Abstract
The construction of Saccharomyces cerevisiae strains that ferment lactose has biotechnological interest, particularly for cheese whey fermentation. A flocculent lactose-consuming S. cerevisiae recombinant expressing the LAC12 (lactose permease) and LAC4 (β-galactosidase) genes of Kluyveromyces lactis was constructed previously but showed poor efficiency in lactose fermentation. This strain was therefore subjected to an evolutionary engineering process (serial transfer and dilution in lactose medium), which yielded an evolved recombinant strain that consumed lactose twofold faster, producing 30% more ethanol than the original recombinant. We identified two molecular events that targeted the LAC construct in the evolved strain: a 1,593-bp deletion in the intergenic region (promoter) between LAC4 and LAC12 and a decrease of the plasmid copy number by about 10-fold compared to that in the original recombinant. The results suggest that the intact promoter was unable to mediate the induction of the transcription of LAC4 and LAC12 by lactose in the original recombinant and that the deletion established the transcriptional induction of both genes in the evolved strain. We propose that the tuning of the expression of the heterologous LAC genes in the evolved recombinant was accomplished by the interplay between the decreased copy number of both genes and the different levels of transcriptional induction for LAC4 and LAC12 resulting from the changed promoter structure. Nevertheless, our results do not exclude other possible mutations that may have contributed to the improved lactose fermentation phenotype. This study illustrates the usefulness of simple evolutionary engineering approaches in strain improvement. The evolved strain efficiently fermented threefold-concentrated cheese whey, providing an attractive alternative for the fermentation of lactose-based media.
At their most basic level, the rules of evolution are remarkably simple: species evolve by means of random variation (via mutation, recombination, or other operators); this process is followed by natural selection, in which the fittest tend to survive and reproduce, propagating their genetic material to future generations (35). Hence, natural selection drives organisms toward adaptation to changing environmental conditions.
Microbial populations and single-celled microorganisms respond in a flexible manner to environmental changes. The genetic basis of adaptation ranges from simple alterations of DNA sequences, such as point mutations, to major events in the genome structure. In a laboratory or in an industrial environment, this adaptation can result in strain instability or can be used as a method to improve biotechnological processes (46).
Classical (nonrecombinant) strain improvement has relied on random mutagenesis and screening procedures and has attained many successes (24). Metabolic engineering introduced the concept of rational design into strain development strategies (reviewed by Nielsen [22]). Inspired by natural evolution, evolutionary engineering exploits evolutionary principles to enhance microbial properties in a biotechnological context, provided the desired phenotype is amenable to direct or indirect selection (35). Evolutionary engineering of whole cells is expected to gain relevance both as a complementary strategy in metabolic engineering for strain development and as a tool to elucidate the molecular basis of desired phenotypes (35).
Many examples of evolutionary engineering approaches have been extensively reviewed by Sauer (35). A number of recent studies on the engineering of Saccharomyces cerevisiae for xylose metabolism (reviewed by Jeffries [14]) highlight the importance of evolutionary engineering strategies to complement rational metabolic engineering efforts. Recombinant strains with the genetic potential to utilize the new substrate (xylose) were subjected to long-term evolution experiments (using chemostats and/or serial batch cultivations), which resulted in improved strains (15, 17, 18, 38, 39). The improvement of an engineered S. cerevisiae strain for glycerol production has also been achieved previously by serial transfer (23).
Lactose is another sugar that cannot be metabolized by wild-type strains of S. cerevisiae. However, the construction of S. cerevisiae strains with the ability to consume lactose has biotechnological interest, particularly for the valorization of cheese whey, a high-level pollutant by-product of dairy industries that contains about 5% (wt/vol) lactose (37).
Kluyveromyces lactis is a naturally lactose-consuming yeast. The GAL-LAC regulon of K. lactis (reviewed in references 29 and 36) and the GAL-MEL regulon of S. cerevisiae (reviewed in references 16 and 20) are closely related. Despite the extensive degree of conservation in this group of genes between the two yeasts, differences have arisen as a result of their evolution in different environments: S. cerevisiae has adapted to glucose, whereas K. lactis has adapted to lactose (29). The ability of K. lactis to assimilate lactose depends on two genes: LAC12, encoding a lactose permease, and LAC4, encoding the enzyme β-galactosidase that catalyzes the hydrolysis of lactose into glucose and galactose. These genes are divergently transcribed from an unusually large intergenic region, which contains four functional upstream activating sites (UASs) that synergistically contribute to the activation of both genes by providing binding sites for the transcriptional activator Lac9p (also known as K. lactis Gal4p), homologous to Gal4p of S. cerevisiae (13). K. lactis GAL-LAC and S. cerevisiae GAL-MEL genes are induced by galactose. Induction also occurs when lactose is the substrate, since intracellular galactose is responsible for triggering induction (for details see, e.g., reference 29).
The development of a high-productivity lactose-fermenting process using recombinant S. cerevisiae has been considered for ethanol production from cheese whey. In this context, a recombinant S. cerevisiae flocculent strain with the ability to express both the LAC4 and LAC12 genes of K. lactis was constructed previously using a 13-kb K. lactis genomic sequence that included the two genes as well as their intergenic region (6). The original recombinant obtained metabolized lactose slowly. After a long-term adaptation experiment, in which the recombinant was cultured in liquid lactose medium, refreshed periodically, the strain was able to consume lactose faster with a higher ethanol yield than before. Here, we describe comparative physiological and genetic studies of the original recombinant and the evolved strain, aiming to identify mechanisms involved in the evolutionary adaptation of the recombinant to lactose. In addition, we show that the evolved recombinant is suitable for efficient alcoholic fermentation of concentrated cheese whey.
MATERIALS AND METHODS
Strains and cultivations.
The bacterial strain used for DNA preparation was Escherichia coli DH5α grown in Luria-Bertani medium (1% casein, 0.5% yeast extract, 0.5% NaCl). Ampicillin at 100 mg liter−1 was used for selection.
The original recombinant S. cerevisiae flocculent strain was named NCYC869-A3/T1 (hereafter referred to as T1), and its construction has been described in detail elsewhere (6). The other recombinant strain evolved from T1, as the result of the adaptation experiment described here, and is referred to as T1-E. These strains express both the LAC4 (β-galactosidase) and LAC12 (lactose permease) genes of K. lactis from an episomal plasmid (pKR1B-LAC4-1) (40).
The auxotrophic strain NCYC869-A3 (MATα FLO1 ura3), which was the host for the construction of T1, is a uracil-deficient mutant of the flocculent wild-type haploid strain S. cerevisiae NCYC869 (MATα FLO1) (6). K. lactis CBS2359 was used for comparative studies. Kluyveromyces marxianus CBS6556 was used as a control in some experiments.
Yeast cultivations were performed with defined mineral medium (41). The concentrations of trace elements and vitamins were doubled. The carbon source (lactose, glucose, or galactose) was autoclaved separately and added after heat sterilization of the medium to a concentration of 20 g·liter−1 (unless otherwise stated). To avoid major drops in pH during cultivation, the medium was supplemented with 100 mM potassium hydrogen phthalate. The initial pH was adjusted to 4.5 with NaOH. The final pH of the cultures in all experiments was higher than 3.7. The cultivations were carried out in Erlenmeyer flasks filled with medium to 40% of the total volume and shaken (150 rpm) at 30°C. Preinocula were grown under the same conditions and used to inoculate the main cultures to an initial optical density at 600 nm (OD600) of 0.05 to 0.15.
