Abstract
An oligonucleotide microarray based on the arrayed-primer extension (APEX) technique has been developed to simultaneously identify pathogenic fungi frequently isolated from invasive and superficial infections. Species-specific oligonucleotide probes complementary to the internal transcribed spacer 1 and 2 (ITS1 and ITS2) region were designed for 24 species belonging to 10 genera, including Candida species (Candida albicans, Candida dubliniensis, Candida famata, Candida glabrata, Candida tropicalis, Candida kefyr, Candida krusei, Candida guilliermondii, Candida lusitaniae, Candida metapsilosis, Candida orthopsilosis, Candida parapsilosis, and Candida pulcherrima), Cryptococcus neoformans, Aspergillus species (Aspergillus fumigatus and Aspergillus terreus), Trichophyton species (Trichophyton rubrum and Trichophyton tonsurans), Trichosporon cutaneum, Epidermophyton floccosum, Fusarium solani, Microsporum canis, Penicillium marneffei, and Saccharomyces cerevisiae. The microarray was tested for its specificity with a panel of reference and blinded clinical isolates. The APEX technique was proven to be highly discriminative, leading to unequivocal identification of each species, including the highly related ones C. parapsilosis, C. orthopsilosis, and C. metapsilosis. Because of the satisfactory basic performance traits obtained, such as reproducibility, specificity, and unambiguous interpretation of the results, this new system represents a reliable method of potential use in clinical laboratories for parallel one-shot detection and identification of the most common pathogenic fungi.
The majority of life-threatening fungal infections are caused by the well-known opportunistic pathogens Candida albicans, Aspergillus fumigatus, and Cryptococcus neoformans, with the latter being associated frequently with severe mycoses in AIDS patients (22, 30). Candidiasis remains the major cause of invasive fungal infections in immunocompromised patients, and in recent years, an impressive increase in mortality rate due to candidemia by non-C. albicans Candida species has been noted (2, 6, 15, 20, 24). It has been suggested that the widespread use of azoles in clinical settings could have contributed to changing the etiology of C. albicans candidemia toward non-C. albicans species, which now account for more than 50% of systemic Candida infections (2, 6, 15). Rapid diagnosis of these mycoses is crucial for prompt management of infection with tailored antifungal treatments. However, conventional laboratory methods for identification of fungal pathogens, though continuously improving, are still time-consuming and therefore often inadequate for ensuring early targeted therapy, especially for uncommon or newly identified fungal species. Unlike what is currently available for bacteria, molecular approaches for the identification of pathogenic fungi have been held back so far due to the lack of a robust sequence data bank. However, several DNA-based methods have been introduced and have improved the identification of fungal pathogens and shortened the time required for their detection (3, 12, 13, 19, 21, 26). Most molecular procedures allow the identification of one or a few species at a time (3, 12, 13), thus resulting in a high cost if all relevant species must be considered. An ideal approach to overcome this limitation is given by the application of DNA microarray technology, which may enable discrimination of a wide range of pathogens in a single assay. The panmicrobial oligonucleotide array developed by Palacios and colleagues (23) was designed mainly to produce a staged strategy for molecular surveillance and discovery of emerging pathogens, as it covers detection of viruses, bacteria, fungi, and parasites. However, in the case of fungi, the panmicrobial chip predominantly allows genus identification rather than fungal species discrimination. It should be noted that rapid identification of pathogenic fungi at the species level is relevant in medical practice, as fungemia and other fungal symptomatic infections are emerging as a leading cause of morbidity and mortality in the general patient population, especially for hospitalized cancer and major surgical patients (15). A DNA microarray specifically developed by Leinberger and colleagues (18) for detecting fungal pathogens enabled discrimination of the 12 most common pathogenic Candida and Aspergillus organisms at the species level, and the array developed by Huang and colleagues (11) enlarged to 20 the number of identified species, which are representative of eight different genera. Nevertheless, neither system encompasses oligonucleotide probes for detection/identification of emerging fungi increasingly reported to be responsible for invasive or other symptomatic infections, such as Candida famata, Candida kefyr, Trichosporon cutaneum, Fusarium solani, and Penicillium marneffei (5, 10, 24, 31); moreover, the probes designed to detect Candida parapsilosis do not differentiate this species from two newly identified and closely related ones, i.e., Candida orthopsilosis and Candida metapsilosis (27), which were recently reported to cause mycoses in humans (14, 28). Oligonucleotide probes effectively enabling simultaneous discrimination of these three species may be useful, since available conventional methods do not allow discrimination of C. parapsilosis from C. orthopsilosis and C. metapsilosis (26, 28).
This report describes the development of an up-to-date oligonucleotide array for the unambiguous identification of 24 fungi, allotted into 10 diverse genera, including (i) species involved in invasive infections and frequently exhibiting a drug-resistant phenotype, such as Candida glabrata, Candida krusei, and Aspergillus terreus (17, 24, 25); (ii) emerging fungal pathogens, such as C. famata, C. kefyr, Trichosporon cutaneum, or molds such as Fusarium solani and Penicillium marneffei; and (iii) the newly defined species C. orthopsilosis and C. metapsilosis. The oligonucleotide probes used in this microarray are complementary to the sequence variation in the internal transcribed spacer 1 and 2 (ITS1 and ITS2) region of each species. Direct labeling of PCR products is not required, and signals for fungal species identification are based on the arrayed-primer extension (APEX) technique, which has been applied successfully to discriminate natural variants of human papillomavirus type 16, to identify germ line mutations in beta-thalassemia, and for single-nucleotide polymorphism genotyping (4, 7, 8, 16). The specificity and reproducibility of the array were validated with reference strains, and its application was tested with blinded clinical isolates.
