Abstract
Previous studies have found that the diversity of begomovirus-associated DNAβ satellites is related to host and geographical origin. In this study, we have cloned and sequenced 20 different isolates of DNAβ molecules associated with Malvastrum yellow vein virus (MYVV) isolated from Malvastrum coromandelianum plants in different geographical locations of Yunnan Province, China. Analyses of their molecular variation indicate that the satellites are clustered together according to their geographical location but that they have only limited sequence diversity. Infectivity tests using infectious clones of MYVV and its associated DNAβ molecule indicate that MYVV DNAβ is indispensable for symptom induction in Nicotiana benthamiana, N. glutinosa, Petunia hybrida, and M. coromandelianum plants. Furthermore, we showed that MYVV interacts functionally with heterologous DNAβ molecules in N. benthamiana plants.
Geminiviruses are a group of plant viruses characterized by a circular single-stranded DNA genome that is encapsidated by the geminate-shaped particles that range in size from 18 nm to 20 nm. Four genera (Mastrevirus, Topocuvirus, Curtovirus, and Begomovirus) of geminiviruses are recognized, but begomoviruses are the most numerous and geographically widespread. Most begomoviruses have a bipartite genome consisting of two similarly sized DNA components, referred to as DNA-A and DNA-B, both of which are essential for infectivity. A smaller number of begomoviruses have only one monopartite genome, equivalent to the DNA-A of bipartite geminiviruses, and appear to lack DNA-B. In recent years, some monopartite begomoviruses such as Ageratum yellow vein virus, Cotton leaf curl Multan virus, and Tomato yellow leaf curl China virus (TYLCCNV) have been shown to be associated with a novel satellite DNAβ molecule. Without the satellite, these begomoviruses are either poorly infectious or not infectious on the hosts from which they were initially isolated (3, 5, 15).
Previous analyses of the diversity of begomovirus-associated DNAβ satellites focused mainly on satellites originating from different hosts (1, 4). In this study, 20 different isolates of DNAβ molecules associated with Malvastrum yellow vein virus (MYVV) isolated from Malvastrum coromandelianum plants in different geographical locations in Yunnan Province, China, were cloned and sequenced. In addition, infectious clones of MYVV and its associated DNAβ molecule were constructed, and we found that the DNAβ molecule is indispensable for symptom induction in Nicotiana benthamiana, N. glutinosa, Petunia hybrida, and M. coromandelianum plants. Furthermore, we demonstrated that MYVV interacts functionally with heterologous DNAβ molecules in N. benthamiana plants.
MATERIALS AND METHODS
Virus sources, DNA extraction, and virus detection.
M. coromandelianum plants showing typical yellow vein symptoms were collected in Chuxiong, Yuxi, and Honghe districts, Yunnan Province, China, between 2003 and 2004 (see Fig. S1 in the supplemental material), and total nucleic acids were extracted from leaf tissue as described previously (21). The MYVV-specific primers Y47F (5′-GAATTCTTTATAGCTGCTAT-3′) and Y47R (5′-GAATTCCTCGACGAGAAAGA-3′) (EcoRI is underlined), designed based on the complete nucleotide sequence of MYVV isolate Y47 (AJ457827), were used to test for the presence of MYVV by PCR. PCR products were purified, cloned, and sequenced.
PCR and sequence determination.
Based on the conserved nucleotide sequence reported for DNAβ, abutting primers β01 (5′-GGTACCACTACGCTACGCAGCAGCC-3′) and β02 (5′-GGTACCTACCCTCCCAGGGGTACAC-3′) were designed and used to amplify full-length DNAβ as described previously (2, 23). PCR products were recovered, purified, and cloned into a pGEM-T Easy vector (Promega, Madison, WI). Sequences were determined using the automated model 3730 DNA sequencing system (Perkin-Elmer, Foster City, CA).
Sequence and phylogenetic analysis.