During the adaptation experiment, yeast was cultivated in semisynthetic lactose (SSlactose) medium, which contained the following (in grams per liter): yeast extract, 1.0; KH2PO4, 5.0; (NH4)2SO4, 2.0; MgSO4·7H2O, 0.4; and lactose, 20 or 50. Fermentation conditions in the 2-liter bioreactor used during the last stages of the adaptation experiment were as follows: temperature, 30°C, and agitation speed, 150 rpm. The pH control was set to 4.0.
For the batch fermentation of threefold-concentrated cheese whey, 600 g of whey powder obtained from a Portuguese dairy was dissolved with 2.2 liters of warm water and the solution was further warmed to boiling temperature. The precipitate was removed by filtering with a cloth followed by centrifugation. A 1.5-liter sample of the cheese whey powder solution was then transferred into the bioreactor and sterilized by autoclaving. Fermentation was performed in a 2-liter benchtop bioreactor (Bioengineering). The temperature was maintained at 30°C, and the pH was maintained at 4.0 by the automatic addition of ammonia. The agitation speed was set at 150 rpm. An airflow rate of 0.1 vvm (where 1 vvm equals 1 standard liter of air per liter of solution per min; adjusted by mass flow control) was applied during the cultivation. The yeast (strain T1-E) for inoculation was grown in a shake flask (containing yeast defined mineral medium with 20 g of lactose·liter−1), as described above.
Adaptation experiment.
The original recombinant (T1) was taken from a −80°C stock and spread onto SSlactose plates. Biomass washed from these plates was used to inoculate test tubes filled with 5 ml of liquid SSlactose medium (containing 20 g of lactose·liter−1). The tubes were incubated overnight at 30°C with gentle agitation (40 rpm), and then the biomass from each tube was transferred into a 250-ml Erlenmeyer flask filled with 50 ml of SSlactose medium (containing 20 g of lactose·liter−1). This flask was incubated at 30°C and 40 rpm for 3 days. From this flask, a 5-ml sample was transferred into another cultivation (under the same conditions) and cultivated for 7 days. Samples taken from this cultivation were spread onto SSlactose plates. Biomass was again washed from these plates to inoculate test tubes filled with 5 ml of liquid SSlactose medium (containing 50 g of lactose·liter−1), the tubes were incubated overnight (at 30°C and 40 rpm), and the contents were used to inoculate a cultivation in 50 ml of SSlactose (containing 50 g of lactose·liter−1). This cultivation was incubated under the same conditions (30°C and 40 rpm). To take advantage of the flocculent properties of the cells, the medium was refreshed periodically: the cultivation broth was decanted, and fresh medium was added to the flocculated cells, which had sedimented to the bottom of the flask. The medium was refreshed 12 times (every 2 to 5 days) over a total of 41 days. The lactose concentration in the fresh medium added was either 20 or 50 g·liter−1. After the 41-day period, 5 ml of the culture was transferred into a subsequent cultivation (to be grown under the same conditions), in which the medium was refreshed two times. This 50-ml cultivation was used to inoculate a 2-liter bioreactor containing 1.5 liters of SSlactose medium. In this fermentation, the 20 g of lactose·liter−1 was consumed by the yeast in about 24 h. Samples from this fermentation were spread onto SSlactose plates. Biomass washed from these plates was again sequentially grown in 5 and 150 ml of SSlactose medium (with incubation at 30°C and 150 rpm) and used for a second fermentation trial in the bioreactor. This time, the yeast consumed the 20 g of lactose·liter−1 in less than 16 h. A third fermentation trial in the bioreactor was done by following the same procedure. In this third fermentation, the yeast consumed 20 g of lactose·liter−1 in about 13 h. Yeast samples were spread onto SSlactose plates. Glycerol stocks from this yeast were stored at −80°C. We named this strain T1-E (evolved T1).
The adaptation experiment involved the growth of the recombinant for >120 generations (given an average of five doublings per batch cultivation in lactose).
Determination of biomass and extracellular metabolites.
The biomass concentration was measured by a dry weight method and/or an absorbance method (at 600 nm [OD600]), essentially as previously described (6).
Lactose, glucose, galactose, ethanol, and glycerol were analyzed by high-performance liquid chromatography using a Chrompack organic acids column. The column was eluted at 60°C with 0.005 M H2SO4 at a flow rate of 0.6 ml·min−1. A refractive-index detector was used.
Flocculation assay.
The flocculation assay was done essentially as described previously (6, 7), except that the absorbance was read at 600 nm (OD600) instead of 620 nm. The sedimentation profiles correspond to the normalized cell concentration (defined as the ratio between actual and initial cell concentrations in the suspension during the assay) plotted against the sedimentation time. The profiles shown correspond to the average of results from two independent assays. In control assays, the cell concentration in the suspension did not change significantly (±4%) over 10 min.
Plasmid retention.
To determine the fraction of plasmid-bearing cells, samples from the yeast culture were washed twice with a 15-g·liter−1 NaCl solution, pH 3.0, to deflocculate cells, appropriate dilutions were prepared with the same solution, and aliquots were spread onto plates containing YPGal (1% yeast extract, 2% peptone, 2% galactose, 2% agar) supplemented with 40 mg of X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside)·liter−1. Cells expressing β-galactosidase, i.e., those that still carry the plasmid, form blue colonies in X-Gal plates. The fraction of plasmid-bearing cells was determined as the ratio between the number of blue colonies and the total number of colonies (blue and white).
β-Galactosidase-specific activity.
Yeast cells were grown to an OD600 of 0.5 to 2.5. A volume of the culture corresponding to 15 to 20 OD600 units was rapidly harvested, and cellular extracts were prepared exactly as described previously (26). The β-galactosidase activity in the extracts was assayed using p-nitrophenyl-β-d-galactopyranoside as the substrate, exactly as described previously (26). The protein concentration in the extracts was measured according to the method of Bradford (2) by using the Bio-Rad protein reagent and ovalbumin as the standard. At least three different dilutions of each extract were assayed for both enzyme activity and protein concentration; the coefficient of variation of each specific activity measurement was <30%. One unit of enzyme activity catalyzes the conversion of 1 nmol of p-nitrophenyl-β-d-galactopyranoside per min at 30°C and pH 7.0. Specific activities were expressed as units per milligram of protein.
General DNA methods.
Standard molecular biology techniques were used basically according to the procedures described by Sambrook and Russell (34). Yeast was transformed by the lithium acetate method (12). E. coli was transformed by electroporation according to protocols from Bio-Rad. Commercial kits (from Qiagen and Sigma) were used for plasmid DNA isolation from E. coli. Enzymes were purchased from different manufacturers and used according to the recommendations.
PCR amplifications.
Ribosomal DNA (rDNA) internal transcribed spacer 1 (ITS1) and ITS1-5.8S rDNA-ITS2 regions were amplified by PCR using universal primers for fungi (42). The primers (FLO11_Fprobe and FLO11_Rprobe) used for PCR screening for the FLO11 gene of S. cerevisiae were described previously (10).
PCR screening for the LAC4 gene was done with primers LAC4_1 (5′-AGGATTTACAGTGGGAG GAT-3′) and LAC4_2 (5′-ATGTCTTGCCTTATTCCTGA-3′). For LAC12, primers LAC12_1 (5′-GGAAGCCTTGAACAGTGATA-3′) and LAC12_2 (5′-AGACCTGCAACCTTACCTCT-3′) were used.
