE) | 5 | 1.4 |
| lr0085 | DQ233699 | Hypothetical protein | 3.3 | 2.5 |
| lr1265 | EU038268 | Multidrug resistance protein (ABC transporter family) | 3.9 | 1.5 |
| lr1291 | AY970991 | Metalloproteinase | 2.8 | 1.6 |
| lr1351 | DQ233687 | Conserved membrane protein of unknown function | 3 | 4.1 |
| lr1516 | DQ219970 | Putative esterase | 7.3 | 2.8 |
| lr1584 | EU038252 | Multidrug resistance protein (major facilitator superfamily) | 1.6 | 2.2 |
| lr1706 | EU038255 | Dps | 2.6 | 2.1 |
| lr1864 | DQ219976 | Clp chaperone (ClpL) | 3.2 | 1.4 |
Changes in boldface were found to be significantly different based on the outlier analysis.
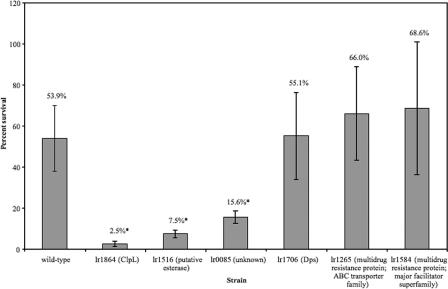
Each of the nine mutants was subjected to treatment with various concentrations of bile to determine which mutants are defective in surviving bile shock. Based on these preliminary experiments, the survival of six of these mutant strains was quantitated and compared to the viability of wild-type cells after 30 min of 0.3% bile exposure; the other three mutants (lr0004, lr1291, and lr1351) showed no defect in bile shock and were not further tested. Using a Student's t test, it was determined that the survival rate after 30 min of bile exposure for three of the mutants (lr1864, lr1516, and lr0085) was significantly different from that of the wild type (P < 0.001). Strains with mutations in dps and the two multidrug resistance transporters (lr1706, lr1584, and lr1265) did not have a survival rate significantly different from wild-type cells (Table 5 and Fig. 2).
TABLE 5.
Effects of 0.3% bile exposure on cell viability and final culture density
| Disrupted gene | GenBank accession no. | Annotation | % Survivala,c | OD600b,c |
|---|---|---|---|---|
| lr0085 | DQ233699 | Unknown | 15.6 ± 2.9* | 1.71 ± 0.03 |
| lr1265 | EU038268 | Multidrug resistance protein (ABC transporter family) | 66 ± 22.8 | 1.04 ± 0.09* |
| lr1516 | DQ219970 | Putative esterase | 7.5 ± 1.9* | 2.06 ± 0.12 |
| lr1584 | EU038252 | Multidrug resistance protein (major facilitator superfamily) | 68.6 ± 32.2 | 0.57 ± 0.03* |
| lr1706 | EU038255 | Dps | 55.1 ± 21.2 | 1.32 ± 0.27 |
| lr1864 | DQ219976 | Clp chaperone (ClpL) | 2.5 ± 1.3* | 1.64 ± 0.15 |
| None | Wild-type cells | 53.9 ± 16 | 1.62 ± 0.21 |
Percent survival 30 min after exposure. Data are presented as mean percent survival ± standard deviation.
Data are presented as mean OD600 ± standard deviation after growth had ceased.
*, P < 0.001 compared with wild type.
FIG. 2.
Comparison of survival rates after 30 min of exposure to 0.3% oxgall for the L. reuteri ATCC 55730 wild type and lr1864 (ClpL), lr1516 (putative esterase), lr0085 (unknown), lr1706 (Dps), lr1265 (multidrug resistance protein in the ABC transporter family), and lr1584 (multidrug resistance protein in the major facilitator superfamily) mutant strains. Cultures were plated onto MRS plates after bile exposure to determine the number of viable cells. Error bars represent standard deviation. *, P < 0.001 compared with wild type.
Mutations in a putative operon encoding a multidrug resistance protein and a hypothetical protein decrease the ability of L. reuteri to adapt in the presence of bile.