Nucleotide sequences were edited using EditSeq (Lasergene package of programs from DNAStar, Inc.) and aligned with the aid of the multiple alignment program Clustal X 1.81 (18). The levels of variation among these sequences were evaluated by two parameters (π and θ) using DnaSP version 4.10.3 (14). Nucleotide sequence diversity (π) was the average number of nucleotide differences per site between two sequences (17), and the population mutation parameter θ was based on the number of segregating sites S (20). A phylogenetic tree from multiple alignments of the 21 sequences was constructed by using the neighbor-joining method calculated with MEGA 3.1 (9). The DNAβ sequence associated with Malvastrum yellow vein Yunnan virus (MYVYNVβ; AJ786712) was used as a root for the comparisons. Bootstrap values were produced from 1,000 replicates.
Construction of infectious clones of MYVV and its associated DNAβ.
The primers Y47F and Y47R were used to amplify the complete nucleotide sequence of MYVV isolate Y47, and the PCR products were cloned into a pGEM-T vector (Promega) to yield the clone T-Y47A-FL. A SalI-EcoRI-digested fragment containing the entire intergenic region was released from T-Y47A-FL and introduced into a PLH9000 binary plasmid, which was kindly provided by Jörg Schubert, Federal Centre for Breeding Research on Cultivated Plants, Germany, to produce PLH9000-Y47A-PL. Then, T-Y47A-FL was digested with EcoRI to obtain a full-length unit of MYVV and inserted into the EcoRI site of PLH9000-Y47A-PL to produce clone PLH9000-Y47-1.9A containing a 1.9-mer partial tandem repeat of MYVV.
The complete monomeric sequence of MYVV DNAβ was amplified using primers β01 and β02, and the fragment was inserted into a pGEM-T Easy vector (Promega) to produce the clone T-Y47β-FL. Subsequently, another copy of complete MYVV DNAβ sequence was amplified using primers β05 (5′-GAAACCACTACGCTACGCAGCAGCC-3′) and β02 to produce the clone T-Y47β-PL. The T-Y47β-FL clone was digested with KpnI and inserted into the unique KpnI site of T-Y47β-PL to produce T-Y47-2β. Then T-Y47-2β was digested with EcoRI and inserted into the binary vector pBinPLUS (19) to produce clone pBinPLUS-Y47-2β, which contains a tandem dimeric repeat of MYVV DNAβ molecules. All the amplified fragments used for construction of infectious clones were sequenced with an automated model 3730 DNA sequencer (Perkin-Elmer), and sequence analysis indicated that no mutations were introduced into the clones.
Agroinoculation of plants.
Agrobacterium tumefaciens strain EHA105 was transformed with PLH9000-Y47-1.9A or pBinPLUS-Y47-2β by electrotransformation, and the transconjugants were selected on kanamycin (50 μg/ml) and streptomycin (50 μg/ml). A. tumefaciens cultures were grown at 28°C for 48 h, after which a 1-ml fine syringe was used to inject 0.2 ml of the culture into the stem or petioles of plants at the six-leaf stage. N. benthamiana, N. glutinosa, and P. hybrida plants were agroinoculated with PLH9000-Y47-1.9A either alone or as a mixture with pBinPLUS-Y47-2β. To test the interaction between MYVV and DNAβ molecules of different origins, N. benthamiana plants were agroinoculated with MYVV and either TYLCCNV DNAβ or Tomato yellow leaf curl Thailand virus (TYLCTHV) DNAβ previously constructed by our laboratory (23; X. P. Zhou, unpublished data). After inoculation, plants were grown in an insect-free greenhouse with supplementary light corresponding to a 16-h day length.
Whitefly transmission.
N. glutinosa plants infected with MYVV alone or in combination with MYVV DNAβ were used as sources of inocula for whitefly transmission. In these experiments, Bemisia tabaci whiteflies were fed on the source plants in closed cages within an insect-free cabinet for an acquisition access period of 3 days. Then, 20 viruliferous adult whiteflies were transferred to each healthy M. coromandelianum plant for a minimum 48-h period of transmission access as described previously (7). After inoculation, insects were removed and plants were sprayed twice weekly with an insecticide and maintained in an insect-free cabinet. Four weeks later, virus infections were evaluated by symptom observation and by PCR using the MYVV-specific primers Y47F and Y47R.
Assay of viral DNA.
