Abstract
Comparative analysis of related biosynthetic gene clusters can provide new insights into the versatility of these pathways and allow the discovery of new natural products. The freshwater cyanobacterium Microcystis aeruginosa NIES298 produces the cytotoxic peptide microcyclamide. Here, we provide evidence that the cyclic hexapeptide is formed by a ribosomal pathway through the activity of a set of processing enzymes closely resembling those recently shown to be involved in patellamide biosynthesis in cyanobacterial symbionts of ascidians. Besides two subtilisin-type proteases and a heterocyclization enzyme, the gene cluster discovered in strain NIES298 encodes six further open reading frames, two of them without similarity to enzymes encoded by the patellamide gene cluster. Analyses of genomic data of a second cyanobacterial strain, M. aeruginosa PCC 7806, guided the discovery and structural elucidation of two novel peptides of the microcyclamide family. The identification of the microcyclamide biosynthetic genes provided an avenue by which to study the regulation of peptide synthesis at the transcriptional level. The precursor genes were strongly and constitutively expressed throughout the growth phase, excluding the autoinduction of these peptides, as has been observed for several peptide pheromone families in bacteria.
Cyanobacteria are known as a prolific source of secondary metabolites exhibiting unique structural features and biological activities. Various cyanobacterial compounds (e.g., curacin A, aeruginosin, and cryptophycin) have great potential for drug development (33, 36). However, due to their adverse effects on higher organisms, several cyanobacterial metabolites are regarded as health-threatening toxins and have caused serious concern among water authorities worldwide. A majority of these compounds, in particular, those that were isolated from planktonic freshwater cyanobacteria belonging to the genera Microcystis, Planktothrix, Nostoc, and Anabaena, can be classified as peptides or possess peptidic substructures often comprising highly modified amino acid moieties. Several of these peptides, such as the hepatotoxin microcystin or the protease inhibitors aeruginosin and anabaenopeptolide, were shown to be produced by nonribosomal peptide synthetase assembly lines (13, 27, 34).
The recent characterization of the patellamide biosynthetic pathway from the as-yet-nonculturable cyanobacterial symbiont of the ascidian Lissoclinum patella has revealed that there are biosynthetic pathways independent from nonribosomal peptide synthetase systems capable of producing modified and cyclic peptide structures in cyanobacteria (29). The patellamide family of cyclic pseudosymmetrical octapeptides is characterized by the presence of thiazole and oxazole moieties. Although nonribosomal biosynthesis was anticipated for the formation of these peptides, heterologous expression of a microcin-like gene cluster discovered in the genome of the symbiotic cyanobacterium Prochloron didemni unambiguously showed that these peptides are produced by a ribosomal pathway (20, 29). In a more recent study, the patellamide biosynthetic pathway could be used as a template for the design of a highly flexible expression platform for the production of libraries of cyclic peptides (7). This experimental breakthrough has revealed the large biotechnological potential of cyanobacterial natural product biosyntheses. An increasing number of further cyclic peptides containing heterocyclic amino acids have recently been isolated from other symbiotic and free-living cyanobacteria, including nostocyclamide (16), tenuecyclamide (1), venturamides (19), dendroamides (25), and microcyclamide (14). The variety of structures is reflected in an equally large variety of bioactivities, such as antibacterial, cytotoxic, and antimalarial activities. Moreover, the investigation of a patellamide-like gene cluster in the bloom-forming organism Trichodesmium erythraeum has guided the discovery of the cyclic heptapeptide trichamide (31).
Small oligopeptides synthesized from ribosomal prepeptides via peptide maturation are also known from bacteria distantly related to cyanobacteria. Two well-studied groups of peptides are the microcins of Escherichia coli and the lanthionine-containing lantibiotics of gram-positive bacteria. Members of both groups exhibit antibiotic activities against other bacteria and were shown to increase the permeability of cell membranes and to inhibit DNA gyrases and RNA polymerases (3, 15). Lantibiotics such as nisin are also used for food preservation (5). Particular microcins share some structural properties with patellamides, such as the cyclic microcin J25 (2) and heterocycle-containing microcin B17 (24). Besides their role in defense against other bacteria, lantibiotics of gram-positive bacteria were shown to play a role in the cell-cell signaling of bacteria (17).
Although the discovery of patellamides has provided first insights into the biosynthesis of microcin-like peptides in cyanobacteria and the distribution and versatility of such peptides in cyanobacteria, their biological function and evolution are still under debate. Investigation of these questions is limited by the number of biosynthetic systems analyzed.
This study therefore aimed to elucidate the molecular basis of the biosynthesis of cyclic hexapeptides of the patellamide family, the microcyclamides produced by the planktonic freshwater genus Microcystis. The analysis could not only provide new lessons on microcin-like biosyntheses in cyanobacteria and the transcriptional dynamics of their genes but could also facilitate the precursor-guided discovery and structural elucidation of a cryptic microcyclamide-like compound.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Microcystis aeruginosa strains PCC 7806 and NIES298 were obtained from the Pasteur Culture Collection of Cyanobacteria (Institut Pasteur, Paris, France) and the National Institute of Environmental Studies (Tsukuba, Japan), respectively. The cyanobacteria were cultivated in BG-11 (26) or Z8 medium (18) under continuous light at 30 μmol photons m−2 s−1 with continuous shaking at 40 rpm at 23°C. For the light experiment, the cells were grown at 18 μmol photons m−2 s−1 until they reached the required cell densities (optical densities at 750 nm of 0.3, 0.5, and 1.0) and then exposed for 2 h to different light intensities as follows: 0 μmol photons m−2 s−1, dark (D); 18 μmol photons m−2 s−1, low light (L); 68 μmol photons m−2 s−1, high light (H); 180 μmol photons m−2 s−1, very high light (VH). Light intensities were measured with a Li-Cor LI250 light meter (Walz, Effeltrich, Germany). Mass cultivation of M. aeruginosa PCC 7806 for purification of microcyclamides was performed with BG-11 medium in 20-liter flasks under continuous aeration. E. coli strain EPI300-T1R was used for fosmid library construction and cultivated according to the suggestions of the manufacturer (Epicenter Technologies, Madison, WI).