When testing the mutant strains for survival in the presence of bile, it was observed that the lr1584 mutant did not adapt to grow in the presence of bile, even after extended incubations (24 h). The final culture density obtained for the lr1584 mutant was found to be threefold lower than that of the wild-type strain. Additional experiments revealed that most of the other mutants were not affected in their ability to adapt and grow in the presence of bile. The exceptions were the lr1265 mutant, which also obtains a lower culture density than the wild type, and the lr1516 mutant, which obtains a slightly higher culture density (Table 5). The decreased ability of the lr1584 and lr1265 mutant strains to adapt to the presence of bile suggests that multidrug resistance efflux pumps play a role in these strains' bile response. Efflux pumps have already been shown to play important roles in the bile response of other bacteria (18, 34, 37).
The lr1584 gene is found in a putative operon with a gene encoding a conserved hypothetical protein, lr1582. lr1582 is also found to be significantly overexpressed in the presence of bile; therefore, we were concerned about the possible polar effects the disruption in lr1584 would have on the downstream gene, lr1582. To distinguish between the effects of the lr1584 mutation and the possible polar effects on lr1582, a separate mutant strain containing a disruption in lr1582 was created and tested for its ability to adapt in the presence of bile. This strain also showed an adaptation defect; the final culture density of the lr1582 mutant was twofold lower than that of wild-type cells. This demonstrates that the adaptation defect seen in the lr1584 mutant cannot be fully explained by polar effects on the downstream gene, lr1582, and suggests that both genes in this operon play a role in L. reuteri's adaptation to bile.
DISCUSSION
The ability of a bacterium to resist bile stress is one of the criteria often used in the selection of a potential probiotic. Bile is a complex mixture of bile acids, phospholipids, proteins, ions, and pigments that has potent antimicrobial properties, particularly against gram-positive bacteria. In this study, we have identified several genes that participate in the ability of Lactobacillus reuteri ATCC 55730 to tolerate bile shock and to resume growth in the presence of bile (bile adaptation).
Stress responses activated in L. reuteri based on gene expression data.
The gene expression data indicate that membrane/cell wall stress, oxidative stress, DNA damage, and protein denaturation occur when L. reuteri is exposed to bile. Several of these pathways have been previously shown to be involved in dealing with various forms of stress in other bacteria. First, the Clp chaperones ClpE and ClpL are induced 15 min after bile addition, as is their known transcriptional regulator in other gram-positive organisms, CtsR. CtsR is a repressor of multiple Clp chaperones in Listeria monocytogenes and Bacillus subtilis and also represses its own expression. Previous work has shown that the induction of the CtsR stress regulon is transient with an initial peak of expression under heat or salt stress that then is reduced after a period of time (32). Consistent with this mode of regulation in other bacteria is the fact that we observed that ctsR and the clp chaperones are overexpressed only during bile shock and not during bile adaptation. clpL was specifically required for L. reuteri to resist bile shock, while the clpE mutant did not survive at rates significantly different from those of wild-type cells. Repeated attempts to construct a mutation in the ctsR gene were unsuccessful. Second, we also observed increased expression of dps, a protein involved in several types of stress adaptation in Escherichia coli, including oxidative stress, irradiation, metal toxicity, heat stress, and pH stress (22, 24). However, disruption of dps in L. reuteri did not significantly affect the organism's ability to survive bile shock or adapt to the presence of bile. Finally, two additional stress response genes and one additional pathway were also induced. A homolog of the gene gls24 (lr2108) was induced; Gls24 was previously identified as a bile-induced protein in Enterococcus faecalis. Subsequent genetic analysis indicated it was required for the ability of E. faecalis to survive bile exposure; however, no molecular function for Gls24 is known (12). The gene lr1346 encodes a homolog of the phage shock transcriptional regulator PspC, which is proposed to be involved in sensing membrane stress during phage infection. Given that bile likely induces membrane stress, it is possible that lr1346 plays a role in bile stress survival. Unfortunately, we were unable to disrupt lr1346 due to limitations of our gene knockout technology. Lastly, genes of the arginine deiminase pathway were specifically induced during bile adaptation. This pathway has been implicated in the ability to resist mild pH shock in bacteria (21).
