Abstract
Candida arabinofermentans PYCC 5603T and Pichia guilliermondii PYCC 3012 were shown to grow well on l-arabinose, albeit exhibiting distinct features that justify an in-depth comparative study of their respective pentose catabolism. Carbon-13 labeling experiments coupled with in vivo nuclear magnetic resonance (NMR) spectroscopy were used to investigate l-arabinose metabolism in these yeasts, thereby complementing recently reported physiological and enzymatic data. The label supplied in l-[2-13C]arabinose to nongrowing cells, under aerobic conditions, was found on C-1 and C-2 of arabitol and ribitol, on C-2 of xylitol, and on C-1, C-2, and C-3 of trehalose. The detection of labeled arabitol and xylitol constitutes additional evidence for the operation in yeast of the redox catabolic pathway, which is widespread among filamentous fungi. Furthermore, labeling at position C-1 of trehalose and arabitol demonstrates that glucose-6-phosphate is recycled through the oxidative pentose phosphate pathway (PPP). This result was interpreted as a metabolic strategy to regenerate NADPH, the cofactor essential for sustaining l-arabinose catabolism at the level of l-arabinose reductase and l-xylulose reductase. Moreover, the observed synthesis of d-arabitol and ribitol provides a route with which to supply NAD+ under oxygen-limiting conditions. In P. guilliermondii PYCC 3012, the strong accumulation of l-arabitol (intracellular concentration of up to 0.4 M) during aerobic l-arabinose metabolism indicates the existence of a bottleneck at the level of l-arabitol 4-dehydrogenase. This report provides the first experimental evidence for a link between l-arabinose metabolism in fungi and the oxidative branch of the PPP and suggests rational guidelines for the design of strategies for the production of new and efficient l-arabinose-fermenting yeasts.
Economic, environmental, and political concerns provided the impetus for expanding research into producing bioethanol from lignocellulosic biomass. d-Xylose and l-arabinose are the predominant sugars in the pentose fraction of hemicellulose hydrolysates. Saccharomyces cerevisiae, the preferred industrial microorganism for use in ethanolic fermentation processes, efficiently ferments hexoses but is unable to metabolize pentoses. Metabolic engineering of S. cerevisiae for the efficient fermentation of pentoses is based on the expression of heterologous genes from organisms naturally able to utilize these sugars. The characterization of d-xylose-utilizing yeasts, like Pichia stipitis (9, 23, 35), was essential for the successful development of recombinant S. cerevisiae strains able to ferment d-xylose (13, 22, 39). In contrast, l-arabinose metabolism in yeast is poorly characterized, as evidenced by the low number of publications on this topic (8, 10, 11, 27, 32, 43). This scarcity of information may hamper the development of an efficient l-arabinose-fermenting strain of S. cerevisiae (2, 21, 31, 44).
In fungi, the catabolic pathways of l-arabinose and d-xylose share most enzymes (1, 6, 10, 45). l-Arabinose and d-xylose are reduced by an unspecific aldose reductase (AR; EC 1.1.1.21) to yield l-arabitol and xylitol, respectively. l-Arabitol is subsequently converted to xylitol in two consecutive redox steps, catalyzed by l-arabitol 4-dehydrogenase (LAD; EC 1.1.1.12) and l-xylulose reductase (LXR; EC 1.1.1.10). Xylitol, an intermediate common to the d-xylose catabolic pathway, is further oxidized to d-xylulose by xylitol dehydrogenase (XDH; EC 1.1.1.9). The phosphorylation by xylulokinase (XK; EC 2.7.1.17) leads to d-xylulose-5-phosphate, an intermediate of the pentose phosphate pathway (PPP) (10). Most aldose and l-xylulose reductases (AR and LXR) are strictly NADPH dependent, with a few also showing a lower affinity for NADH. On the other hand, the l-arabitol and xylitol dehydrogenases (LAD and XDH) require NAD+ as a cofactor. The specificity features of reductases and dehydrogenases with respect to NADPH and NAD+ generate a lack of NAD+ under anaerobic conditions, which is strengthened by the absence of transhydrogenase activity in yeasts (3). NADPH can be regenerated via the oxidative PPP (5, 12, 17, 18) and, possibly, by the action of an NADP+-linked isocitrate dehydrogenase (4).
After screening yeasts for rapid growth and high l-arabinose uptake rates (10), we selected Candida arabinofermentans PYCC 5603T and Pichia guilliermondii PYCC 3012 for comparative characterization and elucidation of l-arabinose catabolic pathways. Both strains have specific high-capacity l-arabinose transporters but displayed distinctly different levels of in vitro enzyme activities for l-arabinose catabolism (10). In terms of substrate regulation, oxygen requirement, and their influences on product formation, both strains showed the capacity to produce ethanol from d-glucose under oxygen-limiting conditions but virtually no d-xylose and l-arabinose fermentation (11). However, C. arabinofermentans, which was considered the best-performing yeast in l-arabinose medium (8), accumulated much less arabitol than P. guilliermondii under oxygen limitation conditions (11), possibly due to a more effective l-arabitol 4-dehydrogenase and/or a higher ability to regenerate NAD+ in the absence of oxygen (10).
In the present work, in vivo 13C nuclear magnetic resonance (NMR) was used to complement the previous biochemical studies and to investigate the in vivo flux distribution through the catabolic pathways of l-arabinose in the two yeast strains, thereby contributing to an understanding of the differences between their respective l-arabinose catabolic pathways. Due to its unique analytical and nondestructive features, NMR is a powerful technique for the elucidation of metabolic pathways. The use of 13C-enriched compounds allows tracing the fate of specific carbon atoms through different metabolic routes (25). Furthermore, positional isotopic information derived from 13C NMR can be used to characterize the flux distribution (7). The work reported here provides new insights into the metabolism of l-arabinose in yeast.
MATERIALS AND METHODS
Strains and maintenance.
Candida arabinofermentans PYCC 5603T and Pichia guilliermondii PYCC 3012 were provided by the Portuguese Yeast Culture Collection (PYCC), Faculty of Sciences and Technology, New University of Lisbon, Caparica, Portugal. Both strains were grown on YPA medium (yeast extract, 10 g liter−1; peptone, 20 g liter−1; agar, 20 g liter−1; l-arabinose, 20 g liter−1) at 25°C and maintained at 4°C.
Growth conditions.
Strains were grown aerobically in mineral defined medium (42) with vitamins, trace elements, and 20 g liter−1 of l-arabinose at 30°C in a 2-liter fermentor (2 liters of culture and continuous air flushing) or in 2-liter shake flasks (400 ml of culture at 180 rpm), depending on the amount of cells needed for the experiments. Inoculum cultures were grown in 100-ml shake flasks (20 ml of culture at 180 rpm) under the same conditions as the 2-liter shake flasks. Inocula were prepared to an initial optical density at 600 nm (OD600) of 0.1 to 0.2. Growth was followed, and cells were harvested during the exponential growth phase (OD600 of 4.0 to 4.4).
In vivo 13C NMR experiments.
