Abstract
The Shigella genus has historically been separated into four species, based on biochemical assays. The classification within each species relies on serotyping. Recently, genome sequencing and DNA assays, in particular the multilocus sequence typing (MLST) approach, greatly improved the current knowledge of the origin and phylogenetic evolution of Shigella spp. The Shigella and Escherichia genera are now considered to belong to a unique genomospecies. Multilocus variable-number tandem-repeat (VNTR) analysis (MLVA) provides valuable polymorphic markers for genotyping and performing phylogenetic analyses of highly homogeneous bacterial pathogens. Here, we assess the capability of MLVA for Shigella typing. Thirty-two potentially polymorphic VNTRs were selected by analyzing in silico five Shigella genomic sequences and subsequently evaluated. Eventually, a panel of 15 VNTRs was selected (i.e., MLVA15 analysis). MLVA15 analysis of 78 strains or genome sequences of Shigella spp. and 11 strains or genome sequences of Escherichia coli distinguished 83 genotypes. Shigella population cluster analysis gave consistent results compared to MLST. MLVA15 analysis showed capabilities for E. coli typing, providing classification among pathogenic and nonpathogenic E. coli strains included in the study. The resulting data can be queried on our genotyping webpage (http://mlva.u-psud.fr). The MLVA15 assay is rapid, highly discriminatory, and reproducible for Shigella and Escherichia strains, suggesting that it could significantly contribute to epidemiological trace-back analysis of Shigella infections and pathogenic Escherichia outbreaks. Typing was performed on strains obtained mostly from collections. Further studies should include strains of much more diverse origins, including all pathogenic E. coli types.
Shigellosis is a widespread disease occurring mainly in developing countries, often in association with poor sanitation, and is responsible for about 600,000 deaths per year in the world, two-thirds of them concerning <10-year-old children (2). The implementation of treatment and, to some extent, control strategies is a significant challenge, especially in Asia, because antibiotic-resistant strains of different species and serotypes have emerged and because the distribution of Shigella species and serotypes is heterogeneous (46). The low infectious dose (10 to 100 cells) allows the organism to spread effectively by contaminated food or water but also by person-to-person contact, and a specific virulence plasmid that encodes a type III secretion system plus the invasion plasmid antigens is responsible for epithelial cell wall invasiveness. Finally, it is worth noting that Shigella, like other bacteria responsible for food and waterborne diseases, has been classified as a potential biological threat agent due to its infection route and environmental stability (38).
Shigella sp. strains are currently classified into four species: S. dysenteriae, S. flexneri, S. boydii, and S. sonnei. Three of them are apparently antigenically heterogeneous, comprising several serotypes, whereas S. sonnei is antigenically homogeneous.
Shigella typing relies on phenotypic characteristics, but discrimination between the four species can be difficult. Serotyping is not always able to provide a correct species identification, due to cross-reactions or the absence of agglutination. New serotypes are regularly discovered (28, 49) and are sometimes found to cross-react with Escherichia coli strains.
In addition, the discriminatory power of phenotypic tools and serotyping is limited and requires the manipulation of the live agent. The introduction of DNA-based molecular typing methods, such as ribotyping (5), plasmid profile analysis, restriction fragment length polymorphism (25, 26), and pulsed-field gel electrophoresis (PFGE) (37), has greatly improved the ability of researchers to discriminate between epidemiologically related and unrelated isolates in outbreaks. In the United States, PulseNet, a network of laboratories implicated in food-borne disease surveillance (42), uses PFGE typing coupled with strict quality control procedures in order to ensure interlaboratory reproducibility, but this approach remains labor-intensive for routine clinical strain typing, so cheaper alternatives are actively being pursued. Multilocus sequence typing (MLST) is a very powerful approach, and it provides a clear view of the population structure (52). However, it is not yet appropriate for the routine, first-line genotyping of a large number of isolates. Recently, PulseNet members acknowledged that multilocus variable-number tandem-repeat (VNTR) analysis (MLVA) is a highly promising typing tool, likely to replace PFGE in the coming years (9). MLVA typing is being actively developed by a number of laboratories, together with the associated Internet-based query tools and databases (3, 6, 17, 18), to genotype several bacterial pathogens, in particular, potential biothreat agents, such as Bacillus anthracis (18), Yersinia pestis (18, 31), Legionella pneumophila (32, 33), Salmonella enterica (36), Brucella spp. (19), Francisella tularensis (11), Burkholderia mallei, and B. pseudomallei (43). MLVA is also increasingly recognized as a future reference technique for bacterial genotyping allowing systematic typing of all isolates for a number of other pathogens of high public health interest (including Mycobacterium tuberculosis [17], Pseudomonas aeruginosa [30, 47], Streptococcus pneumoniae [13], and Staphylococcus aureus [40]) (the technique is reviewed in references 21, 44, and 45).
The present work aimed to set up MLVA for the Shigella genus to facilitate epidemiological follow-up, by providing easy-to-use typing tools, in countries suffering from a recrudescence of Shigella outbreaks. Recently, Liang et al. (20) proposed an MLVA for molecular typing of S. sonnei and explored the capability of a 26-VNTR set for detecting clusters of infection. Because the proposed VNTRs have very short repeat units, this assay requires a high precision of DNA length measurement, as provided, for instance, by microcapillary electrophoresis and fluorescent markers. This is a strong limitation for routine typing because of the cost of such equipment and the associated consumables. In contrast, the present investigation favors markers with larger repeat units in order to provide an assay widely accessible to research and public health laboratories, particularly in developing countries.
MATERIALS AND METHODS
Strains and DNA preparation.
Sixty-eight S. boydii, S. dysenteriae, S. flexneri, and S. sonnei strains and four E. coli strains were obtained from the Centre de Ressources Biologiques de l'Institut Pasteur (CRBIP), the American Type Culture Collection, the Hôpital d'Instruction des Armées (Bégin, France), and the Centre d'Etudes du Bouchet (CEB) strain collections (Table 1). Strains were cultured overnight at 30°C on Trypticase soy agar (reference number 43011; bioMérieux, Marcy L'Etoile, France) before DNA extraction. S. flexneri, S. sonnei, and S. boydii DNA extractions were processed in a biosafety level 2 lab by use of lysozyme, sodium dodecyl sulfate, and proteinase K followed by phenol-chloroform extraction and ethanol precipitation, as described elsewhere (41). S. dysenteriae serotype 1 DNA extraction was done in a biosafety level 3 lab by use of the genomic DNA 500/G extraction kit (Qiagen GmbH, Hilden, Germany), according to the manufacturer's instructions. In several cases, the presence of high quantities of polysaccharides was suspected after lysis and cetyltrimethylammonium bromide extraction was used to remove this excess (41). The average size of the extracted DNA was checked on a 0.8% agarose gel. Nucleic acid concentration and purity were quantified with an ND-1000 UV-Vis spectrophotometer (NanoDrop Technologies, Wilmington, DE).
TABLE 1.