The identification of lr1516, a putative esterase of the serine β-lactamase-like superfamily, as a key enzyme in responding to bile stress suggests these cells are experiencing cell envelope damage upon exposure to bile. lr1516 contains the signature SxxK active site motif associated with these enzymes, which also include the d-alanyl-d-alanine carboxypeptidases. These enzymes are involved in the breakdown and reorganization of peptidoglycan, and thus we expect that lr1516 may play a similar role when adapting to bile and acid stress (42).
Bile adaptation.
Because L. reuteri has been shown to colonize, at least temporarily, the small intestine, we were interested in determining if L. reuteri can thrive in the presence of bile. Our results demonstrate that L. reuteri can sustain growth in the presence of bile concentrations as high as 5%. Interestingly, the data suggest a multidrug resistance transporter (lr1584) is required for this ability to grow in the presence of bile, suggesting that removal of bile or another toxic metabolite from the cytoplasm is required for growth. lr1584 is a member of the EmrB/QacA subfamily of the major facilitator superfamily of multidrug resistance transporters. EmrB has previously been shown to play a role in bile resistance and efflux of bile in Escherichia coli (37).
Interestingly, lr1584 is found in an operon upstream of a conserved hypothetical protein, lr1582; this operon is conserved in many lactic acid bacteria and is overexpressed during exposure of L. reuteri, L. acidophilus, and E. faecalis to bile (25, 35). Due to the limited genetic tools available for use in L. reuteri, we were not completely able to distinguish between the effects of disruption of lr1584 and possible polar effects this disruption may have on the downstream lr1582. A separate mutant strain with a disruption in lr1582 was created and tested for bile adaptation. The lr1582 mutation does result in an adaptation defect, although it is not as severe as the defect found in the lr1584 mutant strain. The final culture density of the lr1584 mutant is approximately threefold lower than that of the wild-type cells, while the final culture density of the lr1582 mutant is approximately twofold lower than that of wild-type cells. This suggests that both genes may contribute to the adaptation defect that we have observed. Further investigation is required to elucidate the specific role of each gene in bile adaptation.
Genes that provide protection in bile stress also protect against acid stress.
We identified three genes in L. reuteri that were induced by bile stress and that significantly reduced their ability to survive bile shock when disrupted. Two of these proteins have recently been shown to be induced by a strong reduction in pH and are necessary for increased survival at low pH (42). Both lr1864 (clpL) and lr1516 (putative esterase) play a role in surviving the initial shock of acid and bile stress.
Indeed, nearly one-third of the genes Wall et al. found to be differentially regulated under acid stress conditions were also altered under bile stress conditions (Table 6) (42). This indicates that once cells experience acid stress in the stomach, many of the important pathways for dealing with bile stress in the small intestine will already be activated.
TABLE 6.
Genes overexpressed or underexpressed during both 15 min of bile exposure and 15 min of acid stressa
| Gene | GenBank accession no. | Annotation |
|---|---|---|
| Overexpressed | ||
| lr0597 | DQ219952 | Thioredoxin domain-containing protein |
| lr0922 | DQ074860 | Extracellular hydrolase |
| lr1139 | AY970988 | Conserved intracellular protein of unknown function |
| lr1191 | DQ219999 | Conserved membrane protein of unknown function |
| lr1468 | DQ219968 | Putative transcriptional regulator |
| lr1515 | DQ074905 | Unknown extracellular protein |
| lr1516 | DQ219970 | Putative esterase |
| lr1797 | DQ219975 | Phosphatidylglycerolphosphatase A and related proteins |
| lr1864 | DQ219976 | ClpL ATPase with chaperone activity |
| lr1937 | DQ219979 | Conserved intracellular protein of unknown function |
| lr1993 | DQ219980 | Putative transcriptional regulator |
| lr2045 | DQ219981 | Phage-associated protein |
| Underexpressed | ||
| lr0190 | DQ219995 | Transcriptional regulator |
| lr0195 | DQ219996 | Putative 5-formyltetrahydrofolate cyclo-ligase |
| lr0382 | AY971000 | Putative branched-chain amino acid transport protein |
| lr0733 | DQ219954 | Conserved intracellular protein of unknown function |
| lr0862 | DQ219957 | Asp-tRNAAsn/Glu-tRNAGln amidotransferase C subunit |
| lr1240 | DQ219962 | Recombinational DNA repair ATPase |
| lr1297 | DQ219963 | Thymidine kinase |
| lr1432 | DQ220005 | Ribosomal protein S1 |
| lr1434 | DQ074902 | Unknown extracellular protein |
| lr1628 | DQ219972 | Conserved intracellular protein, MarZ, of unknown function |
Genes were overexpressed or underexpressed during both 15 min of bile exposure (0.5% oxgall) and 15 min of acid stress (pH 2.7) (42).