Fosmid library construction and screening.
Genomic DNA was prepared from M. aeruginosa PCC 7806 and NIES298 as reported previously (8). DNA fragments of approximately 30 to 40 kb were directly ligated to the pCC1FOS vector (Epicenter Technologies, Madison, WI) by following the manufacturer's instructions. Screening of the library was performed by hybridization under standard conditions (28). The mcaA gene fragment used as a probe was obtained by PCR with primers mcaAdegFw (5′-TTYGGNACYGAAGCNCGNGG-3′) and mcaAdegRv (5′-AGAAGACCAAGAACGAACTTGGCC-3′) at an annealing temperature of 55°C under standard conditions. The DNA probe was radioactively labeled with the HexaLabel kit (Fermentas, St. Leon-Rot, Germany). Colony hybridization was performed in hybridization buffer containing 50% formamide at 42°C according to standard protocols (28).
DNA sequencing and sequence analysis.
DNA sequencing by a primer-walking strategy was performed by SMB GbR (Berlin, Germany) with the Big Dye Terminator cycle sequencing kit (ABI, Foster City, CA). The assembly was performed with the software package Vector NTI (Invitrogen, Karlsruhe, Germany). Analyses of DNA sequences and deduced protein sequences were performed with the NCBI (National Institute for Biotechnology Information, Bethesda, MD) BLAST server (http://www.ncbi.nlm.nih.gov/BLAST/).
RNA isolation and Northern blot analysis.
Cells were harvested by centrifugation at 4°C and homogenized in liquid nitrogen with a mortar and pestle. RNAs were isolated with the Trizol kit (Invitrogen, Karlsruhe, Germany) according to the manufacturer's instructions. DNA blotting and DNA-RNA hybridization were performed as previously described (28).
Extraction and purification of microcyclamides.
Freeze-dried cyanobacterial cells (24.36 g from ca. 90 liters of culture) were extracted with 80% (vol/vol) methanol (MeOH; 960 ml × 2) and MeOH (960 ml). The combined 80% (vol/vol) MeOH and MeOH extracts were concentrated in vacuo. Dried residue was extracted with diethyl ether and H2O. The aqueous layer was concentrated and then extracted with 1-butanol (n-BuOH). The n-BuOH layer was subjected to octyldecyl silane chromatography (YMC GEL ODS A 60-S50, 5 by 28 cm) with aqueous acetonitrile (20% [vol/vol], 40% [vol/vol], and 80% [vol/vol]), MeOH, and CH2Cl2. The 40% and 80% acetonitrile fractions were subjected to reversed-phase column high-performance liquid chromatography (RP-HPLC; Nucleosil 100-5 C18, 21 by 250 mm; 25 to 83% [vol/vol] aqueous acetonitrile containing 0.1% [vol/vol] trifluoroacetic acid [TFA]; flow rate, 10 ml/min; UV detection at 220 nm). Crude fractions containing peptides 1 and 2 were resubjected to RP-HPLC (Nucleosil 100-5 C18, 21 by 250 mm; 25 to 83% [vol/vol] aqueous acetonitrile containing 0.1% [vol/vol] TFA; flow rate, 10 ml/min; UV detection at 220 nm) to yield microcyclamide 7806A (peptide 1; 6.8 mg) and microcyclamide 7806B (peptide 2; 2.1 mg).
Spectroscopic techniques.
The UV spectra were recorded on a Specord 200 spectrophotometer. Optical rotations were measured on a JASCO P-1020 polarimeter. Infrared (IR) spectra were measured on a JASCO FT/IR-4100 spectrometer. 1H and 13C nuclear magnetic resonance (NMR) spectra were obtained with Bruker Avance DPX 300 and DRX 500 in dimethyl sulfoxide-d6 at 30°C. The high-resolution electron spray ionization mass spectra (HR-ESI-MS) were measured on a Finnigan TSQ Quantum Ultra AM.
Ozonolysis and acid hydrolysate.
