Abstract
A 38-year-old patient developed meningitis after a complicated kidney transplantation and organ rejection. Ureaplasma urealyticum was identified as the etiological agent by molecular and microbiological analyses of the cerebrospinal fluid. The patient was successfully treated with doxycycline and chloramphenicol. This is the first report of Ureaplasma urealyticum meningitis in an adult.
CASE REPORT
A 38-year-old man was admitted to the nephrology department of our hospital for kidney transplantation. His medical history showed terminal kidney insufficiency caused by interstitial nephritis and treatment with continuous ambulant peritoneal dialysis for 10 years. On day 11 after transplantation of a kidney into the right fossa iliaca, he suffered from hemorrhagic shock caused by the sudden onset of diffuse arterial bleeding. Mass transfusions and colloidal volume substitution therapy were initiated. After explantation of the kidney graft, the bleeding was stopped, and the patient was admitted to the intensive care unit for further management. Immunosuppressive therapy was discontinued. During the following 8 weeks, several episodes of infection of a residual iliaco-retroperitoneal-perihepatic hematoma with Pseudomonas aeruginosa, Enterococcus faecium, or coagulase-negative staphylococci (mixed cultures), as well as three independent infections of abdominal and liver abscesses caused by Candida albicans, Morganella morganii (extended-spectrum beta-lactamase producer), or Klebsiella pneumoniae, were successfully treated by both surgical intervention and antibiotic therapy. Ventricular drainage was applied after colloid-induced brain edema. An intermittent factor XII deficiency and ventilator-associated pneumonia were successfully treated. The patient was mostly stable when he presented with increasing cephalgias accompanied by a single episode of seizure. Analyses of cerebrospinal fluid (CSF) revealed no conclusive results. Magnetic resonance tomography and computed tomography scans demonstrated a normal configuration of the central nervous system. When clinical signs of meningitis developed 4 weeks later and intubation of the patient was required due to suspected central repression of respiration, a second lumbar puncture was performed. Turbid, xanthochromic CSF was obtained. The constellation of reduced glucose (210 mg/liter), increased lactate (739 mg/liter), protein (2,858 mg/liter), and leucocytosis (mononuclear cells, 810 cells/ml) in the CSF confirmed the suspicion of an infectious etiology (10).
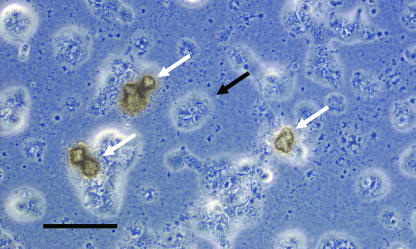
Routine microbiological procedures were performed. No microorganisms on Gram stains, no acid-fast bacteria, and no evidence of meningitis antigens (i.e., Streptococcus pneumoniae or Haemophilus influenzae) could be detected (Wellcogen bacterial antigen kit; Remel, Dartford, United Kingdom), and standard cultures of the CSF for bacterial (i.e., Listeria monocytogenes or Nocardia asteroides) or fungal (i.e., Cryptococcus neoformans) pathogens remained sterile. Mycobacterium tuberculosis complex RNA was not detectable (amplified Mycobacterium tuberculosis direct test; Gen-Probe, San Diego, CA). Molecular techniques were performed as soon as the cultures remained negative for 18 h. From the CSF sediment, a 792-bp fragment was amplified by a broad-range PCR for detection of the bacterial 16S rRNA gene (8). The sequence of the PCR product showed 100% identity to the 16S rRNA gene of Ureaplasma urealyticum (American Type Culture Collection [ATCC] strain 33698; GenBank accession number AF073455). As soon as this surprising PCR result became available, an aliquot of the CSF sediment was spread on solid A7 agar media supplemented with 17 mM urea, 16.7% equine serum, 1.4 mM MnSO4, and 20 units/ml of penicillin (4). After 18 h of anaerobic incubation at 37°C, the growth of typical tiny, brownish, granular cotton wool-like Ureaplasma colonies was observed under the microscope (10 to 20 μm in diameter), confirming the molecular diagnosis of U. urealyticum (Fig. 1). A mycoplasma IST 2 kit (bioMérieux, France) was used for confirmation of strong urealytic activity and antimicrobial susceptibility, and the results were interpreted according to the recommendations of the manufacturer.
FIG. 1.