List of strains used in this study
| Genotype | Genus and species | Serotype or serovar | Collection | Alias | Origina | Cluster | Yr of isolation | Location of isolation | Strain identifier |
|---|---|---|---|---|---|---|---|---|---|
| 1 | Shigella dysenteriae | 5 | CIP 56.17 | CRBIP | 1 | 1956 | Vietnam | Sd#1 | |
| 2 | Shigella dysenteriae | 5 | CIP 57.42 | CRBIP | 1 | 1957 | Tunisia | Sd#2 | |
| 3 | Shigella boydii | 3 | CIP 52.50 | NCTC 9329 | CRBIP | 1 | 1952 | Egypt | Sb#3 |
| 4 | Shigella boydii | 6 | CIP 52.53 | NCTC 9332 | CRBIP | 1 | 1952 | Sb#4 | |
| 5 | Shigella boydii | 10 | CIP 73.2 | CRBIP | 1 | 1973 | Sb#5 | ||
| 6 | Shigella boydii | 1 | NCTC 9327 | CRBIP | 1 | 1938 | Sb#6 | ||
| 7 | Shigella boydii | 1 | CIP 54.73 | CRBIP | 1 | 1954 | Burkina Faso | Sb#7 | |
| 8 | Shigella boydii | 8 | CIP 51.3 | CRBIP | 1 | 1951 | Congo | Sb#8 | |
| 9 | Shigella boydii | 18 | BS512 | Sanger | 1 | AZ | Sb#9 | ||
| 10 | Shigella dysenteriae | 3 | CIP 59.1 | CRBIP | 1 | 1959 | Cameroon | Sd#10 | |
| 11 | Shigella dysenteriae | 3 | CIP 67.58 | CRBIP | 1 | 1967 | Iraq | Sd#11 | |
| 12 | Shigella dysenteriae | 4 | CIP 59.2 | CRBIP | 1 | 1959 | Cameroon | Sd#12 | |
| 13 | Shigella dysenteriae | 4 | CIP 67.59 | CRBIP | 1 | 1967 | Iraq | Sd#13 | |
| 14 | Shigella dysenteriae | 3 | CIP 54.77 | CRBIP | 1 | 1954 | Vietnam | Sd#14 | |
| 15 | Shigella dysenteriae | 11 | CIP 58.16 | NCTC 9349 | CRBIP | 1 | 1958 | Sd#15 | |
| 16 | Shigella dysenteriae | 9 | CIP 58.25 | NCTC 9348 | CRBIP | 1 | 1958 | Sd#16 | |
| 17 | Shigella boydii | 2 | CIP 82.50T | NCTC 12985 | CRBIP | 1 | 1982 | Sb#17 | |
| 18 | Shigella boydii | 4 | CIP 54.76 | CRBIP | 1 | 1954 | Vietnam | Sb#18 | |
| 19 | Shigella boydii | 4 | 227 | ChMPH | 1 | 1950 | China | Sb#19 | |
| 20 | Shigella boydii | 14 | CIP 58.17 | NCTC 9766 | CRBIP | 1 | 1958 | Sb#20 | |
| 21 | Shigella boydii | 14 | CIP 58.18 | NCTC 9765 | CRBIP | 1 | 1958 | Sb#21 | |
| 22 | Shigella flexneri | 6 | CIP 106202 | CRBIP | 1 | 1940 | United Kingdom | Sf#22 | |
| 23 | Shigella boydii | 14 | CIP 53.44 | NCTC 6721 | CRBIP | 1 | 1953 | Sb#23 | |
| 24 | Shigella flexneri | 6 | CIP 55.19 | CRBIP | 1 | 1955 | Vietnam | Sf#24 | |
| 25 | Shigella dysenteriae | 6 | CIP 52.32 | CRBIP | 1 | 1952 | Sd#25 | ||
| 26 | Shigella dysenteriae | 7 | CIP 52.123 | NCTC 9763 | CRBIP | 1 | 1952 | Sd#26 | |
| 27 | Shigella dysenteriae | 7 | CIP 67.60 | CRBIP | 1 | 1967 | Iraq | Sd#27 | |
| 28 | Shigella boydii | 9 | CIP 57.47 | NCTC 9356 | CRBIP | 2 | 1957 | Sb#28a | |
| 28 | Shigella boydii | 9 | CIP 51.4 | NCTC 9355 | CRBIP | 2 | 1951 | Sb#28b | |
| 29 | Shigella boydii | 16 | CIP 67.11 | CRBIP | 2 | 1967 | Hong Kong | Sb#29 | |
| 30 | Shigella boydii | 5 | CIP 56.36 | CRBIP | 2 | 1956 | Ethiopia | Sb#30 | |
| 31 | Shigella boydii | 11 | CIP 51.6 | CRBIP | 2 | 1951 | Sb#31 | ||
| 32 | Shigella boydii | 11 | CIP 56.18 | CRBIP | 2 | 1956 | Vietnam | Sb#32 | |
| 33 | Shigella dysenteriae | 2 | CIP 54.51 | CRBIP | 2 | 1954 | Burkina Faso | Sb#33 | |
| 34 | Shigella dysenteriae | 2 | CIP 54.57 | CRBIP | 2 | 1954 | Vietnam | Sd#34 | |
| 35 | Shigella boydii | 15 | CIP 58.19 | NCTC 9365 | CRBIP | 2 | Sb#35 | ||
| 36 | Shigella boydii | 15 | CIP 58.20 | NCTC 10024 | CRBIP | 2 | 1958 | Sb#36 | |
| 37 | Shigella boydii | 7 | CIP 58.4 | CRBIP | 2 | 1958 | Tunisia | Sb#37 | |
| 38 | Shigella boydii | 7 | CIP 52.54 | NCTC 333 | CRBIP | 2 | 1952 | Sb#38 | |
| 39 | Shigella sonnei | None | CIP 55.56 | CRBIP | 1955 | Sri Lanka | Ss#39a | ||
| 39 | Shigella sonnei | None | CIP 63.10 | CRBIP | 1963 | Ss#39b | |||
| 40 | Shigella sonnei | None | 53G | Sanger | Japan | Ss#40 | |||
| 41 | Shigella sonnei | None | CIP 106345 | CRBIP | 1999 | France | Ss#41 | ||
| 42 | Shigella sonnei | None | CIP 51.44 | CRBIP | 1951 | Ss#42 | |||
| 43 | Shigella sonnei | None | CIP 104223 | ATCC 25931 | CRBIP | 1994 | Panama | Ss#43 | |
| 44 | Shigella sonnei | None | 46 | ChMPH | 1950 | China | Ss#44 | ||
| 45 | Shigella sonnei | None | Bégin | Ss#45 | |||||
| 46 | Shigella sonnei | None | Bégin | Ss#46 | |||||
| 47 | Escherichia coli | Bégin | 2000 | France | Ec#47 | ||||
| 48 | Escherichia coli | CFT073 | Univ. of Wisconsin | 1990 | MD | Ec#48 | |||
| 49 | Escherichia coli | UTI89 | Univ. Goettingen | Ec#49 | |||||
| 50 | Escherichia coli | 536 | Washington Univ. | Ec#50 | |||||
| 51 | Shigella flexneri | 1 | CIP 107169 | CRBIP | 3 | 1995 | France | Sf#51 | |
| 52 | Shigella flexneri | 2 | CIP 106236 | CRBIP | 3 | Sf#52 | |||
| 53 | Shigella flexneri | 4b | CIP 52.43 | NCTC 8336 | CRBIP | 3 | 1947 | United Kingdom | Sf#53 |
| 54 | Shigella flexneri | Y | CIP 52.36 | CRBIP | 3 | 1952 | Sf#54 | ||
| 55 | Shigella flexneri | 1 | CIP 106171 | CRBIP | 3 | Sf#55 | |||
| 56 | Shigella boydii | 12 | CIP 58.23 | NCTC 9772 | CRBIP | 3 | 1958 | Sb#56 | |
| 57 | Shigella flexneri | Untyped | CIP 54.58 | CRBIP | 1954 | Vietnam | Sf#57 | ||
| 58 | Shigella flexneri | Untyped | CIP 106210 | CRBIP | Sf#58 | ||||
| 59 | Shigella flexneri | 2a | 301 | ChMPH | 3 | 1984 | China | Sf#59 | |
| 60 | Shigella flexneri | 2a | CIP 56.19 | CRBIP | 3 | 1956 | Tunisia | Sf#60 | |
| 61 | Shigella flexneri | 2a | ATCC 700930 | Univ. of Wisconsin | 3 | Japan | Sf#61 | ||
| 62 | Shigella flexneri | 2b | CIP 52.39 | CRBIP | 3 | 1952 | Sf#62 | ||
| 63 | Shigella flexneri | 5 | CIP 67.61 | CRBIP | 3 | 1967 | Iraq | Sf#63 | |
| 64 | Shigella flexneri | 5 | 8401 | ChMPH | 3 | Sf#64 | |||
| 65 | Shigella flexneri | 3 | CIP 54.40 | CRBIP | 3 | 1919 | United Kingdom | Sf#65 | |
| 66 | Shigella flexneri | X | CIP 52.34 | CRBIP | 3 | 1952 | Sf#66 | ||
| 67 | Shigella flexneri | 3a | CIP 106211 | CRBIP | 3 | Sf#67 | |||
| 68 | Shigella dysenteriae | 8 | CIP 53.134 | NCTC 9345 | CRBIP | 1953 | Sd#68 | ||
| 69 | Shigella flexneri | 4c | CIP 52.25 | CRBIP | 3 | 1952 | Sf#69 | ||
| 70 | Shigella dysenteriae | 1 | CIP 56.33 | CRBIP | 1956 | Ethiopia | Sd#70 | ||
| 71 | Shigella dysenteriae | 1 | CIP 106200 | CRBIP | 1919 | Sd#71 | |||
| 72 | Shigella dysenteriae | 1 | A147 | Bégin | 1994 | Rwanda | Sd#72 | ||
| 73 | Shigella dysenteriae | 1 | A1-313 | Bégin | 1994 | Rwanda | Sd#73 | ||
| 74 | Shigella dysenteriae | 1 | 197 | ChMPH | 1950 | China | Sd#74a | ||
| 74 | Shigella dysenteriae | 1 | M131649 | Sanger | 1970 | Guatemala | Sd#74b | ||
| 75 | Shigella dysenteriae | 1 | CIP 58.1 | CRBIP | 1958 | Tunisia | Sd#75a | ||
| 75 | Shigella dysenteriae | 1 | CIP 62.17 | CRBIP | 1962 | Cameroon | Sd#75b | ||
| 76 | Escherichia coli | O157:H7 | Sakaï | Osaka Univ. | 1996 | Japan | Ec#76 | ||
| 77 | Escherichia coli | O157:H7 | EDL933 | Univ. of Wisconsin | 1982 | MI | Ec#77 | ||
| 78 | Escherichia coli | K12 | CEB | 1922 | CA | Ec#78a | |||
| 78 | Escherichia coli | K12 | CEB | 1922 | CA | Ec#78b | |||
| 78 | Escherichia coli | MG1655 | Univ. of Wisconsin | 1922 | CA | Ec#78c | |||
| 79 | Escherichia coli | HB101 | CEB | 1922 | CA | Ec#79 | |||
| 80 | Escherichia coli | W3110 | NARA IST | 1922 | CA | Ec#80 | |||
| 81 | Shigella boydii | 13 | CIP 58.21 | NCTC 9361 | CRBIP | 1958 | Sb#81 | ||
| 82 | Shigella boydii | 13 | CIP 58.22 | NCTC 9362 | CRBIP | 1958 | Sb#82 | ||
| 83 | Salmonella enterica | Typhimurium | LT2 | Washington Univ. | 1940s | Se#83 |
Bégin, Hôpital d'Instruction des Armées, Bégin, France; ChMPH, Microbial Genome Center of the Chinese Ministry of Health; CRBIP, Centre de Ressources Biologiques de l'Institut Pasteur; NARA IST, Nara Institute of Science and Technology; Univ., University.