Cells were harvested at a late-exponential growth phase (OD600 of 4.0 to 4.4), centrifuged, washed twice, and suspended in 50 mM KPi buffer (pH 6.0) with 6% (vol/vol) 2H2O and an antifoam agent, to a cell concentration of around 40 mg (dry weight) ml−1. The experiments were performed using a 10-mm NMR tube containing 5 ml of cell suspension or by using a circulating system that couples a bioreactor (50-ml cell suspension) with the NMR tube (29), using a circulation rate of 30 ml min−1. To avoid cell sedimentation and to ensure an adequate supply of gases to the cell suspension, an airlift system was used inside the NMR tube (33). Aerobic conditions were established by bubbling pure oxygen through the cell suspension. Prior to substrate addition, oxygen was bubbled for at least 10 min to allow consumption of internal carbon sources, mainly arabitol. The pH in the bioreactor was controlled at 6.0 by the automatic addition of 2 M NaOH. Dissolved oxygen in cell suspensions was measured by using an oxygen electrode (Hamilton Bonaduz AG, Switzerland). All measurements were performed after calibration by using argon and air for 0% and 100% oxygen saturation, respectively. 13C NMR spectra were acquired sequentially before and after the addition of l-[2-13C]arabinose (20 mM). The labeled metabolites were monitored noninvasively for 60 min. [13C]methanol, in a glass capillary, was used as the concentration standard. After the in vivo experiment, an NMR sample extract was prepared as described previously (28) and used for the quantification of metabolites.
Quantification of intracellular metabolites in living cells by 13C NMR.
Due to the fast pulsing conditions used to acquire in vivo 13C spectra, the conversion of peak areas into concentrations requires the use of correction factors, which were determined for NMR sample extracts spiked with pure compounds. For the α- and β-furanose forms and the α- and β-pyranose forms of l-[2-13C]arabinose, the correction factors were 0.32, 0.30, 0.35, and 0.35, respectively; for C-1, C-2, and C-4 of arabitol, the correction factor was 0.40; for C-1, C-2, and C-3 of trehalose, the correction factors were 0.37, 0.37, and 0.39, respectively. The quantitative kinetic data were calculated from the areas of the respective resonances by applying the respective correction factor and comparing the result with the intensity of the concentration standard. Intracellular metabolite concentrations were calculated by considering a value of 2.0 μl of yeast cell volume per mg (dry weight) (14).
Perchloric extracts of yeast cells.
Cell suspensions of P. guilliermondii PYCC 3012 (five identical samples) were prepared as described previously for in vivo 13C NMR experiments, except that oxygen was bubbled through the cell suspensions for 5 min before 20 mM l-[2-13C]arabinose was added. The metabolism of l-[2-13C]arabinose was stopped at different time points (5, 9, 19, and 39 min) by the addition of cold perchloric acid to a final concentration of 0.6 M. A control sample prior to the addition of l-[2-13C]arabinose was also prepared. The five extracts were kept on ice, with stirring, for 20 min, neutralized with 5 M KOH, and centrifuged. EDTA was added to the supernatant solution to a final concentration of 5 mM. Cell extracts were analyzed by 13C NMR to resolve overlapping resonances and to quantify isotopomer distribution. Furthermore, aliquots were analyzed by high-performance liquid chromatography (HPLC) to determine the total amounts of metabolites.
13C NMR spectroscopy.
All NMR spectra were run at 30°C with a quadruple-nucleus-probe head on a Bruker DRX500 spectrometer. The acquisition of 13C NMR in vivo spectra was performed as previously described (29), except for the number of transients (80, 13C-enriched substrates; 2,500, natural abundance spectroscopy). For the calculation of the correction factors and the metabolite concentrations in cell extracts, 13C NMR spectra were acquired with a 60° flip angle and a recycle delay of 1.5 s (under saturating conditions) or 60.5 s (under relaxed conditions). Carbon chemical shifts are referenced to that of external methanol, which is designated at 49.3 ppm. The assignment of 13C NMR resonances was confirmed by the addition of pure compounds to cell extracts and/or by comparison with spectra of pure compounds. Spectra were analyzed using Topspin (Bruker).
Assessment of intra- and extracellular metabolite distribution.
Intra- and extracellular pools were determined by independent experiments performed with the experimental setup used for in vivo 13C NMR measurements. After l-arabinose (20 mM) was added, 0.6-ml samples were withdrawn at regularly timed intervals and immediately centrifuged, and supernatant solutions were used for the quantification of extracellular metabolites. For the determination of the total concentration of metabolites (intracellular plus extracellular), 1.5-ml samples were taken at 10, 20, 40, and 60 min after l-arabinose was added. Perchloric extractions were performed as described above.
Quantification of metabolites by HPLC.
Supernatant solutions, cell extracts, and in vivo NMR sample extracts were analyzed in a Dionex HPLC system (Dionex, Sunnyvale, CA) equipped with a refractive index detector (LKB, Bromma, Sweden), using an Aminex HPX-87P column (Bio-Rad, Richmond, CA) at 80°C, with bidistilled water as the mobile phase and a flow rate of 0.5 ml min−1. Trehalose, arabinose, ribitol, arabitol, and xylitol were separated after 45 min.
d-Arabitol and l-arabitol identification and relative quantification by chiral GC-MS.
The enantiomeric excess of d- or l-arabitol was determined by using a chiral gas chromatography (GC) column. The arabitol mass composition was confirmed by analyzing mixtures which were converted to their trifluoroacetic esters, as previously described (15). Samples were evaporated to dryness under a stream of nitrogen in 1-ml vials. The pellet was dissolved in dichloromethane (200 μl) and trifluoroacetic anhydride (400 μl), and the mixture was heated at 80°C over 30 min. After samples cooled to room temperature, they were dried under a nitrogen stream and redissolved in dichloromethane. GC-mass spectrometry (GC-MS) analysis was performed using a glass capillary column (30 m by 0.25-mm internal diameter) coated with 2,6-di-O-pentyl-3-trifluoroacetyl gamma-cyclodextrin (Chiraldex G-TA; ASTEC), using a Carlo Erba GC-MS (GC 6000 model; Carlo Erba Vega Series 2, MS QMD 1000). The helium carrier gas flow rate was set at 1.5 ml min−1, and a four-step program was followed: 90°C for 10 min, 0.5°C min−1 up to 103°C, 4°C min−1 up to 180°C, and 7 min at 180°C. The injection port temperature was maintained at 250°C. Mass spectra were measured at an ionizing voltage of 70 eV. The temperatures of the ion source and the transfer line were maintained at 150°C and 220°C, respectively. Spectra of the individual components were identified, using MassLab software (Micromass), by comparison with the spectra of pure compounds. d-Arabitol and l-arabitol were quantified from peak areas.
RESULTS
Metabolism of l-[2-13C]arabinose as monitored by in vivo 13C NMR.