Nucleic acids were isolated from young leaves of N. benthamiana and N. glutinosa plants at 35 days postinoculation as previously described (24), fractionated by 1% agarose gel electrophoresis in TBE buffer (90 mM Tris-borate, 2 mM EDTA, pH 8.3), and then transferred to Hybond-N+ membranes (Amersham Pharmacia, Little Chalfont, Buckinghamshire, England). After alkali denaturation and neutralization, hybridization was detected with randomly labeled probes using a random primer DNA labeling kit (Promega) according to the manufacturer's instructions. A DNA probe specific for MYVV was produced with BamHI digestion of T-Y47A-FL. The MYVV DNAβ probe was amplified from the satellite conserved region (SCR) of MYVV DNAβ using βSCR1 and βSCR2 primers as described previously (22). Hybridization signals were detected by phosphorimaging using a Typhoon 9200 imager (Amersham Pharmacia).
Nucleotide sequence accession numbers.
The complete nucleotide sequences of DNAβ molecules associated with the Yunnan isolates have been submitted to GenBank (accession numbers AJ971459-63, AJ971695-700, and AJ971702-10).
RESULTS AND DISCUSSION
Genomic organization and variation of DNAβ molecules.
PCR analysis using Y47F/Y47R primers specific for MYVV yielded approximately 2.7-kb bands from all 20 samples collected in Yunnan, indicating that they were infected with MYVV. The complete nucleotide sequences of DNAβ molecules associated with these isolates were determined and ranged from 1,339 to 1,358 nucleotides in length. The molecules have common structural features of DNAβ detailed in previous studies (1, 23), including the SCR, which contains a predicted hairpin structure with the loop sequence TAATATT↓AC. Comparisons of the SCRs, which spans 115 bp from 100 bp upstream to 15 bp downstream of the stem-loop, revealed between 97.4% and 100% sequence identity among these isolates. The A-rich regions, which contained 58.3% and 60.5% adenylic acid and ranged from 204 to 206 nucleotides in length, were located between positions 765 and 970 on the virion-sense DNA. These regions have repeated blocks of up to nine consecutive adenylic acid residues. A single conserved βC1 open reading frame encoding a predicted 118-amino-acid product was also located on the complementary-sense strand of viral satellite DNA. These predicted proteins share 92.4% to 100% amino acid sequence identity with each other.
The complete nucleotide sequences of the 20 DNAβ molecules share 93.0% to 99.8% sequence identity. The sequences were most closely related to MYVV DNAβ with the sequence identities ranging from 93.2% to 97.3%, further indicating that these samples were infected by MYVV. The MYVV DNAβ molecules had much less variation than those of TYLCCNV DNAβ (72 to 99%) or Tobacco curly shoot virus DNAβ molecules (83 to 98%) collected in Yunnan Province (23). This is probably due to the fact that TYLCCNV/DNAβ and tobacco curly shoot virus/DNAβ complexes infect a wide variety of hosts, whereas MYVV/DNAβ complexes infect mainly M. coromandelianum.
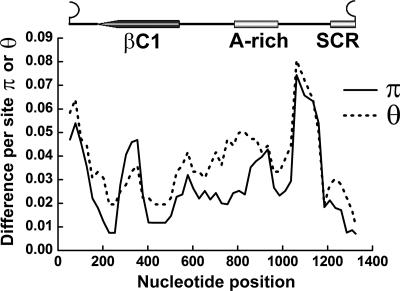
To evaluate the extent of DNAβ variation, statistical analyses regarding nucleotide sequence diversity were carried using the versatile program DnaSP version 4.10.3. The level of intraspecific variation of MYVV DNAβ molecules, including the 20 DNAβ molecules and the DNAβ sequence from MYVV isolate Y47, was estimated by the Watterson parameter θ and the mean pairwise diversity π at all sites along the genome. Using the sliding windows option of DnaSP 4.10.3 software (window length, 100; step size, 25), a noteworthy observation is that there are two regions with a high level of nucleotide diversity located within the βC1 coding region adjacent to the C terminus and the noncoding region between the SCR and the A-rich region (Fig. 1).
FIG. 1.
Sliding window plot showing the distribution of genetic variation estimated by nucleotide diversity (π) and Watterson's parameter (θ) for MYVV DNAβ molecules. The window size is 100 sites wide, with sliding by 50-site increments. The relative positions of the βC1 gene, A-rich region, and SCR of the DNAβ genome are illustrated above the plot in linear DNA format.
