Abstract
Understanding the factors that influence the distribution and abundance of marine diazotrophs is important in order to assess their role in the oceanic nitrogen cycle. Environmental DNA samples from four cruises to the North Atlantic Ocean, covering a sampling area of 0°N to 42°N and 67°W to 13°W, were analyzed for the presence and amount of seven nifH phylotypes using real-time quantitative PCR and TaqMan probes. The cyanobacterial phylotypes dominated in abundance (94% of all nifH copies detected) and were the most widely distributed. The filamentous cyanobacterial type, which included both Trichodesmium and Katagnymene, was the most abundant (51%), followed by group A, an uncultured unicellular cyanobacterium (33%), and gamma A, an uncultured gammaproteobacterium (6%). Group B, unicellular cyanobacterium Crocosphaera, and group C Cyanothece-like phylotypes were not often detected (6.9% and 2.3%, respectively), but where present, could reach high concentrations. Gamma P, another uncultured gammaproteobacterium, was seldom detected (0.5%). Water temperature appeared to influence the distribution of many nifH phylotypes. Very high (up to 1 × 106 copies liter−1) nifH concentrations of group A were detected in the eastern basin (25 to 17°N, 27 to 30°W), where the temperature ranged from 20 to 23°C. The highest concentrations of filamentous phylotypes were measured between 25 and 30°C. The uncultured cluster III phylotype was uncommon (0.4%) and was associated with mean water temperatures of 18°C. Diazotroph abundance was highest in regions where modeled average dust deposition was between 1 and 2 g/m2/year.
Input and removal of fixed nitrogen in the ocean are largely controlled by the microbial processes of dinitrogen fixation and anaerobic ammonia oxidation and denitrification, respectively (23). Global rate estimates of both input and removal processes do not balance and suggest a net loss of fixed nitrogen from oceanic systems (11, 16). One reason for the imbalance of the marine nitrogen budget may be due to the paucity of data for marine diazotrophs. Until recently, the filamentous, nonheterocystous cyanobacterium Trichodesmium and the diatom endosymbiont Richelia were considered the major diazotrophs in the world's oceans (28, 49). However, field measurements and geochemical estimates indicate that these two species together do not fix enough nitrogen to balance the global marine nitrogen budget (3, 20). Methodological advances have led to the discovery of many new marine diazotrophs, and it is now possible to detect diverse diazotrophs by PCR analysis of the nifH gene, which encodes the highly conserved iron-protein subunit of the nitrogenase enzyme (22, 25, 26, 45, 47).
Although our knowledge of nifH sequence diversity from the marine environment has been rapidly expanding (46, 48), the temporal and spatial distributions of the various nifH phylotypes remain poorly characterized (22) and estimates of nifH abundances are rare. Real-time quantitative PCR (qPCR) was successfully used to determine the abundances of two nifH sequences at three stations in the Chesapeake Bay (39) and five nifH phylotypes at Station ALOHA in the Pacific Ocean (9, 10, 38). Diazotrophs in the Sargasso Sea and Amazon River plume have also been investigated using the qPCR technique (13, 17). Although the geographical coverage was limited in each of these studies, patterns in the vertical, temporal, and seasonal distributions of nifH phylotypes could be identified.
The goal of this study was to determine the abundances and distributions of seven different nifH phylotypes over a wide geographical area of the North Atlantic ranging from the equator to 42°N. Gene copy numbers of four cyanobacterial (one filamentous and three unicellular) and two gammaproteobacterial phylotypes and one cluster III nifH phylotype were quantified using qPCR and TaqMan MGB probes, providing estimates of the relative distributions and abundances of these unique diazotroph groups. Our results and those of others (14, 17, 24) suggest that diazotrophy may not be as tightly constrained by geographic location as previously thought.
MATERIALS AND METHODS
Sample collection, nucleic acid extraction, and nifH amplification.
Samples for this study were collected and analyzed from four cruises in the northern Atlantic Ocean. Samples were collected in the spring during the F/S Meteor 60/5 Transient Tracers Revisited cruise (March and April 2004) to the North Atlantic Ocean and the F/S Poseidon 284 cruise (March 2002) to the eastern subtropical Atlantic Ocean. Samples along a transect at 10°N with a second transect to the equator and back were collected during the F/S Meteor 55 cruise (October to November 2002). Samples were collected from the F/S Sonne 152 cruise (November to December 2000) in the western tropical Atlantic basin. All samples were collected using a CTD rosette except for samples from the Sonne 152 cruise and three samples from Meteor 55, which were collected using an overboard pump from a depth of 8 m and a trace-metal clean diaphragm pump, respectively. Poseidon 284 samples were collected from depths ranging from 10 to 30 m. Samples from both Meteor cruises were collected at the surface (5 m) and from one to two depths ranging from 20 to 120 m. After collection, water was filtered onto 0.22-μm Durapore (Millipore) filters under low (2 mbar) vacuum pressure without prescreening to remove large particles. One to two liters of seawater was filtered per sample during the two Meteor and Poseidon cruises. Two to seven liters of seawater was filtered during the Sonne cruise. After filtration, filters were frozen at −80°C until extraction in the lab. Detailed information on some of the Sonne 152, Poseidon 284, and Meteor 55 stations can be found in reference 22. Nutrient samples from the Meteor 55 and 60 and the Poseidon 284 cruises were measured onboard using the methods described in reference 15.
All samples were extracted using the Qiagen DNeasy mini plant kit according to the manufacturer's protocol, except that the DNA was eluted two times with 50 μl of prewarmed PCR-grade water. DNA concentrations were measured using the PicoGreen double-stranded DNA quantitation reagent (Molecular Probes) and a Fluoroscan Ascent microplate reader (Laborsystems) following the manufacturer's protocol. Amplicons of the nifH gene were produced using the previously described primers (45), amplified, and cloned (22).
Primer selection.
Based on nifH sequence information from a subset of the samples (22), primers and TaqMan MGB probes (6-carboxyfluorescein reporter) were selected to target seven diazotroph phylotypes using Primer Express (version 2.0; Applied Biosystems) (Table 1). All primers and probes were checked against the NCBI database with a BLAST search to ensure that they did not contain any near perfect sequence matches, other than the selected phylotypes. Standards for the different phylotypes were obtained by cloning the environmental sequences for each phylotype into Top10 cells. Plasmid extraction and purification was done using the Qiagen plasmid purification kit according to the manufacturer's instructions. Plasmid DNA concentrations were measured with NanoDrop ND-1000 (PeqLab) and diluted to 4 pg μl−1, which corresponds to 107 target sequence copies in a 5-μl volume. Serially diluted plasmid standards for filamentous, unicellular group A, unicellular group B, Cyanothece-like (group C), gammaproteobacterium AO (gamma A), gammaproteobacterium PO (gamma P), and cluster III phylotypes were used to calculate copy numbers in the qPCR assays. The group B and gammaproteobacterial primers and TaqMan probes from our study were very different from those previously reported (10), while the filamentous and group A primers and probes from reference 10 detected a segment of the nifH gene similar to that detected in our study. Although independently designed, the reverse primer and probe for cluster III in our study were almost identical to those described in references 9 and 10. The group C primer and probe set designed for this study selected for a different part of the group C sequence than the set described in Foster et al. (13).