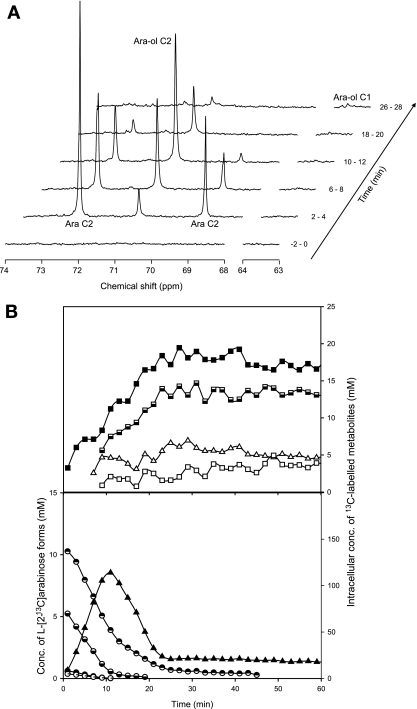
A time series of 13C spectra acquired during the metabolism of 20 mM l-[2-13C]arabinose by nongrowing cell suspensions of C. arabinofermentans or P. guilliermondii are shown in Fig. 1 and 2, respectively. Upon the addition of l-[2-13C]arabinose, resonances due to the α- and β-furanose forms (82.05 and 76.83 ppm, respectively) and the α- and β-pyranose forms (72.49 and 69.11 ppm, respectively) of the sugar were detected and progressively decreased. A strong resonance due to C-2 of arabitol (70.92 ppm) appeared immediately after the substrate addition and increased concomitantly with arabinose consumption. Weak signals assigned to C-1 (63.66 ppm), C-3 (71.14 ppm), C-4 (71.60 ppm), and C-5 (63.56 ppm) of arabitol were also detected. The evolution of the areas of resonances due to C-3 and C-5 of arabitol was similar to that of the strong resonance due to C-2. Moreover, their intensities relative to that of C-2 (approximately 1%) indicated that the C-3 and C-5 signals were due to the naturally abundant 13C in arabitol; i.e., these carbon atoms were not enriched in 13C. The signal due to C-1 of arabitol was composed of a central peak at 63.66 ppm and two satellite peaks separated by about 40 Hz, a typical value for the coupling constant between adjacent carbon atoms. The central peak is due to arabitol singly labeled at C-1, while the satellite peaks are due to the arabitol molecules simultaneously labeled on C-1 and C-2, i.e., [1,2-13C]arabitol. The observed increase for the intensity of these signals showed the progressive accumulation of the isotopomers [1-13C]arabitol and [1,2-13C]arabitol. The weak resonances centered at 93.70, 71.63, and 73.17 ppm also increased in intensity with time and were assigned to C-1, C-2, and C-3 of trehalose (see below for the detailed identification of isotopomers). The resonances due to C-2 of trehalose (centered at 71.63 ppm) overlap the resonance due to C-4 of arabitol (71.60 ppm). Similarly, the α-pyranose form of the l-[2-13C]arabinose resonance (at 72.49 ppm) largely obscured a resonance subsequently assigned to C-2 of xylitol (at 72.54 ppm) in the spectra of cell extracts (see below).
FIG. 1.
l-Arabinose metabolism in suspensions of C. arabinofermentans PYCC 5603T resting cells monitored by in vivo 13C NMR. (A) Sequence of in vivo 13C NMR spectra acquired during the metabolism of l-[2-13C]arabinose (20 mM) at 30°C and under aerobic conditions. Each spectrum represents 2 min of acquisition. Cells were grown in aerobic batch cultures with l-arabinose (20 g liter−1), harvested at exponential growth phase, and suspended in 50 mM phosphate buffer (pH 6.0) to a concentration of around 40 g (dry weight) liter−1. Ara, arabinose; Ara-ol, arabitol. (B) Time course for the consumption of l-[2-13C]arabinose and evolution of the intracellular metabolite pools. Metabolite concentrations were obtained from in vivo 13C NMR data. Symbols: circles, l-arabinose (◓, α-l-[2-13C]arabinopyranoside; ◒, β-l-[2-13C]arabinopyranoside; •, α-l-[2-13C]arabinofuranoside; ○, β-l-[2-13C]arabinopyranoside); triangles, arabitol (▵, C-1 labeled; ▴, C-2 labeled); squares, trehalose (□, C-1 labeled; ▪, C-2 labeled;, C-3 labeled).
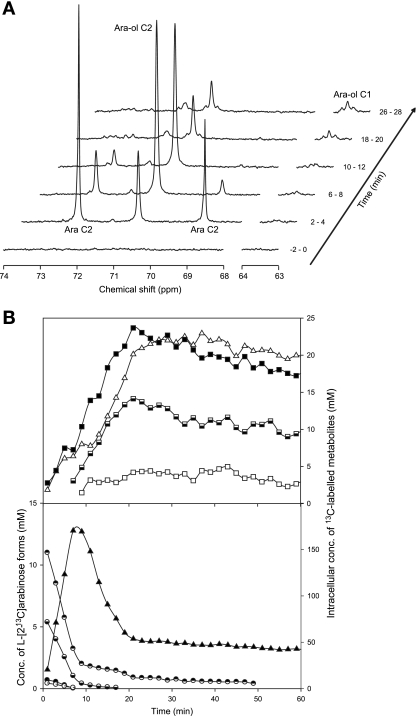
FIG. 2.
l-Arabinose metabolism in suspensions of P. guilliermondii PYCC 3012 resting cells monitored by in vivo 13C NMR. (A) Sequence of in vivo 13C NMR spectra acquired during the metabolism of l-[2-13C]arabinose (20 mM) at 30°C and under aerobic conditions. Each spectrum represents 2 min of acquisition. Cells were grown in aerobic batch cultures with l-arabinose (20 g liter−1), harvested at exponential growth phase, and suspended in 50 mM phosphate buffer (pH 6.0) to a concentration of around 40 g (dry weight) liter−1. Ara, arabinose; Ara-ol, arabitol. (B) Time course for the consumption of l-[2-13C]arabinose and the evolution of the intracellular metabolite pools. Metabolite concentrations were obtained from in vivo 13C NMR data. Circles, l-arabinose (◓, α-l-[2-13C]arabinopyranoside; ◒, β-l-[2-13C]arabinopyranoside; •, α-l-[2-13C]arabinofuranoside; ○, β-l-[2-13C]arabinopyranoside); triangles, arabitol (▵, C-1 labeled; ▴, C-2 labeled); squares, trehalose (□, C-1 labeled; ▪, C-2 labeled;, C-3 labeled).
P. guilliermondii consumed l-[2-13C]arabinose at a rate 1.5-fold higher than C. arabinofermentans (58 ± 3 μmol min−1 g [dry weight]−1 versus 41 ± 2 μmol min−1 g [dry weight]−1, respectively) (Fig. 1B and 2B). The level of arabitol labeled on C-2 increased with time, reaching maximal intracellular concentrations of 170 and 114 mM in P. guilliermondii and C. arabinofermentans, respectively, at around 10 min after the substrate addition. An increase in the signals due to trehalose C-2/arabitol C-4 and trehalose C-3 was also observed with both yeasts (Fig. 1B and 2B). The buildup of label on C-1 of arabitol was concomitant with the reduction of arabitol labeled on C-2. The concentration of arabitol labeled on C-1 was 3.5-fold higher in P. guilliermondii, whereas the concentration of trehalose labeled on C-1 was similar for the two strains examined.
Analysis of 13C isotopomers in perchloric acid extracts.
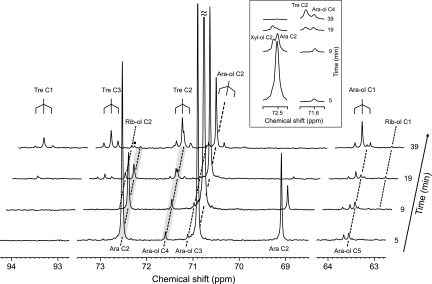
Resonance overlapping and low signal-to-noise ratios hindered a detailed characterization of 13C isotopomers in the 13C NMR spectra of living cells. Therefore, we resorted to the analysis of perchloric acid extracts of P. guilliermondii obtained at different time points during the metabolism of l-[2-13C]arabinose in parallel experiments. The narrower spectral lines allowed a better separation between overlapping peaks. Moreover, whenever resonances were not satisfactorily resolved, the areas of individual resonances were determined using standard deconvolution algorithms from Topspin (Bruker). Cell extracts were prepared at an early stage of substrate consumption (5 min), at the time of maximum arabitol accumulation (9 min), at the onset of l-[2-13C]arabinose depletion (19 min), and finally at 39 min after substrate addition (Fig. 2B). 13C NMR spectra of the corresponding extracts of P. guilliermondii are shown in Fig. 3.
FIG. 3.
13C NMR spectra of cell extracts obtained at different time points during the metabolism of l-[2-13C] arabinose (20 mM) in P. guilliermondii cells, under aerobic conditions, at 30°C. Cells were grown in aerobic batch cultures with l-arabinose (20 g liter−1), harvested at exponential growth phase, and suspended in 50 mM phosphate buffer (pH 6.0) to a concentration of 38 g (dry weight) liter−1. Ara, arabinose; Ara-ol, arabitol; Xyl-ol, xylitol; Tre, trehalose; Rib-ol, ribitol; ▪, Tre C-5. The inset shows magnifications of the shaded areas.