Microcyclamides 7806A and 7806B (each 0.1 mg) were dissolved in CH2Cl2 (3 ml) and ozonized at −78°C for 20 min, respectively. After removal of the solvent in vacuo, the resulting residue was suspended in 6 M HCl and heated at 105°C overnight. To the reaction residue, which was removed from the solvent by reduced pressure, 1 M NaHCO3 (100 μl) and 1-fluoro-2,4-dinitrophenyl-5-l-alanine-amide (l-FDAA; 50 μl of 10 mg ml−1 in acetone) were added and the reaction mixture was incubated at 50°C for 1 h. The reaction mixture was quenched by 2 N HCl (50 μl) and diluted with 50% (vol/vol) acetonitrile (200 μl). l-FDAA derivatives were analyzed by RP-HPLC with Cosmosil 5 C18-MS, 4.6 by 250 mm, a flow rate of 1 ml min−1, UV detection at 340 nm, and a gradient time program of buffer A (0.1% [vol/vol] TFA) and buffer B (100% acetonitrile) of buffer A/B at 80:20 to buffer A/B at 65:35 in 30 min to buffer A/B at 45:55 in 20 min to buffer A/B at 0.5:99.5 in 1 min. The retention times (minutes) of standard amino acids were as follows: l-Thr/allo Thr, 16.61; d-allo Thr, 18.90; d-Thr, 21.54; l-Ala, 23.33; d-Ala, 28.09; l-Val, 33.82; d-Val, 40.61; l-Ile/allo Ile, 39.84; d-Ile/allo Ile, 45.50; peptide 1, l-Thr/allo Thr, 16.61; l-Ala, 23.34; d-Ala, 28.14; l-Val, 33.86; d-Ile/allo Ile, 45.50; peptide 2, l-Thr/allo Thr, 16.59; l-Ala, 23.32; l-Val, 33.85; d-Ile/allo Ile, 45.62. To separate the l-FDAA-Thr derivatives, different RP-HPLC conditions, consisting of Cosmosil 5 C18-MS (4.6 by 250 mm), a 1-ml-min−1 flow rate, UV detection at 340 nm, and a gradient time program of buffer A (0.1% [vol/vol] TFA) and buffer B (100% acetonitrile) of buffer A/B at 80:20 for 5 min to buffer A/B at 70:30 in 10 min to buffer A/B at 0.5:99.5 in 5 min, were used. The retention times (minutes) of standard amino acids were as follows: l-Thr, 17.68; l-allo Thr, 17.92; d-allo Thr, 19.22; d-Thr, 20.66; peptide 1, l-Thr, 17.66; peptide 2, l-Thr, 17.62. To separate the l-FDAA-Ile derivatives, different RP-HPLC conditions were used, i.e., Phenomenex Synergi 4 μ Fusion-RP80, 4.6 by 250 mm, a flow rate of 1 ml min−1, UV detection at 340 nm, and 60% (vol/vol) buffer A (0.1% [vol/vol] TFA)-40% (vol/vol) buffer B (100% acetonitrile). The retention times (minutes) of standard amino acids were as follows: l-allo Ile, 16.70; l-Ile, 17.37; d-allo Ile, 24.05; d-Ile, 24.69; peptide 1, d-Ile, 24.43; peptide 2, d-Ile, 24.53.
Microcyclamide 7806A (peptide 1).
Microcyclamide 7806A is a colorless, amorphous powder with the following characteristics: UV (MeOH) λmax, 204 (ɛ, 20,900) nm; [α]20D, −43.1° (c 0.1, MeOH); IR (ATR), 2,965, 1,748, 1,661, 1,597, 1,543, 1,301, 1,198, 1,135, 1,029, 833, and 797 cm−1; HR-ESI-MS (positive), m/z 517.2228 [M+H]+ (calculated for C24H33O5N6S, 517.2233).
Microcyclamide 7806B (peptide 2).
Microcyclamide 7806B is a colorless, amorphous powder with the following characteristics: UV (MeOH) λmax, 204 (ɛ, 25,500) nm; [α]20D, −120.4° (c 0.1, MeOH); IR (ATR), 2,967, 1,780, 1,652, 1,599, 1,514, 1,378, 1,206, 1,161, 1,104, 924, 888, and 797 cm−1; HR-ESI-MS (positive), m/z 535.2318 [M+H]+ (calculated for C24H35O6N6S, 535.2339).
Nucleotide sequence accession number.
The sequence of the mca gene cluster and flanking regions has been deposited in the EMBL database under accession number AM774406.
RESULTS AND DISCUSSION
Identification of the microcyclamide biosynthesis genes.
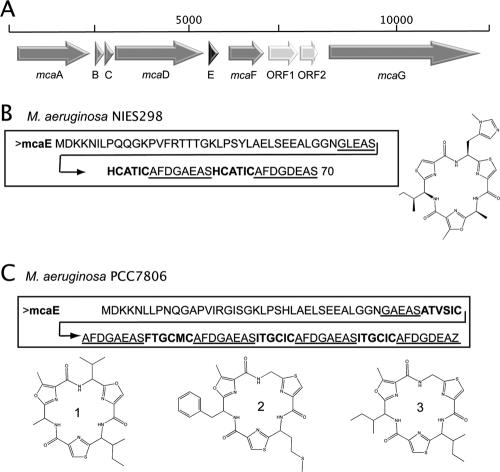
The striking structural similarity of microcyclamide to patellamides (14) prompted us to search for a patellamide-like pathway in the microcyclamide-producing strain M. aeruginosa NIES298. To detect orthologous genes in strain NIES298, we therefore designed a set of degenerated primers based on the patA sequence from P. didemni (29) and the corresponding triA sequence from Trichodesmium erythraeum ISM101 (31). As a result, we obtained a PCR product exhibiting 79% and 68% identity to the patA and triA genes, respectively. In order to determine the entire biosynthetic gene cluster in Microcystis, a genomic fosmid library of M. aeruginosa NIES298 was constructed and hybridized with the Microcystis patA-like PCR product as a probe. Sequencing of the positive clones led to the identification of a patellamide-like gene cluster consisting of nine genes (Fig. 1A). The putative precursor protein encoded by the cluster comprised two copies of the amino acid sequence HCATIC, as expected from the composition and order of the amino acids in the microcyclamide molecule (Fig. 1B). The cluster could therefore unambiguously be assigned to microcyclamide biosynthesis, and the genes were designated mca. Seven of the mca genes resemble those of the patellamide biosynthesis cluster, although the top-scoring BLAST hits for five of these genes were annotated as hypothetical proteins in the genome of the mat-forming cyanobacterium Lyngbya sp. strain PCC 8106 (Table 1). The mca gene cluster is flanked by two genes encoding a putative transposase and a restriction-modification enzyme (not shown).
FIG. 1.
Microcyclamide gene cluster in M. aeruginosa. (A) Schematic representation of the mca biosynthetic gene cluster in strains NIES298 and PCC 7806. Genes with similarity to patellamide biosynthesis genes in P. didemni are in gray. The precursor protein is highlighted in black. The light gray arrows represent ORFs that could not directly be assigned to microcyclamide biosynthesis. (B) McaE sequence of M. aeruginosa NIES298. The bold sequences show the product-coding sequences. The proposed start and stop cyclization sequences are underlined. The structure of microcyclamide from NIES298 is shown on the right. (C) McaE sequence. The bold sequences show the product-coding sequences. The proposed start and stop cyclization sequences are underlined. Predicted microcyclamide structures in PCC 7806 are shown below.