TABLE 1.
qPCR primers and TaqMan probes designed for this study
| Type | Sequence (position) ofa:
|
Reference sequence | ||
|---|---|---|---|---|
| Primer
|
Probe | |||
| Reverse | Forward | |||
| Group A | TCAGGACCACCGGACTCAAC (146-127) | TAGCTGCAGAAAGAGGAACTGTAGAAG (50-76) | TAATTCCTGGCTATAACAAC (98-117) | AF059627.1 |
| Filamentous | GCAAATCCACCGCAAACAAC (275-256) | TGGCCGTGGTATTATTACTGCTATC (165-189) | AAGGAGCTTATACAGATCTA (206-225) | AY896367.1 |
| Group B | TCAGGACCACCAGATTCTACACACT (146-122) | TGCTGAAATGGGTTCTGTTGAA (54-75) | CGAAGACGTAATGCTC (87-102) | AY896454.1 |
| Group C | GGTATCCTTCAAGTAGTACTTCGTCTAGCT (112-83) | TCTACCCGTTTGATGCTACACACTAA (1-26) | AAACTACCATTCTTCACTTAGCAG (32-55) | AY896461.1 |
| Gamma A | AACAATGTAGATTTCCTGAGCCTTATTC 321-294 | TTATGATGTTCTAGGTGATGTG (240-266) | TTGCAATGCCTATTCG (275-290) | AY896371.1 |
| Gamma P | CATCGCGAAACCACCACATAC (282-262) | TTGTGCAGGTCGTGGTGTAATC (159-180) | CCTATGACGAAGACCTAGAC (212-231) | AY896428.1 |
| Cluster III | GCAGACCACGTCACCCAGTAC (267-247) | ACCTCGATCAACATGCTCGAA (175-195) | CCTGGACTACGCGTTC (225-240) | AY896461.1 |
All positions and sequences relate to the 324-base nifH segment length (5′→3′).
qPCR assays and detection limits.
All qPCRs were run on an ABI Prism 7000 (Applied Biosystems) using the default cycling program, but increasing the number of cycles to 45. The program's cycling conditions were 2 min at 50°C, 10 min at 95°C, and 45 cycles of 95°C for 15 s, followed by 1 min at 60°C. The specificity of the qPCR primer and probe sets was confirmed by testing for cross-reactivity and sensitivity against each standard diluted to 104 and 105 nifH copies. DNA amplification signals were detected only for the homologous phylotype. It is important to note that the filamentous primer and probe set was designed for Trichodesmium, but due to the high similarity in the filamentous nifH sequences it amplified Katagnymene equally well. Additionally, the sensitivity of the primers and probes in a mixed-DNA sample was tested in combinations of serially diluted standards mixed in ratios ranging from 107:101 to 101:107 copies of the specific and unspecific standard. There were no significant changes in the linear regressions, indicating that the presence of various amounts of closely related DNA sequences does not affect the quantification of a specific phylotype. However, the detection limit of the filamentous primer and probe set was 100 filamentous nifH copies when mixed with 107 group A nifH copies, a situation that was not observed in any samples.
qPCR mixtures contained 1× TaqMan PCR buffer (Applied Biosystems), 100 nM TaqMan probe, 5 pmol/μl each of the forward and reverse primers, 400 ng/μl bovine serum albumin (BSA), 3 μl PCR water, and 5 μl of either standard or environmental sample (which corresponded to 1 to 6 ng environmental DNA, with an average of 2 ng). To test environmental samples for PCR inhibition, serial dilutions of several samples from each cruise were run. The results indicated different levels of inhibition in almost all samples tested. Also, additions of environmental DNA to standards resulted in an underestimation of the true copy number. Dilution of the samples partially reduced PCR inhibition but affected the detection limit. Inhibition of PCR is a common problem with environmental samples and has been reported before in other studies, which estimated the degree of the inhibition from qPCR efficiency without relieving the inhibition (13). Addition of BSA to the reaction consistently relieved the inhibition without affecting the standard curves or detection limit (data not shown). We therefore routinely added BSA to all qPCR mixtures. Environmental DNA samples were run in triplicate. The coefficient of variation for the cycle threshold (CT) value of all samples was 1.77% (n = 251). Standards were serially diluted (107 to 101 copies) and run in duplicate. The prepared serial dilutions were stored at 4°C and were stable for about 1 month. Standard curves from stored standard serial dilutions were highly reproducible and varied daily by an average CT of 0.7. In contrast, freeze-thaw cycles of samples and standards resulted in 10-fold decrease in copy numbers (results not shown). To avoid this, extracted DNA was frozen in aliquots and thawed only once for qPCR determination.
No-template controls were run in duplicate for each primer and probe set and were undetectable after 45 cycles, thus setting the theoretical detection limit of our assay mixture to one nifH copy. However, the realized detection limit depends on the amount of seawater filtered per sample, elution volume after extraction, and the amount of sample loaded. In this study, the amount of seawater filtered determined the detection limits and these are 20, 50, and 75 copies liter−1 of seawater for Sonne, Poseidon, Meteor 55, and Meteor 60 samples, respectively. Thus, in our assays, samples with a CT value below 36, which corresponded to <100 copies liter−1, were considered detectable but not quantifiable. To minimize potential differences between qPCRs run on different days, a sample was quantified with all primer and probe sets on 1 day and the same standard serial dilutions were used for all samples. The ABI 7000 system SDS software (version 1.2.3) with RQ study application was used to calculate linear regressions of CT versus log10 nifH standard copy numbers. PCR efficiencies were calculated using the formula E = 10−1/slope − 1 (1). Average PCR efficiencies were 96.5% ± 2% (n = 37; R2 = 0.999) for filamentous, 94.9% ± 1% (n = 39; R2 = 0.999) for group A, 96.1% ± 2% (n = 22; R2 = 0.997) for group B, 89.1% ± 6% (n = 24; R2 = 0.998) for group C, 95.3% ± 2% (n = 24; R2 = 0.998) for gamma A, 100% ± 2% (n = 14, R2 = 0.997) for gamma P, and 100% ± 1% (n = 22; R2 = 0.994) for cluster III. Statistical analyses (Kruskal-Wallis and Mann-Whitney U tests) were performed with Statistica (version 6). P values of <0.05 were considered significant. Maps of the distribution and abundance of nifH phylotypes in the North Atlantic were generated with Ocean Data View (37).
RESULTS
Sampling and DNA concentrations.