The time courses of the l-[2-13C]arabinose and [2-13C]arabitol pools were similar to those observed directly with living cells (Fig. 2A). The presence of 13C-13C couplings due to adjacent carbon atoms is clearly observed for C-1 and C-2 of arabitol and for C-1, C-2, and C-3 of trehalose, all showing a similar resonance pattern that consists of a central peak due to singly labeled molecules and a doublet of resonances (1JCC around 40 Hz), indicating double labeling. Furthermore, it was confirmed that the broad peak at about 72.5 ppm in the in vivo spectra included contributions from resonances due to C-2 of xylitol (72.54 ppm) and C-2 of α-l-arabinopyranoside (72.49 ppm) by the addition of the suspected compounds (Fig. 3, see expanded regions of the spectra). Additionally, the resonance at 71.60 ppm was composed of two components, one due to the central signal of trehalose C-2 (71.63 ppm) and the other due to arabitol C-4 (71.60 ppm). The three signals due to trehalose C-2 (isotopomers [2-13C]trehalose and [2,3-13C]trehalose) appeared only at the latest stages of metabolism (19 and 39 min), while the intensity of the resonance due to arabitol C-4 was relatively constant over time (Fig. 3, inset). It is worth noting that resonances due to trehalose C-4 (expected at 70.3 ppm) were not detected, showing that the 13C present in trehalose due to natural abundance is below the detection limit of our method. On the other hand, a weak resonance assigned to C-5 of trehalose (at 72.66 ppm) was observed. The two doublets centered at 71.63 ppm (C-2 of trehalose) and 73.17 ppm (C-3 of trehalose) have similar intensities and were assigned to [2,3-13C]trehalose (time point, 19 min). The degree of 13C enrichment on C-1 of trehalose (at 93.7 ppm) was lower than on C-2 or C-3; moreover, weak satellite signals due to trehalose doubly labeled on C-1 and C-2 were visible only in the last spectrum (time point, 39 min). A peak at 72.72 ppm seen in the spectra of extracts at 19 and 39 min was assigned as C-2 of ribitol on the basis of the comparison between the chemical shift and the spectra of the pure compound.
The fractional 13C enrichment of specific carbons of arabitol, trehalose, xylitol, and ribitol was determined (Table 1). For this purpose, all extracts were also analyzed by HPLC to determine total metabolite concentrations (arabinose, arabitol, trehalose, xylitol, and ribitol). The amount of label quantified under a certain resonance in the 13C NMR spectra of cell extracts was divided by the total amount of compound to yield the fractional enrichment. Values above 1% (naturally abundant 13C) indicate labeling originating from the metabolism of l-[2-13C]arabinose. As expected, the label appears primarily on C-2 of arabitol, but at the latest stages of metabolism, C-1 and C-4 of arabitol also become labeled. Likewise, the enrichment of trehalose and ribitol C-1 occurred only at the onset of l-arabinose exhaustion. The presence of unlabeled metabolite pools justify a total enrichment value lower than 100% and the relative increase in the fractional enrichment of arabitol and xylitol.
TABLE 1.
Time courses for enrichment of metabolitesa
| Metabolite | Time (min) |
13C fractional enrichment (%)
|
||||
|---|---|---|---|---|---|---|
| C-1 | C-2 | C-3 | C-4 | C-5 | ||
| Arabitol | 5 | 0.8 | 26.1 | 0.6 | 0.9 | 0.8 |
| 9 | 0.8 | 37.0 | 0.4 | 1.0 | 0.9 | |
| 19 | 1.8 | 28.1 | 0.4 | 1.4 | 0.7 | |
| 39 | 9.7 | 21.0 | 0.7 | 3.9 | 0.8 | |
| Xylitol | 5 | 25.1 | ||||
| 9 | 35.4 | |||||
| 19 | 37.1 | |||||
| 39 | ||||||
| Ribitol | 5 | |||||
| 9 | 32.7 | |||||
| 19 | 6.9 | 23.2 | ||||
| 39 | 8.2 | 18.8 | ||||
| Trehalose | 5 | |||||
| 9 | ||||||
| 19 | 8.1 | 28.1 | 23.5 | |||
| 39 | 9.6 | 29.7 | 20.7 | 1.5 | ||
Time courses are shown for 13C fractional enrichment of arabitol, xylitol, ribitol, and trehalose in P. guilliermondii cell extracts obtained during the metabolism of l-[2-13C]arabinose (20 mM) under aerobic conditions at 30°C. Values result from the combination of HPLC total quantification and 13C NMR information for 13C labeling.
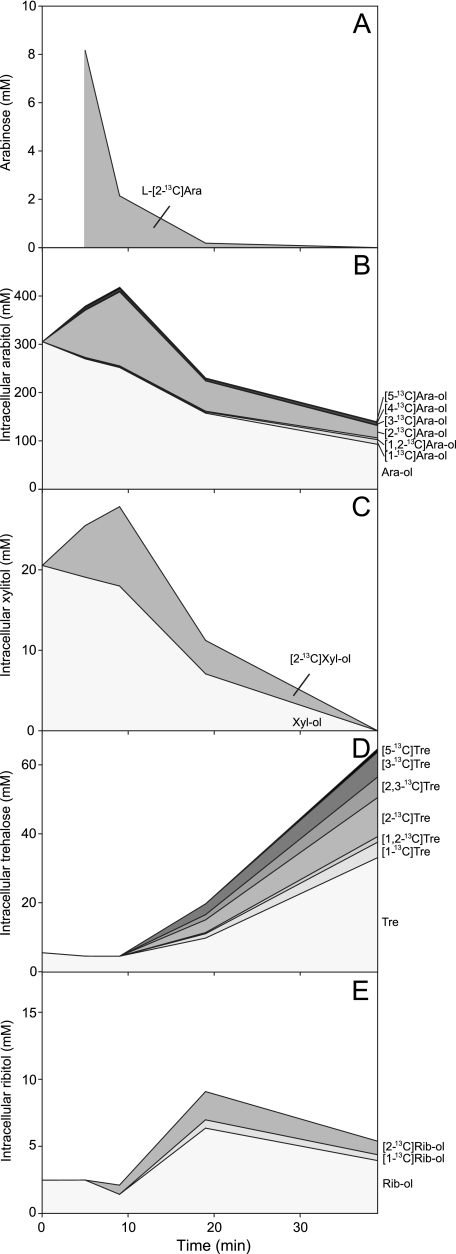
The time dependence of the absolute concentrations of the major isotopomers of arabitol, xylitol, ribitol, trehalose, and arabinose is plotted in Fig. 4. The large size of the initial unlabeled arabitol pool is noteworthy. The intermediate xylitol showed labeling time courses parallel to those of arabitol. Finally, the label appeared on ribitol and trehalose only at the latest stages of metabolism.
FIG. 4.
Evolution of metabolite isotopomeric pools as assessed in cell extracts obtained during l-[2-13C]arabinose (20 mM) metabolism in P. guilliermondii cells under aerobic conditions at 30°C. The data were obtained by combining 13C NMR information on 13C labeling with quantification of total pools by HPLC. (A) l-[2-13C]arabinose (Ara); (B) arabitol (Ara-ol); (C) xylitol (Xyl-ol); (D) trehalose (Tre); and (E) ribitol (Rib-ol).
Arabitol accumulates intracellularly during l-arabinose metabolism.