MATERIALS AND METHODS
Strains.
The yeast and mold reference isolates used in this study were obtained from the American Type Culture Collection (ATCC), Deutsche Sammlung von Mikroorganismen und Zellkulturen (DSMZ), and Centraalbureau voor Schimmelcultures (CBS) (Table 1). A selected panel of 20 clinical fungal isolates, previously identified with conventional laboratory tests (API32 ID and a Vitek 2 advanced colorimetry semiautomated system [bioMerieux, Marcy l'Etoile, France] for yeast identification and microscopic examination of reproductive structures for hyphomycetes), were provided by the Unità Operativa di Microbiologia, Ospedale Universitario, Pisa, Italy. The isolates (Table 2) were blindly submitted to the DNA microarray identification system. All of the isolates were maintained on Sabouraud agar (yeasts) or potato dextrose agar (molds) (Liofilchem S.R.L., TE, Italy) for the duration of the study. Genomic DNAs from representative bacterial reference strains (Bacillus subtilis ATCC 6633, Escherichia coli SCS 110 [ATCC], and Mycobacterium bovis bacillus Calmette-Guérin strain Pasteur [bioMerieux, Lyon, France]) were used as negative controls in PCR experiments.
TABLE 1.
Fungal species tested and oligonucleotides used with the APEX technique
| Species | GenBank accession no. | Probe | Probe sequence |
|---|---|---|---|
| Candida albicans ATCC 10231 | X71088 | CA1 | GGCCCAGCCTGCCGCCAGAGGTCTAAACT |
| CA2 | ACCAATTTTTTATCAACTTGTCACACCAGATTATTAC | ||
| Candida dubliniensis CBS 8500 | AJ249484 | CD1 | AAACTTACAACCAAATTTTTTATAAACTTGTCACGAGA |
| CD2 | CCTGCCGCCAGAGGACATAAACTTACAACCAAAT | ||
| CD3 | CAATACGACTTGGGTTTGCTTGAAAGATGA | ||
| CD4 | GCTTGACAATGGCTTAGGTGTAACCAAAAACA | ||
| Candida famata DSMZ 70590 | AB053101 | CFA1 | CTAGAAATAGTTTGGGCCAGAGGTTTACTGAAC |
| CFA2 | AAACCTTACACACAGTGTTTTTTGTTATTACAAGAAC | ||
| Candida glabrata DSMZ 11226 | AY208055 | CG1 | GCTCGGAGAGAGACATCTCTGGGGAGGACCAGTG |
| CG2 | AAAGGAGGTGTTTTATCACACGACTCGACACTTTC | ||
| Candida guilliermondii ATCC 6260 | L47110 | CGU1 | TGATACAGAACTCTTGCTTTGGTTTGGCCTAGAGA |
| CGU2 | GGGCCAGAGGTTTAACAAAACACAATTTAATTA | ||
| Candida kefyr CBS 1970 | L47107 | CKE1 | AGCTCGTCTCTCCAGTGGACATAAACACAAACAATAT |
| CKE2 | CTCTGCTATCAGTTTTCTATTTCTCATCCTAAACACAA | ||
| CKE3 | GATTATGAATGAATAGATTGCTGGGGGAATCGTC | ||
| CKE4 | CAATTCGTGGTAAGCTTGGGTCATAGAGACTCA | ||
| Candida krusei ATCC 6258 | L47113 | CKR1 | CGCAGAGTTGGGGGAGCGGAGCGGACGACG |
| Candida lusitaniae DSMZ 70102 | AF172262 | CAL1 | AAAATTTAATTTTTTTGTTCGCAAAAACAATGTG |
| CAL2 | TGTTGAAAGTTTTGATATTAAGAAATTCGAATAAAAAAA | ||
| Candida metapsilosis ATCC 96144 | AJ698049 | CM1 | TAAACTCAACCAAATTTTTATTTAATTGTCAACTTGAT |
| CM2 | TAGGAGAAGGTTGCTTAACTGCAATCCTTTTCTTTC | ||
| Candida orthopsilosis ATCC 96139 | AJ698048 | CO1 | CCAGAGATTAAACTCAACCAAATTTTATTTAAGTCAAC |
| CO2 | CGTTGTTGAAAGTTTTGACTATTAGTTAATCAGTTGAC | ||
| Candida parapsilosis ATCC 22019 | AF287909 | CPA1 | GAATGAAAAGTGCTTAACTGCATTTTTTCTTACACA |
| CPA2 | GGGGCCTGCCAGAGATTAAACTCAACCAAA | ||
| Candida pulcherrima DSMZ 70336 | AF235809 | CPU1 | CACCCTTTTAGGCACAAACTCTAAATCTTAACCG |
| Canadida tropicalis DSMZ 11953 | L47112 | CT1 | TATTGAACAAATTTCTTTGGTGGCGGGAGCAATCC |
| CT2 | GCAATCCTACCGCCAGAGGTTATAACTAAACCAAACT | ||
| Cryptococcus neoformans DSMZ 11959 | AY208079 | CRN1 | GGCACGTTTTACACAAACTTCTAAATGTAATGAATG |