A total of 141 DNA samples were collected from four cruises to the northern Atlantic Ocean. Sixty-four samples were collected in surface waters, 41 from depths of 20 to 80 m, and 36 from depths below 80 m. Water temperatures from all samples ranged from 15 to 30°C, with the coldest (15 to 23°C) and highest (25 to 29°C) temperatures recorded during the Meteor 60 and Meteor 55 expeditions, respectively. All samples were collected in waters with a salinity of 34.6 to 37.2 ppt, with the exception of samples from four stations located in the Amazon River plume, where surface water salinities ranged between 31.2 and 33.4 ppt. NO3 concentrations ranged from 0 to 25 μM; however, 65% of the samples were collected from waters with 0 to 1 μM NO3. PO4 concentrations ranged from 0 to 1.5 μM, and 75% of the samples had concentrations of 0.25 μM PO4 or less. DNA concentrations ranged from 16 ng liter−1 to 640 ng liter−1 (average, 132 ng liter−1). The lower DNA concentrations generally came from samples collected at depth. The Sonne and Meteor 55 samples collected with an overboard pump had on average a higher DNA concentration (237 ng liter−1) than samples collected with a CTD rosette (114 ng liter−1). No other trends were observed between DNA concentrations and location, depth, or time of year.
Comparison of clone libraries versus qPCR.
Cyanobacterial phylotypes were the most abundantly detected diazotrophs (93.8% of the total nifH copies detected), with the majority being the filamentous phylotype (Table 2). The group A phylotype was the next most abundant, followed by gamma A, which was lower by 1 order of magnitude. The gamma P and cluster III phylotypes made up about 1% of all nifH sequences detected. These results reflect the relative abundance of phylotypes in clone libraries (22), except that rare groups detected at low abundance with qPCR were overestimated in pooled clone libraries. In direct comparisons of qPCR data and clone libraries from the same station, more diazotroph phylotypes were detected with qPCR, thus demonstrating the higher sensitivity of this technique, especially for the detection of rare phylotypes. For 10 of the samples, which contained low diversity (two to three phylotypes present), the relative abundance of phylotypes from the cloned library accurately (5% difference) reflected abundances obtained with qPCR, except that groups with low copy numbers were sometimes absent from the clone libraries. However, results from qPCR and clone libraries diverged greatly for samples containing high (more than three phylotypes present) nifH diversity, with minor phylotypes completely absent from the clone library and large over- or underestimates of more dominant groups based on data from clone libraries. Results showed that qPCR provided a highly sensitive and quantitative estimate of phylotype abundance, in contrast with clone libraries, which can only show relative abundances at best (Table 2).
TABLE 2.
Comparison of clone libraries and qPCR data given as percentages of each phylotype from the total number of either clones or phylotypes detected
| Phylotype | % of clones recovereda | % of nifH copies detected from:
|
|
|---|---|---|---|
| Samples with clone librariesb | All samplesc | ||
| Filamentous | 57.4 | 71.1 | 53.1 |
| Group A | 26.9 | 23.9 | 34.1 |
| Group B | 1.4 | 0.3 | 4.3 |
| Group C | 3.6 | 1.2 | 2.3 |
| Gamma A | 5.9 | 2.8 | 5.3 |
| Gamma P | 2.5 | 0.7 | 0.6 |
| Cluster III | 2.2 | 0.05 | 0.4 |
A total of 357 clones was obtained from 23 cloned samples.
A total of 4.9 × 106 nifH copies liter−1 was detected in samples with a corresponding clone library.
A total of 1.1 × 107 nifH copies liter−1 was detected in all 142 samples.
Distributions of nifH phylotypes.
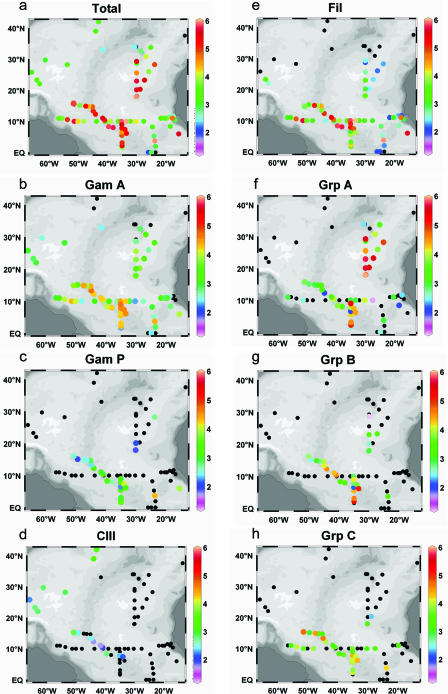
Diazotroph phylotypes were detected throughout the northern Atlantic Ocean (Fig. 1a). In general, very low to undetectable nifH copy numbers were observed along the equator and at latitudes higher than 30°N. The highest nifH copy numbers were measured in Poseidon samples between 18 to 30°N and 30 to 23°W; however, the lowest diversity was also observed at this location. High copy numbers and high diversity were detected in Sonne samples between 50 and 34°W and 2 and 15°N. The filamentous, group A, and gamma A phylotypes were the most commonly detected phylotypes and were distributed throughout the North Atlantic (Fig. 1b, e, and f). Both gammaproteobacterial and cluster III phylotypes were only detectable at low copy numbers (Fig. 1b to d). In contrast to the other phylotypes, the cluster III distribution reached as far as 40°N (Fig. 1d). This rare phylotype was detectable in Sonne and Meteor 60 samples, but only west of 33°W. Copy numbers ranged from the detection limit to 1,600 cluster III nifH copies liter−1, with the highest copy numbers measured at northern stations where the spring bloom was in progress (29). The gamma P phylotype occurred mainly in Sonne samples, and occasionally in Poseidon samples and the Meteor 55 samples, which were collected using a pump. In contrast, the gamma A nifH phylotype was detectable in most samples from all cruises, and ranged between 103 (Sonne) and 104 (Meteor 55) nifH copies liter−1.
FIG. 1.
Total nifH copies liter−1 detected at each station (a) and nifH copies liter−1 of each phylotype in panels b to h were plotted using Ocean Data View (37). Black dots indicate samples with concentrations below the detection limit. Note that the color scale is a log scale. Fil, filamentous; Gam, gamma; Grp, group; CIII, cluster III.
Filamentous diazotroph nifH copy numbers were found north of the equator to about 15°N (Fig. 1e) and reached the highest abundances between 52°W and 35°W, though areas of high abundances were also present in the eastern part of the North Atlantic basin (up to 106 copies liter−1). Others have also observed higher abundances of Trichodesmium in the western Atlantic basin (12) than in the eastern basin. The filamentous phylotype was found at low copy numbers between 20°N and 30°N, and it was undetectable along the northern cruise track, except at the two southernmost stations (approximately 22°N).
Unicellular cyanobacterial diazotroph copy numbers were more variable than filamentous copy numbers. Group A unicellular cyanobacterial phylotypes were present throughout the study area and reached high concentrations (up to 106 group A nifH copies liter−1) (Fig. 1f). No group A nifH copies were detectable in the northwestern quadrant of the Atlantic Ocean (west of 30°W and north 17°N) during spring. Group A copy numbers were moderately high (9 × 103 to 2 × 104 group A nifH copies liter−1) between the equator and 15°N but reached the highest abundances between 30°W to 23°W and 30°N to 17°N (Poseidon cruise track), where copy numbers ranged from 4 × 105 to 106 copies liter−1. Group C nifH copy numbers were low to undetectable throughout the study area, except for some Sonne samples. Group B unicellular phylotypes were mainly found in Sonne samples, as well, but at higher concentrations: 3 × 104 group B nifH copies liter−1 as opposed 1 × 104 group C nifH copies liter−1 (Fig. 1g and h). No quantifiable group B or group C phylotypes were detected north of 20°N. The group B phylotype accounted for 6.9% of the total nifH copies detected in our samples, despite being detected at only 27 stations.