Earlier studies have shown that arabitol accumulates extracellularly only under oxygen-limiting conditions with C. arabinofermentans and P. guilliermondii cultures growing on l-arabinose (11). Therefore, the location of arabitol detected in living cells by 13C NMR (Fig. 1 and 2) was determined. The quantification of arabitol in supernatant solutions and in total cell extracts (as described in Material and Methods) showed that arabitol was exclusively intracellular. Upon the addition of l-arabinose during the in vivo NMR experiments, the oxygen level in cell suspensions of C. arabinofermentans and P. guilliermondii dropped to 0 and 20%, respectively, despite continuous sparging with pure oxygen at a flow rate of approximately 250 ml min−1. Nongrowing cells of C. arabinofermentans did not excrete arabitol under oxygen-limited conditions, and the intracellular arabitol concentration amounted to 95 mM. A higher intracellular level of arabitol (177 mM) was detected in P. guilliermondii, even though the dissolved oxygen remained at 20% in this case. Intracellular arabitol accumulation was also determined for C. arabinofermentans and P. guilliermondii cultures growing aerobically with l-arabinose. Cells were sampled at the exponential phase and treated immediately with perchloric acid, and arabitol was determined by HPLC in the resulting cell extracts. The intracellular arabitol concentration in C. arabinofermentans and P. guilliermondii were 108 mM and 410 mM, respectively. Oxygenation of cell suspensions at 30°C led to arabitol consumption. In C. arabinofermentans, the intracellular concentration decreased from 108 to 75 mM in the first 5 min, remaining approximately constant thereafter. In comparison, concentrations of 319 mM and 151 mM were found in P. guilliermondii after 5 and 15 min in the presence of oxygen, respectively.
d-Arabitol and l-arabitol ratios at different stages of l-arabinose metabolism.
The analysis of l-[2-13C]arabinose metabolism by NMR showed the production of labeled ribitol. Since the production of ribitol and d-arabitol from d-xylose has been proposed as a potential redox sink (16), we suspected that d-arabitol could also be a product of l-arabinose metabolism. Therefore, to obtain a comprehensive picture of the pathways involved in l-arabinose metabolism in yeasts, it was deemed important to discriminate between arabitol enantiomers. Enantiomeric resolution is not achieved by techniques such as standard HPLC or NMR. Therefore, the d-arabitol/l-arabitol ratio was determined by chiral GC-MS. The enantiomeric excess was determined with several cell extracts of C. arabinofermentans and P. guilliermondii (at 10, 20, 40 and 60 min after arabinose addition) (Table 2). Significant differences were observed between the two yeasts. During l-arabinose consumption, l-arabitol was the only enantiomer found in P. guilliermondii. However, d-arabitol was detected in a 1:1 ratio at later stages of metabolism. Conversely, considerable amounts of d-arabitol were observed in C. arabinofermentans before l-arabinose exhaustion. Moreover, in the latter, the l-arabitol pool was virtually depleted within 20 min and d-arabitol accounted for the great majority of total arabitol monitored thereafter.
TABLE 2.
Time courses for l- and d-arabitol concentrationsa
| Strain | Sampling time (min) | Concn (%) of metabolite ± SD
|
|
|---|---|---|---|
| l-Arabitol | d-Arabitol | ||
| C. arabinofermentans | 10 | 84.1 ± 0.9 | 15.9 ± 0.9 |
| PYCC 5603T | 20 | 11.3 ± 5.4 | 88.7 ± 5.4 |
| 40 | 11.5 ± 0.6 | 88.5 ± 0.6 | |
| 60 | 11.6 ± 2.3 | 88.4 ± 2.3 | |
| P. guilliermondii | 10 | 100.0 ± 0.0 | ND |
| PYCC 3012 | 20 | 100.0 ± 0.0 | ND |
| 40 | 51.5 ± 1.7 | 48.5 ± 1.7 | |
| 60 | 51 ± 15 | 49 ± 15 | |
Time courses are shown for l- and d-arabitol concentrations ± standard deviations (SD) as measured by chiral GC-MS during l-arabinose (20 mM) metabolism under aerobic conditions at 30°C. l-Arabinose was added at time zero. ND, not detected.
Model of l-[2-13C]arabinose metabolism in yeast.
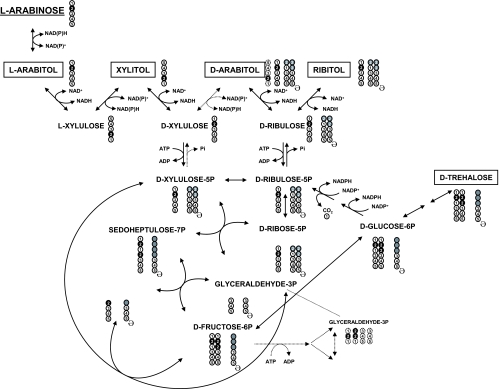
Based on the combination of data obtained by 13C NMR, HPLC, and chiral GC-MS, a metabolic scheme of l-[2-13C]arabinose metabolism in yeast was constructed. The reactions, metabolites, and qualitative isotopomer distributions are depicted in Fig. 5. The detection of resonances due to l-[2-13C]arabitol and [2-13C]xylitol shows that l-[2-13C]arabinose is metabolized through the redox catabolic pathway. The resulting d-[2-13C]xylulose is either phosphorylated to d-[2-13C]xylulose-5-P and/or converted to d-[4-13C]arabitol. The detection of only small amounts of [4-13C]arabitol indicates that the direct conversion of d-xylulose to d-arabitol (Fig. 5) does not occur or has a minor contribution. In fact, the formation of [4-13C]arabitol results most likely from glyceraldehyde-3-P labeled on C-2 and back flux (see below for experimental evidence). The definite production of d-arabitol (identified by GG-MS) and the large excess of [2-13C]arabitol compared to [4-13C]arabitol (as assessed by 13C NMR) strongly indicates that the production of d-arabitol occurs preferentially through dephosphorylation of d-ribulose-5-P, an isomer of d-xylulose-5-P (Fig. 5). Detection of [2-13C]ribitol further supports this hypothesis.
FIG. 5.
Schematic representation of l-arabinose metabolism and pattern of carbon distribution as determined by 13C NMR, HPLC, and chiral GC-MS. l-[2-13C]arabinose is metabolized through the redox catabolic pathway to d-[2-13C]xylulose-5-P. d-Xylulose-5-P proceeds through the PPP or is further converted to d-ribulose, yielding d-arabitol and ribitol. The reactions through the PPP yield fructose-6-P labeled in C-2 and C-3. C-3 labeling is specifically obtained with the carbon transfer (two carbons) from d-xylulose-5-P to d-erythrose-4-P. Fructose-6-P, labeled in C-2 and C-3, either enters the glycolytic pathway or is isomerized to glucose-6-P, the precursor of trehalose. In addition, glucose-6-P follows the oxidative PPP with decarboxylation and conversion to d-ribulose-5-P. The cycle is closed with the decarboxylation, and new species labeled in C-1 are produced (arabitol and trehalose). Extra cycles will maintain the same pattern of carbon labeling. The circles represent individual carbon atoms, numbered according to their position in the original l-[2-13C]arabinose. For each metabolite, filled circles represent 13C labeling. Gray circles represent all carbon positions that can become 13C enriched after carbon recycling (ə;) when l-[2-13C]arabinose is metabolized. Dashed lines indicate putative pathways.