MLVA setup.
A multiple comparison of Shigella genome sequences available at the onset of the study was done through the Microorganism Tandem Repeats database (3, 6), accessible as a Web service (http://minisatellites.u-psud.fr/). The methods previously described (3, 18) and the genome sequence data for strains S. flexneri 2457T (GenBank accession no. AE005174) (48), S. flexneri 301 (GenBank accession no. NC_004337) (10), S. sonnei 46 (GenBank accession no. NC_007384), S. dysenteriae 197 (GenBank accession no. CP000034), and S. boydii 227 (GenBank accession no. NC_007613) (50) were used to identify presumably polymorphic tandem repeats (TRs).
Loci with repeat units longer than 9 bp were favored in order to facilitate allele repeat number calling by a variety of DNA amplicon length estimation devices (24) and as a complement to a previous investigation (20). Selected primers pairs were validated in silico with available genome sequence data for Shigella and E. coli strains by using the multiple-PCR-primer BLAST Web service (http://minisatellites.u-psud.fr/) which provides the expected size and sequence of the PCR products. The resulting in silico typing data were integrated into the MLVA.
Some VNTR loci of E. coli O157:H7 described by Keys et al. (12) are present in the Shigella genus genome sequences. Five of them (O157-11, O157-13, O157-25, O157-33, and O157-34), selected for their relative ease of use (repeat unit size, >6 bp) or high rate of polymorphism, were evaluated in this study.
VNTR amplification and genotyping.
PCRs were performed as previously described (18) and using an annealing temperature of 60°C. DNA from the S. flexneri 2457T strain was used as an internal control to ensure high-quality size assignments as described previously (36). The PCR products were run on agarose gels, stained with ethidium bromide, visualized under UV, and photographed as previously described (19).
Sequence-based typing.
To allow comparison between MLVA and MLST assays, a region of the thrC gene described by Pupo et al. that carries enough information to discriminate the collection in the clusters previously described (35) was selected. Primers were designed with Primer3 software (39) and led to a 351-bp-long amplicon. This region was amplified in all strains of the present collection, and PCR products were sent to MWG-BIOTECH AG (Ebersberg, Germany) for sequencing.
Data analysis.
Gel electrophoresis images were analyzed by using the Bionumerics software package, version 5.0 (Applied-Maths, Sint-Martens-Latem, Belgium), as previously described (19). The number of repeats in each allele was deduced from the amplicon size. The resulting data were analyzed as a character data set with Bionumerics software. Clustering analysis was done by using the categorical parameter and the unweighted-pair group method with arithmetic averages (UPGMA) coefficient. The same weight was given to large and small numbers of differences in the repeats at each locus. The categorical parameter was also used to calculate the minimum spanning tree (MST). MST is a convenient complementary tool to cluster multiple isolates and visualize the relative diversity within different lineages. Polymorphism was quantified by the Hunter-Gaston diversity index (HGDI) (7). MLST sequences published by Pupo et al. (35) were imported into the Bionumerics program.
RESULTS
The in silico multiple-genome comparison of five genomes identified 32 TRs (repeat unit size, >9 bp; repeat sequence conservation, >80%) predicted to show at least two alleles among the five available genomes. One of the 32 loci was previously investigated (22). Five VNTR loci described by Keys et al. (12) for MLVA typing of E. coli O157:H7 were added to this panel owing to their presence and their polymorphism in both Shigella spp. and E. coli O157:H7. All VNTR loci considered herein are located on the chromosome. Specific primers for these VNTRs were checked using an initial selection of 12 bacterial strains (1 S. dysenteriae strain, 4 S. flexneri strains, 2 S. sonnei strains, 4 S. boydii strains, and 1 E. coli strain) and led to the elimination of 18 markers (8 failed to amplify DNA satisfactorily and 10 did not show polymorphism) (see Table S1 in the supplemental material). Analysis of the collection of 72 Shigella strains (Table 1) eliminated four additional markers that amplified less than 90% of the strains (see Table S1 in the supplemental material), leading to a final set of 15 markers (Table 2), including 3 from the panel described by Keys et al.
TABLE 2.
List of TRs selected
| Locusa | TR | S. flexneri 2457T chromosome location (positions) | Primer sequence
|
Product | Reference | |
|---|---|---|---|---|---|---|
| Upper | Lower | |||||
| flex0880_39bp_301bp_2U | ms06 | 880507-880807 | AAACGGGAGAGCCGGTTATT | TGTTGGTACAACGGCTCCTG | Cell division protein | This report |
| flex0881_39bp_626bp_7U | CVN001 (ms07) | 881365-881990 | GTCAGTTCGCCCAGACACAG | CGGTGTCAGCAAATCCAGAG | Cell division protein | 22 |
| flex1282_179bp_535bp_2U | ms09 | 1282058-1282592 | GTGCCATCGGGCAAAATTAG | CCGATAAGGGAGCAGGCTAGT | This report | |
| flex1724_96bp_766bp_5U | ms11 | 1724780-1725545 | GAAACAGGCCCAGGCTACAC | CTGGCGCTGGTTATGGGTAT | Electron transport | This report |
| flex2172_23bp_123bp_2U | ms18 | 2172858-2172980 | AACAAGAATATGAAAAATCATAGCAC | TTGTAAATTCGGTCATGATAATAATTC | complex protein RnfC | This report |
| flex2786_141bp_646bp_4U | ms21 | 2786095-2786740 | GCTGATGGCGAAGGAGAAGA | GGGAGTATGCGGTCAAAAGC | This report | |
| flex2917_9bp_117bp_7U | ms22 | 2917680-2917796 | TGGATACCCGATAACGCAGT | CCTGAAGAATCAACAAGGCTTTA | Prepilin peptidase- | This report |
| flex3171_375bp_1334bp_3U | ms23 | 3171829-3173162 | GCTCCGCTGATTGACTCCTT | CGGTTGCTCGACCACTAACA | dependent protein C | This report |
| flex3237_78bp_316bp_2U | ms24 | 3237723-3238038 | CGTTTAGCCGAGGATGAAGC | TGTGCGTTAGCAAGCCATTC | Hypothetical protein | This report |
| flex3406_17bp_340bp_3U | ms25 | 3406473-3406812 | CCGTACAAGCTGCTGATGCT | TGCGTACATCGTCTCGAACA | Hypothetical protein | This report |
| flex3450_18bp_134bp_2U | ms26 | 3450707-3450840 | TTCTTCTCCAGGTCTACAAAAATC | GCAACAAAACGACATCTTTACCTG | This report | |
| flex4573_101bp_456bp_2U | ms32 | 4573218-4573673 | TGAGATTGCCGAAGTGTTGC | AACTGGCGGCGTTTATCAAG | Hypothetical protein | This report |
| flex3996_6bp_220bp_6U | O157-11 | 3776930-3777149 | GATGCTGGAAAAACTGATGCAGACTCGCGT | GACCGGCAATCATCGGGCCAACCA | Uroporphyrinogen III methylase | 12 |
| flex3659_9bp_121bp_2U | O157-13 | 4114040-4114160 | GCAGCAAACGCCACAGTACCCATGCC | GTAGGTCATCTGCCGTGGTTCGAGCGCT | Hypothetical protein | 12 |
| flex4448_16bp_176bp_1U | O157-33 | 4440283-4440458 | GTGAAGGATAAGCTGCATTTGTCAGTGATGTCCGAAG | GCCTGACGCTAAAGATAAAGAAGAAAGCGTCGCG | Hypothetical protein | 12 |
Locus names were created as follows: four letters for the species strain used as a reference, four digits for genome location in the sequenced reference strain, TR size, locus size in the reference strain, and number of TR in the reference strain, separated by underscore sign.