The intergenic region between genes LAC4 and LAC12 (LACIR) was amplified using the primers LACIR1 (5′-GCAATCTATTTTCGTGAACC-3′) and LACIR2 (5′-CCCAAAGTGTC TTTATGCTC-3′). Primer LACIR1 is complementary to nucleotides 42 to 61 of the coding sequence of LAC4 (25). Primer LACIR2 is complementary to nucleotides 58 to 77 of the coding sequence of LAC12 (4).
RNA extraction.
Yeast cells for total RNA isolation were grown to an OD600 of 0.5 to 0.7. A volume of the culture corresponding to 15 OD600 units was rapidly centrifuged (3,000 rpm at 4°C for 3 min), and the cell pellets were immediately frozen in liquid nitrogen and stored at −80°C. The cells were mechanically disrupted using a ball mill (Mikro-Dismembrator S; B. Braun Biotech International). Total RNA was extracted using an RNeasy mini kit (Qiagen). The extracted RNA was quantified using the Bioanalyzer 2100 with the RNA 6000 Nano LabChip kit (Agilent) and the ND-1000 UV-visible light spectrophotometer (NanoDrop Technologies). The absence of contaminant DNA in RNA preparations was verified using RNA as a template in real-time PCR assays (see below).
Quantitative real-time RT-PCR.
One microgram of total RNA was transcribed into cDNA in a 20-μl reaction mixture by using the iScript cDNA synthesis kit (Bio-Rad). The mRNA levels were analyzed by quantitative reverse transcription-PCR (RT-PCR) using the Bio-Rad MyIQ real-time PCR system. Each sample was tested in duplicate in a 96-well plate (Bio-Rad, CA). The reaction mix (a 25-μl final volume) consisted of 12.5 μl of Sybr green 2× supermix (Bio-Rad), 2.5 μl of each primer (at a 250 nM final concentration), 2.5 μl of H2O, and 5 μl of a 1/10 dilution of the cDNA preparation. The thermocycling program consisted of one hold at 95°C for 4 min, followed by 40 cycles of 10 s at 95°C and 45 s at 56°C. Melting-curve data were then collected to verify PCR specificity and the absence of primer dimers. Oligonucleotides for quantitative PCR (Table 1) were designed using Beacon Designer 2.0 software (PREMIER Biosoft International). The PCR efficiency of each primer pair was evaluated by the dilution series method using sample cDNA as the template.
TABLE 1.
Oligonucleotides used for quantitative RT-PCR
| Oligonucleotide | Sequence | Amplicon size (bp) | Efficiencya |
|---|---|---|---|
| ACT1_QPCR_F1bis | ATTATATGTTTAGAGGTTGCTGCTTTGG | 285 | 1.98 |
| ACT1_QPCR_R1bis | CAATTCGTTGTAGAAGGTATGATGCC | ||
| Bla_QPCR_F13 | CATTTCCGTGTCGCCCTTATTCCC | 147 | 2.11 |
| Bla_QPCR_R159 | CTTACCGCTGTTGAGATCCAGTTCG | ||
| LAC12_QPCR_F742 | GGTCTTGTGTGTATATTTGGTTGGTTAATCCC | 246 | 1.97 |
| LAC12_QPCR_R987 | TGCTCTGTACCTATCCGATCTCGTTCTG | ||
| LAC4_QPCR_F1733 | GTGGCTTTATCTGGGAATGGGCAAATC | 278 | 2.03 |
| LAC4_QPCR_R2010 | CAATAAGTGGTCTGTCGTAATGAAGTCGTG |
Efficiency (E) was determined using the formula E = 10(−1/slope), with “slope” being the slope of the standard curve that was obtained from serial dilution of sample cDNA.
Relative expression levels were determined using the Δ threshold cycle method, which takes into account differences in primer pair amplification efficiencies and yields more accurate data than the 2ΔΔ threshold cycle method. For standardization, the results were expressed as ratios of target gene expression to reference gene expression, the reference gene being genome-borne ACT1 or a plasmid-borne bla gene.
RESULTS
Adaptation.
The original recombinant strain (T1) was able to metabolize lactose, but rather slowly. Moreover, the flocculation performance of the recombinant was poor compared to that of the host strain S. cerevisiae NCYC869-A3 (6). Hence, the adaptation of the recombinant strain to growth on lactose was attempted. The adaptation experiment consisted of a serial transfer-dilution strategy using flasks subjected to gentle shaking (40 rpm; see details in Materials and Methods). This strategy was designed to keep the recombinant growing in lactose for many generations (>120), as well as to select for flocculent cells. Therefore, in some stages of the process, the medium was simply refreshed periodically in the same cultivation flask: the cultivation broth was decanted and fresh medium was added to the flocculated cells, which had sedimented to the bottom of the flask. The yeast cells recovered at the end of the process presented improved lactose fermentation performance compared to T1 cells. We now consider these evolved cells to belong to an independent strain, which was named T1-E.
The evolved strain culture was tested for possible contamination, particularly with Kluyveromyces yeasts. ITS regions located on the rRNA gene clusters are highly variable, allowing for species distinction even within the Saccharomyces genus (21). Restriction fragment length polymorphism analysis (with the enzymes EcoRI, HaeIII, HindIII, HinfI, MseI, PstI, RsaI, and TaqI) of the PCR-amplified ITS1-5.8S rDNA-ITS2 region generated identical profiles for the recombinants T1 and T1-E and for the host strain S. cerevisiae NCYC869. Moreover, PCR amplification of the ITS1 region resulted in a smaller product for K. marxianus (ca. 350 bp) than for S. cerevisiae (ca. 450 bp) (data not shown). In an independent test, PCR screening for the S. cerevisiae FLO11 gene yielded positive results for strains T1, T1-E, and S. cerevisiae NCYC869 but negative results for K. lactis CBS2359 and K. marxianus CBS6556 (data not shown). These results confirmed that strains T1 and T1-E are S. cerevisiae, excluding the possibility of contamination during the adaptation experiment.
Physiological characterization of the recombinants.