The subsequent processing of d-[2-13C]xylulose-5-P through the reactions of the PPP yields fructose-6-P, singly labeled on C-2 and doubly labeled on C-2 and C-3, and unlabeled glyceraldehyde-3-P. The C-2/C-3 labeling in fructose-6-P is specifically obtained with carbon transfer (two carbons) from d-[2-13C]xylulose-5-P to [1-13C]erythrose-4-P. The isotopomers [2-13C]fructose-6-P and [2,3-13C]fructose-6-P can be processed through the glycolytic pathway, producing glyceraldehyde-3-P labeled primarily on C-2 or isomerized to glucose-6-P, the precursor of trehalose. The detection of [2-13C]trehalose and [2,3-13C]trehalose by 13C NMR fully supports the proposed pathway. In addition, glucose-6-P can follow the oxidative PPP with decarboxylation and conversion to d-ribulose-5-P. The decarboxylation generates new species labeled on C-1 as indicated by the detection of resonances due to [1-13C]arabitol, [1-13C]ribitol, and [1-13C]trehalose.
The pattern of carbon labeling is not considerably affected by further cycles of decarboxylation. However, back flux from C-2-labeled glyceraldehyde-3-P to fructose-6-P can lead to C-5 enrichment, as observed for 13C enrichment of trehalose C-5 and arabitol C-4 (Table 1 and Fig. 5), the latter originating from decarboxylation through the oxidative PPP.
The presence of carbon reserves of (unlabeled) arabitol can give rise to all possible combinations of C-1 and C-2 labeling in d-arabitol and ribitol (four isotopomers each) and of C-1, C-2, and C-3 labeling in trehalose (eight possible isotopomers). This explains the observation of singly labeled trehalose (see, for example, the resonance due to [3-13C]trehalose as shown in Fig. 2).
DISCUSSION
The increased interest in the utilization of hemicellulose-containing materials has drawn considerable attention to the metabolism of pentoses in yeasts. Fermentation of l-arabinose is a very rare feature among yeasts, and l-arabinose metabolism is poorly characterized. Intending to fill this gap in knowledge, we compared two natural l-arabinose-utilizing yeasts, C. arabinofermentans PYCC 5603T and P. guilliermondii PYCC 3012 (10), selected in a previous search for rapid growth and high-capacity l-arabinose influx rates. These yeasts have been characterized in terms of their physiology, including the influences of sugar and oxygen in product formation (11), in l-arabinose transport, and in in vitro enzyme activities at the early steps of l-arabinose catabolism (10). Since these studies did not provide answers concerning the distribution of metabolic fluxes, which is important for understanding the incapacity to ferment l-arabinose and the differences between the two yeasts in terms of metabolite production, 13C NMR was used to trace the fate of l-[2-13C]arabinose through branching metabolic pathways.
Consumption of l-[2-13C]arabinose was faster in resting cells of P. guilliermondii (58 ± 3 μmol min−1 g [dry weight]−1) than in C. arabinofermentans (41 ± 2 μmol min−1 g [dry weight]−1) (Fig. 1 and 2). Similar relative values were found for l-arabinose consumption during fully aerobic and exponentially growing batch cultures in 5 g/liter (33 mM) l-arabinose: approximately 63 μmol min−1 g (dry weight)−1 in P. guilliermondii compared to 55 μmol min−1 g (dry weight)−1 in C. arabinofermentans, values obtained from similar maximum specific growth rates (μmax) and relative biomass yields obtained from media with an initial concentration of 5 g/liter (33 mM) l-arabinose (11) by applying the equation μmax = biomass yield × specific substrate consumption rate. These results are consistent with the higher transport capacity of P. guilliermondii (10). In particular, the initial uptake rates at 20 mM l-arabinose (1.36 mmol min−1 g [dry weight]−1 and 0.49 mmol min−1 g [dry weight]−1 for P. guilliermondii and C. arabinofermentans, respectively) (10) are about 20- and 10-fold higher than the l-arabinose consumption rates in resting cells at the same substrate concentration, suggesting that transport does not limit l-arabinose catabolism in these yeasts.
Contrary to what was expected from the presence of arabitol in the external medium with an actively growing population in l-arabinose under oxygen-limited conditions (11), all arabitol produced by resting cells was intracellular. We first hypothesized that the accumulation of arabitol resulted from the impairment in NAD+ regeneration by the respiratory chain, since it coincided with oxygen depletion (11). This explanation appears to suit C. arabinofermentans, since oxygen disappeared during l-arabinose utilization in the NMR experiments, despite high fluxes of oxygen supply. However, in P. guilliermondii, mechanisms other than those associated with oxygen limitation need to be invoked, as arabitol accumulated when oxygen was still detectable during l-arabinose utilization by resting cells. Results from the present work, together with data on enzyme kinetics (10), point to a bottleneck at the level of LAD. It should be noted that the l-arabitol consumption rate by P. guilliermondii resting cells (0.32 ± 0.07 mmol min−1 g [dry weight]−1) was of the same order of magnitude as the LAD activity determined for crude cell extracts (10) (approximately 0.40 mmol min−1 g [dry weight]−1 for an arabitol concentration of 177 mM). On the other hand, the same enzyme activity in C. arabinofermentans (approximately 1.94 mmol min−1 g [dry weight]−1 for an arabitol concentration of 95 mM) far exceeds (10-fold) the l-arabitol consumption rate measured by in vivo NMR (0.20 ± 0.01 mmol min−1 g [dry weight]−1). The l-arabitol accumulation observed for C. arabinofermentans during growth is likely to be related to insufficient XK activity (10), the enzyme catalyzing the first irreversible step in the l-arabinose catabolic pathway. As to the intracellular accumulation of arabitol in resting cells, without secretion to the external medium, we have no explanation thus far. Polyol efflux has been thoroughly investigated in S. cerevisiae, namely the mechanisms involved in glycerol accumulation and release, which are controlled by the Fps1 transporter (26, 30, 36, 37). The Zygosaccharomyces rouxii Fps1p homologue ZrFps1p was found to be involved in glycerol and d-arabitol efflux (38), but d-arabitol accumulated intracellularly in concentrations up to around 0.5 M in response to osmotic stress. What prevents the efflux of l-arabitol concentrations as high as 0.4 M in l-arabinose-grown P. guilliermondii resting cells warrants further study.
Besides l-arabitol, other polyols (xylitol, d-arabitol, and ribitol) were derived from l-[2-13C]arabinose. Chiral GC-MS was used to quantify the two stereoisomers of arabitol, which cannot be differentiated by NMR. The detection of l-arabitol and xylitol, both labeled on C-2, confirms that the metabolism of l-arabinose in these yeasts proceeds through the redox catabolic pathway first identified in filamentous fungi (6, 45) and is in line with results of previously reported in vitro enzyme assays (10). A primary route for the biosynthesis of d-arabitol and ribitol was identified by the observation of isotopic enrichment at position C-2 of their carbon backbones: d-xylulose is converted to d-xylulose-5-P, which is isomerized to d-ribulose-5-P, followed by dephosphorylation to ribulose, the precursor of the polyols, as hypothesized by Jeppsson et al. (16). The direct conversion of d-xylulose to d-arabitol, proposed as an alternative pathway in pentose utilization (19, 20, 34), would lead to d-[4-13C]arabitol, a compound detected only in trace amounts. The occurrence of poor enrichment in C-4 most likely stems from the utilization of [5-13C]fructose-6-P (which is produced from glyceraldehyde-3-P labeled on C-2), a hypothesis further supported by the detection of [5-13C]trehalose. The production of trehalose, d-arabitol, and ribitol may act as a sink to restore inorganic phosphate (Pi) levels (26). The need to balance the level of sugar phosphates and Pi will reduce the glycolytic flux.