TABLE 1.
Deduced functions of ORFs in the microcyclamide biosynthetic gene clusters
| Protein and strain | No. of amino acids | Protein identification no. | Deduced function | Protein with similar sequence | % Identity/similarity (no. of amino acids) |
|---|---|---|---|---|---|
| McaA | |||||
| Nies298 | 657 | CAO82081 | Subtilisin-like protease | PatA; Prochloron didemni | 68/78 (702) |
| PCC 7806 | 657 | CAP64335 | 69/78 (702) | ||
| McaB | |||||
| Nies298 | 83 | CAO82082 | Conserved hypothetical protein | PatB; Prochloron didemni | 72/79 (83) |
| PCC 7806 | 83 | CAP64336 | 69/78 (83) | ||
| McaC | |||||
| Nies298 | 80 | CAO82083 | Conserved hypothetical protein | PatC; Prochloron didemni | 53/67 (70) |
| PCC 7806 | 80 | CAP64337 | 50/65 (72) | ||
| McaD | |||||
| Nies298 | 776 | CAO82084 | Adenylation/heterocyclization | PatD; Prochloron didemni | 77/86 (785) |
| PCC 7806 | 776 | CAP64338 | 77/86 (785) | ||
| McaE | |||||
| Nies298 | 74 | CAO82085 | Microcyclamide precursor protein | Patellamide precursor protein; Prochloron didemni | 68/78 (69) |
| PCC 7806 | 97 | CAP64339 | 60/74 (70) | ||
| McaF | |||||
| Nies298 | 321 | CAO82086 | Conserved hypothetical protein | PatF; Prochloron didemni | 52/69 (311) |
| PCC 7806 | 321 | CAP74572 | 54/70 (312) | ||
| McaG | |||||
| Nies298 | 1351 | CAO82089 | Thiazoline oxidase/subtilisin-like protease | PatG; Prochloron didemni | 71/81 (729) |
| PCC 7806 | 1313 | CAP64342 | 71/81 (729) | ||
| ORF1 | |||||
| Nies298 | 267 | CAO82087 | Unknown | Hypothetical; Lyngbya sp. strain PCC 8106 | 90/94 (267) |
| PCC 7806 | 267 | CAP64340 | 91/95 (267) | ||
| ORF2 | |||||
| Nies298 | 115 | CAO82088 | Unknown | PatG; Prochloron didemni | 54/70 (37) |
| PCC 7806 | 115 | CAP64341 | 54/70 (37) |
In silico analysis of the mca gene cluster of M. aeruginosa NIES298.
Table 1 summarizes the results of the analysis of the mca region (13 kb). The order of the mcaA to -G genes in M. aeruginosa NIES298 is congruent with the order of the patellamide biosynthesis genes in P. didemni. The deduced gene product of mcaA is 68% identical to PatA from P. didemni and encodes a multidomain enzyme comprising a putative subtilisin-like protease domain at the N terminus. No conserved motif was detected in the C-terminal part of McaA. Subtilisin proteases are frequently involved in the posttranslational tailoring of peptide pheromones in gram-positive bacteria (30, 35). Accordingly, the PatA protease was predicted to be responsible for the processing of the patellamide precursor peptide in P. didemni (29). McaA is expected to play a similar role in strain NIES298. McaB and McaC are 72% and 53% identical to PatB and PatC, respectively. Both proteins do not show significant similarity to characterized proteins in the database and were shown to be dispensable for patellamide biosynthesis in E. coli (7). A PatB homolog is nevertheless encoded in all patellamide-like gene clusters and may thus be related to the functional role of the cyclic peptides.
McaD is 77% identical to the PatD protein from P. didemni that comprises two domains. Like PatD, McaD shows low similarity to MccB from microcin biosynthesis (11) and to a possible hydrolase, SagD, from Streptomyces iniae (10). McaD is therefore proposed to be involved in the heterocyclization of cysteine, threonine, and/or serine into thiazoline and oxazoline rings. The putative microcyclamide precursor McaE partially differs from the corresponding ortholog in P. didemni (Table 1). The leader sequence of 49 amino acids and the proposed cyclization signals surrounding the peptide-coding region (GAEAS…AFD, Fig. 1) resemble the corresponding parts of the patellamide precursor. However, whereas the patellamide precursor in P. didemni encodes two different patellamides, two copies of the same peptide are encoded in M. aeruginosa NIES298. Another feature of the microcyclamide precursor peptide that diverges from the patellamide precursor is the presence of a double-glycine motif at the C-terminal part of the leader sequence, suggesting possible transport through the inner cell wall, since this motif is known to play a role in peptide translocation in different groups of bacteria (22).
McaF shares 52% identity with PatF from P. didemni. The protein was shown to be essential for patellamide biosynthesis but is not encoded in the trichamide gene cluster (7, 31). As the protein does not contain any conserved domain motifs, no functional role in microcyclamide biosynthesis could be deduced. A PatG homolog, McaG, is encoded at the 3′ end of the microcyclamide cluster. This putative protein shows similarity to McaA and to the subtilisin-like family of proteases. Furthermore, the N-terminal domain reveals characteristic features of NAD(P)H oxidoreductases. Similar to its homolog in P. didemni, the McaG protein is predicted to play a role in oxidizing thiazoline rings to the thiazole oxidation state in the patellamide biosynthetic pathway (29).
Two open reading frames (ORFs) present in the microcyclamide gene cluster are not part of the trichamide and patellamide gene clusters. The first one encodes a protein of 267 amino acids that shows no significant similarity to any in the database except a hypothetical protein encoded in the respective peptide gene cluster from Lyngbya. The second ORF encodes a small protein of 115 amino acids with partial similarity to McaG, suggesting that this gene could represent a pseudogene (Table 1).