| Saccharomyces cerevisiae DSMZ 70449 | AJ544253 | SCE1 | CCAAACGGTGAGAGATTTCTGTGCTTTTGTTA |
| Trichosporon cutaneum CBS 2466 | AF444325 | TRC1 | GTTTCTTAATGGATTGGATTTGGGCGCTGCCAG |
| Aspergillus fumigatus DSMZ 819 | AB051071 | AFU1 | CTGTTCTGAAAGTATGCAGTCTGAGTTGATTATCG |
| AFU2 | GAAGACCCCAACATGAACGCTGTTCTGAAAG | ||
| Aspergillus terreus DSMZ 826 | AF078896 | AST1 | ACCTCCCACCCGTGACTATTGTACCTTGTTGCT |
| AST2 | CCCTGTTCTGAAAGCTTGCAGTCTGAGTGTGA | ||
| Epidermophyton floccosum CBS 214.63 | AF168130 | EPF1 | CGAAATCTCCATAGGTGGTTCAGTCTGAGCG |
| EPF2 | TCCCCCTTCTCTCTGAATGCTGGACGGTG | ||
| EPF3 | ATCCCCCGTTCCACCGGGAGAGGAGAAAGG | ||
| Fusarium solani CBS 208.29 | AF129105 | FSO1 | GCCGCAGCTTCCATCGCGTAGTAGCTAACACC |
| FSO2 | GCGGGCACACGCCGTCCCCCAAATACAG | ||
| Microsporum canis CBS 113480 | AF168127 | MCA1 | CGGGGAGGTTGCGGGCGGCGAGGGGTGCC |
| MCA2 | CTCGCCGGAGGATTACTCTGGAAAACACACTC | ||
| MCA3 | TGTGTGATGGACGACCGTCCCCCCTCCCCAG | ||
| MCA4 | CGTCCCCCCTCCCCAGTAACCACCCACCGC | ||
| Penicillium marneffei CBS 334.59 | AB049129 | PEM1 | TGATGAAGATGGACTGTCTGAGTACCATGAAAA |
| PEM2 | CGCCCTGTGAACCCTGATGAAGATGGACTG | ||
| PEM3 | GAAGCGCCCTGTGAACCCTGATGAAGATGGAC | ||
| PEM4 | AAGCACGGCTTGTGTGTTGGGTGTGGTCCC | ||
| Trichophyton rubrum CBS 286.30 | AF168123 | TRU1 | GACCGACGTTCCATCAGGGGTGAGCAGACG |
| TRU2 | CGCCCGCCGGAGGACAGACACCAAGAAAAAA | ||
| TRU3 | AGAGCCGTCCGGCGGGCCCCTTCTGGGAGCC | ||
| TRU4 | CTGTCAGTCTGAGCGTTTAGCAAGCACAATCAG | ||
| TRU5 | CTAGGCGAATGGGCAGCCAATTCAGCGCCC | ||
| Trichophyton tonsurans CBS 120.30 | AB166667 | TRT1 | AGGGCCAAACGTCCGTCAGGGGTGAGCAGATG |
| TRT2 | CAGGATAGGGCCAAACGTCCGTCAGGGG |
TABLE 2.
Selected clinical fungal isolates blindly run to validate the DNA array-based identification system
| Isolate | Site of isolation | Identification by conventional testsa,b | Array identificationb |
|---|---|---|---|
| CF1 | Balanopreputial swab | Candida albicans | Candida albicans |
| CF2 | Bronchial aspirate | Candida tropicalis | Candida tropicalis |
| CF3 | Anal swab | Candida guilliermondii | Candida guilliermondii |
| CF4 | Anal swab | Candida glabrata | Candida glabrata |
| CF5 | Sputum | Candida albicans | Candida albicans |
| CF6 | Sputum | Candida albicans | Candida albicans |
| CF7 | Sputum | Candida dubliniensis | Candida dubliniensis |
| CF8 | Urine | Candida parapsilosis | Candida orthopsilosis |
| CF9 | Drainage | Candida albicans | Candida albicans |
| CF10 | Feces | Candida tropicalis | Candida tropicalis |
| CF11 | Scalp | Microsporum canis | Microsporum canis |
| CF12 | Oral swab | Candida famata | Candida tropicalis |
| CF13 | Feces | Candida dubliniensis | Candida dubliniensis |
| CF14 | Nail | Trichophyton rubrum | Trichophyton rubrum |
| CF15 | Bronchial aspirate | Candida tropicalis | Candida tropicalis |
| CF16 | Oral swab | Candida glabrata | Candida glabrata |
| CF17 | Scalp | Trichophyton tonsurans | Trichophyton tonsurans |
| CF18 | Vaginal swab | Candida parapsilosis | Candida metapsilosis |
| CF19 | Blood | Candida parapsilosis | Candida parapsilosis |
| CF20 | Balanopreputial swab | Candida lusitaniae | Candida lusitaniae |
API32 ID and Vitek 2 advanced colorimetry for yeasts and microscopic examination of reproductive structures for molds.
Data in bold show differences between conventional and array identification results.
Genomic DNA extraction.