In an attempt to identify what factors influence the distribution of diazotrophs, we considered sampling depth, dissolved inorganic nutrient concentrations, mixed layer depth, water temperature, geographical distribution, and Fe fertilization via mineral dust deposition. Comparison of paired surface (up to 30 m deep) and deep (30 to 200 m) samples from 37 stations indicated that overall total diazotroph abundance decreased with depth (Mann-Whitney U test; P = 0.004). More specifically, the filamentous, gamma A, and group C phylotype concentrations were significantly higher in the surface samples (Mann-Whitney U test; P < 0.001). There were a few stations where the group A phylotype was detected both at the surface and at depth; however, the majority of group A phylotypes were detected between 10 and 30 m. Cluster III and gamma P concentrations appeared to increase with depth, but this result was not statistically significant (Table 3).
TABLE 3.
Effect of sampling depth and NO3 concentration on the abundance of nifH phylotypesa
| Typeb | % of nifH phylotype copies detected
|
|||
|---|---|---|---|---|
| Surface samples | Depth samples | <0.5 μM NO3 | >0.5 μM NO3 | |
| Filamentous*† | 99.2 | 0.8 | 99.5 | 0.5 |
| Group A† | 30.7 | 69.3 | 97.5 | 2.5 |
| Group C*† | 90.5 | 9.5 | 96.5 | 3.5 |
| Gamma A*† | 89.0 | 11.0 | 95.8 | 4.2 |
| Gamma P | 0.0 | 100.0 | 89.8 | 10.2 |
| Cluster III | 24.6 | 75.4 | 40.2 | 59.8 |
| Total*† | 94.1 | 5.9 | 97.9 | 2.1 |
Data are given as the percentage of either nifH phylotype copies detected in surface and depth samples (columns 1 and 2) or nifH phylotypes recovered in waters with less than or greater than 0.5 μM NO3. Surface samples (up to 30 m deep) and corresponding depth samples (from 30 to 200 m) were available for 37 stations, and no group B phylotypes were detected in these samples (columns 1 and 2). NO3 measurements were available for 112 samples.
Phylotypes whose concentrations were significantly different (P < 0.05, Mann-Whitney U test) in samples collected at depth than in surface samples are identified by *, while those significantly affected by NO3 concentrations are identified by †.
Significant relationships were also observed between nifH distribution and residual NO3 (Table 3) and PO4 concentrations (results not shown), with most phylotypes being almost completely restricted to NO3 concentrations less than 0.5 μM. One clear exception was the cluster III phylotype, which showed no significant preference for low NO3 waters (Table 3). Mixed-layer depth showed no direct relationship with specific nifH phylotypes (results not shown).
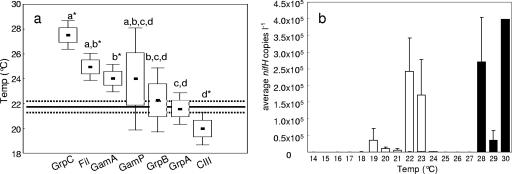
The influence of temperature on the distribution of nifH phylotypes was investigated by calculating the mean water temperature for each phylotype in samples containing greater than 100 nifH copies liter−1. The mean temperature for each phylotype ranged from 28°C for group C to 18°C for cluster III (Fig. 2a). We observed that most phylotypes were associated with narrow temperature ranges except for gamma P, the least abundant phylotype, which appeared to be poorly constrained by temperature. The group C phylotype was detected in very warm waters, while gamma A, gamma P, and group B were detected mainly in waters of about 24°C. Temperature means for group C, filamentous, gamma A, and cluster III were significantly different from the mean water temperature of all samples. Interestingly, cluster III was the only phylotype whose mean detection temperature was below the mean of all samples. The clearest associations between temperature and nifH phylotypes were for the filamentous and group A phylotypes, which were detected in significant quantities at average temperatures of 26°C and 20.9°C, respectively (Fig. 2a). Although the filamentous and group A phylotypes were detected throughout the temperature range, the highest copy numbers were detected in samples with water temperatures from 28 to 30°C and 19 to 24°C, respectively (Fig. 2b). This suggests that these two dominant groups may have different temperature optima.
FIG. 2.
Box-whisker plots (black dots, mean; boxes, standard error; bars, 95% confidence interval) show the temperatures where >100 nifH copies liter−1 were detected for all phylotypes in panel a. The mean temperature of all samples (22°C) and the associated standard error are indicated by a black line and dashed line, respectively. Phylotype temperature means which are significantly different from 22°C (P < 0.05, Kruskal-Wallis test) are identified by asterisks. Letters signify groups of phylotypes whose temperature means are not significantly different from another group (Mann-Whitney U test). Average filamentous and group A nifH copies liter−1 detected at sample temperatures rounded to the nearest whole degree are displayed in panel b. Error bars indicate the standard error.
Distribution by geographical areas.
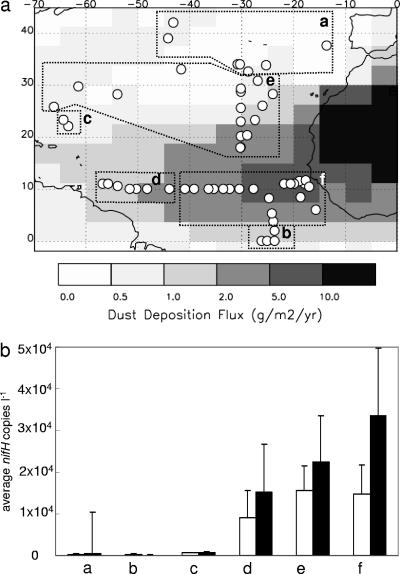
The large geographical coverage of our data prompted us to analyze the distributions and abundances of the nifH phylotypes in six regions (Fig. 3a), defined as a function of the basic water characteristics, temperature and salinity, of the surface water sample (see Fig. S1 in the supplemental material). Average nifH gene copy numbers for the stations within regions A, B, and C, corresponding to areas north of 35°N, the equator, and the Sargasso Sea respectively, contained very low abundances of potential diazotrophs (Fig. 3b). In contrast, the highest average surface and total nifH gene copy numbers were detected in regions E and F (Fig. 1a and 3b).
FIG. 3.
Separation of stations according to geographical location (a) and physical characteristics (see the supplemental material). Station locations are overlaid on a map of predicted annual dust deposition. The average number of nifH copies liter−1 and standard error from all samples (clear bars) and only surface samples (black bars) in each region are plotted in panel b. Region letters correspond to the panels in Fig. 4. Sonne 152 samples were not plotted because no temperature and salinity data were available.