The microcyclamide gene cluster does not code for a protein possessing methyltransferase domain signatures that could be responsible for the methylation of histidine in microcyclamide (Fig. 1B). Therefore, it remains unclear whether one of the uncharacterized enzymes in the gene cluster or factors encoded in trans provide the required methylation activity.
Taken together, a detailed analysis of the microcyclamide gene cluster in M. aeruginosa NIES298 has revealed the expected similarity but also clear differences from the recently described patellamide and trichamide gene clusters. Our work provides the first evidence for the biosynthesis of a cyclic hexapeptide from a ribosomal precursor in cyanobacteria.
Identification of a similar peptide gene cluster in M. aeruginosa PCC 7806.
Although M. aeruginosa NIES 298 is the only strain of the genus Microcystis that was previously shown to produce a microcyclamide-like structure, the genetic potential to produce this family of cyclic peptides may be far more widespread than expected. In order to test this hypothesis, the genome database (AM778843 to AM778958) of a second strain, M. aeruginosa PCC 7806, was searched for the presence of similar genes. Blastp analysis revealed a gene cluster encoding proteins with more than 90% identity to the nine proteins encoded in NIES298 (Fig. 1C and Table 1), suggesting the capability of strain PCC 7806 to produce a microcyclamide-like peptide. Detailed analysis of the precursor peptide McaE in strain PCC 7806 revealed an almost identical leader peptide and the same cyclization signals as in M. aeruginosa NIES298. However, the peptide-coding sequences differ. Whereas the precursor in NIES298 contains two peptide-coding regions, the corresponding protein in strain PCC 7806 encodes four (Fig. 1C). The entire sequence of the microcyclamide gene cluster of M. aeruginosa PCC 7806 is available under accession number AM931579. Using microcyclamide from NIES298 as a model, three different hexapeptides containing thiazole and oxazole rings could be predicted (Fig. 1C). The accurate prediction of chemical structures produced by cryptic pathways can guide the structure-based detection and elucidation of the corresponding compounds (12). Therefore, a reanalysis of the metabolite spectrum produced by M. aeruginosa PCC 7806 aimed at the detection of novel microcyclamide-like compounds was initiated.
Isolation of new microcyclamides and structural elucidation.
M. aeruginosa PCC 7806 is well known to produce microcystins (6) and cyanopeptolins (21). However, the analysis of the n-BuOH fraction from freeze-dried M. aeruginosa PCC 7806 cells by RP-HPLC revealed the presence of at least four different peptide types (see Fig. S1 in the supplemental material). Whereas microcystins and cyanopeptolins could be assigned on the basis of their specific UV maxima at 238 and 280 nm, respectively, two of the putative peptide peaks did not show any specific UV absorbance maxima but rather a spectrum similar to that of microcyclamide from M. aeruginosa NIES298 (14).
The molecular formula of microcyclamide 7806A (peptide 1) was established as C24H32O5N6S by the HR-ESI-MS and NMR spectral data (see Table S1 in the supplemental material). The 1H NMR spectrum supported the peptidic nature of peptide 1 from three doublet amide protons (δ 8.05, 8.21, and 8.32). The 1H-1H correlation spectroscopy spectrum suggested that peptide 1 contained an Ile-like unit (5-NH to H-9), a Val-like unit (14-NH to H-16/17), a Thr-like unit (H-20 to H-21), and an Ala-like unit (23-NH to H-24) (see Fig. S1 in the supplemental material). These units were further identified as thiazole-isoleucinyl, oxazole-valyl, and methyloxazoline-alanyl units on the basis of the heteronuclear multiple quantum correlation and heteronuclear multiple bond correlation (HMBC) (see Fig. S2 in the supplemental material). The sequence of the three units was determined by the HMBC correlations, i.e., H-5 to C-10, 14-NH to C-18, and 23-NH to C-1 (see Fig. S2 in the supplemental material). The amino acid sequence of peptide 1 could be deduced as ATVSIC, corresponding to the first precursor peptide of McaE of strain M. aeruginosa PCC 7806 (Fig. 2).
FIG. 2.
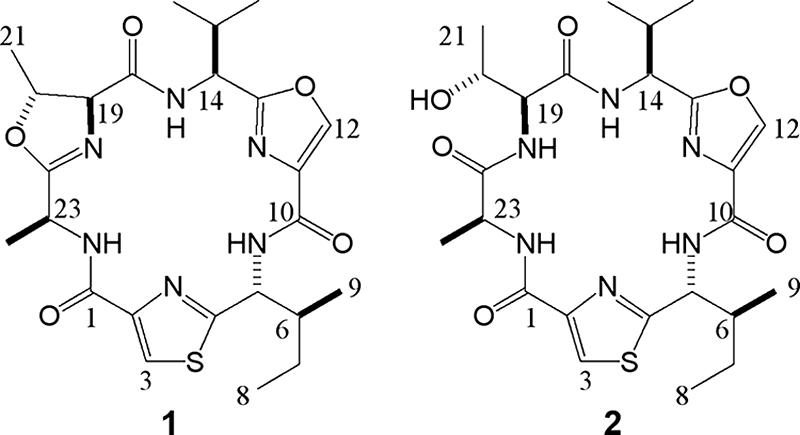
Structures of microcyclamides 7806A (peptide 1) and 7806B (peptide 2) from M. aeruginosa PCC 7806 as determined by NMR and MS analyses.