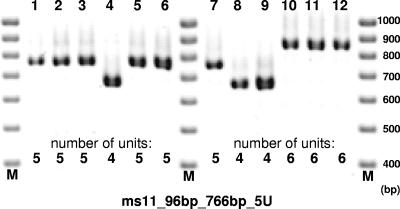
The sizes of the amplification products for the sequenced strain S. flexneri 2457T were as expected for each marker, and DNA from this strain was used as the reference in all analyses (Fig. 1). Four VNTR loci yielded an allele assignation for all Shigella and Escherichia strains investigated, while 11 others yielded no PCR product in some strains in spite of repeated attempts, suggesting locus absence and/or sequence polymorphism at priming sites.
FIG. 1.
Illustration of the MLVA assay setup. The PCR products of an amplification using ms06 primers were loaded on an agarose gel, electrophoresed, and stained with ethidium bromide. Lanes M show a 100-bp-ladder molecular weight marker. Lanes 1 and 7 correspond to the S. flexneri strain 2457T; lanes 2 to 6 and lanes 8 to 12 correspond to strains from our assay. The image illustrates how the number of units can be directly deduced by manual reading. The marker name below the gel provides the repeat unit size of the TR, the expected PCR product size in the S. flexneri 2457T genome, and the corresponding number of units in the S. flexneri 2457T genome.
Table 3 highlights characteristics and diversity of the 15 selected VNTR loci. Ten loci are located within coding or putative coding regions, and five are intergenic. Repeat unit lengths ranged from 6 to 375 base pairs. These VNTRs were also tested in silico by using the 17 available sets of genomic sequence data of Shigella, E. coli, and Salmonella enterica serovar Typhimurium (Table 1).
TABLE 3.
Diversity indexes calculated for MLVA panels and individual markers in the four Shigella species and E. colia
| Locus | Alias | Total (n = 89)
|
E. coli (n = 11)
|
S. boydii (n = 26)
|
S. dysenteriae (n = 23)
|
S. flexneri (n = 19)
|
S. sonnei (n = 9)
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. of genotypes | HGDI | No. of genotypes | HGDI | No. of genotypes | HGDI | No. of genotypes | HGDI | No. of genotypes | HGDI | No. of genotypes | HGDI | ||
| ms09 | 8 | 0.65 | 4 | 0.75 | 5 | 0.69 | 3 | 0.53 | 3 | 0.4 | 2 | 0.22 | |
| ms11 | 5 | 0.51 | 3 | 0.59 | 2 | 0.41 | 4 | 0.74 | 2 | 0.22 | 1 | 0.00 | |
| ms18 | 4 | 0.27 | 1 | 0.00 | 2 | 0.14 | 2 | 0.47 | 3 | 0.23 | 1 | 0.00 | |
| ms22 | 6 | 0.53 | 2 | 0.33 | 3 | 0.22 | 2 | 0.36 | 5 | 0.67 | 1 | 0.00 | |
| ms23 | 5 | 0.40 | 3 | 0.56 | 2 | 0.21 | 4 | 0.25 | 2 | 0.51 | 1 | 0.00 | |
| ms24 | 4 | 0.35 | 2 | 0.18 | 3 | 0.22 | 1 | 0.00 | 2 | 0.38 | 1 | 0.00 | |
| ms25 | 6 | 0.38 | 1 | 0.00 | 3 | 0.22 | 2 | 0.09 | 4 | 0.71 | 1 | 0.00 | |
| ms26 | 5 | 0.68 | 2 | 0.51 | 4 | 0.54 | 4 | 0.64 | 2 | 0.38 | 2 | 0.50 | |
| ms32 | 6 | 0.56 | 3 | 0.69 | 4 | 0.65 | 3 | 0.32 | 1 | 0 | 1 | 0.00 | |
| O157-13 | 5 | 0.42 | 2 | 0.33 | 4 | 0.65 | 3 | 0.37 | 2 | 0.12 | 1 | 0.00 | |
| O157-33 | 4 | 0.50 | 2 | 0.33 | 1 | 0.00 | 2 | 0.47 | 1 | 1 | 2 | 0.50 | |
| Panel 1 | 53 | 0.98 | 8 | 0.89 | 14 | 0.95 | 14 | 0.93 | 15 | 0.96 | 3 | 0.64 | |
| ms06 | 6 | 0.77 | 2 | 0.51 | 6 | 0.77 | 6 | 0.79 | 2 | 0.52 | 1 | 0.00 | |
| CNV-001 | ms07 | 11 | 0.78 | 4 | 0.75 | 7 | 0.70 | 4 | 0.66 | 4 | 0.42 | 1 | 0.00 |
| ms21 | 7 | 0.76 | 4 | 0.75 | 3 | 0.69 | 5 | 0.68 | 3 | 0.59 | 2 | 0.56 | |
| Panels 1 and 2 | 65 | 0.99 | 9 | 0.95 | 19 | 0.97 | 18 | 0.98 | 16 | 0.98 | 3 | 0.64 | |
| O157-11 (panel 3) | 19 | 0.93 | 5 | 0.85 | 15 | 0.96 | 6 | 0.81 | 10 | 0.92 | 7 | 0.94 | |
| Total for MLVA-15 | 83 | 1.00 | 9 | 0.95 | 25 | 1.00 | 21 | 0.99 | 19 | 1.00 | 8 | 0.97 | |
n, number of strains.
The HGDI (7) of each VNTR was calculated for the complete collection and for each species. Three different panels of VNTR loci (panel 1 included 11 loci, panel 2 included 3 loci, and panel 3 included 1 locus) were defined according to their HGDI value and were subsequently combined in a composite data set using three different levels of weight (individual marker weight of 20, 10, or 1) along the line previously proposed for Brucella MLVA (1). The rationale for this approach is that the diversity index indirectly reflects mutation rate and homoplasy level at each locus. Markers with a higher homoplasy level have a lower phylogenetic value. Panel 1 included 11 VNTRs (individual weights of 20) with HGDIs below 0.75 (ms09, ms11, ms18, ms22, ms23, ms24, ms25, ms26, ms32, O157-13, and O157-33), panel 2 included 3 VNTRs (individual weights of 10) with HGDIs ranging from 0.75 to 0.9 (ms06, CNV-001, and ms21), and panel 3 is limited to the hypervariable VNTR, O157-11 (individual weight of 1), with an HGDI higher than 0.9.
Diversity indexes differ within the genus Shigella and the species E. coli (Table 3). As expected, the least variable group of organisms is the S. sonnei genus, with an HGDI of 0.64 when the highly variable locus O157-11 is not included (MLVA14, comprising panels 1 and 2). Locus O157-11 alone has an HGDI of 0.94 for S. sonnei and leads to a general HGDI of 0.97 for this species. S. flexneri, S. boydii, and S. dysenteriae show much greater MLVA14 diversity, correlated with the higher number of serotypes (12, 15, and 10, respectively) in these species. Regarding E. coli, the limited number of strains included in the study does not allow comparison with the others.