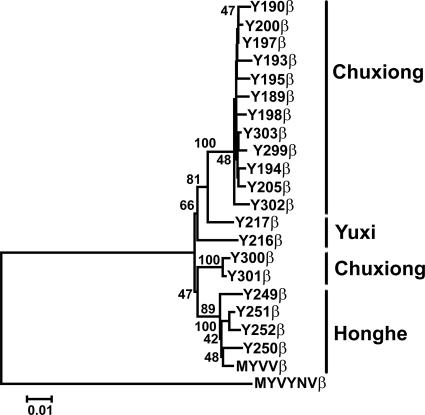
A rooted phylogenetic tree of DNAβ molecules was created by the neighbor-joining method using MEGA version 3.1 (9). The tree delineated two subpopulations: subgroup 1 isolates from Chuxiong and Yuxi districts and subgroup 2 isolates from the Honghe district (Fig. 2). However, there is an exception in the tree where DNAβ molecules from Y300 (Y300β) and Y301 (Y301β) should cluster into subgroup 1 according to geographical location but instead form a relatively independent cluster. Sequence comparison among DNAβ molecules from Y300, Y301, Y302, and Y303, which were taken from the same area in Chuxiong, shows that the major sequence differences are point mutations.
FIG. 2.
Rooted neighbor-joining tree constructed based on the full-length DNAβ sequences. Sequences of the DNAβs obtained from the above samples are named according to their associated begomovirus: thus, Y189β refers to DNAβ from isolate Y189. The sequence of MYVYNV DNAβ (MYVYNVβ) was used as the root. The statistical significance of the branches was calculated by bootstrap analysis with 1,000 replicates of the data. Numbers at the nodes indicate the percentages of identical branches obtained by bootstrapping. Only bootstrap values greater than 40% are indicated.
Infectivity and symptoms induced by MYVV and its associated DNAβ.
To investigate the biological role of DNAβ, MYVV (PLH9000-Y47-1.9A) was agroinoculated into plants either alone or together with MYVV DNAβ (pBinPLUS-Y47-2β). MYVV alone was systemically infectious in N. benthamiana, N. glutinosa, and P. hybrida plants, but no symptoms were visible in these plants (Fig. 3C, E, and G). In contrast, when these plants were infected with MYVV and MYVV DNAβ, the coinoculated plants developed downward curling of the leaves associated with yellow vein and leaf crinkling (Fig. 3D, F, and H). Agroinoculation of either MYVV alone or together with MYVV DNAβ was not successful in M. coromandelianum plants, and so whitefly transmission assays were carried out to fulfill Koch's postulates. In these transmission tests, M. coromandelianum plants infected with MYVV and MYVV DΝΑβ displayed yellow vein symptoms similar to those observed in the field (Fig. 3B). However, plants exposed to whiteflies that had fed only on MYVV-infected plants displayed no symptoms, although MYVV could be detected by PCR with MYVV-specific primers (Fig. 3A).
FIG. 3.
Symptoms induced by MYVV alone or in the presence of MYVV DNAβ (MYVVβ) in M. coromandelianum at 45 days after whitefly transmission (A and B) and in N. benthamiana (C and D), N. glutinosa (E and F), or P. hybrida (G and H) at 35 days after agroinoculation.
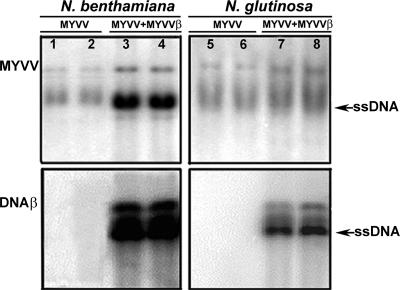
Southern hybridization analysis of N. benthamiana and N. glutinosa plants agroinoculated with both MYVV and MYVV DNAβ showed that both MYVV and MYVV DNAβ were present (Fig. 4, lanes 3, 4, 7, and 8). N. benthamiana and N. glutinosa plants coinfected with MYVV and MYVV DNAβ accumulated more viral DNA than did those infected with MYVV alone (Fig. 4). Also, N. benthamiana has much more intense bands than does N. glutinosa whether the plants were infected with MYVV alone or with combinations of MYVV DNAβ (Fig. 4).