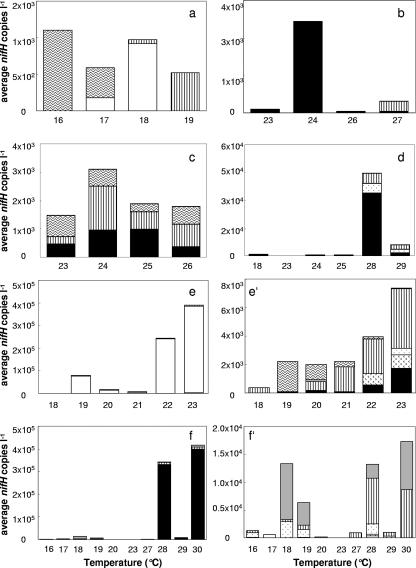
For each geographical area, average nifH concentrations of individual phylotypes were plotted versus temperature (Fig. 4). Region A (northerly stations) was characterized by low surface water temperatures (<19°C) and relatively high salinity (>36.2 ppt). This area of very low nifH gene copy numbers (Fig. 3 and 4a) contained only the cluster III, group A, and gamma A phylotypes. Of the eight samples measured in region B (equator), only one contained abundances well above the detection limit (Fig. 4b). Here, diversity was low and only the filamentous and a few gamma A phylotypes were detected. Stations in the Sargasso Sea were sampled in the spring time (region C, Fig. 4c), exhibited deep mixed layers, and contained low nifH concentrations. The three phylotypes found there consisted of the rare cluster III phylotype as well as the gamma A and filamentous phylotypes. These phylotypes were homogeneously distributed throughout the stations, even at depths up to 160 m. Region D contained samples that were influenced by the Amazon River, where surface water temperatures were greater than 28°C and diazotroph concentrations, especially the filamentous phylotype, were higher in the surface samples (Fig. 4d).
FIG. 4.
Average concentrations of nifH copies liter−1 by sample temperature (rounded to the nearest whole degree) in each geographic location are shown, with panel letters corresponding to the regions in Fig. 3. ▪, filamentous; □, group A,  , group B;
, group B;  , group C; ▥, gamma A;
, group C; ▥, gamma A;  , gamma P; and
, gamma P; and  , cluster III. The numbers of samples for regions a, b, c, d, e, and f are 21, 8, 6, 40, 105, and 96, respectively. Panels e′ and f′ show the average numbers of nifH copies liter−1 from panels e and f without the dominant group A and filamentous phylotypes, respectively.
, cluster III. The numbers of samples for regions a, b, c, d, e, and f are 21, 8, 6, 40, 105, and 96, respectively. Panels e′ and f′ show the average numbers of nifH copies liter−1 from panels e and f without the dominant group A and filamentous phylotypes, respectively.
Total nifH concentrations were relatively similar in regions E and F (Fig. 3b), but different groups were dominant (Fig. 4e and f′). Region E samples were characterized by surface water temperatures of 20 to 23°C and salinity of 36.8 to 37 ppt. This area was dominated by the group A phylotype. The cluster III phylotype was also detected, but only in the samples from the western half of this region. In contrast, the filamentous phylotype dominated region F, where surface water temperatures were greater than 27°C and the salinity was 34.7 to 35.6 ppt.
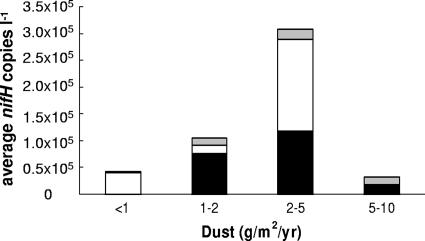
The amount of dust deposition may play a role in determining the abundance and distribution of diazotrophs in a certain region. In Fig. 3a, the stations are plotted in relation to the annual dust deposition. Samples were then grouped according to estimated annual dust deposition (Fig. 5). The highest diazotroph concentrations were detected where the dust deposition was between 2 and 5 g m−2 year−1 (Fig. 5). The geographical regions with the lowest number of nifH copies detected (i.e., all of region A and most of region B samples) were located in the modeled dust deposition area with <1 g m−2 year−1. Only the cluster III phylotype was detected more often in regions with low dust deposition (Fig. 1d and 5).
FIG. 5.
Average concentrations of nifH copies liter−1 for filamentous (▪), group A (□), and all other phylotypes combined (gray square) as a function of estimated annual dust deposition are shown. Average temperatures of each region were 22.3°C (5 to 10 g m−2 year−1), 24.5°C (2 to 5 g m−2 year−1), 26.8°C (1 to 2 g m−2 year−1), and 19.3°C (<1 g m−2 year−1). The dust deposition map was provided by I. Tegen, using the data from reference 42.
DISCUSSION
Assessing relative diversity and phylotype abundances from qPCR data.
Although extensive sequencing of clone libraries generated from endpoint PCR products has the potential to uncover rare and new phylotypes, amplification biases can occur from preferential primer binding, the use of different types of Taq polymerases (32), nonselection by a primer set (30), and multiple target gene copies. For this reason, it is well recognized that endpoint PCR techniques are not suitable for quantitative analysis of genetically diverse phylotypes from natural microbial communities. Prior knowledge of the nifH gene diversity from the study area (22) allowed us to target and quantitatively estimate the abundance of the dominant phylotypes, avoiding amplification biases by using specific primer and probe sets. We found that an approach combining both sequence analyses from clone libraries and TaqMan-based qPCR assays was needed for accurately assessing both diversity and abundances of diazotrophs in a given environment.
The presence of multiple gene copies in the genome of an organism as well as multiple genome copies per cell can affect the extrapolation of qPCR results to estimates of cell abundance. It is known that some diazotrophs, such as Clostridium pasteurianum, have multiple copies of the nifH gene (35). Others, such as Trichodesmium erythraeum and Crocosphaera watsonii, have only one copy of the nifH gene per genome. As there is no available genome information for the uncultured phylotypes and little information on the variability of genome copies per cell for all nifH phylotypes, the concentrations of nifH genes presented in this study should be considered an upper limit of the cell density. Given the limited dissolved P and N supply in the euphotic zone of the oligotrophic North Atlantic (27, 44), the presence of multiple genome copies per cell is unlikely. In contrast, this may be a consideration for diazotrophs located below the euphotic zone, where nutrient levels are high (26).
Diazotrophic communities of the Atlantic Ocean.
Our results on nifH phylotype distribution emphasize the importance of Trichodesmium in the tropical Atlantic Ocean, complementing other studies (6), as filamentous nifH sequences made up 51% of all detected sequences. Filamentous concentrations were similar to Trichodesmium cell counts (7). Although the distribution of Trichodesmium exhibits a latitudinal trend with increasing abundances in southerly latitudes, seasonality may also play an important role in explaining the distribution patterns of this phylotype in our study. Nitrogen fixation rates have been reported to be highest during the late summer and autumn and lowest in the winter in the area above 10°N and west of 40°W (6) The two cruises with the highest filamentous copy numbers (Sonne and Meteor 55) were during the fall. The Meteor 60 cruise was during the spring, the season corresponding to the lowest Trichodesmium abundances in this latitudinal range (31).