Comparison of multiple genomic studies of bile stress in lactic acid bacteria.
Multiple genomic studies have now been completed that identified genes important for bile tolerance in different species of Lactobacillus and Enterococcus (5, 6, 25, 35). Although different culture conditions, types of bile, and species were used for these studies, which resulted in a limited overlap in the genes identified in these studies, there are some common themes that have emerged. In both Lactobacillus plantarum and Lactobacillus acidophilus, several genes involved in the reorganization of the cell envelope were induced. Thus, dealing with membrane stress is a common theme that has emerged from these three studies. In addition, the operon containing lr1584 (multidrug resistance transporter) and lr1582 (unknown function) is also conserved as an operon in L. acidophilus and Enterococcus faecalis. Interestingly, both homologs in E. faecalis and L. acidophilus are also overexpressed when cells are exposed to bile, indicating the function of this operon in bile adaptation may be conserved in other lactic acid bacteria (25, 35). Lastly, clp proteases were identified in L. acidophilus as being upregulated by bile stress, as we found with clpL in our study, further supporting that protein denaturation is one stress being encountered by bile-treated cells.
One common finding that is not easily explained is the reduction in gene expression of recF, which encodes a protein that participates in the repair of DNA damage during active DNA replication. Since bile has been implicated in generating DNA damage, on the surface it seems that a reduction in the expression of RecF would not be productive. However, recent evidence indicates that RecF is predominantly utilized in DNA repair at replication forks during active growth. The reduction in recF expression may simply indicate a reduction in growth rate in the presence of bile.
Although there were significant similarities in the transcriptional profiles between the E. faecalis, L. acidophilus, L. plantarum, and L. reuteri responses to bile treatment, overall there was much more discordance in the data than similarities. The lack of concordance may be due to the different physiological strategies utilized by these organisms to adapt to bile stress. In addition, the differences in experimental strategy (the type of bile used, the way bile was administered, and for how long) likely also played a significant role in the differences that were noted. Bron et al. exposed L. plantarum to 0.1% porcine bile on plates and looked at the response to 3 days of exposure, while Pfeiler et al. conducted their L. acidophilus experiments in liquid media containing 0.5% oxgall with 30 min of exposure (6, 25). The use of purified bile acids or different sources of bile will also have different effects on cell physiology (3).
Bile has been implicated as a potential signaling molecule that would indicate to a bacterium that it had entered the small intestine. Such a signal could serve to stimulate the organism to adapt its physiology to optimize growth and survival in the GI tract. Several candidates from these experiments have now been identified in vitro. Future work in relevant animal models will determine if the strategies uncovered here are important for survival in vivo.
Supplementary Material
Acknowledgments
We thank Eamonn Connolly for helpful discussions and access to the L. reuteri ATCC 5730 genome and Eric Hufner and Christian Hertel for assistance with generating mutants. We also thank Jeff Landgraf in the Research Technology Support Facility at Michigan State University for assistance with microarray production.
This work was supported in part by funding support to R.A.B. from the Michigan State Center for Microbial Pathogenesis and the Rackham Foundation.