FIG. 4.
Southern blot analysis of nucleic acids extracted from infected N. benthamiana and N. glutinosa plants agroinoculated with MYVV alone (lanes 1, 2, 5, and 6) or in the presence of MYVV DNAβ (MYVVβ) (lanes 3, 4, 7, and 8) at 35 days after inoculation. Equal amounts of nucleic acid (20 μg) were loaded in each lane. Blots were probed with either MYVV (top) or MYVV DNAβ (bottom). The positions of single-stranded DNA (ssDNA) forms are indicated.
The results above confirmed that MYVV is a monopartite begomovirus associated with a satellite DNAβ, which is necessary for symptom induction, and that DNAβ can enhance the accumulation level of viral DNA in infected plants.
Interaction between MYVV and other DNAβ molecules.
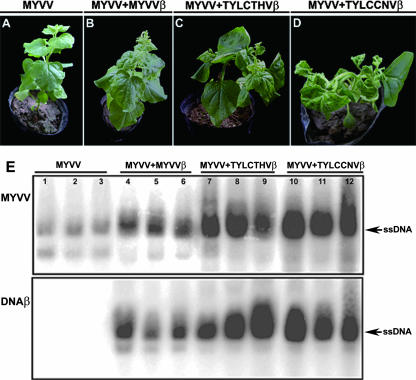
The ability of MYVV to act as a helper virus to replicate heterologous DNAβ molecules was evaluated by agrobacterium-mediated coinoculation of N. benthamiana plants with MYVV and TYLCTHV DNAβ or TYLCCNV DNAβ. N. benthamiana seedlings systemically infected with MYVV and either TYLCTHV DNAβ or TYLCCNV DNAβ developed typical systemic symptoms that were distinguishable from those induced by MYVV and MYVV DNAβ (Fig. 5A to D). N. benthamiana plants coinoculated with MYVV and TYLCTHV DNAβ showed downward leaf curling and stem distortion (Fig. 5C). However, when agroinoculated with MYVV and TYLCCNV DNAβ, N. benthamiana plants developed more accentuated downward leaf curling and stem distortion symptoms accompanied by enations on the undersides of the leaves (Fig. 5D). These symptoms were substantially more severe than those induced by MYVV and MYVV DNAβ (Fig. 5B).
FIG. 5.
Symptoms and Southern blot analyses in N. benthamiana plants induced by MYVV and different DNAβ molecules at 35 days postinoculation. (A to D) Plants were agroinoculated with MYVV alone (A) or in the presence of MYVV DNAβ (B), TYLCTHV DNAβ (C), or TYLCCNV DNAβ (D). (E) Southern blot analyses of nucleic acids extracted from agroinoculated N. benthamiana plants with MYVV alone (lanes 1 to 3) or in the presence of MYVV DNAβ (MYVVβ, lanes 4 to 6), TYLCTHV DNAβ (TYLCTHVβ, lanes 7 to 9), or TYLCCNV DNAβ (TYLCCNVβ, lanes 10 to 12). Equal amounts of nucleic acid (20 μg) were loaded in each lane. Blots were probed with either MYVV (top) or MYVV DNAβ (bottom). The positions of the single-stranded DNA (ssDNA) forms are indicated to the right of the blot.
The relative amounts of MYVV and of the homologous and heterologous DNAβs in the MYVV-infected and -coinfected N. benthamiana plants were compared by Southern hybridization analysis. All of the heterologous DNAβ isolates could be detected in systemically symptomatic leaves, indicating that they can replicate and spread systemically in association with MYVV. However, viral DNA concentrations in the plants were variable. Viral DNA levels in N. benthamiana plants coinoculated with MYVV and TYLCCNV DNAβ were similar to those in plants coinoculated with MYVV and TYLCTHV DNAβ (Fig. 5E, lanes 5 and 7 to 12). In contrast, the relative concentrations of viral DNA in N. benthamiana plants coinoculated with MYVV and MYVV DNAβ were nearly an order of magnitude lower than concentrations in those inoculated with the heterologous DNAβ molecules (Fig. 5E). These results indicate that MYVV can interact functionally with homologous and heterologous DNAβ and that the variation of viral DNA amounts in infected plants correlates well with the severity of symptoms.