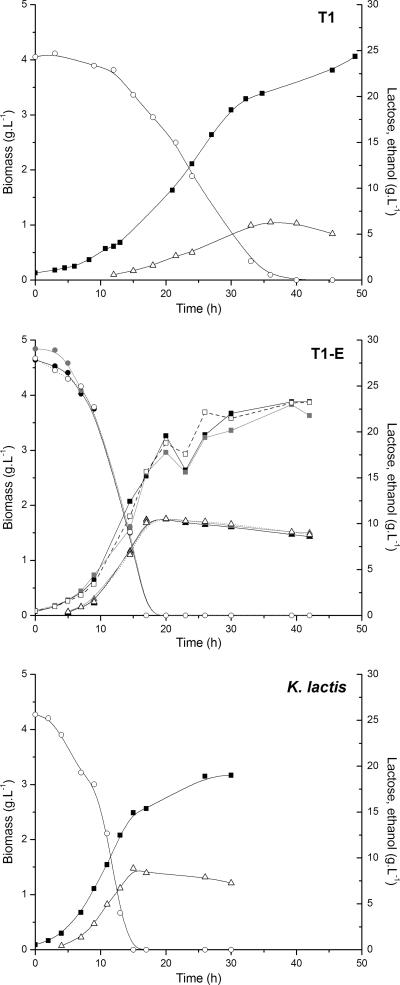
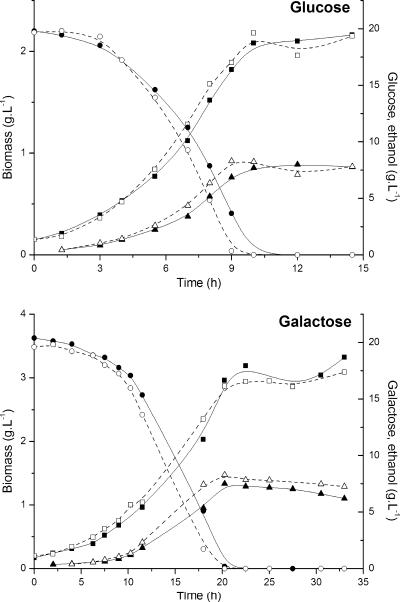
Three single-colony isolates of the evolved strain showed identical fermentation profiles (Fig. 1; Table 2), indicating that the T1-E population was homogenous. The lactose fermentation performance of the evolved strain was considerably improved compared to that of the original recombinant. T1-E fermented lactose twice as fast as the original recombinant T1, presenting a higher growth rate as well as 30% higher ethanol production, which indicated that the flux of carbon (lactose) through the fermentative pathway occurred at a higher level in T1-E (Fig. 1; Table 2). The differences in lactose fermentation between the evolved recombinant and the K. lactis wild-type strain were small, though K. lactis grew faster and produced less ethanol (Fig. 1; Table 2). Conversely, the two recombinant strains behaved similarly in cultivations with glucose and galactose (the two products of lactose hydrolysis) (Fig. 2; Table 2).
FIG. 1.
Data from lactose cultivations with strains T1, T1-E, and K. lactis CBS2359. For T1-E, data from triplicate cultivations with single-colony isolates are shown. Concentrations of lactose (circles), ethanol (triangles), and biomass (squares) were monitored during shake flask cultivations (as described in Materials and Methods).
TABLE 2.
Comparison of fermentation parameters of strains T1 and T1-E in lactose, glucose, and galactose cultivations, as well as K. lactis CBS2359 in lactosea
| Sugar | Strain | μ (h−1) | Xfinal (g liter−1) | Emax (g liter−1) | Eyield (%) |
|---|---|---|---|---|---|
| Lactose | T1 | 0.14 ± 0.01 | 3.48 ± 0.09 | 7.08 ± 0.79 | 53 ± 5 |
| T1-E | 0.21 ± 0.01 | 2.81 ± 0.09 | 10.52 ± 0.04 | 69 ± 1 | |
| K. lactis | 0.28 | 2.56 | 8.86 | 65 | |
| Glucose | T1 | 0.32 ± 0.03 | 2.14 ± 0.03 | 8.19 ± 0.19 | 81 ± 2 |
| T1-E | 0.34 ± 0.03 | 2.13 ± 0.03 | 8.05 ± 0.26 | 80 ± 2 | |
| Galactose | T1 | 0.15 ± 0.01 | 2.94 ± 0.13 | 7.10 ± 0.40 | 69 ± 4 |
| T1-E | 0.15 ± 0.01 | 2.94 ± 0.00 | 7.65 ± 0.64 | 76 ± 7 |
Cultivations were done as described in Materials and Methods. The initial lactose concentration was 25 g liter−1; the initial glucose and galactose concentrations were 20 g liter−1. μ, specific growth rate; Xfinal: final biomass concentration (when sugar from the medium was exhausted); Emax, maximum ethanol concentration; Eyield, ethanol conversion yield (percentage of the theoretical value). Data for T1 and T1-E are means ± ranges of results from duplicate independent cultivations, except data for T1-E in lactose, which correspond to the means ± standard deviations of results from triplicate cultivations with single-colony isolates.
FIG. 2.
Data from glucose and galactose cultivations with strains T1 and T1-E. Concentrations of glucose or galactose (circles), ethanol (triangles), and biomass (squares) were monitored during shake flask cultivations (as described in Materials and Methods). Solid symbols and solid lines correspond to T1 cultures; hollow symbols and dotted lines correspond to T1-E cultures.
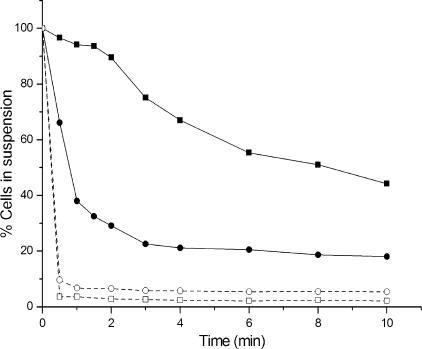
The adaptation experiment was also successful in the selection of cells with improved flocculation, which is an interesting property for the development of fermentation processes (5). T1-E flocculated earlier and formed much bigger flocs than T1, as could be easily observed by visual inspection of the cultivation flasks. Sedimentation profiles of lactose-grown T1 and T1-E cells are shown in Fig. 3. When the culture reached an OD600 of about 2, nearly all T1-E cells showed the ability to flocculate. The flocculation performance of T1 was much weaker, although that of stationary-phase cells improved compared to that of growing cells. Similar differences in the flocculation behavior between the two strains were also observed in glucose and galactose cultures.
FIG. 3.
Sedimentation profiles of T1 and T1-E cells grown in lactose. Cells were harvested during growth at an OD600 of about 2 (T1-E, □; T1, ▪) or at the stationary phase after 50 h of cultivation (T1-E, ○; T1, •).
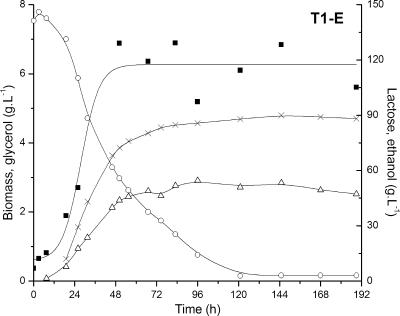
The evolved strain T1-E was tested for the fermentation of cheese whey. Cheese whey powder solution was used in order to get threefold-concentrated whey (corresponding to about 150 g of lactose·liter−1). The recombinant was able to grow and flocculate in cheese whey powder solution and consumed nearly all the lactose in about 120 h, producing 55 g of ethanol·liter−1 (which corresponds to 70% of the theoretical conversion yield) and 5 g·of glycerol liter−1 (Fig. 4). In a shake flask cultivation with buffered defined mineral medium containing 140 g of lactose·liter−1, T1-E was able to consume lactose completely in only 36 h, producing 63 g of ethanol·liter−1 (84% of the theoretical yield).
FIG. 4.
Fermentation of threefold-concentrated cheese whey by the evolved recombinant T1-E. Concentrations of lactose (○), ethanol (▵), biomass (▪), and glycerol (×) were monitored during batch fermentation in a 2-liter bioreactor (as described in Materials and Methods).
Plasmid retention.
Under positive selection pressure conditions, i.e., with lactose as the sole carbon source, recombinant cells need to carry the plasmid in order to retain their capacity to grow on lactose. However, in glucose and in galactose cultivations, there was no selective pressure to prevent plasmid loss. Therefore, we counted the fraction of cells expressing β-galactosidase (blue colonies in plates supplemented with X-Gal), which corresponds to the fraction of cells still carrying the plasmid. The vast majority of the cells in T1-E cultures contained the plasmid (96 and 93% in glucose and galactose, respectively). In contrast, only 74 and 27% of T1 cells in glucose and galactose cultures, respectively, maintained the construct. For both T1 and T1-E, plasmid retention patterns were similar in yeast samples harvested during growth phase (sampling at 7 h of growth in glucose or 6 to 11 h in galactose) and stationary phase (28 h in glucose and 30 to 50 h in galactose).