Although filamentous phylotypes were dominant in our study, the group A unicellular phylotype was responsible for over a third of our total nifH copies, suggesting that this group may periodically contribute significantly to N2 fixation in the northern Atlantic. The gamma A phylotype, although detected at low concentrations, was found in almost all samples and may also play an important role globally due its widespread distribution. It has already been demonstrated that gammaproteobacteria are cosmopolitan (8), and an nifH gammaproteobacterial sequence 99% similar to the gamma A target sequence of this study was present at all stations in an Arabian Sea study (4). Taken together, these results suggest that the widespread distribution of nifH phylotypes belonging to the gammaproteobacterial group warrants further study of its role in the global nitrogen cycle.
The presence of cluster III nifH phylotypes in oxygenated low light waters with detectable to high nitrate is puzzling. In our study, the highest concentrations of cluster III phylotypes were detected during the spring bloom at stations characterized by a deep mixed layer and high nitrate concentrations (29), suggesting that high nitrate conditions do not select against all diazotrophs (20). Even Trichodesmium, which in our study was abundant only in oligotrophic waters, could retain some ability to fix nitrogen when grown in cultures with high nitrate concentrations (18). Diverse cluster III nifH transcripts, distantly related (<60%) to the ones described here, have also been reported in well-oxygenated surface waters of the Mediterranean Sea (24). It was suggested that cluster III diazotrophs from oxygenated waters may be either facultative anaerobes or associated with zooplankton (24). Photosynthetic, diazotrophic, purple sulfur bacteria have been reported in association with copepods and have been found to fix nitrogen (34). Although prefiltered (10 μm) samples preclude zooplankton as the host of cluster III phylotypes in some studies (9), a zooplankton-cluster III association cannot be ruled out in the present study. Whether cluster III contributes significantly to global nitrogen fixation remains an open question requiring further work.
To date there have been few other studies which have looked at the distribution of oceanic diazotrophs quantitatively. Published concentrations of diazotroph phylotypes (9, 10) measured by real-time qPCR were comparable to those obtained in this study. Church et al. (9) reported the group A phylotype as being the most numerous (2 × 105 copies liter−1) in upper waters and the cluster III phylotype abundant in dim waters (100 to 1,000 copies liter−1). The situation was different in the Atlantic Ocean, with some stations dominated by the filamentous phylotype and cluster III often undetectable even at depth. Concentrations reported by Foster et al. (13) of the filamentous, group A, group B, and group C phylotypes were comparable to those detected in region D. We did not observe any nifH sequences related to the diatom-diazotroph symbiont Richelia in our clone libraries and therefore did not quantify these groups, although our samples were collected from waters with higher salinities and lower Si concentrations (0 to 1 μM) than those where Foster et al. (13) observed the diatom-diazotroph association. The Sargasso Sea has been considered an area where nitrogen fixation can be important (31); however, we detected low nifH copy numbers in this area (region C), a finding shared by others (17).
In our study, filamentous and group A were the dominant phylotypes whose distributions were well separated as a function of temperature (Fig. 2). The filamentous and group A phylotypes were dominant at stations with water temperatures ranging between 28 and 30°C and 22 and 23°C, respectively. Similarly, group A dominated at the ALOHA time series site when the water temperature ranged between 23 and 24°C (9, 22). The direct effects of temperature on the physiology of an organism can be confounded by the role temperature plays in water column stratification, light regimens, and nutrient availability. In the case of Trichodesmium, the association with high water temperature reflects a direct effect on nitrogen fixation and growth rates, with the optima for both physiological processes occurring between 24 and 30°C (5, 40). The physiological reasons for this high temperature preference are not completely understood (41). A combination of factors such as lower dissolved O2 concentration and a higher respiration rate at high temperatures may favor Trichodesmium growth by facilitating O2 scavenging and thereby protecting nitrogenase against irreversible O2 damage. A change from 30°C to 20°C waters would result in a 20% increase in dissolved O2 concentration and in a twofold reduction in respiration rates. Except for a few sparse field observations, there are currently no data for the other nifH phylotypes primarily because there are no culture isolates. It is therefore not possible at this stage to establish the effects of temperature on organism physiology for the other diazotrophic groups, although there is a need to carry out laboratory experiments with a group B isolate, C. watsonii.
The detection of nifH in a DNA sample gives an estimate of the potential for diazotrophy within the microbial community. Whether any of the detected diazotrophs were actively fixing dinitrogen cannot be determined from our results, as only information on nifH DNA copy numbers is presented here. However, dinitrogen fixation rates ranging from 3.7 to 255 μmol m−2 day−1 were measured during the same cruises (27, 43), indicating that some of the nifH phylotypes were actively fixing N2.
nifH distribution relative to dust deposition.
Iron supply has been shown to limit diazotrohic growth (33) and, in combination with phosphate, nitrogen fixation (27). This limitation is probably because diazotrophs can have an iron requirement up to 10 times greater than that of other photoautotrophs (21, 40) which is needed for the synthesis of the nitrogenase enzyme. Iron is supplied to the Atlantic Ocean mainly through deposition of aeolian dust, originating from the Sahara (19). The concentration of nifH phylotypes was higher in regions which receive an estimated annual dust deposition of 2 to 5 g m−2 year−1 than in the area with 5 to 10 g m−2 year−1. Trichomes were indeed visible in water samples and on filters in the area with 2 to 5 g m−2 year−1. Although the western coast of Africa receives the largest amount of dust, seasonal upwelling brings colder temperatures and higher macronutrient concentrations, which most likely favor groups other than diazotrophs, thus potentially explaining why nifH abundance was considerably lower in the area with dust deposition of 5 to 10 g m−2 year−1. Dust particle size may also play a role in determining the bioavailability of Fe, with heavier particles deposited more quickly and sinking faster than finer particles (2). Similar to our findings, Voss et al. (43) found that nitrogen fixation rates were correlated with dissolved iron concentrations during the Meteor 55 cruise. In contrast, a study focused on nitrogen fixation by Trichodesmium in the central North Atlantic found no correlation of N2 fixation rates with dissolved iron (36). The authors concluded that Trichodesmium was not iron limited in their study because of high dissolved iron concentrations observed in April, a month of high dust deposition. The cruise track from their study was mainly located within the area with a modeled annual dust deposition of 2 to 5 g/m2/year, covering a fraction of the geographical area presented in our study. Whether temperature, geographical location, residual NO3, or amount of dust deposition plays a larger role in determining the composition of diazotrophs requires further study.
Conclusions.
Our study presents a snapshot of nifH phylotype distribution and abundance covering a large geographical area of the North Atlantic and expands the known distribution range of diazotrophs. The dominant phylotypes filamentous and group A had different temperature optima, indicating that these two groups may occupy different niches. Although most nifH phylotypes were associated with warm waters with very low NO3 concentration, the presence of the cluster III phylotypes at relatively high latitudes and high NO3 concentrations was unexpected. Data on nitrogenase activity or nifH expression will be needed to confirm that these nifH phylotypes actively fix dinitrogen in all of the locations where they were detected. There is currently no information on the seasonal distribution and diurnal activity of most nifH phylotypes, with the notable exception of Trichodesmium (10). Our study suggests that atmospheric mineral dust deposition and temperature are important factors determining the distribution and abundance of the various nifH phylotypes. However, confirmation of our observations will require a coupled seasonal analysis of the nifH phylotype distribution and mineral dust deposition in the North Atlantic.