The structure of microcyclamide 7806B (peptide 2) exhibited a 1H NMR spectrum almost identical to that of peptide 1, except for low field protons (7.3 ppm to 9.3 ppm). Further inspection by means of HR-ESI-MS revealed that the molecular mass of peptide 2 was increased by 18 mmu (H2O) compared to that of peptide 1. The detailed analysis of the 13C NMR spectrum of peptide 2 indicated that C-18 (δ 169.8), C-19 (δ 59.6), C-20 (δ 66.8), C-21 (δ 20.5), and C-23 (δ 51.3) of peptide 2 were clearly different from C-18 (δ 166.2), C-19 (δ 55.0), C-20 (δ 69.7), C-21 (δ 17.1), and C-23 (δ 47.7) in the methyloxazoline-alanyl unit of peptide 1, providing evidence for the presence of a threonyl-alanyl unit in peptide 2. The 1H-1H correlation spectroscopy, heteronuclear multiple quantum correlation, and HMBC results supported the structure of peptide 2 as a variant of peptide 1 without threonine heterocyclization (Fig. 2; see Fig. S1 in the supplemental material).
The absolute stereochemistry of peptides 1 and 2 was determined by HPLC analysis of the l-FDAA (9) derivatives of the acid hydrolysates after ozonolysis of peptides 1 and 2. Detection of l-Thr and l-Val derivatives indicated that C-14, C-19, and C-20 were in the S, S, and R configurations, respectively. Although both l- and d-Ala (ca. 50/50) derivatives of peptide 1 were detected, only l-Ala was detected in peptide 2. It was concluded that d-Ala in peptide 1 could be the result of racemization during the process of ozonolysis or acid hydrolysis. Finally, d-Ile was detected in both derivatives and C-5 and C-6 were in the R and S configurations, respectively. In conclusion, peptide 1 was identified as cyclo-L-A-mOzl-L-V-Ozl-D-I-Tzl and peptide 2 was identified as cyclo-L-A-L-T-L-V-Ozl-D-I-Tzl (Fig. 2). Milne et al. (23) proposed that epimerization of single amino acids within patellamides occurs spontaneously and is interdependent with macrocyclization of the linear prepeptide. This may also explain the fact that different patellamide and cyclic hexapeptide variants were shown to differ in the stereochemistry of individual amino acids (1, 4, 14, 16). The amount of the two microcyclamides per unit of dry cell weight was estimated as 0.29% (wt/wt).
Both of the microcyclamide structures detected in M. aeruginosa PCC 7806 could be assigned to the first precursor of McaE. The fact that no microcyclamides corresponding to the other precursors comprised in McaE of PCC 7806 were found might be due to detection problems. Alternatively, the three remaining precursors of McaE of strain PCC 7806 are not correctly processed or unstable. The generation of mutants of the microcyclamide pathway that could have facilitated the identification of unknown microcyclamide variants failed in several independent experiments, probably because of the extensive restriction barrier of strains of the genus Microcystis (32).
Microcyclamide function.
Microcyclamide isolated from M. aeruginosa NIES298 exhibited moderate cytotoxic activity against P388 murine leukemia cells (14), suggesting a possible antibiotic function of the metabolites. To further confirm this assumption for a second Microcystis strain, the two microcyclamides identified in the course of this study were tested against HeLa cells. However, both peptides did not show any inhibitory activity. Microcyclamides 7806A and 7806B were also negative in standard antiproliferative, antibacterial, and antifungal assays (data not shown). These results argue against a general antibiotic function of microcyclamides. The different activities observed for individual cyclamide structures could be incidental and may not reflect the true biological function of the cyclic peptides (1, 14, 16, 19, 25). The striking diversity of the cyclic heptapeptide variants suggests that microcyclamides could potentially be involved in communication and self-recognition of Microcystis ecotypes. In order to get insights into a possible communication role for the cyclic peptides, we initiated transcriptional studies of microcyclamide biosynthesis genes. Several of the peptide bacteriocins of gram-positive bacteria like subtilin, nisin, and other lantibiotics are primarily produced in the late growth phase and act as autoinducers of their own biosynthetic genes and other target genes (17). To elucidate the expression profile of microcyclamide genes in M. aeruginosa NIES298, we performed DNA hybridizations with RNA prepared from cells incubated under different light intensities and collected at different growth phases (Fig. 3). Hybridization with an mcaE probe showed similar transcript levels, whatever the cell growth phase or light intensity. From these data, it seems unlikely that microcyclamides act as autoinducers of their biosynthesis genes, as this should have been reflected by cell density-dependent expression.
FIG. 3.
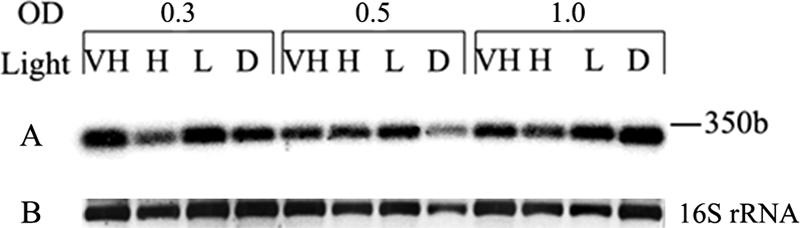
Transcription analysis of the mcaE precursor RNA from M. aeruginosa NIES298. Total RNA isolated from cells incubated under different light intensities and collected at different cell densities (OD, optical density at 750 nm). D, dark (0 μmol photons m−2 s−1); L, low light (16 μmol photons m−2 s−1); H, high light (68 μmol photons m−2 s−1); VH, very high light (180 μmol photons m−2 s−1). (A) Autoradiogram of DNA-RNA hybridization with an mcaE probe. (B) Agarose gel picture of the 16S rRNA stained with ethidium bromide under UV light. b, bases.
The size of the strong transcript of about 350 bases indicates transcription of the precursor RNA independent from the genes encoding microcyclamide-processing enzymes.
Conclusion.