Footnotes
Published ahead of print on 1 February 2008.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Båth, K., S. Roos, T. Wall, and H. Jonsson. 2005. The cell surface of Lactobacillus reuteri ATCC 55730 highlighted by identification of 126 extracellular proteins from the genome sequence. FEMS Microbiol. Lett. 253:75-82. [DOI] [PubMed] [Google Scholar]
- 2.Begley, M., C. G. M. Gahan, and C. Hill. 2002. Bile stress response in Listeria monocytogenes LO28: adaptation, cross-protection, and identification of genetic loci involved in bile resistance. Appl. Environ. Microbiol. 68:6005-6012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Begley, M., C. G. Gahan, and C. Hill. 2005. The interaction between bacteria and bile. FEMS Microbiol. Rev. 29:625-651. [DOI] [PubMed] [Google Scholar]
- 4.Britton, R. A., P. Eichenberger, J. E. Gonzalez-Pastor, P. Fawcett, R. Monson, R. Losick, and A. D. Grossman. 2002. Genome-wide analysis of the stationary-phase sigma factor (sigma-H) regulon of Bacillus subtilis. J. Bacteriol. 184:4881-4890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bron, P. A., M. Marco, S. M. Hoffer, E. Van Mullekom, W. M. de Vos, and M. Kleerebezem. 2004. Genetic characterization of the bile salt response in Lactobacillus plantarum and analysis of responsive promoters in vitro and in situ in the gastrointestinal tract. J. Bacteriol. 186:7829-7835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bron, P. A., D. Molenaar, W. M. de Vos, and M. Kleerebezem. 2006. DNA micro-array-based identification of bile-responsive genes in Lactobacillus plantarum. J. Appl. Microbiol. 100:728-738. [DOI] [PubMed] [Google Scholar]
- 7.Casas, I., and W. Dobrogosz. 2000. Validation of the probiotic concept: Lactobacillus reuteri confers broad-spectrum protection against disease in humans and animals. Microb. Ecol. Health Dis. 12:247-285. [Google Scholar]
- 8.Derré, I., G. Rapoport, K. Devine, M. Rose, and T. Msadek. 1999. ClpE, a novel type of HSP100 ATPase, is part of the CtsR heat shock regulon of Bacillus subtilis. Mol. Microbiol. 32:581-593. [DOI] [PubMed] [Google Scholar]
- 9.Dunne, C., L. Murphy, S. Flynn, L. O'Mahony, S. O'Halloran, M. Feeney, D. Morrissey, G. Thornton, G. Fitzgerald, C. Daly, B. Kiely, E. M. Quigley, G. C. O'Sullivan, F. Shanahan, and J. K. Collins. 1999. Probiotics: from myth to reality. Demonstration of functionality in animal models of disease and in human clinical trials. Antonie Leeuwenhoek 76:279-292. [PubMed] [Google Scholar]
- 10.FAO. 2001. Health and nutritional properties of probiotics in food including powder milk with live lactic acid bacteria: report of a joint FAO/WHO expert consultation on evaluation of health and nutritional properties of probiotics in food including powder milk with live lactic acid bacteria. Food and Agriculture Organization, Rome, Italy.
- 11.Frees, D., K. Savijoki, P. Varmanen, and H. Ingmer. 2007. Clp ATPases and ClpP proteolytic complexes regulate vital biological processes in low GC, Gram-positive bacteria. Mol. Microbiol. 63:1285-1295. [DOI] [PubMed] [Google Scholar]
- 12.Giard, J.-C., A. Rince, H. Capiaux, Y. Auffray, and A. Hartke. 2000. Inactivation of the stress- and starvation-inducible gls24 operon has a pleiotrophic effect on cell morphology, stress sensitivity, and gene expression in Enterococcus faecalis. J. Bacteriol. 182:4512-4520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gunn, J. S. 2000. Mechanisms of bacterial resistance and response to bile. Microbes Infect. 2:907-913. [DOI] [PubMed] [Google Scholar]
- 14.Hofman, A. 1998. Bile secretion and the enterohepatic circulation of bile acids, p. 937-948. In M. Feldman, B. Scharschmidt, and M. Sleisenger (ed.), Sleisengers and Fordtran's gastrointestinal and liver disease, 6th ed. W. B. Saunders, Co., Philadelphia, PA.