By use of the MLVA14 assay (panels 1 and 2), the 89 strains (72 DNA samples tested and 17 sets of sequence data) were differentiated into 65 genotypes. When the O157-11 VNTR is used, the discriminatory power of the assay is significantly increased, with 83 genotypes numbered 1 to 83 in the dendrogram produced (Fig. 2). With the low relative weight given in the clustering analysis to this very highly variable marker, the two clusters are highly similar. All strains are identified by species acronym and genotype number (e.g., Sd#01 or Sf#70). Several strains sharing the same genotype will be additionally differentiated by a letter (e.g., a, b, etc.).
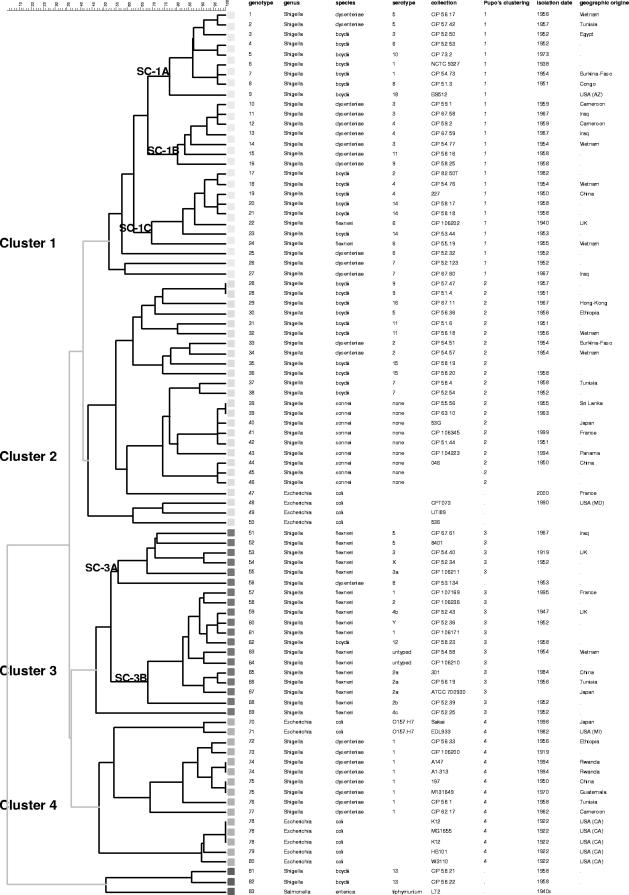
FIG. 2.
Dendrogram with all the typed strains, including the ones corresponding to sequenced strains that were included in the data set by their allele numbers as calculated theoretically from the expected amplicon sizes of all 15 loci based on their genome sequences. Clustering analysis was done using the categorical and UPGMA options. The columns indicate the genotype number, the genus, the species, the serotype, the strain identification number (collection), the cluster number as defined by Pupo et al., and the year and place of isolation.
Cluster analysis shows that most of the Shigella strains fall into four main clusters. Sb#81 and Sb#82 stand as outliers, Se#83 being an outgroup. Sb#81 and Sb#82 belong to the S. boydii serotype 13 (B13 in reference 35) and have an MLVA15 profile very different from those of all the other Shigella strains, with seven cases lacking amplification and two alleles observed only in these strains. Two strains of S. sonnei, two strains of S. boydii, three strains of E. coli, and two pairs of S. dysenteriae strains share the same genotype. Additional markers might allow further differentiation of Shigella strains, supposed to have been isolated in different places at different times (20). The HGDIs, however, are high and, by adding in the cluster analysis markers discarded previously, the strains remain undistinguishable (data not shown), supporting the idea that they are in all likelihood epidemiologically related.
To estimate the validity of the clustering observed by VNTR typing, selected strains were analyzed by sequence typing using published MLST data (35). Strains provided amplification products of the expected size, regarding the segment of the thrC gene. Sequences were imported and aligned in the Bionumerics program, and a cluster analysis was produced (see Table S2 in the supplemental material). A single discrepancy between clusters inferred from known genera and serotypes on one hand and sequence analysis on the other hand was observed due to the incongruence of phylogeny at the different loci. S. dysenteriae CIP 53.134 (D8 in reference 35) fell within cluster 3 when only the thrC sequence data were analyzed. It is the trpC-trpB sequence used in the MLST assay, in particular, which excludes that strain from cluster 3. Subsequently, all strains from the collection for which the thrC locus was investigated were characterized by their relevance to the clusters classified by Pupo et al., as indicated in Fig. 2.
This sequence analysis and comparison with published sequence data show that three of these clusters do correspond to the clusters classified by Pupo et al., and they are numbered accordingly in Fig. 2. Cluster 1 grouped S. boydii serotypes 1 to 4, 6, 8, 10, 14, and 18, S. dysenteriae serotypes 3 to 7, 9, and 11, and S. flexneri serotype 6. Cluster 1 was the most diverse, grouping representatives of all Shigella species except S. sonnei. This cluster could be divided into three subclusters, i.e., SC-1A, SC-1B, and SC-1C. Strains sharing the same serotype are usually discriminated by MLVA15 typing.
Cluster 2 comprises S. boydii serotypes 5, 7, 9, 11, 15, and 16, S. dysenteriae serotype 2, and all S. sonnei serotypes. The two S. boydii serotype 9 displayed the same MLVA15 pattern. The S. sonnei strains are located into two branches, one with the two representatives of S. boydii serotype 7. Three uropathogenic E. coli strains are included in cluster 2, together with an E. coli strain isolated from a patient at Hôpital d'Instruction des Armées, Bégin, France (Ec#47).
Cluster 3 groups all S. flexneri strains (with the exception of S. flexneri serotype 6, located in cluster 1), S. boydii serotype 12, and S. dysenteriae serotype 8 (Sd#56). Sd#56 is weakly associated with subcluster 3A together with S. flexneri serotypes 3, 3a, 5, and X. A second subcluster, tentatively called 3B (Fig. 2), comprises S. boydii serotype 12 and S. flexneri serotypes 1, 2, 2a, 2b, 4b, and Y. S. flexneri serotype 4c is more distantly related. Each strain has a unique genotype, although several serotypes were represented by more than one strain.
Consequently, and with the exception of Sd#56 (Sd8), the composition of the clusters is in remarkable agreement with the report of Pupo et al., although the underlying approaches are quite different (35).
We propose here to define a fourth cluster. Cluster 4 would include all the S. dysenteriae serotype 1 strains, five representatives of the E. coli K-12 strain (including two genomic sequences), and the two E. coli O157:H7 strains included in the study. The E. coli O157:H7 representatives are located in a quite distinct branch, slightly closer to the S. dysenteriae serotype 1 branch than to the E. coli K-12 branch. The eight S. dysenteriae serotype 1 strains are separated into six genotypes, showing the discriminative power of the VNTR panel. Among those strains, two were isolated from patients during an outbreak in a refugee camp in Rwanda in 1994. They show exactly the same MLVA15 pattern. Three of the E. coli K-12 strains were wild-type K-12 and two were derivatives (14). The three K-12 wild types share exactly the same genotype, while derivatives present different patterns at locus ms22, ms32, or O157-11. The O157:H7 strains exhibited two differences among them, at loci ms11 and O157-11.
In very rare instances, a given allele is strongly associated with a specific cluster, at least in the limited collection of strains investigated here. The three-repeat-unit allele at locus ms06 is observed only in cluster 2, and locus O157-33 has more than one repeat unit only in clusters 2 and 4 (associated with the E. coli strains investigated here).
ms25 and ms26 each gave rise to a very large amplicon in two strains and four strains, respectively, and these amplicons correspond to alleles with more than 60 repeat units (compared to the usual allele size ranges of one to three and two to four repeat units, respectively) (see Table S2 in the supplemental material). The six alleles were sequenced. The ms25 sequencing revealed that two different insertion sequences (ISs) were present, IS629 and IS2 for Sf#22 and Sd#26, respectively. IS629 is a member of the IS3 family of transposable elements. The ms26 allele sequencing led to the finding that only one IS, known as IS630, was present in this TR (27), and it was inserted at the same position and orientation in the four strains Sd#33, Sd#34, Sb#35, and Sb#36, confirming the close relationship between the four strains already suggested by MLVA15 clustering, in spite of a different species assignment.