Supplementary Material
Acknowledgments
We thank the captains and crews of the F/S Meteor and Poseidon and chief scientists D. Wallace (Meteor 55 and 60 expeditions) and P. Kähler (Poseidon 284). We thank I. Tegen for providing the dust deposition map and F. Malien for nutrient analyses. We thank two anonymous reviewers for constructive comments.
This work was supported by DFG grants RO 2138/4-1 and RO 2138/5-1 to J.L.
Footnotes
Published ahead of print on 1 February 2008.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Atallah, Z. K., J. Bae, S. H. Jansky, D. I. Rouse, and W. R. Stevenson. 2007. Multiplex real-time quantitative PCR to detect and quantify Verticillium dahliae colonization in potato lines that differ in response to verticillium wilt. Am. Phytopathol. Soc. 97:865-872. [DOI] [PubMed] [Google Scholar]
- 2.Baker, A. R., and T. D. Jickells. 2006. Mineral particle size as a control on aerosol iron solubility. Geophys. Res. Lett. 33:.
- 3.Berman-Frank, I., P. Lundgren, and P. Falkowski. 2003. Nitrogen fixation and photosynthetic oxygen evolution in cyanobacteria. Res. Microbiol. 154:157-164. [DOI] [PubMed] [Google Scholar]
- 4.Bird, C., J. Martinez Martinez, A. G. O'Donnell, and M. Wyman. 2005. Spatial distribution and transcriptional activity of an uncultured clade of planktonic diazotrophic γ-proteobacteria in the Arabian Sea. Appl. Environ. Microbiol. 71:2079-2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Breitbarth, E., A. Oschlies, and J. La Roche. 2006. Physiological constraints on the global distribution of Trichodesmium—effect of temperature on diazotrophy. Biogeosciences. www.biogeosciences.net/3/1/2006.
- 6.Capone, D., J. Burns, J. P. Montoya, A. Subramaniam, C. Mahaffey, T. Gunderson, A. F. Michaels, and E. J. Carpenter. 2005. Nitrogen fixation by Trichodesmium spp.: an important source of new nitrogen to the tropical and subtropical North Atlantic Ocean. Global Biogeochem. Cycles. doi: 10.1029/2004GB002331. [DOI]
- 7.Carpenter, E. J., A. Subramaniam, and D. Capone. 2004. Biomass and primary productivity of the cyanobacterium Trichodesmium spp. in the tropical N Atlantic Ocean. Deep-Sea Res. 51:173-203. [Google Scholar]
- 8.Cho, J.-C., and S. J. Giovannoni. 2004. Cultivation and growth characteristics of a diverse group of oligotrophic marine Gammaproteobacteria. Appl. Environ. Microbiol. 70:432-440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Church, M. J., B. D. Jenkins, D. M. Karl, and J. P. Zehr. 2005. Vertical distributions of nitrogen-fixing phylotypes at Stn ALOHA in the oligotrophic North Pacific Ocean. Aquat. Microb. Ecol. 38:3-14. [Google Scholar]
- 10.Church, M. J., C. M. Short, B. D. Jenkins, D. M. Karl, and J. P. Zehr. 2005. Temporal patterns of nitrogenase (nifH) gene expression in the oligotrophic North Pacific Ocean. Appl. Environ. Microbiol. 71:5362-5370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Codispoti, L. A., J. A. Brandes, J. P. Christensen, A. H. Devol, S. W. A. Naqvi, H. W. Paerl, and T. Yoshinari. 2001. The oceanic fixed nitrogen and nitrous oxide budgets: moving targets as we enter the anthropocene? Sci. Mar. 62:85-105. [Google Scholar]
- 12.Davis, C. S., and D. J. McGillicuddy, Jr. 2006. Transatlantic abundance of the N2-fixing colonial cyanobacterium Trichodesmium. Science 312:1517-1520. [DOI] [PubMed] [Google Scholar]
- 13.Foster, R. A., A. Subramaniam, C. Mahaffey, E. J. Carpenter, D. Capone, and J. P. Zehr. 2007. Influence of the Amazon River plume on distributions of free-living and symbiotic cyanobacteria in the western tropical north Atlantic Ocean. Limnol. Oceanogr. 52:517-532. [Google Scholar]
- 14.Fulweiler, R. W., S. W. Nixon, B. A. Buckley, and S. L. Granger. 2007. Reversal of the net dinitrogen gas flux in coastal marine sediments. Nature 448:180-182. [DOI] [PubMed] [Google Scholar]
- 15.Grasshoff, K., M. Ehrhardt, and K. Kremling. 1983. Methods of seawater analysis. Springer-Verlag, New York, NY.
- 16.Gruber, N., and J. L. Sarmiento. 1997. Global patterns of marine nitrogen fixation and denitrification. Global Biogeochem. Cycles 11:235-266. [Google Scholar]
- 17.Hewson, I., P. H. Moisander, K. M. Achilles, C. A. Carlson, B. D. Jenkins, E. Mondragon, A. E. Morrison, and J. P. Zehr. 2007. Characteristics of diazotrophs in surface to abyssopelagic waters of the Sargasso Sea. Aquat. Microb. Ecol. 46:15-30. [Google Scholar]
- 18.Holl, C. M., and J. P. Montoya. 2005. Interactions between nitrate uptake and nitrogen fixation in continuous cultures of the marine diazotroph Trichodesmium (Cyanobacteria). J. Phycol. 41:1178-1183. [Google Scholar]
- 19.Jickells, T. D., Z. S. An, K. K. Andersen, A. Baker, G. Bergametti, N. Brooks, J. J. Cao, P. W. Boyd, R. A. Duce, K. A. Hunter, H. Kawahata, N. Kubilay, J. La Roche, P. S. Liss, N. Mahowald, J. M. Prospero, A. J. Ridgwell, I. Tegen, and O. Torres. 2005. Global iron connections between desert dust, ocean biogeochemistry, and climate. Science 308:67-71. [DOI] [PubMed] [Google Scholar]
- 20.Karl, D., A. Michaels, B. Bergman, D. Capone, E. Carpenter, R. Letelier, F. Lipschultz, H. Paerl, D. Sigman, and L. Stal. 2002. Dinitrogen fixation in the world's oceans. Biogeochemistry 57:47-98. [Google Scholar]
- 21.Kustka, A., E. J. Carpenter, and S. A. Sanudo-Wilhelmy. 2002. Iron and marine nitrogen fixation: progress and future directions. Res. Microbiol. 153:255-262. [DOI] [PubMed] [Google Scholar]
- 22.Langlois, R. J., J. La Roche, and P. A. Raab. 2005. Diazotrophic diversity and distribution in the tropical and subtropical Atlantic Ocean. Appl. Environ. Microbiol. 71:7910-7919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mahaffey, C., R. G. Williams, G. A. Wolff, N. Mahowald, W. Anderson, and M. Woodward. 2003. Biogeochemical signatures of nitrogen fixation in the eastern North Atlantic. Geophys. Res. Lett. 30:1300. [Google Scholar]
- 24.Man-Aharonovich, D., N. Kress, E. B. Zeev, I. Berman-Frank, and O. Beja. 2007. Molecular ecology of nifH genes and transcripts in the eastern Mediterranean Sea. Environ. Microbiol. 9:2354-2363. [DOI] [PubMed] [Google Scholar]