Characterization of the LAC region in the plasmid isolated from both recombinants.
To search for mutations that could explain the differences observed in the lactose fermentation phenotypes of the two recombinants, the plasmid bearing the K. lactis LAC12-LAC4 construct was isolated from the two strains and recloned into E. coli. Restriction analyses with the enzymes EcoRI, BglII, XbaI, BamHI, and PstI (data not shown) revealed a deletion of 1,300 to 1,600 bp in the LACIR on the plasmid isolated from T1-E, while the restriction pattern of the plasmid isolated from T1 was identical to that of the plasmid pKR1B-LAC4-1 used for transformation. PCR amplification of the LACIR in T1 and T1-E resulted in single bands (data not shown) and confirmed the deletion in T1-E.
The LACIR of the T1-E plasmid was therefore sequenced and compared with the same region in the recently published K. lactis genome (GenBank accession no. CR382122) (9). A deletion of 1,593 bp was mapped between positions −2108 and −516 (inclusive), +1 referring to the adenosine in the LAC4 initiation codon (Fig. 5). Furthermore, a single nucleotide mutation was found at position −2422 which replaced the adenosine in the K. lactis genome sequence with a cytosine in the T1-E sequence. This substitution is interesting because it leads to a putative binding site for the transcription factor Gal4p: CGGCCCACGCAGACCCG (the A-to-C mutation occurred in the underlined position). We found the same substitution in the LACIR borne on the plasmid isolated from T1. However, we do not know whether this putative UAS (pU) (Fig. 5) was functional in any of the strains.
FIG. 5.
Deletion identified in the LAC12-LAC4 intergenic region of the plasmid isolated from the evolved strain (T1-E). The 1,593-bp deletion (open box) was mapped between positions −516 and −2108, +1 referring to the adenosine in the LAC4 initiation codon. The functional UAS elements (U) present in the LAC promoter (13) are represented by vertical black bars and correspond to the 17-bp consensus sequence 5′-CGG(N5)A/T(N5)CCG-3′ (in the figure, the central position of the consensus sequence at each UAS is indicated in parentheses). The gray vertical bar represents an additional putative UAS (pU) found in T1 and T1-E (see the text for details).
LAC12 and LAC4 coding sequences in the T1-E plasmid were determined to be 100% identical to the published sequences for these genes (4, 9, 25).
Transformation of the host strain with the plasmid isolated from T1-E did not yield Lac+ transformants.
Several attempts were made to transform S. cerevisiae strains NCYC869 (the wild type) and NCYC869-A3 (a uracil-deficient mutant) with the plasmid isolated from T1-E (with the deletion). Although transformants resistant to the antibiotic G418 (used for selection) were obtained, these transformants were unable to grow on lactose. It should be stressed that the same result happened in control experiments using the original plasmid pKR1B-LAC4-1. We reasoned that genetic alterations in the T1 genome may have contributed to obtaining the Lac+ phenotype, in addition to the cloning of the K. lactis LAC genes. Therefore, we used the cured strain T1 (a plasmid-free colony selected after nonselective sequential growth in glucose) as the host for transformation. With this host, we again obtained transformants resistant to G418 (up to 150 mg·liter−1). Some of the G418-resistant transformants (>10%) gave a positive PCR signal for the genes LAC4 and LAC12, both in the transformations with the plasmid isolated from T1-E and in those with pKR1B-LAC4-1. Surprisingly, none of the transformants could grow on lactose (plates or liquid medium) or showed β-galactosidase activity (as judged by color screening in YPGal plates supplemented with X-Gal).
LAC4 and LAC12 expression levels and estimation of relative plasmid copy numbers.
We reasoned that the deletion in the promoter region may have altered the expression of one or both genes. Thus, we compared the levels of LAC4 and LAC12 transcripts in strains T1 and T1-E by using quantitative real-time RT-PCR, as well as β-galactosidase activity.
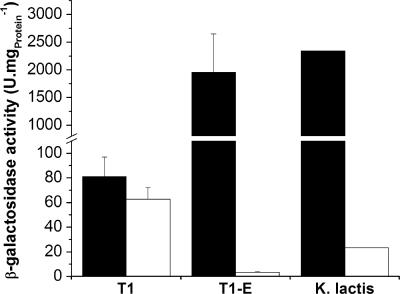
The LAC4 and LAC12 mRNA levels were initially normalized to ACT1 transcript levels. In lactose cultures (inducing conditions), the LAC4 mRNA level in T1-E was about 2.2-fold higher than that in T1 (Table 3). Nevertheless, the specific β-galactosidase activity measured in T1-E was 24-fold higher than that in T1 and was essentially similar to that in K. lactis CBS2359 (Fig. 6). Conversely, the LAC12 transcript level in T1-E was about 2.2-fold lower than that in T1 (Table 3). In glucose cultures (noninducing and repressing conditions), the levels of both LAC4 and LAC12 transcripts in T1-E were about 10-fold lower than those in T1 (Table 3). This result could be attributed to the decrease of plasmid copy number in the evolved strain (discussed below). The decrease in the LAC4 transcript level may account for the lower level of β-galactosidase activity observed in T1-E glucose cultures than in T1 cultures (Fig. 6).
TABLE 3.
Relative expression of bla, LAC4, and LAC12 genes during growth of T1 and T1-E in lactose and in glucose (after normalization with ACT1)a
| Gene | % Expression during growth in lactose of:
|
% Expression during growth in glucose of:
|
||
|---|---|---|---|---|
| T1 | T1-E | T1 | T1-E | |
| bla | 100 ± 1 | 8 ± 0 | 50 ± 5 | 6 ± 1 |
| LAC4 | 100 ± 4 | 225 ± 6 | 69 ± 2 | 7 ± 2 |
| LAC12 | 100 ± 3 | 45 ± 5 | 66 ± 5 | 5 ± 2 |
Expression levels were determined by quantitative real-time RT-PCR, as described in Materials and Methods. Expression levels of bla, LAC4, and LAC12 were normalized using ACT1 as the reference gene. T1 cells grown in lactose were used as the calibrator sample (the expression was set at 100%). Results shown are means ± ranges of results from duplicate biological cultivations. Each sample (one sample from each biological duplicate) was analyzed in duplicate, and the coefficient of variation (after normalization) between the results for these technical duplicates was <30%.
FIG. 6.
β-Galactosidase activities of strains T1, T1-E, and K. lactis CBS2359. Cultivations were done as described in Materials and Methods with either 2% lactose (solid bars) or 2% glucose (hollow bars). Cell extracts were prepared from growing cells (OD600, 0.5 to 2.5). Error bars (for T1 and T1-E) represent standard deviations of results from three (lactose) or four (glucose) independent cultivations. β-Galactosidase activity was not detected in the supernatants from any of the cultivations.