Genomic DNAs were extracted from yeasts or molds grown in Sabouraud dextrose broth (yeast) or on potato dextrose agar (molds) (Liofilchem S.R.L., TE, Italy). DNAs were extracted from yeast samples as previously described (28). Briefly, yeast cells were harvested in stationary phase, while mold suspensions were prepared by collecting conidia from 72-h potato dextrose agar plate colonies washed with 5 ml of 0.1% Tween 20 (Sigma, St. Louis, MO)-sterile saline. Both yeast and conidia were lysed by vortexing the pellet for 3 min with 0.3 g glass beads (0.45- to 0.52-mm diameter; Sigma) in 200 μl of lysis buffer (100 mM Tris-HCl, pH 8.0, 2% [vol/vol] Triton X-100, 1% [wt/vol] sodium dodecyl sulfate, and 1 mM EDTA) and 200 μl of 1:1 (vol/vol) phenol-chloroform. After the pellet was vortexed, 200 μl of TE (10 mM Tris-HCl, pH 8.0, 1 mM EDTA) was added to the lysate; the mixture was microcentrifuged at full speed for 10 min, and the aqueous phase was transferred to a new tube. DNA was precipitated by the addition of 1 ml of ethanol. Samples were centrifuged, and the pellet was suspended in 400 μl of TE containing 100 μg RNase (Sigma). The mixture was incubated for 1 h at 37°C and subsequently treated with proteinase K (5 μl of a 20-mg/ml stock solution; Sigma) for 1 h at 65°C and for 30 min at 72°C. Phenol-chloroform treatment was repeated, and then DNA was precipitated with 1 ml of isopropanol and 10 μl of 4 M ammonium acetate, dried, and dissolved in 50 μl of TE, pH 8.0.
ITS amplification.
The universal fungal primers ITS1 and ITS4 described by White et al. (29) (Sigma-Genosys, Steinheim, Germany) were used to amplify the noncoding ITS regions (ITS1 and ITS2) as well as the 5.8S rRNA gene, positioned between the ITS regions. Reaction mixtures (20 μl) contained fungal DNA (100 ng), 0.4 μl primers (5 μM), 2 μl of deoxynucleoside triphosphate mix (2 mM dATP, 2 mM dCTP, 2 mM dGTP, 1.7 mM dTTP, and 0.03 mM dUTP), 2 μl magnesium-free buffer (10×; Solis Biodine, Tartu, Estonia), 2 μl MgCl2 (25 mM; Solis Biodine), and 0.2 μl Hot Fire DNA polymerase (5 U/μl; Solis Biodine). A negative control, which consisted of an equal volume of water replacing the DNA template, was included in all PCR experiments. Conditions for PCR amplification were as follows: 94°C for 15 min (hot start); 35 cycles of denaturation at 95°C (1 min), annealing at 56°C (30 s), and elongation at 72°C (75 s); and a final extension step at 72°C (10 min). Amplified DNA products were separated by electrophoresis in a 1% agarose gel containing ethidium bromide (0.5 mg/ml); the running buffer was TAE (40 mM Tris acetate [pH 8.0], 1 mM EDTA). A 100-bp DNA ladder was used as a molecular size marker (Promega). DNA bands were visualized by UV transillumination. The universal fungal primers used were tested versus representative bacterial (n = 3) and human genomic DNA preparations.
Chip probe design.
5′ C-6 aminolinker-modified oligonucleotides (C-6 oligonucleotides) were designed by matching a region of about 35 bp within the ITS1 amplification products to the sequences of ITS deposited in GenBank (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=Nucleotide&itool=toolbar) (accession numbers are given in Table 1). Each C-6 oligonucleotide was designed in order to exclusively add a uracyl (fluorescein-ddUTP) at its 3′ end during the single-base extension reaction. All of the C-6 oligonucleotides were synthesized by MWG Biotech (Ebersberg, Germany) and spotted onto silanized slides by Asperbio (Tartu, Estonia), as previously reported (1, 9). Preliminary experiments were performed by designing a large number of oligonucleotides for each species in order to test them for the ability to identify and discriminate the 24 fungal species. Details of the probes used in these experiments are provided in Table 1. For each fungal species, sets of oligonucleotides were selected among those previously designed on the basis of their discriminatory power and lack of cross-reactivity (Table 1).
Purification, hybridization of PCR products on the chip, and signal detection.
PCR products were purified and concentrated using Millipore Y30 columns (7), and their sizes were reduced by fragmentation to improve hybridization with the arrayed oligonucleotides. To this end, Y30 column eluate samples (15 μl) were treated with 1 U uracil N-glycosylase (Epicenter Technologies, Madison, WI) and 1 U shrimp alkaline phosphatase (Amersham Biosciences, Milwaukee, WI), and the mixtures were incubated at 37°C for 1.5 h and at 95°C for 30 min. At this temperature, DNA with abasic sites is denatured and fragmented, whereas uracil N-glycosylase and shrimp alkaline phosphatase are inactivated. Aliquots of the treated amplicons (9 μl) were added to a reaction mixture containing fluorescein-ddUTP, cyanine 3-ddATP, ROX-ddCTP, cyanine 5-ddGTP (4 × 50 pmol), 10× buffer, and 4 U of Thermo Sequenase (Amersham Biosciences, Uppsala, Sweden) diluted in the provided dilution buffer to give a final volume of 20 μl. Mixtures were quickly placed onto the spotted slide, which was previously washed with 100 mM NaOH and rinsed with distilled water at 95°C, and incubated at 58°C for 10 min. Slides were washed again with distilled water at 95°C to remove both traces of nonhybridized PCR products and nonincorporated labeled dideoxynucleoside triphosphates. A drop of SlowFade Light antifade reagent (Molecular Probes, Eugene, OR) was then added to prevent signal fading. Slides were imaged by a Genorama-003 four-color detector (Asper Biotech, Tartu, Estonia). Fluorescence intensities at each position were measured and converted to base calls according to Genorama image analysis and genotyping software (Asper Biotech, Tartu, Estonia).
Interpretation of results and identification criteria.