The number of cryptic biosynthetic pathways is increasing steadily, first and foremost through microbial genome sequencing projects. Bioinformatic predictions have been shown to be particularly useful for the bioassay-independent discovery of natural products. Microcyclamides produced by bloom-forming freshwater cyanobacteria provide another example of the success of genomic mining strategies. It can be speculated that similar approaches could help in elucidating other and diverse cyclamide structures produced by Microcystis strains. Some of them may prove useful for medical applications, whereas others could give us some insights into the functional role of the peptides.
Supplementary Material
Acknowledgments
We thank SMB GbR for fosmid sequencing and Cyanobiotech GmbH for mass cultivation of M. aeruginosa PCC 7806. We also thank Hans-Martin Dahse for cytotoxic activity assays and Uta Wohlfeld for antimicrobial and antifungal assays at the Leibniz Institute for Natural Product Research and Infection Biology. We acknowledge L. Frangeul and A. Lepelletier from the Pasteur Genopole and A. M. Castets (Unité des Cyanobactéries, Institut Pasteur), who were involved in the genome sequencing project financed by the Institut Pasteur, the Ministère de l'Education Nationale, de la Recherche et de la Technologie, and the Centre National de la Recherche Scientifique (URA2172).
This work was supported by a grant of the German-Israeli Foundation (GIF) to E.D.
Footnotes
Published ahead of print on 1 February 2008.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Banker, R., and S. Carmeli. 1998. Tenuecyclamides A-D, cyclic hexapeptides from the cyanobacterium Nostoc spongiaeforme var. tenue. J. Nat. Prod. 61:1248-1251. [DOI] [PubMed] [Google Scholar]
- 2.Blond, A., J. Peduzzi, C. Goulard, M. J. Chiuchiolo, M. Barthelemy, Y. Prigent, R. A. Salomon, R. N. Farias, F. Moreno, and S. Rebuffat. 1999. The cyclic structure of microcin J25, a 21-residue peptide antibiotic from Escherichia coli. Eur. J. Biochem. 259:747-755. [DOI] [PubMed] [Google Scholar]
- 3.Breukink, E. 2006. A lesson in efficient killing from two-component lantibiotics. Mol. Microbiol. 61:271-273. [DOI] [PubMed] [Google Scholar]
- 4.Degnan, B. M., C. J. Hawkins, M. F. Lavin, E. J. McCaffrey, D. L. Parry, and D. J. Watters. 1989. Novel cytotoxic compounds from the ascidian Lissoclinum bistratum. J. Med. Chem. 32:1354-1359. [DOI] [PubMed] [Google Scholar]
- 5.Delves-Broughton, J., P. Blackburn, R. J. Evans, and J. Hugenholtz. 1996. Applications of the bacteriocin, nisin. Antonie van Leeuwenhoek 69:193-202. [DOI] [PubMed] [Google Scholar]
- 6.Dierstein, R., I. Kaiser, J. Weckesser, U. Matern, W. A. König, and R. Krebber. 1990. Two closely related peptide toxins in axenically grown Microcystis aeruginosa PCC-7806. Syst. Appl. Microbiol. 13:86-91. [Google Scholar]
- 7.Donia, M. S., B. J. Hathaway, S. Sudek, M. G. Haygood, M. J. Rosovitz, J. Ravel, and E. W. Schmidt. 2006. Natural combinatorial peptide libraries in cyanobacterial symbionts of marine ascidians. Nat. Chem. Biol. 2:729-735. [DOI] [PubMed] [Google Scholar]
- 8.Franche, C., and T. Damerval. 1988. Tests on nif probes and DNA hybridizations. Methods Enzymol. 167:803-808. [Google Scholar]
- 9.Fujii, K., Y. Yahashi, T. Nakano, S. Imanishi, S. F. Baldia, and K. Harada. 2002. Simultaneous detection and determination of the absolute configuration of thiazole-containing amino acids in a peptide. Tetrahedron 58:6873-6879. [Google Scholar]
- 10.Fuller, J. D., A. C. Camus, C. L. Duncan, V. Nizet, D. J. Bast, R. L. Thune, D. E. Low, and J. C. S. De Azavedo. 2002. Identification of a streptolysin S-associated gene cluster and its role in the pathogenesis of Streptococcus iniae disease. Infect. Immun. 70:5730-5739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.González-Pastor, J. E., J. L. San Millan, M. A. Castilla, and F. Moreno. 1995. Structure and organization of plasmid genes required to produce the translation inhibitor microcin C7. J. Bacteriol. 177:7131-7140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gross, H. 2007. Strategies to unravel the function of orphan biosynthesis pathways: recent examples and future prospects. Appl. Microbiol. Biotechnol. 75:267-277. [DOI] [PubMed] [Google Scholar]
- 13.Ishida, K., G. Christiansen, W. Y. Yoshida, R. Kurmayer, M. Welker, N. Valls, J. Bonjoch, C. Hertweck, T. Börner, T. Hemscheidt, and E. Dittmann. 2007. Biosynthesis and structure of aeruginoside 126A and 126B, cyanobacterial peptide glycosides bearing a 2-carboxy-6-hydroxyoctahydroindole moiety. Chem. Biol. 14:565-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ishida, K., H. Nakagawa, and M. Murakami. 2000. Microcyclamide, a cytotoxic cyclic hexapeptide from the cyanobacterium Microcystis aeruginosa. J. Nat. Prod. 63:1315-1317. [DOI] [PubMed] [Google Scholar]
- 15.Jack, R. W., and G. Jung. 2000. Lantibiotics and microcins: polypeptides with unusual chemical diversity. Curr. Opin. Chem. Biol. 4:310-317. [DOI] [PubMed] [Google Scholar]
- 16.Jüttner, F., A. K. Todorova, N. Walch, and W. von Philipsborn. 2001. Nostocyclamide M: a cyanobacterial cyclic peptide with allelopathic activity from Nostoc 31. Phytochemistry 57:613-619. [DOI] [PubMed] [Google Scholar]
- 17.Kleerebezem, M., L. E. Quadri, O. P. Kuipers, and W. M. de Vos. 1997. Quorum sensing by peptide pheromones and two-component signal-transduction systems in gram-positive bacteria. Mol. Microbiol. 24:895-904. [DOI] [PubMed] [Google Scholar]
- 18.Kotai, J. 1972. Instructions for preparation of modified nutrient solution Z8 for algae. Publication B-11/69. Norwegian Institute for Water Research, Oslo, Norway.