The global differences in levels of LAC4 and LAC12 mRNAs, as calculated by normalization to the expression levels of an endogenous gene (ACT1) (Table 3), may be due to differential regulation of transcription and/or variation of gene copy number, i.e., plasmid copy number, between the two strains. Therefore, to verify if our results were biased by a difference in LAC gene copy number, we estimated the relative plasmid copy numbers in the two recombinants by quantifying the transcript levels of the plasmid-borne bla (β-lactamase) gene. bla confers resistance to ampicillin on E. coli and is known to be expressed in S. cerevisiae (28). Assuming that there is no differential regulation of this gene in T1 and T1-E, the relative levels of bla expression reflect the difference in the average numbers of plasmid copies per cell in the cultures. The level of bla expression in T1 was higher than that in T1-E; specifically, it was 12-fold higher in lactose cultures and 8-fold higher in glucose cultures (Table 3). However, in glucose cultures, only the fraction of plasmid-bearing cells (74 and 96% for T1 and T1-E, respectively, as described above) was expressing the bla gene. Hence, considering only the plasmid-bearing cells, bla expression in T1 was 11-fold higher than that in T1-E in glucose cultures, and this difference was therefore similar to the difference observed in lactose. These results for bla expression were obtained in independent duplicate biological experiments, both with lactose and with glucose, indicating that the differences observed were not due to random variation in plasmid copy number between cultivations. Furthermore, T1 showed a higher level of resistance to the antibiotic G418 (conferred on yeast by the plasmid-encoded bacterial kanamycin resistance gene). T1 grew well on plates supplemented with 50 mg of G418·liter−1, while only a few small T1-E colonies could be observed. In plates with 75 mg of G418·liter−1, T1 was the only strain still able to grow. Altogether, these results indicate that the plasmid copy number in the evolved strain was lower (approximately 10-fold) than that in the original recombinant.
The LAC4 and LAC12 mRNA levels were therefore normalized to bla transcript levels, which eliminates the effect of plasmid copy number variation so that the results represent only the effect of differential regulation of the transcription of the LAC genes in T1 and T1-E. As illustrated by the results shown in Table 4, LAC4 and LAC12 induction factors (the ratio between expression in lactose and that in glucose) for T1-E were 26 and 7, respectively, whereas the induction factor for T1 was 0.7 for both genes. These results show that lactose activated the transcription of both genes in T1-E, although the induction factor was higher for LAC4 than for LAC12. On the other hand, lactose did not induce the expression of the LAC genes in T1, which is supported by the observation that the β-galactosidase activities of T1 in lactose and in glucose cultures were similar (Fig. 6). We propose that the intact promoter was not able to mediate activation of the transcription of the LAC genes in T1 (no induction by lactose), whereas the 1,593-bp deletion in the promoter region contributed to lactose activation in T1-E.
TABLE 4.
Relative expression of LAC4 and LAC12 genes during growth of T1 and T1-E in lactose and in glucose (after normalization with bla)a
| Gene | % Expression during growth in lactose of:
|
% Expression during growth in glucose of:
|
||
|---|---|---|---|---|
| T1 | T1-E | T1 | T1-E | |
| LAC4 | 100 ± 3 | 2,677 ± 211 | 138 ± 17 | 103 ± 12 |
| LAC12 | 100 ± 4 | 531 ± 30 | 133 ± 21 | 76 ± 19 |
Expression levels were determined by quantitative real-time RT-PCR, as described in Materials and Methods. Expression levels of LAC4 and LAC12 were normalized using bla as the reference gene. T1 cells grown in lactose were used as the calibrator sample (the expression was set at 100%). Results shown are means ± ranges of results from duplicate biological cultivations. Each sample (one sample from each biological duplicate) was analyzed in duplicate, and the coefficient of variation (after normalization) between the results for these technical duplicates was <30%.
DISCUSSION
A recombinant S. cerevisiae flocculent strain with the genetic potential to utilize lactose was constructed previously (6). Using a simple evolutionary engineering process consisting of serial transfer and dilution in lactose medium for >120 generations, we succeeded in isolating an evolved recombinant that fermented lactose twofold faster than the original recombinant T1, presenting a higher ethanol yield and improved flocculation.
A similar process of adaptation had already been attempted before with the recombinant T1, which resulted in a comparable improvement of the lactose fermentation phenotype. That adapted strain was successfully used in long-term continuous lactose fermentations (7, 8). However, the strain lost its apparently adapted phenotype after storage at −80°C. When cultures stored at −80°C were regrown, the limitation on lactose consumption (the T1 phenotype) was again observed (for details, see reference 6). Presumably, the outcome of that former adaptation experiment was a heterogeneous population containing adapted and nonadapted cells, and during the storage and regrowth phase nonadapted cells were consistently selected. The evolved strain described here (T1-E) resulted from an independent adaptation experiment, and its fermentative characteristics were maintained after storage at −80°C.
The evolved strain T1-E presented an improved lactose fermentation phenotype but otherwise performed identically to the original recombinant T1 in the fermentation of glucose and galactose. This result suggested the occurrence of mutations in lactose-specific genes, rather than in pathways affecting sugar metabolism in general. Accordingly, we identified two major molecular events that specifically targeted the lactose metabolism system in T1-E. The first event was a 1,593-bp deletion in the LACIR in the plasmid isolated from the evolved recombinant. The second event was a reduction of the plasmid copy number by about 10-fold in T1-E compared to that in the nonevolved strain T1. Moreover, our transcriptional analysis revealed that lactose strongly induced the expression of LAC4 and LAC12 in the evolved strain but that in the original recombinant, these genes were expressed at the same levels in lactose and in glucose cultures. This finding indicates that the K. lactis LAC promoter was unable to mediate the induction of the LAC genes by lactose in S. cerevisiae and that the 1,593-bp deletion in the LACIR apparently restored induction. It is noteworthy that the K. lactis LACIR contains four functional UASs, which are binding sites for the trans-activator Lac9p, the K. lactis homologue of S. cerevisiae Gal4p. Two UAS elements are located in front of each of the genes at almost symmetrical positions (Fig. 5), and it has been proposed previously that LAC4- and LAC12-proximal sites interact to achieve the maximal expression of both genes simultaneously in K. lactis (13).
In our recombinant strains, the trans-activator is S. cerevisiae Gal4p, which substitutes for the K. lactis Lac9p. Lac9p and Gal4p bind to the same DNA consensus sequence: 5′-CGG(N5)A/T(N5)CCG-3′ (13, 19). GAL4 can complement the transcriptional activation function of LAC9 in K. lactis (27), and conversely, LAC9 complements the gal4 mutation in S. cerevisiae (33, 43). However, Gal4p did not exactly mimic Lac9p function, and vice versa. Even though Gal4p is functionally analogous to Lac9p, specific features of each of the proteins (which share only three regions of significant homology, accounting for about 30% of the amino acids), as well as their cellular concentrations, seem to have regulatory relevance (29, 44, 45, 47). Thus, a reasonable interpretation of our results is that Gal4p cannot effectively replace Lac9p in the LAC promoter. We propose that the deletion in the LACIR altered the architecture of the promoter, giving rise to a structure favorable for transcriptional activation by the S. cerevisiae Gal4p. The removal of sequences involved in transcription repression mechanisms existing in the deleted region cannot be excluded.
Unexpectedly, the copy number of the plasmid bearing the LAC construct in T1-E was lower than that in T1. We reasoned that the transformant originally selected (T1) owed its ability to grow on lactose to its high plasmid content. Considering that under the control of the intact K. lactis LAC promoter the LAC genes were expressed at low (noninduced) levels, it is conceivable that only transformants with a high dosage of these genes would be able to sustain viability and growth on lactose. Furthermore, during T1 lactose cultivations, cells with higher plasmid copy numbers would be selected, since these would have a clear advantage. In fact, Sreekrishna and Dickson (40) found that only the transformants that had integrated the LAC genes at high copy numbers (15 to 25 tandem copies) were able to grow on lactose.