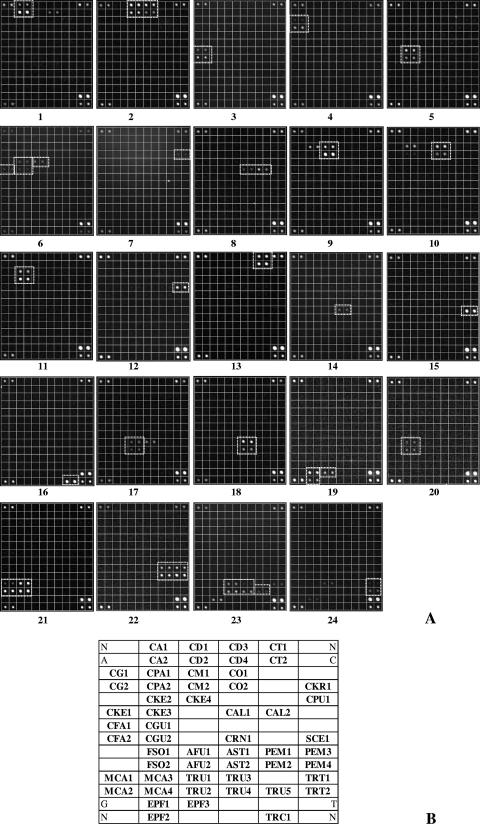
For species identification, the following two criteria were required: (i) both APEX probes had to give a signal and (ii) the extension had to yield a signal on the U channel, as the probes were designed to extend uridine only. Although A, G, and C signals are not expected to be incorporated in the APEX reaction, ddA, ddC, and ddG were also included in the reaction mixture, thus providing a further specificity control to detect any unspecific extensions. Moreover, to ensure quality control, the following strategies were adopted: (i) DNA samples were randomly processed by personnel who were blinded to the strain typed, (ii) each APEX oligonucleotide was spotted in duplicate, and (iii) internal positive controls were included in the microarray (Fig. 1) to verify that the intensities of the four channels (A, C, U, and G) were equilibrated.
FIG. 1.
(A) Representative snapshots from Genorama Basecaller software (Asperbio, Tartu, Estonia), showing the identification of fungal species by the chip. Cy3 fluorescence images (U channel) show the following identification panels: 1, Candida albicans; 2, C. dubliniensis; 3, C. famata; 4, C. glabrata; 5, C. guilliermondii; 6, C. kefyr; 7, C. krusei; 8, C. lusitaniae; 9, C. metapsilosis; 10, C. orthopsilosis; 11, C. parapsilosis; 12, C. pulcherrima; 13, C. tropicalis; 14, Cryptococcus neoformans; 15, Saccharomyces cerevisiae; 16, Trichosporon cutaneum; 17, Aspergillus fumigatus; 18, A. terreus; 19, Epidermophyton floccosum; 20, Fusarium solani; 21, Microsporum canis; 22, Penicillium marneffei; 23, Trichophyton rubrum; and 24, T. tonsurans. No signals were detected in the Cy5 channel (i.e., the non-U channel, which was not displayed for brevity). (B) Layout of chip capture probes (for probe sequences, see Table 1). Control probes are positioned at the corners of each panel.
Confirmation of C. parapsilosis and related species identification by the array.
The two-step DNA-based identification test described by Tavanti et al. (27) was used to confirm the identification of C. parapsilosis, C. metapsilosis, and C. orthopsilosis isolates. Briefly, a 716-bp fragment of the SADH (secondary alcohol dehydrogenase) gene was amplified, purified, and digested with BanI. The different digestion patterns were used to identify the three species, since the C. parapsilosis, C. metapsilosis, and C. orthopsilosis SADH amplicons contain 1, 3, and 0 BanI restriction sites, respectively.
RESULTS
Probe design and array setup.
An up-to-date DNA microarray system was developed for the unambiguous identification of 24 fungal species of clinical relevance (Table 1). Oligonucleotides were designed on the basis of the available ITS1 and/or ITS2 sequences (for accession numbers, see Table 1). The amplicons obtained for each species included the region carrying ITS1, the 5.8S rRNA gene, and ITS2 and varied in size from 375 bp (Candida pulcherrima) to 880 bp (Candida glabrata) (data not shown). A check test was performed by testing the specificity of the universal fungal primers used versus bacterial (three different strains) and human genomic DNA preparations. No amplification product was detected for any nonfungal genomic DNA template (data not shown). A pilot study was undertaken with a redundant number of oligonucleotide probes in order to evaluate probe reliability, discrimination power, and lack of cross-reactivity, which would potentially lead to misinterpretation of the results. The data obtained with this pilot chip led to an accurate selection of a reduced number of probes. Sets of C-6 oligonucleotides, ranging from 1 to 5 (Table 1), were proven to unambiguously identify the 24 fungal species and were used to validate their application for detecting/identifying fungal reference strains and blinded clinical isolates.
Validation of the DNA-based chip with reference strains.
The results presented in Fig. 1 were obtained from experiments repeated at least three times using samples containing amplicons generated from different preparations of genomic DNAs of reference species, for a total of 72 slides. As shown in Fig. 1A, the array system gave an unequivocal response for each of the 24 reference species considered. The layout of the chip allows rapid and accurate species identification (Fig. 1B): most of the species (n = 13) were identified by two oligonucleotide probes, replicated twice for a total of four spots, with the exception of Epidermophyton floccosum, detected with three replicated probes (Fig. 1A, slide 19); Candida dubliniensis, C. kefyr, Penicillium marneffei, and Microsporum canis, detected with a set of four replicated probes (Fig. 1A, slides 2, 6, 21, and 22) for a total of eight spots; and Trichophyton rubrum, detected with five replicated probes (Fig. 1A, slide 23).