- 25.Mazard, S. L., N. J. Fuller, K. M. Orcutt, O. Bridle, and D. J. Scanlan. 2004. PCR analysis of the distribution of unicellular cyanobacterial diazotrophs in the Arabian Sea. Appl. Environ. Microbiol. 70:7355-7364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mehta, M. P., D. A. Butterfield, and J. A. Baross. 2003. Phylogenetic diversity of nitrogenase (nifH) genes in deep-sea and hydrothermal vent environments of the Juan de Fuca ridge. Appl. Environ. Microbiol. 69:960-970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mills, M. M., C. Ridame, M. Davey, J. La Roche, and R. J. Geider. 2004. Iron and phosphorus co-limit nitrogen fixation in the eastern tropical North Atlantic. Nature 429:292-294. [DOI] [PubMed] [Google Scholar]
- 28.Montoya, J. P., C. M. Holl, J. P. Zehr, A. Hansen, T. A. Villareal, and D. Capone. 2004. High rates of N2 fixation by unicellular diazotrophs in the oligotrophic Pacific Ocean. Nature 430:1027-1031. [DOI] [PubMed] [Google Scholar]
- 29.Moore, C. M., M. Mills, A. Milne, R. J. Langlois, E. P. Achterberg, K. Lochte, R. Geider, and J. La Roche. 2006. Iron limits primary productivity during spring bloom development in the central North Atlantic. Global Change Biol. 12:626-634. [Google Scholar]
- 30.Nuebel, U., F. Garcia-Pichel, M. Kühl, and G. Muyzer. 1999. Quantifying microbial diversity: morphotypes, 16S rRNA genes, and carotenoids of oxygenic phototrophs in microbial mats. Appl. Environ. Microbiol. 65:422-430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Orcutt, K. M., F. Lipschulz, K. Gundersen, R. Arimoto, A. F. Michaels, A. H. Knap, and J. R. Gallon. 2001. A seasonal study of the significance of N2 fixation by Trichodesmium spp. at the Bermuda Atlantic Time-series Study (BATS) site. Deep-Sea Res. Part II 48:1583-1608. [Google Scholar]
- 32.Osborn, A. M., E. R. B. Moore, and K. N. Timmis. 2000. An evaluation of terminal-restriction fragment length polymorphism (T-RFLP) analysis for the study of microbial community structure and dynamics. Environ. Microbiol. 2:39-50. [DOI] [PubMed] [Google Scholar]
- 33.Paerl, H. W., L. E. Prufert-Bebout, and C. Guo. 1994. Iron-simulated N2 fixation and growth in natural and cultured populations of the planktonic marine cyanobacterium Trichodesmium spp. Appl. Environ. Microbiol. 60:1044-1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Proctor, L. 1997. Nitrogen-fixing, photosynthetic, anaerobic bacteria associated with pelagic copepods. Aquat. Microb. Ecol. 12:105-113. [Google Scholar]
- 35.Rosado, A., G. F. Duarte, L. Seldin, and J. D. van Elsas. 1998. Genetic diversity of nifH gene sequences in Paenibacillus azotofixans strains and soil samples analyzed by denaturing gradient gel electrophoresis of PCR-amplified gene fragments. Appl. Environ. Microbiol. 64:2770-2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sañudo-Wilhelmy, S. A., A. B. Kustka, C. J. Gobler, D. A. Hutchins, M. Yang, K. Lwiza, J. Burns, D. G. Capone, J. A. Raven, and E. J. Carpenter. 2001. Phosphorus limitation of nitrogen fixation by Trichodesmium in the central Atlantic Ocean. Nature 411:66-69. [DOI] [PubMed] [Google Scholar]
- 37.Schlitzer, R. 2004. Ocean Data View, 2.0 ed. AWI, Bremerhaven, Germany.
- 38.Short, S. M., and J. P. Zehr. 2005. Quantitative analysis of nifH genes and transcripts from aquatic environments. Methods Enzymol. 397:380-394. [DOI] [PubMed] [Google Scholar]
- 39.Short, S. M., B. D. Jenkins, and J. P. Zehr. 2004. Spatial and temporal distribution of two diazotrophic bacteria in the Chesapeake Bay. Appl. Environ. Microbiol. 70:2186-2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Staal, M., S. te Lintel Hekkert, P. Herman, and L. J. Stal. 2002. Comparison of models describing light dependence of N2 fixation in heterocystous cyanobacteria. Appl. Environ. Microbiol. 68:4679-4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Staal, M., F. J. R. Meysman, and L. J. Stal. 2003. Temperature excludes N2-fixing heterocystous cyanobacteria in the tropical oceans. Nature 425:504-507. [DOI] [PubMed] [Google Scholar]
- 42.Tegen, I., M. Werner, S. P. Harrison, and K. Kohfeld. 2004. Relative importance of climate and land use in determining present and future global soil dust emission. Geophys. Res. Lett. doi: 10.1029/2003GL019216. [DOI]
- 43.Voss, M., P. Croot, K. Lochte, M. Mills, and I. Peeken. 2004. Patterns of nitrogen fixation along 10°N in the tropical Atlantic. Geophys. Res. Lett. do1: 10.1029/2004GL020127. [DOI]
- 44.Wu, J., W. Sunda, E. A. Boyle, and D. M. Karl. 2000. Phosphate depletion in the western North Atlantic Ocean. Science 289:759-762. [DOI] [PubMed] [Google Scholar]
- 45.Zani, S., M. T. Mellon, J. L. Collier, and J. P. Zehr. 2000. Expression of nifH genes in natural microbial assemblages in Lake George, New York, detected by reverse transcriptase PCR. Appl. Environ. Microbiol. 66:3119-3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zehr, J. P., B. D. Jenkins, S. M. Short, and G. F. Steward. 2003. Nitrogenase gene diversity and microbial community structure: a cross-system comparison. Environ. Microbiol. 5:539-554. [DOI] [PubMed] [Google Scholar]
- 47.Zehr, J. P., M. T. Mellon, and S. Zani. 1998. New nitrogen-fixing microorganisms detected in oligotrophic oceans by amplification of nitrogenase (nifH) genes. Appl. Environ. Microbiol. 64:3444-3450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zehr, J. P., J. P. Montoya, B. D. Jenkins, I. Hewson, E. Mondragon, C. M. Short, M. J. Church, A. Hansen, and D. M. Karl. 2007. Experiments linking nitrogenase gene expression to nitrogen fixation in the North Pacific subtropical gyre. Limnol. Oceanogr. 52:169-183. [Google Scholar]
- 49.Zehr, J. P., and B. B. Ward. 2002. Nitrogen cycling in the ocean: new perspectives on processes and paradigms. Appl. Environ. Microbiol. 68:1015-1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.