Under oxygen limitation conditions, only trace amounts of ethanol were produced from l-[2-13C]arabinose by l-arabinose-grown resting cells of both of the yeasts studied, whereas cells grown under the same conditions (i.e., on l-arabinose) were able to produce considerable amounts of ethanol from [1-13C]glucose (data not shown). These results indicate that it is not due to the absence of pyruvate decarboxylase and alcohol dehydrogenase that l-arabinose-grown cells could not ferment l-arabinose. We propose that the low glycolytic flux from l-arabinose is associated with a metabolic shift to circumvent NADPH and NAD+ limitations in the early steps of pentose utilization. The reductases in l-arabinose metabolism usually require NADPH, which cannot be regenerated in the oxidation of the resulting polyols via the NAD+-dependent l-arabitol and xylitol dehydrogenases. Because transhydrogenases are absent in yeast (3), the strategies to overcome these redox limitations deviate the carbon flux from glycolysis: NADPH regeneration is achieved by carbon recycling through the oxidative PPP, demonstrated in this work by the detection of C-1-labeled arabitol, ribitol, and trehalose; NAD+ can be regenerated by d-arabitol and ribitol production, as well as in the respiratory chain. The recycling through the oxidative PPP has been proposed as a way to recycle NADPH in Candida utilis (5) and Candida (Pichia) guilliermondii (12) and in recombinant S. cerevisiae (17, 18) during d-xylose utilization. However, earlier attempts to demonstrate recycling by resorting to specifically labeled xylose coupled to NMR experiments failed (24, 40, 41). The level of recycling through the oxidative PPP is apparently higher in P. guilliermondii than in C. arabinofermentans, as indicated by the higher amounts of [1-13C]arabitol. The dual cofactor specificity of LXR and XDH (10), although with a clear preference for NADPH and NAD+, respectively, may alleviate the requirement for NADPH in C. arabinofermentans and consequently diminish the need of carbon recycling for NADPH regeneration. The higher biomass yield in C. arabinofermentans (0.50 g [dry weight]−1 versus 0.43 g [dry weight]−1 in P. guilliermondii) determined during aerobic growth in batch cultures with 5 g/liter l-arabinose (11) further supports this hypothesis.
The cofactor imbalance in the early steps of l-arabinose catabolism, aggravated in comparison to that of d-xylose utilization and remaining unsolved under anaerobic conditions, probably explains the natural nonoccurrence of l-arabinose fermentation in yeast. The LAD and, possibly, the XK steps appear as potentially useful targets for improvement. The present work sheds light into l-arabinose metabolism in yeasts, providing rational guidelines for the design of strategies for the production of new and efficient l-arabinose-fermenting yeasts.
Acknowledgments
This work was funded in part by EU project NILE (FP6-019882).
C.F. held a Ph.D. fellowship (SFRH/BD/6794/2001) from Fundação para a Ciência e a Tecnologia, Portugal.
Footnotes
Published ahead of print on 1 February 2008.
REFERENCES
- 1.Barnett, J. A. 1976. The utilization of sugars by yeasts. Adv. Carbohydr. Chem. Biochem. 32:125-234. [DOI] [PubMed] [Google Scholar]
- 2.Becker, J., and E. Boles. 2003. A modified Saccharomyces cerevisiae strain that consumes l-arabinose and produces ethanol. Appl. Environ. Microbiol. 69:4144-4150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bruinenberg, P. M., R. Jonker, J. P. van Dijken, and W. A. Scheffers. 1985. Utilization of formate as an additional energy source by glucose-limited chemostat cultures of Candida utilis CBS 621 and Saccharomyces cerevisiae CBS 8066: evidence for the absence of transhydrogenase activity in yeasts. Arch. Microbiol. 142:302-306. [Google Scholar]
- 4.Bruinenberg, P. M., J. P. van Dijken, and W. A. Scheffers. 1983. An enzymic analysis of NADPH production and consumption in Candida utilis. J. Gen. Microbiol. 129:965-971. [DOI] [PubMed] [Google Scholar]
- 5.Bruinenberg, P. M., J. P. van Dijken, and W. A. Scheffers. 1984. Production and consumption of NADPH and NADH during growth of Candida utilis on xylose. Antonie van Leeuwenhoek 50:81-82. [Google Scholar]
- 6.Chiang, C., and S. G. Knight. 1960. A new pathway of pentose metabolism. Biochim. Biophys. Res. Commun. 3:554-559. [DOI] [PubMed] [Google Scholar]
- 7.De Graaf, A. A. 2000. Metabolic flux analysis of Corynebacterium glutamicum, p. 506-555. In K. Schügerl and K. H. Bellgardt (ed.), Bioreaction engineering. Springer Verlag, Berlin, Germany.
- 8.Dien, B. S., C. P. Kurtzman, B. C. Saha, and R. J. Bothast. 1996. Screening for l-arabinose fermenting yeasts. Appl. Biochem. Biotechnol. 57-58:233-242. [PubMed] [Google Scholar]
- 9.du Preez, J. C., B. van Driessel, and B. A. Prior. 1989. Effect of aerobiosis on fermentation and key enzyme levels during growth of Pichia stipitis, Candida shehatae and Candida tenuis on d-xylose. Arch. Microbiol. 152:143-147. [Google Scholar]
- 10.Fonseca, C., R. Romão, H. Rodrigues de Sousa, B. Hahn-Hägerdal, and I. Spencer-Martins. 2007. l-Arabinose transport and catabolism in yeast. FEBS J. 274:3589-3600. [DOI] [PubMed] [Google Scholar]
- 11.Fonseca, C., I. Spencer-Martins, and B. Hahn-Hägerdal. 2007. l-Arabinose metabolism in Candida arabinofermentans PYCC 5603T and Pichia guilliermondii PYCC 3012: influence of sugar and oxygen on product formation. Appl. Microbiol. Biotechnol. 75:303-310. [DOI] [PubMed] [Google Scholar]
- 12.Granström, T., H. Ojamo, and M. Leisola. 2001. Chemostat study of xylitol production by Candida guilliermondii. Appl. Microbiol. Biotechnol. 55:36-42. [DOI] [PubMed] [Google Scholar]
- 13.Ho, N. W., Z. Chen, and A. P. Brainard. 1998. Genetically engineered Saccharomyces yeast capable of effective cofermentation of glucose and xylose. Appl. Environ. Microbiol. 64:1852-1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Höfer, M., and P. C. Misra. 1978. Evidence for a proton/sugar symport in the yeast Rhodotorula gracilis (glutinis). Biochem. J. 172:15-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hui, M., S. W. Cheung, M. L. Chin, K. C. Chu, R. C. Chan, and A. F. Cheng. 2004. Development and application of a rapid diagnostic method for invasive Candidiasis by the detection of d-/l-arabinitol using gas chromatography/mass spectrometry. Diagn. Microbiol. Infect. Dis. 49:117-123. [DOI] [PubMed] [Google Scholar]
- 16.Jeppsson, H., N. J. Alexander, and B. Hahn-Hägerdal. 1995. Existence of cyanide-insensitive respiration in the yeast Pichia stipitis and its possible influence on product formation during xylose utilization. Appl. Environ. Microbiol. 61:2596-2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jeppsson, M., B. Johansson, B. Hahn-Hägerdal, and M. F. Gorwa-Grauslund. 2002. Reduced oxidative pentose phosphate pathway flux in recombinant xylose-utilizing Saccharomyces cerevisiae strains improves the ethanol yield from xylose. Appl. Environ. Microbiol. 68:1604-1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jeppsson, M., B. Johansson, P. R. Jensen, B. Hahn-Hägerdal, and M. F. Gorwa-Grauslund. 2003. The level of glucose-6-phosphate dehydrogenase activity strongly influences xylose fermentation and inhibitor sensitivity in recombinant Saccharomyces cerevisiae strains. Yeast 20:1263-1272. [DOI] [PubMed] [Google Scholar]