Despite the fact that for five species (Candida krusei, C. pulcherrima, Cryptococcus neoformans, Saccharomyces cerevisiae, and Trichosporon cutaneum) only one probe among the originally designed probes was found to be suitable for application in the final version of the identification array, the results obtained for these species provided clear and reproducible identification for each of them (Fig. 1A, slides 7, 12, 14, 15, and 16, respectively). In addition, the microarray was proven to be highly discriminative, being able to distinguish closely related species, such as C. orthopsilosis, C. metapsilosis, and C. parapsilosis as well as Aspergillus fumigatus and A. terreus. As expected, the results obtained for the “psilosis” complex (Fig. 1A, slides 9 to 11) indicated that a moderate level of cross-reactivity still exists with the probes used in the final version of the chip: indeed, when C. metapsilosis (slide 9) or C. orthopsilosis (slide 10) was detected by the system, one of the C. parapsilosis probes (CPA1) (Table 1) gave a simultaneous signal, whereas when C. parapsilosis was identified, no other probes produced a positive signal (Fig. 1, slide 11). However, the low level of cross-reactivity observed within the “psilosis” species did not prevent their unambiguous identification. As already observed by Leinberger et al. (18), detection of Aspergillus fumigatus led to the appearance of a faint signal where one probe designed for A. terreus was positioned (Fig. 1A, slide 17). This cross-reactivity may be explained by the close genetic relatedness of these species; nevertheless, it does not prevent the correct identification of the two Aspergillus species (Fig. 1A, slides 17 and 18). Penicillium marneffei was included in the panel of fungi identifiable by the system since it has increasingly been associated with opportunistic infections (31) and it shares morphological traits with the more clinically relevant organism Aspergillus fumigatus. As shown in Fig. 1A, slide 22, P. marneffei was clearly identified by eight characteristic spots (four probes) and did not give any cross-reaction with either A. fumigatus or A. terreus (Fig. 1A, slides 17 and 18, respectively). Among the three dermatophyte genera included in the array, Microsporum canis was identified by a clear and unique signal (four probes) (Fig. 1A, slide 21), while a weak level of cross-reactivity was observed, as expected, between the two species of the Trichophyton genus, i.e., T. rubrum (Fig. 1A, slide 23) and T. tonsurans (Fig. 1A, slide 24). When the latter species were present in an independent tested sample, one of the probes for Epidermophyton floccosum also gave a faint signal with T. rubrum (Fig. 1A, slides 23 and 24), while E. floccosum identification (Fig. 1A, slide 19) did not produce any cross-reactive signal. Therefore, despite the fact that faint aspecific signals were observed within related species, the identification of all dermatophytes was clear and unambiguous for any of the species tested.
Blinded test of a panel of clinical fungal isolates.
In separate experiments, a panel of clinical fungal isolates, including yeasts and molds, were blindly submitted to the array identification system. These isolates were previously identified with conventional laboratory tests by the Unità Operativa di Microbiologia, Ospedale Universitario, Pisa, Italy. As reported in Table 2, 17 of the isolates were unambiguously identified as the same species as those obtained by conventional methods (data not shown). Two clinical isolates identified as Candida parapsilosis (Table 2) (CF8 and CF18) were detected as C. orthopsilosis (CF8) and C. metapsilosis (CF18) by the array system. Molecular identification of these two isolates was then performed by BanI digestion of the SADH amplicon (27), and the digestion profiles confirmed the results obtained with the array (data not shown). Isolate CF12, identified as Candida famata (Table 2), turned out to be C. tropicalis on the basis of the spot position on the array; the conventional identification was therefore repeated, confirming that the array identification was indeed correct and that mislabeling of the isolate plate may have occurred.
DISCUSSION
This paper describes the setup of an up-to-date version of a DNA microarray-based system for the identification of the most common fungal pathogens in humans, encompassing 24 different species belonging to 10 genera. Despite the fact that a panmicrobial oligonucleotide array was recently proposed for the diagnosis of infectious diseases (23), it appears to be better suited for molecular surveillance of infectious agents in high-throughput/reference laboratories than in small mycology units. Indeed, the use of the panmicrobial chip may supersede the effective needs of the local diagnostic routine and may have high running costs. Moreover, with this array, the use of fungal probes designed based on the 18S rRNA sequence does not provide enough discriminatory power to identify each single species of the 73 fungal genera included in the chip.
The microarray we propose utilizes oligonucleotide probes complementary to the ITS region (ITS1 or ITS2) of each species and is based on the APEX technique. The present work describes the first application of APEX to the field of medical mycology. The technique proposed here relies both on specific hybridization between probes and their targets, like conventional microarray-based methods (11, 18, 23), and on the specific extension of a single base at the 3′ side of the anchored probe. In other words, APEX provides a further level of analysis and thus, at least theoretically, should give more clear-cut results and reduced misclassification of fungal species than do hybridization-only array-based methods.
Optimization of the detection system was a crucial point for ensuring the reproducibility, specificity, and sensitivity of the results obtained; in many cases, it was necessary to reduce the number of probes originally designed and spotted on the pilot chip (such as those for Candida krusei) due to cross-reaction among species, which could possibly cause misinterpretation of results. The optimized microarray detection system was proven to be highly discriminative, and in its final layout, it allowed the unequivocal identification of each of the 24 species, including yeasts and hyphomycetes. Despite a very low level of cross-reactivity still detectable in the identification of genetically related species, the signals obtained provided effective discrimination of the “psilosis” group, Aspergillus, and dermatophyte species tested.