- 15.Law, J., G. Buist, A. Haandrikman, J. Kok, G. Venema, and K. Leenhouts. 1995. A system to generate chromosomal mutations in Lactococcus lactis which allows fast analysis of targeted genes. J. Bacteriol. 177:7011-7018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Le Breton, Y., A. Mazé, A. Hartke, S. Lemarinier, Y. Auffray, and A. Rincé. 2002. Isolation and characterization of bile salts-sensitive mutants of Enterococcus faecalis. Curr. Microbiol. 45:434-439. [DOI] [PubMed] [Google Scholar]
- 17.Leverrier, P., D. Dimova, V. Pichereau, Y. Auffray, P. Boyaval, and G. Jan. 2003. Susceptibility and adaptive response to bile salts in Propionibacterium freudenreichii: physiological and proteomic analysis. Appl. Environ. Microbiol. 69:3809-3818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin, J., O. Sahin, L. O. Michel, and Q. Zhang. 2003. Critical role of multidrug efflux pump CmeABC in bile resistance and in vivo colonization of Campylobacter jejuni. Infect. Immun. 71:4250-4259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maguin, E., P. Duwat, T. Hege, D. Ehrlich, and A. Gruss. 1992. New thermosensitive plasmid for gram-positive bacteria. J. Bacteriol. 174:5633-5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marchler-Bauer, A., and S. H. Bryant. 2004. CD-Search: protein domain annotations on the fly. Nucleic Acids Res. 32:W327-W331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marquis, R. E., G. R. Bender, D. R. Murray, and A. Wong. 1987. Arginine deiminase system and bacterial adaptation to acid environments. Appl. Environ. Microbiol. 53:198-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martinez, A., and R. Kolter. 1997. Protection of DNA during oxidative stress by the nonspecific DNA-binding protein Dps. J. Bacteriol. 179:5188-5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Metchnikoff, E. 1907. Lactic acid as inhibiting intestinal putrefaction, p. 161-183. In The prolongation of life: optimistic studies. W. Heinemann, London, United Kingdom.
- 24.Nair, S., and S. E. Finkel. 2004. Dps protects cells against multiple stresses during stationary phase. J. Bacteriol. 186:4192-4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pfeiler, E. A., M. A. Azcarate-Peril, and T. R. Klaenhammer. 2007. Characterization of a novel bile-inducible operon encoding a two-component regulatory system in Lactobacillus acidophilus. J. Bacteriol. 189:4624-4634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reid, G. 1999. The scientific basis for probiotic strains of Lactobacillus. Appl. Environ. Microbiol. 65:3763-3766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reuter, G. 2001. The Lactobacillus and Bifidobacterium microflora of the human intestine: composition and succession. Curr. Issues Intest. Microbiol. 2:43-53. [PubMed] [Google Scholar]
- 28.Rolfe, R. D. 2000. The role of probiotic cultures in the control of gastrointestinal health. J. Nutr. 130:396S-402S. [DOI] [PubMed] [Google Scholar]
- 29.Russell, W. M., and T. R. Klaenhammer. 2001. Efficient system for directed integration into the Lactobacillus acidophilus and Lactobacillus gasseri chromosomes via homologous recombination. Appl. Environ. Microbiol. 67:4361-4364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Savino, F., E. Pelle, E. Palumeri, R. Oggero, and R. Miniero. 2007. Lactobacillus reuteri (American Type Culture Collection strain 55730) versus Simethicone in the treatment of infantile colic: a prospective randomized study. Pediatrics 119:e124-e130. [DOI] [PubMed] [Google Scholar]
- 31.Saxelin, M., S. Tynkkynen, T. Mattila-Sandholm, and W. M. de Vos. 2005. Probiotic and other functional microbes: from markets to mechanisms. Curr. Opin. Biotechnol. 16:204-211. [DOI] [PubMed] [Google Scholar]
- 32.Schumann, W., M. Hecker, and T. Msadek. 2002. Regulation and function of heat-inducible genes in Bacillus subtilis, p. 359-368. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to cells. ASM Press, Washington, DC.