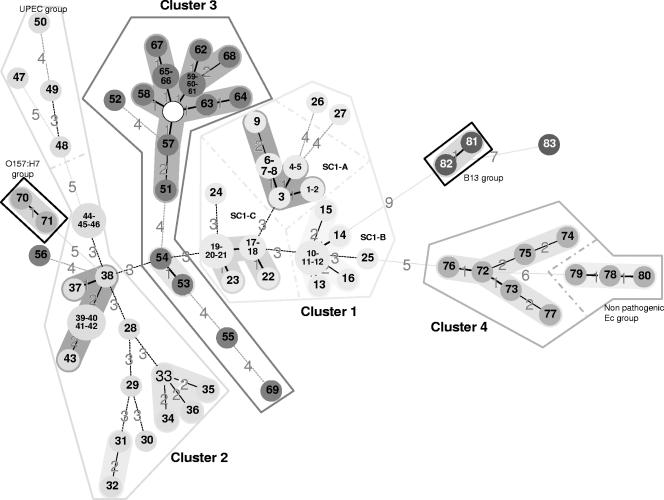
As a complementary analysis, an MST analysis was performed. MST analysis is a convenient tool to cluster multiple isolates and illustrate the relative diversity within different lineages (Fig. 3). This kind of analysis is applicable to categorical data sets. The creation of hypothetical types further minimizes the summed distance of all branches of the tree. The MST was drawn without locus O157-11, which is too variable to address correct relatedness within different lineages, especially since this analysis does not currently allow the assignment of different relative weights to markers. In cluster 1, the three main subclusters are conserved, with only minor changes. The two S. dysenteriae serotype 7 strains, Sd#26 and Sd#27, are located in the vicinity of subcluster A but interspaced by Sb#04 and Sb#05. The composition of cluster 2, with S. sonnei and S. boydii serotype 7 in the same branch, is also found by this approach. Sd#56 is not included in cluster 3, which is in accordance with results obtained from other molecular methods and reflects the absence of amplification at several loci, which is taken into account in this analysis. The only change in cluster 4 is the location of E. coli O157:H7 representatives that are far from other members of the cluster and weakly linked to cluster 2.
FIG. 3.
Shigella population modeling. The number in each circle indicates the corresponding genotype identified in Fig. 2. The empty circle indicates a hypothetical genotype (not present in the population analyzed) created to minimize overall distances between neighboring genotypes. The distance between neighboring genotypes is expressed as the number of allelic changes and is outlined by different shapes of lines: a hatched bold line indicates one change, a full gray line indicates two changes, a black dotted line indicates three changes, and a gray dotted line indicates more than four changes. Gray hatched lines are used to separate subgroups inside clusters. The tree calculation was made without locus O157-11, owing the location of different genotypes in the same circle.
DISCUSSION
The Shigella genus has been classified into species and serotypes for a long time and for practical reasons, but MLST (15, 16, 34, 52) and genome comparisons (51) have emphasized the strong genetic homogeneity between the different species. Most importantly, MLST has further strongly suggested that in some instances, the current species designations are not compatible with the actual phylogeny of the Shigella genus. It is now considered more likely that Shigella sp. strains, as well as enteroinvasive E. coli strains, are derived from multiple origins within the species E. coli (50).
The purpose of the present study was to develop an MLVA assay for Shigella and evaluate if the resulting data would be in agreement with the complex concept of the Shigella-E. coli genomospecies, as revealed by MLST data. We investigated TR loci predicted to be polymorphic by comparing five Shigella genome sequences. The selection of 15 VNTRs allowed us to discriminate 89 strains, covering the large majority of known diversity in Shigella species and several E. coli strains (77 Shigella spp., 11 E. coli strains, and 1 S. enterica strain), into 83 genotypes. Strains sharing the same serotype are rarely from the same genotype, with difference ranging from one marker (Sd#1 and Sd#2, of S. dysenteriae serotype 5) to five markers (Sd#26 and Sd#27, of S. dysenteriae serotype 7). On the other hand, the two strains of S. dysenteriae serotype 1 isolated in Rwanda in 1994 during a dysentery outbreak in refugee camps are identical.
Through MLVA15 analysis, four main clusters were defined, together with some outliers. Among them, the strains of S. boydii serotype 13 were clearly distinguished from the other Shigella strains. Some cases lacking amplification, as well as unique sizes for several loci, illustrate the wide distance between S. boydii serotype 13 and other Shigella representatives. According to Hyma et al. (8), S. boydii serotype 13 is closely related to a different Escherichia species, E. albertii. It supports the hypothesis of an ancient separation of lineages, S. boydii serotype 13, as well as S. enterica LT2, being a clear outgroup. This finding also corroborated the results of MLST investigations by Pupo et al. (35) and Lan et al. (15).
In order to be able to compare our MLVA15 clustering data with published MLST data, part of the thrC gene was sequenced. In this way, the strains used in this study could be assigned to the different clusters defined by Pupo et al. (35), and it very clearly appears that MLVA15 clustering provides the same view of the Shigella population structure as MLST. Recently, Yang et al. (52) conducted another MLST analysis, based on 23 housekeeping genes. The greater number of genes analyzed allowed them to define three subclusters inside cluster 1. MLVA15 clustering reveals the same fine substructure. Such striking similarity demonstrates the potential usefulness of MLVA for Shigella typing. While the general patterns are highly similar, there are some differences between MLST and MLVA approaches. In the present study, the nine S. sonnei strains are assigned to cluster 2, close to S. boydii serotype 7. Yang et al. (52) placed the single S. sonnei investigated close to cluster 1 by MLST. The S. sonnei strain investigated by Escobar-Paramo et al. (4) was found to be closer to cluster 2 and cluster 3 by analysis of four other housekeeping genes. Pupo et al. (35) placed the single S. sonnei strain they analyzed far from the three clusters. Thus, the definitive location of S. sonnei strains remains undefined. Further MLST analysis of S. sonnei might eventually permit us to revisit the relative evolutionary position of this species.
Considering that Shigella clusters are now believed to have independently arisen several times from E. coli species while acquiring pathogenicity factors (16, 35, 52), the underrepresentation of E. coli investigated here led to the representation of small groups of E. coli strains arising from Shigella clusters, while, in fact, the situation would be the opposite. Such analysis should be done by including a Shigella collection in a larger E. coli collection to obtain a general overview of E. coli/Shigella sp. genomospecies (such work is in progress).
Combining UPGMA cluster analysis and MST provides an overview of Shigella intraspecies relationships. In agreement with previous investigations, we can infer from this combination that Shigella taxonomy is of little phylogenetic value. This is illustrated for instance by S. flexneri serotype 6 being associated with cluster 1 by MLVA or MLST, whereas the other S. flexneri serotypes are assigned to cluster 3 (together with the two serologically untyped S. flexneri strains). Four strains clustered together by MLVA share the same rare IS insertion event at the same position, while they are assigned to different species, reinforcing the fact that current classification does not reflect the genetic relatedness within the Shigella genus. It is now believed that Shigella organisms arose several times within E. coli species (35, 52), leading to three clusters not related to the currently recognized Shigella species taxonomy based upon biochemical and serological characteristics. The three main clusters are also identified here with a fourth corresponding essentially to S. dysenteriae serotype 1.
This study augments the results relative to discrimination power, specificity, and sensitivity of the MLVA approach to the Shigella species. Moreover, the MLVA15 panel shows some capabilities to be applied to E. coli for typing purposes, with a wider range of use than considered before in several published studies, mainly focused on Shiga toxin-producing E. coli (12, 23, 29). Another MLVA assay was validated for O157:H7 outbreak detection (12). It showed high discrimination between the E. coli O157 strains and appeared to have equal sensitivity to that of PFGE and specificity superior to that of PFGE. Lindstedt et al. (22) described a seven-TR panel designed to type the ECOR collection and pathogenic E. coli species. Several Shigella strains were included in the study and typed. However, all serotypes were not available, and in one case, the TR was absent, and in another, the TR was monomorphic. Our panel primarily intended to cover all Shigella species, and its satisfactory performance indicates also good capabilities to discriminate between nonpathogenic E. coli, uropathogenic E. coli, and enterohemorrhagic E. coli strains.