- 19.Linington, R. G., J. Gonzalez, L. D. Urena, L. I. Romero, E. Ortega-Barria, and W. H. Gerwick. 2007. Venturamides A and B: antimalarial constituents of the Panamanian marine cyanobacterium Oscillatoria sp. J. Nat. Prod. 70:397-401. [DOI] [PubMed] [Google Scholar]
- 20.Long, P. F., W. C. Dunlap, C. N. Battershill, and M. Jaspars. 2005. Shotgun cloning and heterologous expression of the patellamide gene cluster as a strategy to achieving sustained metabolite production. ChemBioChem 6:1760-1765. [DOI] [PubMed] [Google Scholar]
- 21.Martin, C., L. Oberer, T. Ino, W. A. Konig, M. Busch, and J. Weckesser. 1993. Cyanopeptolins, new depsipeptides from the cyanobacterium Microcystis sp. PCC 7806. J. Antibiot. (Tokyo) 46:1550-1556. [DOI] [PubMed] [Google Scholar]
- 22.Michiels, J., G. Dirix, J. Vanderleyden, and C. Xi. 2001. Processing and export of peptide pheromones and bacteriocins in gram-negative bacteria. Trends Microbiol. 9:164-168. [DOI] [PubMed] [Google Scholar]
- 23.Milne, B. F., P. F. Long, A. Starcevic, D. Hranueli, and M. Jaspars. 2006. Spontaneity in the patellamide biosynthetic pathway. Org. Biomol. Chem. 4:631-638. [DOI] [PubMed] [Google Scholar]
- 24.Milne, J. C., A. C. Eliot, N. L. Kelleher, and C. T. Walsh. 1998. ATP/GTP hydrolysis is required for oxazole and thiazole biosynthesis in the peptide antibiotic microcin B17. Biochemistry 37:13250-13261. [DOI] [PubMed] [Google Scholar]
- 25.Ogino, J., R. E. Moore, G. M. Patterson, and C. D. Smith. 1996. Dendroamides, new cyclic hexapeptides from a blue-green alga. Multidrug-resistance reversing activity of dendroamide A. J. Nat. Prod. 59:581-586. [DOI] [PubMed] [Google Scholar]
- 26.Rippka, R., J. Deruelles, J. B. Waterby, M. Herdmann, and R. T. Stanier. 1979. Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J. Gen. Microbiol. 111:1-61. [Google Scholar]
- 27.Rouhiainen, L., L. Paulin, S. Suomalainen, H. Hyytiainen, W. Buikema, R. Haselkorn, and K. Sivonen. 2000. Genes encoding synthetases of cyclic depsipeptides, anabaenopeptilides, in Anabaena strain 90. Mol. Microbiol. 37:156-167. [DOI] [PubMed] [Google Scholar]
- 28.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 29.Schmidt, E. W., J. T. Nelson, D. A. Rasko, S. Sudek, J. A. Eisen, M. G. Haygood, and J. Ravel. 2005. Patellamide A and C biosynthesis by a microcin-like pathway in Prochloron didemni, the cyanobacterial symbiont of Lissoclinum patella. Proc. Natl. Acad. Sci. USA 102:7315-7320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schnell, N., K. D. Entian, U. Schneider, F. Gotz, H. Zahner, R. Kellner, and G. Jung. 1988. Prepeptide sequence of epidermin, a ribosomally synthesized antibiotic with four sulphide-rings. Nature 333:276-278. [DOI] [PubMed] [Google Scholar]
- 31.Sudek, S., M. G. Haygood, D. T. Youssef, and E. W. Schmidt. 2006. Structure of trichamide, a cyclic peptide from the bloom-forming cyanobacterium Trichodesmium erythraeum, predicted from the genome sequence. Appl. Environ. Microbiol. 72:4382-4387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takahashi, I., D. Hayano, M. Asayama, F. Masahiro, M. Watahiki, and M. Shirai. 1996. Restriction barrier composed of an extracellular nuclease and restriction endonuclease in the unicellular cyanobacterium Microcystis sp. FEMS Microbiol. Lett. 145:107-111. [DOI] [PubMed] [Google Scholar]
- 33.Tan, L. T. 2007. Bioactive natural products from marine cyanobacteria for drug discovery. Phytochemistry 68:954-979. [DOI] [PubMed] [Google Scholar]
- 34.Tillett, D., E. Dittmann, M. Erhard, H. von Döhren, T. Börner, and B. A. Neilan. 2000. Structural organization of microcystin biosynthesis in Microcystis aeruginosa PCC7806: an integrated peptide-polyketide synthetase system. Chem. Biol. 7:753-764. [DOI] [PubMed] [Google Scholar]
- 35.van der Meer, J. R., J. Polman, M. M. Beerthuyzen, R. J. Siezen, O. P. Kuipers, and W. M. De Vos. 1993. Characterization of the Lactococcus lactis nisin A operon genes nisP, encoding a subtilisin-like serine protease involved in precursor processing, and nisR, encoding a regulatory protein involved in nisin biosynthesis. J. Bacteriol. 175:2578-2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Welker, M., and H. von Döhren. 2006. Cyanobacterial peptides—nature's own combinatorial biosynthesis. FEMS Microbiol. Rev. 30:530-563. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.