Pseudorecombination is one of the most important sources of viral genetic variation. In bipartite begomoviruses, the two components share a common region containing repeat sequences (iteron). When the replication-associated protein (Rep) initiates transreplication of DNA-B, Rep specifically recognizes the iteron. This recognition mechanism is extremely sequence specific (6) and is usually confined to a particular begomovirus species, although interspecific replication can occur if the iterons are conserved (16). Our results further indicate that begomoviruses can support heterologous DNAβ replication. The replication of DNA-A and DNA-B genomic components of different bipartite begomoviruses is highly isolate and sequence specific, compared to the interactions between DNAβ and its helper virus (11, 13).
So far, three types of begomoviruses have been identified that are distinguishable by their genome composition. One group consists of bipartite begomoviruses that contain DNA-A and DNA-B components; a second group is composed of monopartite begomoviruses, which lack a component equivalent to DNA-B; and the third group is represented by monopartite begomoviruses that harbor a satellite DNAβ molecule in infected plants (1, 4, 12). The monopartite begomoviruses associated with DNAβ molecules can be divided into two types according to whether or not their DNAβ molecules are indispensable for induction of symptoms. In the majority of these begomoviruses, DNAβ molecules are required for induction of typical disease symptoms. The viral DNA components are either poorly infectious or not infectious without DNAβ in the host from which they were initially isolated, although cloned viral DNA genomes have been shown to be infectious in the experimental host N. benthamiana (3, 8, 15). In the remaining minority, the associated DNAβ molecules do not appear to play a key role in symptom induction and their relationship with the helper virus is less well understood. These classes of monopartite begomoviruses may represent intermediates between the majority type and true monopartite begomoviruses that have a single genomic component similar to the DNA-A component of bipartite begomoviruses (10). Based on our results, it is evident that MYVV belongs to the majority type.
Supplementary Material
Acknowledgments
This research work was supported by the National Natural Science Foundation of China (grant no. 30471137) and the National Key Basic Research and Development Program (2006CB101903).
Footnotes
Published ahead of print on 1 February 2008.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Briddon, R. W., S. E. Bull, I. Amin, A. M. Idris, S. Mansoor, I. D. Bedford, P. Dhawan, N. Rishi, S. S. Siwatch, A. M. Abdel-Salam, J. K. Brown, Y. Zafar, and P. G. Markham. 2003. Diversity of DNAβ, a satellite molecule associated with some monopartite begomoviruses. Virology 312:106-121. [DOI] [PubMed] [Google Scholar]
- 2.Briddon, R. W., S. E. Bull, S. Mansoor, I. Amin, and P. G. Markham. 2002. Universal primers for the PCR-mediated amplification of DNAβ: a molecule associated with some monopartite begomoviruses. Mol. Biotechnol. 20:315-318. [DOI] [PubMed] [Google Scholar]
- 3.Briddon, R. W., S. Mansoor, I. D. Bedford, M. S. Pinner, K. Saunders, J. Stanley, Y. Zafar, K. A. Malik, and P. G. Markham. 2001. Identification of DNA components required for induction of cotton leaf curl disease. Virology 285:234-243. [DOI] [PubMed] [Google Scholar]
- 4.Bull, S. E., W. S. Tsai, R. W. Briddon, P. G. Markham, J. Stanley, and S. K. Green. 2004. Diversity of begomovirus DNAβ satellites of non-malvaceous plants in east and south east Asia—brief report. Arch. Virol. 149:1193-1200. [DOI] [PubMed] [Google Scholar]