As suggested previously (45), the proposed combination of well-selected independent polymorphic TR loci is highly discriminatory and provides a relevant clustering compared with the currently accepted classification. The panel of 15 markers has been divided into three panels according to the polymorphism of each locus. It is important to keep in mind that the very high discriminatory power of some markers usually results in a very high homoplasy level. For this reason, such markers must be given a lower weight when similarity matrices are calculated. Here, the different weights proposed for each panel were empirically determined, but it is very likely that in future MLVA developments, special attention will be given to the optimization of these coefficients. For this to be done, much larger collections of strains, with detailed epidemiological data, will need to be typed first. It can only be hoped that in the future, funding bodies will support international consortiums of laboratories aimed at producing high-quality data of this kind. VNTR typing could then be widely accessible for research and public health laboratories, particularly in developing countries, where the majority of cases of Shigella occur, since this method is highly suitable for sharing results and for the generation of databases as previously demonstrated (http://mlva.u-psud.fr).
Supplementary Material
Acknowledgments
We thank Sabrina Gausson and Claudette Simoes-Miranda for technical help. We thank Jean-Didier Cavallo (Hôpital d'Instruction des Armées, Bégin, France) for providing some of the strains used in this study. O. Gorgé also acknowledges Rozenn Bernard and Philippe Le Flèche for many useful discussions and ideas.
Work on the typing of collections of dangerous pathogens and the making of reference genotype databases was supported by Délégation Générale pour l'Armement (PEA02-36-01).
Footnotes
Published ahead of print on 23 January 2008.
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1.Al Dahouk, S., P. Le Flèche, K. Nockler, I. Jacques, M. Grayon, H. C. Scholz, H. Tomaso, G. Vergnaud, and H. Neubauer. 2007. Evaluation of Brucella MLVA typing for human brucellosis. J. Microbiol. Methods 69137-145. [DOI] [PubMed] [Google Scholar]
- 2.Chaignat, C. 2004. Shigellosis, p. 527-531. In D. L. Heyman (ed.), Control of communicable diseases manual, 18th ed. American Public Health Association, Washington, DC.
- 3.Denoeud, F., and G. Vergnaud. 2004. Identification of polymorphic tandem repeats by direct comparison of genome sequence from different bacterial strains: a web-based resource. BMC Bioinformatics 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Escobar-Paramo, P., C. Giudicelli, C. Parsot, and E. Denamur. 2003. The evolutionary history of Shigella and enteroinvasive Escherichia coli revised. J. Mol. Evol. 57140-148. [DOI] [PubMed] [Google Scholar]
- 5.Faruque, S. M., K. Haider, M. M. Rahman, A. R. Abdul Alim, Q. S. Ahmad, M. J. Albert, and R. B. Sack. 1992. Differentiation of Shigella flexneri strains by rRNA gene restriction patterns. J. Clin. Microbiol. 302996-2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grissa, I., P. Bouchon, C. Pourcel, and G. Vergnaud. On-line resources for bacterial micro-evolution studies using MLVA or CRISPR typing. Biochimie, in press. [DOI] [PubMed]
- 7.Hunter, P. R., and M. A. Gaston. 1988. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J. Clin. Microbiol. 262465-2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hyma, K. E., D. W. Lacher, A. M. Nelson, A. C. Bumbaugh, J. M. Janda, N. A. Strockbine, V. B. Young, and T. S. Whittam. 2005. Evolutionary genetics of a new pathogenic Escherichia species: Escherichia albertii and related Shigella boydii strains. J. Bacteriol. 187619-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hyytia-Trees, E., S. C. Smole, P. A. Fields, B. Swaminathan, and E. M. Ribot. 2006. Second generation subtyping: a proposed PulseNet protocol for multiple-locus variable-number tandem repeat analysis of Shiga toxin-producing Escherichia coli O157 (STEC O157). Foodborne Pathog. Dis. 3118-131. [DOI] [PubMed] [Google Scholar]
- 10.Jin, Q., Z. Yuan, J. Xu, Y. Wang, Y. Shen, W. Lu, J. Wang, H. Liu, J. Yang, F. Yang, X. Zhang, J. Zhang, G. Yang, H. Wu, D. Qu, J. Dong, L. Sun, Y. Xue, A. Zhao, Y. Gao, J. Zhu, B. Kan, K. Ding, S. Chen, H. Cheng, Z. Yao, B. He, R. Chen, D. Ma, B. Qiang, Y. Wen, Y. Hou, and J. Yu. 2002. Genome sequence of Shigella flexneri 2a: insights into pathogenicity through comparison with genomes of Escherichia coli K12 and O157. Nucleic Acids Res. 304432-4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johansson, A., J. Farlow, P. Larsson, M. Dukerich, E. Chambers, M. Bystrom, J. Fox, M. Chu, M. Forsman, A. Sjostedt, and P. Keim. 2004. Worldwide genetic relationships among Francisella tularensis isolates determined by multiple-locus variable-number tandem repeat analysis. J. Bacteriol. 1865808-5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Keys, C., S. Kemper, and P. Keim. 2005. Highly diverse variable number tandem repeat loci in the E. coli O157:H7 and O55:H7 genomes for high-resolution molecular typing. J. Appl. Microbiol. 98928-940. [DOI] [PubMed] [Google Scholar]
- 13.Koeck, J. L., B. M. Njanpop-Lafourcade, S. Cade, E. Varon, L. Sangare, S. Valjevac, G. Vergnaud, and C. Pourcel. 2005. Evaluation and selection of tandem repeat loci for Streptococcus pneumoniae MLVA strain typing. BMC Microbiol. 566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuhnert, P., J. Nicolet, and J. Frey. 1995. Rapid and accurate identification of Escherichia coli K-12 strains. Appl. Environ. Microbiol. 614135-4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lan, R., M. C. Alles, K. Donohoe, M. B. Martinez, and P. R. Reeves. 2004. Molecular evolutionary relationships of enteroinvasive Escherichia coli and Shigella spp. Infect. Immun. 725080-5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lan, R., and P. R. Reeves. 2002. Escherichia coli in disguise: molecular origins of Shigella. Microbes Infect. 41125-1132. [DOI] [PubMed] [Google Scholar]
- 17.Le Flèche, P., M. Fabre, F. Denoeud, J. L. Koeck, and G. Vergnaud. 2002. High resolution, on-line identification of strains from the Mycobacterium tuberculosis complex based on tandem repeat typing. BMC Microbiol. 237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Le Flèche, P., Y. Hauck, L. Onteniente, A. Prieur, F. Denoeud, V. Ramisse, P. Sylvestre, G. Benson, F. Ramisse, and G. Vergnaud. 2001. A tandem repeats database for bacterial genomes: application to the genotyping of Yersinia pestis and Bacillus anthracis. BMC Microbiol. 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Le Flèche, P., I. Jacques, M. Grayon, S. Al Dahouk, P. Bouchon, F. Denoeud, K. Nockler, H. Neubauer, L. A. Guilloteau, and G. Vergnaud. 2006. Evaluation and selection of tandem repeat loci for a Brucella MLVA typing assay. BMC Microbiol. 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liang, S. Y., H. Watanabe, J. Terajima, C. C. Li, J. C. Liao, S. K. Tung, and C. S. Chiou. 2007. Multilocus variable-number tandem repeat analysis for molecular typing of Shigella sonnei. J. Clin. Microbiol. 453574-3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lindstedt, B. A. 2005. Multiple-locus variable number tandem repeats analysis for genetic fingerprinting of pathogenic bacteria. Electrophoresis 262567-2582. [DOI] [PubMed] [Google Scholar]
- 22.Lindstedt, B. A., L. T. Brandal, L. Aas, T. Vardund, and G. Kapperud. 2007. Study of polymorphic variable-number of tandem repeats loci in the ECOR collection and in a set of pathogenic Escherichia coli and Shigella isolates for use in a genotyping assay. J. Microbiol. Methods 69197-205. [DOI] [PubMed] [Google Scholar]