- 33.Simons, L. A., S. G. Amansec, and P. Conway. 2006. Effect of Lactobacillus fermentum on serum lipids in subjects with elevated serum cholesterol. Nutr. Metabol. Cardiovasc. Dis. 16:531-535. [DOI] [PubMed] [Google Scholar]
- 34.Sleator, R. D., H. H. Wemekamp-Kamphuis, C. G. Gahan, T. Abee, and C. Hill. 2005. A PrfA-regulated bile exclusion system (BilE) is a novel virulence factor in Listeria monocytogenes. Mol. Microbiol. 55:1183-1195. [DOI] [PubMed] [Google Scholar]
- 35.Solheim, M., Å. Aakra, H. Vebo, L. Snipen, and I. F. Nes. 2007. Transcriptional responses of Enterococcus faecalis V583 to bovine bile and sodium dodecyl sulfate. Appl. Environ. Microbiol. 73:5767-5774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Talarico, T. L., I. A. Casas, T. C. Chung, and W. J. Dobrogosz. 1988. Production and isolation of reuterin, a growth inhibitor produced by Lactobacillus reuteri. Antimicrob. Agents Chemother. 32:1854-1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thanassi, D. G., L. W. Cheng, and H. Nikaido. 1997. Active efflux of bile salts by Escherichia coli. J. Bacteriol. 179:2512-2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tubelius, P., V. Stan, and A. Zachrisson. 2005. Increasing work-place healthiness with the probiotic Lactobacillus reuteri: a randomised, double-blind placebo-controlled study. Environ. Health 4:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Uicker, W. C., L. Schaefer, and R. A. Britton. 2006. The essential GTPase RbgA (YlqF) is required for 50S ribosome assembly in Bacillus subtilis. Mol. Microbiol. 59:528-540. [DOI] [PubMed] [Google Scholar]
- 40.Usman and A. Hosono. 2000. Effect of administration of Lactobacillus gasseri on serum lipids and fecal steroids in hypercholesterolemic rats. J. Dairy Sci. 83:1705-1711. [DOI] [PubMed] [Google Scholar]
- 41.Valeur, N., P. Engel, N. Carbajal, E. Connolly, and K. Ladefoged. 2004. Colonization and immunomodulation by Lactobacillus reuteri ATCC 55730 in the human gastrointestinal tract. Appl. Environ. Microbiol. 70:1176-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wall, T., K. Bath, R. A. Britton, H. Jonsson, J. Versalovic, and S. Roos. 2007. The early response to acid shock in Lactobacillus reuteri involves the ClpL chaperone and a putative cell wall-altering esterase. Appl. Environ. Microbiol. 73:3924-3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wall, T., S. Roos, K. Jacobsson, A. Rosander, and H. Jonsson. 2003. Phage display reveals 52 novel extracellular and transmembrane proteins from Lactobacillus reuteri DSM 20016(T). Microbiology 149:3493-3505. [DOI] [PubMed] [Google Scholar]
- 44.Walter, J., P. Chagnaud, G. W. Tannock, D. M. Loach, F. Dal Bello, H. F. Jenkinson, W. P. Hammes, and C. Hertel. 2005. A high-molecular-mass surface protein (Lsp) and methionine sulfoxide reductase B (MsrB) contribute to the ecological performance of Lactobacillus reuteri in the murine gut. Appl. Environ. Microbiol. 71:979-986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weizman, Z., G. Asli, and A. Alsheikh. 2005. Effect of a probiotic infant formula on infections in child care centers: comparison of two probiotic agents. Pediatrics 115:5-9. [DOI] [PubMed] [Google Scholar]
- 46.Wenus, C., R. Goll, E. B. Loken, A. S. Biong, D. S. Halvorsen, and J. Florholmen. 14 March 2007, posting date. Prevention of antibiotic-associated diarrhoea by a fermented probiotic milk drink. Eur. J. Clin. Nutr. doi: 10.1038/sj.ejcn.1602718. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.