Furthermore, the system was validated by using a selected panel of clinical fungal isolates previously identified with conventional laboratory tests, which were blindly submitted to the array identification system. The majority of the isolates were detected unambiguously in perfect concordance with the results obtained with the laboratory tests. In two cases, the microarray resulted in more discriminative results than did conventional biochemical tests, suggesting that this system could efficaciously flank the diagnostic routine, providing a rapid and sensitive tool for the identification of closely related species such as C. orthopsilosis, C. metapsilosis, and C. parapsilosis or aspergilli.
The chip we have developed is currently used in our clinical laboratory in parallel with conventional identification procedures. Our aims are both to widen the collection of clinically significant fungi run with the array system and to confirm its performance traits, such as reproducibility, specificity, and unequivocal interpretation of the results. Indeed, high-throughput processing of clinical samples will be necessary to validate the system by testing fungal isolates undergoing selective pressure during the complex interactions occurring within the infected host.
Acknowledgments
This study was supported by research grant 2005068754 from the Italian Ministero dell'Istruzione, dell'Università e della Ricerca (MIUR) and by an Associazione Italiana Ricerca sul Cancro (AIRC) regional grant.
Footnotes
Published ahead of print on 26 December 2007.
REFERENCES
- 1.Auffray, C., C. Mundy, and A. Metspalu. 2000. DNA arrays: methods and applications: report on HUGO Meeting, Tartu, Estonia, 23-26 May, 1999. Eur. J. Hum. Genet. 8236-238. [DOI] [PubMed] [Google Scholar]
- 2.Bedini, A., C. Venturelli, C. Mussini, G. Guaraldi, M. Codeluppi, V. Borghi, F. Rumpianesi, F. Barchiesi, and R. Esposito. 2006. Epidemiology of candidaemia and antifungal susceptibility patterns in an Italian tertiary-care hospital. Clin. Microbiol. Infect. 1275-80. [DOI] [PubMed] [Google Scholar]
- 3.Bolehovska, R., L. Pliskova, V. Buchta, J. Cerman, and P. Hamal. 2006. Detection of Aspergillus spp. in biological samples by real-time PCR. Biomed. Pap. Med. Fac. Univ. Palacky Olomouc Czech Repub. 150245-248. [DOI] [PubMed] [Google Scholar]
- 4.Campa, D., S. Zienolddiny, H. Lind, D. Ryberg, V. Skaug, F. Canzian, and A. Haugen. 2007. Polymorphisms of dopamine receptor/transporter genes and risk of non-small cell lung cancer. Lung Cancer 5617-23. [DOI] [PubMed] [Google Scholar]
- 5.Chang, S. E., K. J. Kim, W. S. Lee, J. H. Choi, K. J. Sung, K. C. Moon, and J. K. Koh. 2003. A case of Trichosporon cutaneum folliculitis and septicaemia. Clin. Exp. Dermatol. 2837-38. [DOI] [PubMed] [Google Scholar]
- 6.Colombo, A. L., T. Guimaraes, L. R. Silva, L. P. de Almeida Monfardini, A. K. Cunha, P. Rady, T. Alves, and R. C. Rosas. 2007. Prospective observational study of candidemia in Sao Paulo, Brazil: incidence rate, epidemiology, and predictors of mortality. Infect. Control Hosp. Epidemiol. 28570-576. [DOI] [PubMed] [Google Scholar]
- 7.Gemignani, F., S. Landi, A. Chabrier, A. Smet, I. Zehbe, F. Canzian, and M. Tommasino. 2004. Generation of a DNA microarray for determination of E6 natural variants of human papillomavirus type 16. J. Virol. Methods 11995-102. [DOI] [PubMed] [Google Scholar]
- 8.Gemignani, F., C. Perra, S. Landi, F. Canzian, A. Kurg, N. Tonisson, R. Galanello, A. Cao, A. Metspalu, and G. Romeo. 2002. Reliable detection of beta-thalassemia and G6PD mutations by a DNA microarray. Clin. Chem. 482051-2054. [PubMed] [Google Scholar]
- 9.Guo, Z., R. A. Guilfoyle, A. J. Thiel, R. Wang, and L. M. Smith. 1994. Direct fluorescence analysis of genetic polymorphisms by hybridization with oligonucleotide arrays on glass supports. Nucleic Acids Res. 225456-5465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ha, Y. S., S. F. Covert, and M. Momany. 2006. FsFKS1, the 1,3-beta-glucan synthase from the caspofungin-resistant fungus Fusarium solani. Eukaryot. Cell 51036-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang, A., J. W. Li, Z. Q. Shen, X. W. Wang, and M. Jin. 2006. High-throughput identification of clinical pathogenic fungi by hybridization to an oligonucleotide microarray. J. Clin. Microbiol. 443299-3305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iwen, P. C., A. G. Freifeld, T. A. Bruening, and S. H. Hinrichs. 2004. Use of a panfungal PCR assay for detection of fungal pathogens in a commercial blood culture system. J. Clin. Microbiol. 422292-2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kardjeva, V., R. Summerbell, T. Kantardjiev, D. Devliotou-Panagiotidou, E. Sotiriou, and Y. Graser. 2006. Forty-eight-hour diagnosis of onychomycosis with subtyping of Trichophyton rubrum strains. J. Clin. Microbiol. 441419-1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kocsube, S., M. Toth, C. Vagvolgyi, I. Doczi, M. Pesti, I. Pocsi, J. Szabo, and J. Varga. 2007. Occurrence and genetic variability of Candida parapsilosis sensu lato in Hungary. J. Med. Microbiol. 56190-195. [DOI] [PubMed] [Google Scholar]