- 19.Jin, Y. S., J. Cruz, and T. W. Jeffries. 2005. Xylitol production by a Pichia stipitis d-xylulokinase mutant. Appl. Microbiol. Biotechnol. 68:42-45. [DOI] [PubMed] [Google Scholar]
- 20.Jin, Y. S., S. Jones, N. Q. Shi, and T. W. Jeffries. 2002. Molecular cloning of XYL3 (d-xylulokinase) from Pichia stipitis and characterization of its physiological function. Appl. Environ. Microbiol. 68:1232-1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karhumaa, K., B. Wiedemann, B. Hahn-Hägerdal, E. Boles, and M. F. Gorwa-Grauslund. 2006. Co-utilization of l-arabinose and d-xylose by laboratory and industrial Saccharomyces cerevisiae strains. Microb. Cell Fact. 5:18-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kötter, P., and M. Ciriacy. 1993. Xylose fermentation by Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 38:776-783. [Google Scholar]
- 23.Ligthelm, M. E., B. A. Prior, and J. C. du Preez. 1988. The oxygen requirements of yeasts for the fermentation of d-xylose and d-glucose to ethanol. Appl. Microbiol. Biotechnol. 28:63-68. [Google Scholar]
- 24.Ligthelm, M. E., B. A. Prior, J. C. du Preez, and V. Brandt. 1988. An investigation of d-{1-13C} xylose metabolism in Pichia stipitis under aerobic and anaerobic conditions. Appl. Microbiol. Biotechnol. 28:293-296. [Google Scholar]
- 25.London, R. E. 1988. 13C labeling in studies of metabolic regulation. Prog. Nucl. Magn. Res. Spectrosc. 20:337-383. [Google Scholar]
- 26.Luyten, K., J. Albertyn, W. F. Skibbe, B. A. Prior, J. Ramos, J. M. Thevelein, and S. Hohmann. 1995. Fps1, a yeast member of the MIP family of channel proteins, is a facilitator for glycerol uptake and efflux and is inactive under osmotic stress. EMBO J. 14:1360-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McMillan, J. D., and B. L. Boynton. 1994. Arabinose utilization by xylose-fermenting yeasts and fungi. Appl. Biochem. Biotechnol. 45:569-584. [DOI] [PubMed] [Google Scholar]
- 28.Neves, A. R., A. Ramos, H. Costa, I. I. van Swam, J. Hugenholtz, M. Kleerebezem, W. de Vos, and H. Santos. 2002. Effect of different NADH oxidase levels on glucose metabolism by Lactococcus lactis: kinetics of intracellular metabolite pools determined by in vivo nuclear magnetic resonance. Appl. Environ. Microbiol. 68:6332-6342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Neves, A. R., A. Ramos, M. C. Nunes, M. Kleerebezem, J. Hugenholtz, W. M. de Vos, J. Almeida, and H. Santos. 1999. In vivo nuclear magnetic resonance studies of glycolytic kinetics in Lactococcus lactis. Biotechnol. Bioeng. 64:200-212. [DOI] [PubMed] [Google Scholar]
- 30.Oliveira, R., F. Lages, M. Silva-Graça, and C. Lucas. 2003. Fps1p channel is the mediator of the major part of glycerol passive diffusion in Saccharomyces cerevisiae: artefacts and re-definitions. Biochim. Biophys. Acta 1613:57-71. [DOI] [PubMed] [Google Scholar]
- 31.Richard, P., R. Verho, M. Putkonen, J. Londesborough, and M. Penttilä. 2003. Production of ethanol from l-arabinose by Saccharomyces cerevisiae containing a fungal l-arabinose pathway. FEMS Yeast Res. 3:185-189. [DOI] [PubMed] [Google Scholar]
- 32.Saha, B. C., and R. J. Bothast. 1996. Production of l-arabitol from l-arabinose by Candida entomaea and Pichia guilliermondii. Appl. Microbiol. Biotechnol. 45:299-306. [Google Scholar]
- 33.Santos, H., and D. L. Turner. 1986. 13C and proton NMR studies of horse cytochrome c. Assignment and temperature dependence of methyl resonances. FEBS Lett. 194:73-77. [DOI] [PubMed] [Google Scholar]
- 34.Shi, N. Q., K. Prahl, J. Hendrick, J. Cruz, P. Lu, J. Y. Cho, S. Jones, and T. Jeffries. 2000. Characterization and complementation of a Pichia stipitis mutant unable to grow on d-xylose or l-arabinose. Appl. Biochem. Biotechnol. 84-86:201-216. [DOI] [PubMed] [Google Scholar]
- 35.Skoog, K., and B. Hahn-Hägerdal. 1990. Effect of oxygenation on xylose fermentation by Pichia stipitis. Appl. Environ. Microbiol. 56:3389-3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sutherland, F. C., F. Lages, C. Lucas, K. Luyten, J. Albertyn, S. Hohmann, B. A. Prior, and S. G. Kilian. 1997. Characteristics of Fps1-dependent and -independent glycerol transport in Saccharomyces cerevisiae. J. Bacteriol. 179:7790-7795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tamás, M. J., K. Luyten, F. C. Sutherland, A. Hernandez, J. Albertyn, H. Valadi, H. Li, B. A. Prior, S. G. Kilian, J. Ramos, L. Gustafsson, J. M. Thevelein, and S. Hohmann. 1999. Fps1p controls the accumulation and release of the compatible solute glycerol in yeast osmoregulation. Mol. Microbiol. 31:1087-1104. [DOI] [PubMed] [Google Scholar]
- 38.Tang, X. M., G. Kayingo, and B. A. Prior. 2005. Functional analysis of the Zygosaccharomyces rouxii Fps1p homologue. Yeast 22:571-581. [DOI] [PubMed] [Google Scholar]
- 39.Tantirungkij, M., N. Nakashima, T. Seki, and T. Yoshida. 1993. Construction of xylose-assimilating Saccharomyces cerevisiae. J. Ferment. Bioeng. 75:83-88. [Google Scholar]
- 40.Taylor, K. B., M. J. Beck, D. H. Huang, and T. T. Sakai. 1990. The fermentation of xylose: studies by carbon-13 nuclear magnetic resonance spectroscopy. J. Ind. Microbiol. 6:29-41. [Google Scholar]
- 41.van Zyl, C., B. A. Prior, S. G. Kilian, and E. V. Brandt. 1993. Role of d-ribose as a cometabolite in d-xylose metabolism by Saccharomyces cerevisiae. Appl. Environ. Microbiol. 59:1487-1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Verduyn, C., E. Postma, W. A. Scheffers, and J. P. van Dijken. 1992. Effect of benzoic acid on metabolic fluxes in yeasts - a continuous-culture study on the regulation of respiration and alcoholic fermentation. Yeast 8:501-517. [DOI] [PubMed] [Google Scholar]
- 43.Verho, R., M. Putkonen, J. Londesborough, M. Penttilä, and P. Richard. 2004. A novel NADH-linked l-xylulose reductase in the l-arabinose catabolic pathway of yeast. J. Biol. Chem. 279:14746-14751. [DOI] [PubMed] [Google Scholar]
- 44.Wisselink, H. W., M. J. Toirkens, B. M. Rosario Franco, A. A. Winkler, J. P. van Dijken, J. T. Pronk, and A. J. van Maris. 2007. Engineering of Saccharomyces cerevisiae for efficient anaerobic alcoholic fermentation of l-arabinose. Appl. Environ. Microbiol. 73:4881-4891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Witteveen, C. F. B., R. Busink, P. Vandevondervoort, C. Dijkema, K. Swart, and J. Visser. 1989. l-Arabinose and d-xylose catabolism in Aspergillus niger. J. Gen. Microbiol. 135:2163-2171. [Google Scholar]