Our results suggest that the tuning of the expression levels of the LAC genes in the evolved recombinant was accomplished by a fine molecular interplay between the decreased copy numbers of both genes and the different levels of transcriptional induction of LAC4 and LAC12 by lactose resulting from a large deletion in the LACIR. Changes in gene copy number (1, 3), as well as mutations in cis regulatory sequences, have been identified previously as genetic events involved in yeast adaptive responses to environmental stresses. Recently, Fidalgo et al. (11) found that a 111-bp deletion within a repression region of the FLO11 gene promoter (possibly the largest S. cerevisiae promoter, with about 3 kb), which significantly increased the expression of FLO11, was determined to confer to wine “flor” yeasts the ability to float (an adaptive mechanism of these yeasts to gain direct access to oxygen).
Besides these two identified mechanisms, we cannot exclude the existence of additional mutations related to the improvement of the lactose fermentation phenotype. Surprisingly, transforming the original host or cured (plasmid-free) T1 strain with the plasmid isolated from T1-E did not yield Lac+ transformants. Our results emphasize that lactose utilization in S. cerevisiae is a complex trait, which is not easily achieved by the transfer of the LAC genes. To our knowledge, only our previous work (6) and the work of Sreekrishna and Dickson (40) have addressed the construction of Lac+ S. cerevisiae recombinants by cloning the K. lactis LAC genes under the control of the endogenous promoter (the LACIR). Sreekrishna and Dickson (40) obtained Lac+ transformants only when using indirect selection (first selecting for G418 resistance and then for growth on lactose) and when the LAC construct integrated in 15 to 25 tandem copies. The strategy for the construction of our original recombinant T1 also involved indirect selection (for ura mutation complementation and for β-galactosidase activity in galactose plates containing X-Gal) using cotransformation with two different plasmids and yielded only 2 (out of 1,212) transformants with a stable Lac+ phenotype (for details, see reference 6).
The engineering of S. cerevisiae for lactose utilization has been addressed over the last 20 years by different strategies (for a recent review, see reference 30). However, most of the strains obtained displayed undesirable characteristics (such as genetic instability or problems derived from the use of glucose-galactose mixtures) or were ineffective for ethanol production, as is the case for other S. cerevisiae strains expressing both LAC4 and LAC12 K. lactis genes (31, 32). Our evolved recombinant T1-E was able to ferment efficiently high concentrations of lactose (in particular, those in concentrated cheese whey) into ethanol, which together with the flocculent properties of the strain provides decisive advantages over other engineered S. cerevisiae strains for application in high-cell-density fermentation processes. The present study illustrates the usefulness of simple evolutionary engineering approaches in the improvement of genetically engineered strains that display poor efficiency.
Acknowledgments
We thank Carla Oliveira for performing the adaptation experiment and John Londesborough for useful discussions and support.
P.M.R.G. acknowledges support from the Fundação para a Ciência e a Tecnologia, Portugal (grant SFRH/BD/13463/2003). Grant support at the J.F. lab was from Reseau National des Genopole and ANR Blan 2005. The Institute for Biotechnology and Bioengineering-Centre of Biological Engineering lab acknowledges financial support from the Fundação para a Ciência e a Tecnologia, Portugal (project ProBioethanol PTDC/BIO/66151/2006).
Footnotes
Published ahead of print on 28 January 2008.
REFERENCES
- 1.Adams, J. 2004. Microbial evolution in laboratory environments. Res. Microbiol. 155:311-318. [DOI] [PubMed] [Google Scholar]
- 2.Bradford, M. M. 1976. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 3.Brown, C. J., K. M. Tood, and R. F. Rosenzweig. 1998. Multiple duplications of yeast hexose transport genes in response to selection in a glucose-limited environment. Mol. Biol. Evol. 15:931-942. [DOI] [PubMed] [Google Scholar]
- 4.Chang, Y., and R. C. Dickson. 1988. Primary structure of the lactose permease gene from the yeast Kluyveromyces lactis: presence of an unusual transcript structure. J. Biol. Chem. 263:16696-16703. [PubMed] [Google Scholar]
- 5.Domingues, L., A. A. Vicente, N. Lima, and J. A. Teixeira. 2000. Applications of yeast flocculation in biotechnological processes. Biotechnol. Bioprocess Eng. 5:288-305. [Google Scholar]
- 6.Domingues, L., J. A. Teixeira, and N. Lima. 1999. Construction of a flocculent Saccharomyces cerevisiae fermenting lactose. Appl. Microbiol. Biotechnol. 51:621-626. [DOI] [PubMed] [Google Scholar]
- 7.Domingues, L., M. M. Dantas, N. Lima, and J. A. Teixeira. 1999. Continuous ethanol fermentation of lactose by a recombinant flocculating Saccharomyces cerevisiae strain. Biotechnol. Bioeng. 64:692-697. [DOI] [PubMed] [Google Scholar]
- 8.Domingues, L., N. Lima, and J. A. Teixeira. 2001. Alcohol production from cheese whey permeate using genetically modified flocculent yeast cells. Biotechnol. Bioeng. 72:507-514. [DOI] [PubMed] [Google Scholar]
- 9.Dujon, B., D. Sherman, G. Fischer, P. Durrens, S. Casaregola, et al. 2004. Genome evolution in yeasts. Nature 430:35-44. [DOI] [PubMed] [Google Scholar]
- 10.Dyk, D., I. S. Pretorius, and F. F. Bauer. 2005. Mss11p is a central element of the regulatory network that controls FLO11 expression and invasive growth in Saccharomyces cerevisiae. Genetics 169:91-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fidalgo, M., R. R. Barrales, J. I. Ibeas, and J. Jiménez. 2006. Adaptive evolution by mutations in the FLO11 gene. Proc. Natl. Acad. Sci. USA 103:11228-11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gietz, D., A. St. Jean, R. A. Woods, and R. H. Schiestl. 1992. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 20:1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gödecke, A., W. Zachariae, A. Arvanitidis, and K. D. Breunig. 1991. Coregulation of the Kluyveromyces lactis lactose permease and β-galactosidase genes is achieved by interaction of multiple LAC9 binding sites in a 2.6 kbp divergent promoter. Nucleic Acids Res. 19:5351-5358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jeffries, T. W. 2006. Engineering yeasts for xylose metabolism. Curr. Opin. Biotechnol. 17:320-326. [DOI] [PubMed] [Google Scholar]
- 15.Jin, Y., H. Alper, Y. Yang, and G. Stephanopoulos. 2005. Improvement of xylose uptake and ethanol production in recombinant Saccharomyces cerevisiae through an inverse metabolic engineering approach. Appl. Environ. Microbiol. 71:8249-8256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnston, M. 1987. A model fungal gene regulatory mechanism: the GAL genes of Saccharomyces cerevisiae. Microbiol. Rev. 51:458-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuyper, M., A. A. Winkler, J. P. van Dijken, and J. T. Pronk. 2004. Minimal metabolic engineering of Saccharomyces cerevisiae for efficient anaerobic xylose fermentation: a proof of principle. FEMS Yeast Res. 4:655-664. [DOI] [PubMed] [Google Scholar]