- 15.Krcmery, V., and A. J. Barnes. 2002. Non-albicans Candida spp. causing fungaemia: pathogenicity and antifungal resistance. J. Hosp. Infect. 50243-260. [DOI] [PubMed] [Google Scholar]
- 16.Landi, S., F. Gemignani, L. Gioia-Patricola, A. Chabrier, and F. Canzian. 2003. Evaluation of a microarray for genotyping polymorphisms related to xenobiotic metabolism and DNA repair. BioTechniques 35816-827. [DOI] [PubMed] [Google Scholar]
- 17.Lass-Florl, C., K. Griff, A. Mayr, A. Petzer, G. Gastl, H. Bonatti, M. Freund, G. Kropshofer, M. P. Dierich, and D. Nachbaur. 2005. Epidemiology and outcome of infections due to Aspergillus terreus: 10-year single centre experience. Br. J. Haematol. 131201-207. [DOI] [PubMed] [Google Scholar]
- 18.Leinberger, D. M., U. Schumacher, I. B. Autenrieth, and T. T. Bachmann. 2005. Development of a DNA microarray for detection and identification of fungal pathogens involved in invasive mycoses. J. Clin. Microbiol. 434943-4953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luo, G., and T. G. Mitchell. 2002. Rapid identification of pathogenic fungi directly from cultures by using multiplex PCR. J. Clin. Microbiol. 402860-2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marr, K. A. 2004. Invasive Candida infections: the changing epidemiology. Oncology (Williston Park) 189-14. [PubMed] [Google Scholar]
- 21.Martin, C., D. Roberts, M. van Der Weide, R. Rossau, G. Jannes, T. Smith, and M. Maher. 2000. Development of a PCR-based line probe assay for identification of fungal pathogens. J. Clin. Microbiol. 383735-3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McNeil, M. M., S. L. Nash, R. A. Hajjeh, M. A. Phelan, L. A. Conn, B. D. Plikaytis, and D. W. Warnock. 2001. Trends in mortality due to invasive mycotic diseases in the United States, 1980-1997. Clin. Infect. Dis. 33641-647. [DOI] [PubMed] [Google Scholar]
- 23.Palacios, G., P. L. Quan, O. J. Jabado, S. Conlan, D. L. Hirschberg, Y. Liu, J. Zhai, N. Renwick, J. Hui, H. Hegyi, A. Grolla, J. E. Strong, J. S. Towner, T. W. Geisbert, P. B. Jahrling, C. Buchen-Osmond, H. Ellerbrok, M. P. Sanchez-Seco, Y. Lussier, P. Formenty, M. S. Nichol, H. Feldmann, T. Briese, and W. I. Lipkin. 2007. Panmicrobial oligonucleotide array for diagnosis of infectious diseases. Emerg. Infect. Dis. 1373-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pfaller, M. A., D. J. Diekema, D. L. Gibbs, V. A. Newell, J. F. Meis, I. M. Gould, W. Fu, A. L. Colombo, and E. Rodriguez-Noriega. 2007. Results from the ARTEMIS Disk Global Antifungal Surveillance study, 1997 to 2005: an 8.5-year analysis of susceptibilities of Candida species and other yeast species to fluconazole and voriconazole determined by CLSI standardized disk diffusion testing. J. Clin. Microbiol. 451735-1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pfaller, M. A., S. A. Messer, L. Boyken, S. Tendolkar, R. J. Hollis, and D. J. Diekema. 2004. Geographic variation in the susceptibilities of invasive isolates of Candida glabrata to seven systemically active antifungal agents: a global assessment from the ARTEMIS Antifungal Surveillance Program conducted in 2001 and 2002. J. Clin. Microbiol. 423142-3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pryce, T. M., S. Palladino, D. M. Price, D. J. Gardam, P. B. Campbell, K. J. Christiansen, and R. J. Murray. 2006. Rapid identification of fungal pathogens in BacT/ALERT, BACTEC, and BBL MGIT media using polymerase chain reaction and DNA sequencing of the internal transcribed spacer regions. Diagn. Microbiol. Infect. Dis. 54289-297. [DOI] [PubMed] [Google Scholar]
- 27.Tavanti, A., A. D. Davidson, N. A. Gow, M. C. Maiden, and F. C. Odds. 2005. Candida orthopsilosis and Candida metapsilosis spp. nov. to replace Candida parapsilosis groups II and III. J. Clin. Microbiol. 43284-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tavanti, A., L. A. Hensgens, E. Ghelardi, M. Campa, and S. Senesi. 2007. Genotyping of Candida orthopsilosis clinical isolates by amplification fragment length polymorphism reveals genetic diversity among independent isolates and strain maintenance within patients. J. Clin. Microbiol. 451455-1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.White, T., T. Bruns, S. Lee, and J. Taylor. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics, p. 315-322. In M. Innis, D. Gelfand, J. Sninsky, and T. White (ed.), PCR protocols: a guide to methods and application. Academic Press, San Diego, CA.
- 30.Wingard, J. R. 2005. The changing face of invasive fungal infections in hematopoietic cell transplant recipients. Curr. Opin. Oncol. 1789-92. [DOI] [PubMed] [Google Scholar]
- 31.Xi, L., X. Xu, W. Liu, X. Li, Y. Liu, M. Li, J. Zhang, and M. Li. 2007. Differentially expressed proteins of pathogenic Penicillium marneffei in yeast and mycelial phases. J. Med. Microbiol. 56298-304. [DOI] [PubMed] [Google Scholar]