- 23.Lindstedt, B. A., E. Heir, E. Gjernes, T. Vardund, and G. Kapperud. 2003. DNA fingerprinting of Shiga-toxin producing Escherichia coli O157 based on multiple-locus variable-number tandem-repeats analysis (MLVA). Ann. Clin. Microbiol. Antimicrob. 212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lista, F., G. Faggioni, S. Valjevac, A. Ciammaruconi, J. Vaissaire, C. le Doujet, O. Gorgé, R. De Santis, A. Carattoli, A. Ciervo, A. Fasanella, F. Orsini, R. D'Amelio, C. Pourcel, A. Cassone, and G. Vergnaud. 2006. Genotyping of Bacillus anthracis strains based on automated capillary 25-loci multiple locus variable-number tandem repeats analysis. BMC Microbiol. 633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Litwin, C. M., A. L. Storm, S. Chipowsky, and K. J. Ryan. 1991. Molecular epidemiology of Shigella infections: plasmid profiles, serotype correlation, and restriction endonuclease analysis. J. Clin. Microbiol. 29104-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu, P. Y., Y. J. Lau, B. S. Hu, J. M. Shyr, Z. Y. Shi, W. S. Tsai, Y. H. Lin, and C. Y. Tseng. 1995. Analysis of clonal relationships among isolates of Shigella sonnei by different molecular typing methods. J. Clin. Microbiol. 331779-1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matsutani, S., H. Ohtsubo, Y. Maeda, and E. Ohtsubo. 1987. Isolation and characterization of IS elements repeated in the bacterial chromosome. J. Mol. Biol. 196445-455. [DOI] [PubMed] [Google Scholar]
- 28.Melito, P. L., D. L. Woodward, J. Munro, J. Walsh, R. Foster, P. Tilley, A. Paccagnella, J. Isaac-Renton, J. Ismail, and L. K. Ng. 2005. A novel Shigella dysenteriae serovar isolated in Canada. J. Clin. Microbiol. 43740-744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Noller, A. C., M. C. McEllistrem, A. G. Pacheco, D. J. Boxrud, and L. H. Harrison. 2003. Multilocus variable-number tandem repeat analysis distinguishes outbreak and sporadic Escherichia coli O157:H7 isolates. J. Clin. Microbiol. 415389-5397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Onteniente, L., S. Brisse, P. T. Tassios, and G. Vergnaud. 2003. Evaluation of the polymorphisms associated with tandem repeats for Pseudomonas aeruginosa strain typing. J. Clin. Microbiol. 414991-4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pourcel, C., F. Andre-Mazeaud, H. Neubauer, F. Ramisse, and G. Vergnaud. 2004. Tandem repeats analysis for the high resolution phylogenetic analysis of Yersinia pestis. BMC Microbiol. 422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pourcel, C., Y. Vidgop, F. Ramisse, G. Vergnaud, and C. Tram. 2003. Characterization of a tandem repeat polymorphism in Legionella pneumophila and its use for genotyping. J. Clin. Microbiol. 411819-1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pourcel, C., P. Visca, B. Afshar, S. D'Arezzo, G. Vergnaud, and N. K. Fry. 2007. Identification of variable-number tandem repeat sequences in Legionella pneumophila and development of an optimized MLVA typing scheme. J. Clin. Microbiol. 451190-1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pupo, G. M., D. K. Karaolis, R. Lan, and P. R. Reeves. 1997. Evolutionary relationships among pathogenic and nonpathogenic Escherichia coli strains inferred from multilocus enzyme electrophoresis and mdh sequence studies. Infect. Immun. 652685-2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pupo, G. M., R. Lan, and P. R. Reeves. 2000. Multiple independent origins of Shigella clones of Escherichia coli and convergent evolution of many of their characteristics. Proc. Natl. Acad. Sci. USA 9710567-10572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ramisse, V., P. Houssu, E. Hernandez, F. Denoeud, V. Hilaire, O. Lisanti, F. Ramisse, J. D. Cavallo, and G. Vergnaud. 2004. Variable number of tandem repeats in Salmonella enterica subsp. enterica for typing purposes. J. Clin. Microbiol. 425722-5730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ribot, E. M., M. A. Fair, R. Gautom, D. N. Cameron, S. B. Hunter, B. Swaminathan, and T. J. Barrett. 2006. Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157:H7, Salmonella, and Shigella for PulseNet. Foodborne Pathog. Dis. 359-67. [DOI] [PubMed] [Google Scholar]
- 38.Rotz, L. D., A. S. Khan, S. R. Lillibridge, S. M. Ostroff, and J. M. Hughes. 2002. Public health assessment of potential biological terrorism agents. Emerg. Infect. Dis. 8225-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rozen, S., and H. Skaletsky. 2000. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132365-386. [DOI] [PubMed] [Google Scholar]
- 40.Sabat, A., J. Krzyszton-Russjan, W. Strzalka, R. Filipek, K. Kosowska, W. Hryniewicz, J. Travis, and J. Potempa. 2003. New method for typing Staphylococcus aureus strains: multiple-locus variable-number tandem repeat analysis of polymorphism and genetic relationships of clinical isolates. J. Clin. Microbiol. 411801-1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sambrook, J., and D. Russell. 2001. Molecular cloning, a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 42.Swaminathan, B., T. J. Barrett, S. B. Hunter, and R. V. Tauxe. 2001. PulseNet: the molecular subtyping network for foodborne bacterial disease surveillance, United States. Emerg. Infect. Dis. 7382-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.U'Ren, J. M., J. M. Schupp, T. Pearson, H. Hornstra, C. L. Friedman, K. L. Smith, R. R. Daugherty, S. D. Rhoton, B. Leadem, S. Georgia, M. Cardon, L. Y. Huynh, D. DeShazer, S. P. Harvey, R. Robison, D. Gal, M. J. Mayo, D. Wagner, B. J. Currie, and P. Keim. 2007. Tandem repeat regions within the Burkholderia pseudomallei genome and their application for high resolution genotyping. BMC Microbiol. 723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van Belkum, A. 2007. Tracing isolates of bacterial species by multilocus variable number of tandem repeat analysis (MLVA). FEMS Immunol. Med. Microbiol. 4922-27. [DOI] [PubMed] [Google Scholar]
- 45.Vergnaud, G., and C. Pourcel. 2006. Multiple locus VNTR (variable number of tandem repeat) analysis (MLVA), p. 83-104. In E. Stackebrandt (ed.), Molecular identification, systematics and population structure of prokaryotes, 1st ed. Springer-Verlag, New York, NY.
- 46.von Seidlein, L., D. R. Kim, M. Ali, H. Lee, X. Wang, V. D. Thiem, G. Canh do, W. Chaicumpa, M. D. Agtini, A. Hossain, Z. A. Bhutta, C. Mason, O. Sethabutr, K. Talukder, G. B. Nair, J. L. Deen, K. Kotloff, and J. Clemens. 2006. A multicentre study of Shigella diarrhoea in six Asian countries: disease burden, clinical manifestations, and microbiology. PLoS Med. 3e353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vu-Thien, H., G. Corbineau, K. Hormigos, B. Fauroux, H. Corvol, A. Clement, G. Vergnaud, and C. Pourcel. 2007. Multiple-locus variable-number tandem-repeat analysis for longitudinal survey of sources of Pseudomonas aeruginosa infection in cystic fibrosis patients. J. Clin. Microbiol. 453175-3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wei, J., M. B. Goldberg, V. Burland, M. M. Venkatesan, W. Deng, G. Fournier, G. F. Mayhew, G. Plunkett III, D. J. Rose, A. Darling, B. Mau, N. T. Perna, S. M. Payne, L. J. Runyen-Janecky, S. Zhou, D. C. Schwartz, and F. R. Blattner. 2003. Complete genome sequence and comparative genomics of Shigella flexneri serotype 2a strain 2457T. Infect. Immun. 712775-2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Woodward, D. L., C. G. Clark, R. A. Caldeira, R. Ahmed, G. Soule, L. Bryden, H. Tabor, P. Melito, R. Foster, J. Walsh, L. K. Ng, G. B. Malcolm, N. Strockbine, and F. G. Rodgers. 2005. Identification and characterization of Shigella boydii 20 serovar nov., a new and emerging Shigella serotype. J. Med. Microbiol. 54741-748. [DOI] [PubMed] [Google Scholar]
- 50.Yang, F., J. Yang, X. Zhang, L. Chen, Y. Jiang, Y. Yan, X. Tang, J. Wang, Z. Xiong, J. Dong, Y. Xue, Y. Zhu, X. Xu, L. Sun, S. Chen, H. Nie, J. Peng, J. Xu, Y. Wang, Z. Yuan, Y. Wen, Z. Yao, Y. Shen, B. Qiang, Y. Hou, J. Yu, and Q. Jin. 2005. Genome dynamics and diversity of Shigella species, the etiologic agents of bacillary dysentery. Nucleic Acids Res. 336445-6458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang, J., L. Chen, J. Yu, L. Sun, and Q. Jin. 2006. ShiBASE: an integrated database for comparative genomics of Shigella. Nucleic Acids Res. 34D398-D401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang, J., H. Nie, L. Chen, X. Zhang, F. Yang, X. Xu, Y. Zhu, J. Yu, and Q. Jin. 2007. Revisiting the molecular evolutionary history of Shigella spp. J. Mol. Evol 6471-79. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.