- 18.Kuyper, M., M. J. Toirkens, J. A. Diderich, A. A. Winkler, J. P. van Dijken, and J. T. Pronk. 2005. Evolutionary engineering of mixed-sugar utilization by a xylose-fermenting Saccharomyces cerevisiae strain. FEMS Yeast Res. 5:925-934. [DOI] [PubMed] [Google Scholar]
- 19.Leonardo, J. M., S. M. Bhairi, and R. C. Dickson. 1987. Identification of upstream activator sequences that regulate induction of the β-galactosidase gene in Kluyveromyces lactis. Mol. Cell. Biol. 7:4369-4376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lohr, D., P. Venkov, and J. Zlatanova. 1995. Transcriptional regulation in the yeast GAL gene family: a complex genetic network. FASEB J. 9:777-787. [DOI] [PubMed] [Google Scholar]
- 21.McCullough, M. J., K. V. Clemons, J. H. McCusker, and D. A. Stevens. 1998. Intergenic transcribed spacer PCR ribotyping for differentiation of Saccharomyces species and interspecific hybrids. J. Clin. Microbiol. 36:1035-1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nielsen, J. 2001. Metabolic engineering. Appl. Microbiol. Biotechnol. 55:263-283. [DOI] [PubMed] [Google Scholar]
- 23.Overkamp, K. M., B. M. Bakker, P. Kötter, M. A. H. Luttik, J. P. van Dijken, and J. T. Pronk. 2002. Metabolic engineering of glycerol production in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 68:2814-2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parekh, S., V. A. Vinci, and R. J. Strobel. 2000. Improvement of microbial strains and fermentation processes. Appl. Microbiol. Biotechnol. 54:287-301. [DOI] [PubMed] [Google Scholar]
- 25.Poch, O., H. L'Hote, V. Dallery, F. Debeaux, R. Fleer, and R. Sodover. 1992. Sequence of the Klyuveromyces lactis β-galactosidase: comparison with prokaryotic enzymes and secondary structure analysis. Gene 118:55-63. [DOI] [PubMed] [Google Scholar]
- 26.Ribeiro, O., A. K. Gombert, J. A. Teixeira, and L. Domingues. 2007. Application of the Cre-loxP system for multiple gene disruption in the yeast Kluyveromyces marxianus. J. Biotechnol. 131:20-26. [DOI] [PubMed] [Google Scholar]
- 27.Riley, M. I., J. E. Hopper, S. A. Johnston, and R. C. Dickson. 1987. GAL4 of Saccharomyces cerevisiae activates the lactose-galactose regulon of Kluyveromyces lactis and creates a new phenotype: glucose repression of the regulon. Mol. Cell. Biol. 7:780-786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roggenkamp, R., B. Kustermann-Kuhn, and C. P. Hollenberg. 1981. Expression and processing of bacterial β-lactamase in the yeast Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 78:4466-4470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rubio-Texeira, M. 2005. A comparative analysis of the GAL genetic switch between not-so-distant cousins: Saccharomyces cerevisiae versus Kluyveromyces lactis. FEMS Yeast Res. 5:1115-1128. [DOI] [PubMed] [Google Scholar]
- 30.Rubio-Texeira, M. 2006. Endless versatility in the biotechnological applications of Kluyveromyces LAC genes. Biotechnol. Adv. 24:212-225. [DOI] [PubMed] [Google Scholar]
- 31.Rubio-Texeira, M., J. I. Castrillo, A. C. Adam, U. O. Ugalde, and J. Polaina. 1998. Highly efficient assimilation of lactose by a metabolically engineered strain of Saccharomyces cerevisiae. Yeast 14:827-837. [DOI] [PubMed] [Google Scholar]
- 32.Rubio-Texeira, M., M. Arévalo-Rodríguez, J. L. Lequerica, and J. Polaina. 2000. Lactose utilization by Saccharomyces cerevisiae strains expressing Kluyveromyces lactis LAC genes. J. Biotechnol. 84:97-106. [DOI] [PubMed] [Google Scholar]
- 33.Salmeron, J. M., Jr., and S. A. Johnston. 1986. Analysis of the Kluyveromyces lactis positive regulatory gene LAC9 reveals functional homology to, but sequence divergence from, the Saccharomyces cerevisiae GAL4 gene. Nucleic Acids Res. 14:7767-7781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 35.Sauer, U. 2001. Evolutionary engineering of industrially important microbial phenotypes. Adv. Biochem. Eng. Biotechnol. 73:129-169. [DOI] [PubMed] [Google Scholar]
- 36.Schaffrath, R., and K. D. Breunig. 2000. Genetics and molecular physiology of the yeast Kluyveromyces lactis. Fungal Genet. Biol. 30:173-190. [DOI] [PubMed] [Google Scholar]
- 37.Siso, M. I. G. 1996. The biotechnological utilization of cheese whey: a review. Bioresour. Technol. 57:1-11. [Google Scholar]
- 38.Sonderegger, M., and U. Sauer. 2003. Evolutionary engineering of Saccharomyces cerevisiae for anaerobic growth on xylose. Appl. Environ. Microbiol. 69:1990-1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sonderegger, M., M. Jeppsson, C. Larsson, M. Gorwa-Grauslund, E. Boles, L. Olsson, I. Spencer-Martins, B. Hahn-Hägerdal, and U. Sauer. 2004. Fermentation performance of engineered and evolved xylose-fermenting Saccharomyces cerevisiae strains. Biotechnol. Bioeng. 87:90-98. [DOI] [PubMed] [Google Scholar]
- 40.Sreekrishna, K., and R. C. Dickson. 1985. Construction of strains of Saccharomyces cerevisiae that grow on lactose. Proc. Natl. Acad. Sci. USA 82:7909-7913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Verduyn, C., E. Postma, W. A. Scheffers, and J. P. van Dijken. 1992. Effect of benzoic acid on metabolic fluxes in yeasts: a continuous-culture study on the regulation of respiration and alcoholic fermentation. Yeast 8:501-517. [DOI] [PubMed] [Google Scholar]
- 42.White, T. J., T. Bruns, S. Lee, and J. Taylor. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics, p. 315-322. In M. A. Innis, D. H. Gelfang, J. J. Sninsky, and T. J. White (ed.), PCR protocols: a guide to methods and applications. Academic Press, San Diego, CA.
- 43.Wray, L. V., Jr., M. M. Witte, R. C. Dickson, and M. I. Riley. 1987. Characterization of a positive regulatory gene, LAC9, that controls induction of the lactose-galactose regulon of Kluyveromyces lactis: structural and functional relationships to GAL4 of Saccharomyces cerevisiae. Mol. Cell. Biol. 7:1111-1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zachariae, W., and K. D. Breunig. 1993. Expression of the transcriptional activator LAC9 (KlGAL4) in Kluyveromyces lactis is controlled by autoregulation. Mol. Cell. Biol. 13:3058-3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zachariae, W., P. Kuger, and K. D. Breunig. 1993. Glucose repression of lactose/galactose metabolism in Kluyveromyces lactis is determined by the concentration of the transcriptional activator LAC9 (KlGal4). Nucleic Acids Res. 21:69-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zelder, O., and B. Hauer. 2000. Environmental directed mutations and their impact on industrial biotransformation and fermentation processes. Curr. Opin. Microbiol. 3:248-251. [DOI] [PubMed] [Google Scholar]
- 47.Zenke, F. T., W. Zachariae, A. Lunkes, and K. D. Breunig. 1993. Gal80 proteins of Kluyveromyces lactis and Saccharomyces cerevisiae are highly conserved but contribute differently to glucose repression of the galactose regulon. Mol. Cell. Biol. 13:7566-7576. [DOI] [PMC free article] [PubMed] [Google Scholar]