- 5.Cui, X. F., X. R. Tao, Y. Xie, C. M. Fauquet, and X. P. Zhou. 2004. A DNAβ associated with Tomato yellow leaf curl China virus is required for symptom induction. J. Virol. 78:13966-13974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fontes, E. P. B., P. A. Eagle, P. S. Sipe, V. A. Luckow, and L. Hanley-Bowdoin. 1994. Interaction between a geminivirus replication protein and origin DNA is essential for viral replication. J. Biol. Chem. 269:8459-8465. [PubMed] [Google Scholar]
- 7.Jiu, M., X. P. Zhou, and S. S. Liu. 2006. Acquisition and transmission of two begomoviruses by the B and a non-B biotype of Bemisia tabaci from Zhejiang, China. J. Phytopathol. 154:587-591. [Google Scholar]
- 8.Jose, J., and R. Usha. 2003. Bhendi yellow vein mosaic disease in India is caused by association of a DNAβ satellite with a begomovirus. Virology 305:310-317. [DOI] [PubMed] [Google Scholar]
- 9.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5:150-163. [DOI] [PubMed] [Google Scholar]
- 10.Li, Z. H., Y. Xie, and X. P. Zhou. 2005. Tobacco curly shoot virus DNAβ is not necessary for infection but intensifies symptoms in a host-dependent manner. Phytopathology 95:902-908. [DOI] [PubMed] [Google Scholar]
- 11.Lin, B., S. A. A. Behjatnia, I. B. Dry, J. W. Randles, and M. A. Rezaian. 2003. High-affinity Rep-binding is not required for the replication of a geminivirus DNA and its satellite. Virology 305:353-363. [DOI] [PubMed] [Google Scholar]
- 12.Mansoor, S., R. W. Briddon, Y. Zafar, and J. Stanley. 2003. Geminivirus disease complexes: an emerging threat. Trends Plant Sci. 8:128-134. [DOI] [PubMed] [Google Scholar]
- 13.Mansoor, S., Y. Zafar, and R. W. Briddon. 2006. Geminivirus disease complexes: the threat is spreading. Trends Plant Sci. 11:209-212. [DOI] [PubMed] [Google Scholar]
- 14.Rozas, J., J. C. Sanchez-DelBarrio, X. Messeguer, and R. Rozas. 2003. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19:2496-2497. [DOI] [PubMed] [Google Scholar]
- 15.Saunders, K., I. D. Bedford, R. W. Briddon, P. G. Markham, S. M. Wong, and J. Stanley. 2000. A unique virus complex causes Ageratum yellow vein disease. Proc. Natl. Acad. Sci. USA 97:6890-6895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saunders, K., N. Salim, V. R. Mali, V. G. Malathi, R. Briddon, P. G. Markham, and J. Stanley. 2002. Characterization of Sri Lankan cassava mosaic virus and Indian cassava mosaic virus: evidence for acquisition of a DNA B component by a monopartite begomovirus. Virology 293:63-74. [DOI] [PubMed] [Google Scholar]
- 17.Tajima, F. 1983. Evolutionary relationship of DNA sequences in finite populations. Genetics 105:437-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Engelen, F. A., J. W. Molthoff, A. J. Conner, J. P. Nap, A. Pereira, and W. J. Stiekema. 1995. pBINPLUS: an improved plant transformation vector based on pBIN19. Transgenic Res. 4:288-290. [DOI] [PubMed] [Google Scholar]
- 20.Watterson, G. A. 1975. On the number of segregating sites in genetical models without recombination. Theor. Popul. Biol. 7:256-276. [DOI] [PubMed] [Google Scholar]
- 21.Xie, Y., X. P. Zhou, Z. H. Li, Z. K. Zhang, and G. X. Li. 2002. Identification of a novel DNA molecule associated with tobacco leaf curl virus. Chin. Sci. Bull. 47:1273-1276. [Google Scholar]
- 22.Xiong, Q., S. Fan, J. Wu, and X. Zhou. 2007. Ageratum yellow vein China virus is a distinct begomovirus species associated with a DNAβ molecule. Phytopathology 97:405-411. [DOI] [PubMed] [Google Scholar]
- 23.Zhou, X. P., Y. Xie, X. R. Tao, Z. K. Zhang, Z. H. Li, and C. M. Fauquet. 2003. Characterization of DNAβ associated with begomoviruses in China and evidence for co-evolution with their cognate viral DNA-A. J. Gen. Virol. 84:237-247. [DOI] [PubMed] [Google Scholar]
- 24.Zhou, X. P., Y. Xie, Z. K. Zhang, Y. J. Qi, and J. J. Wu. 2001. Molecular characterization of a novel defective DNA isolated from tobacco tissues infected with tobacco leaf curl virus. Acta Virol. 45:45-50. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.