Microcolonies (white arrows) of U. urealyticum on A7 agar after 18 h of anaerobic incubation at 37°C. Note the cellular debris of CSF granulocytes in the background (black arrow). Shown is a phase contrast microscopy image at ×800 magnification (scale bar, 25 μm).
The hitherto antibacterial therapy consisting of meropenem (1 g every 8 h [q8h]) and tobramycin (200 mg q8h) was immediately supplemented with doxycycline (200 mg q24h) and moxifloxacin (400 mg q24h), while intermittent antimycobacterial therapy was discontinued. Since the patient's condition did not improve within 72 h and the Ureaplasma isolate tested resistant to ciprofloxacin (MIC, >2 mg/liter) and intermediately susceptible to ofloxacin (1 mg/liter < MIC < 4 mg/liter), moxifloxacin was replaced by chloramphenicol (1 g q6h), which is well known for good effectiveness against Ureaplasma (6) and which should accumulate in high concentrations in CSF. Within 48 h, the patient responded to this therapy. His neurological conditions improved rapidly, his CSF cell counts dropped, and he no longer required oxygen supplementation. In parallel, the concentrations of interleukin-6 (IL-6), IL-8, and neopterin in the CSF dropped significantly, from 12.5 ng/ml, 2.8 ng/ml, and 1.2 mmol/liter, respectively, to undetectable levels. Growth of U. urealyticum, as well as its DNA, was no longer detectable in the CSF. Urethral swabs screened for U. urealyticum were negative. The patient received a total dose of 28 g of chloramphenicol. He recovered completely from his complicated course of meningitis and did not suffer any permanent neurological damage.
In the light of the unusual manifestation of adult meningitis, we further characterized the Ureaplasma isolate designated CM239 by analyzing the multiple-banded antigen (MBA) gene according to the method of Kong et al. (5). The MBA gene sequence was identical to that of U. urealyticum genotype C (biovar 2), which comprises serovars 4, 12, and 13. We also determined the sequences of the quinolone resistance-determining regions of the gyrA (298-bp), gyrB (271-bp), and parC (269-bp) genes (3). In comparison to the reference sequences from the Ureaplasma parvum genome (accession numbers AE002107 and AE002143), the sequences showed a high number of silent-base mismatches, with 24 for gyrA, 18 for gyrB, and 24 for parC. In addition, one amino acid exchange was found in GyrA (D95E; Escherichia coli numbering) and three in ParC (Y56H, A123T, and A134T). No amino acid exchange was found in GyrB.
After organ transplantation, meningitis is a life-threatening complication in the immunosuppressed host and may be caused by several opportunistic pathogens, including Listeria monocytogenes, gram-negative rods, Cryptococcus neoformans, Aspergillus fumigatus, Toxoplasma gondii, and Nocardia asteroides, and progressive dementia may be a sequela of JC polyomavirus or other viruses (7). We report the first case of adult U. urealyticum meningitis, which developed 10 weeks after a complicated kidney transplantation with organ rejection.
Ureaplasma is well known as the agent of non-gonococcal urethritis as well as perinatal infections, including neonatal pneumonia, bacteremia, and meningitis (11). U. urealyticum was categorized into 14 serotypes and divided into two biovars. Recently, biovar 1, including serotypes 1, 3, 6, and 14, was designated a separate species, Ureaplasma parvum, whereas biovar 2, which includes the remaining 10 serotypes, retained the U. urealyticum designation (9). Using the MBA sequence-typing method, our isolate was identified as U. urealyticum serotype 4, 12, or 13. Some studies that addressed the question of the differential pathogenicity of Ureaplasma serotypes found a higher prevalence of serotype 4 in non-gonococcal urethritis (summarized in reference 11). However, it appears that many serotypes are invasive and that antigen variability and host factors are more-important determinants for mycoplasmal infections than different serotypes.
The possibility that the transplant itself or one of the numerous blood products which initially had to be transfused were contaminated with undetectable numbers of U. urealyticum cannot be excluded. However, it seems more likely that after explantation of the kidney graft, the iliaco-retroperitoneal-perihepatic hematoma was superinfected with U. urealyticum as part of the resident urogenital flora of the patient. Since the antibiotics applied at that time to eliminate M. morganii, P. aeruginosa, and E. faecium did not target this uncommon type of cell wall-free microorganism, it caused a slowly progressing systemic infection, which finally spread hematogenously into the brain. Unfortunately, the urethral swabs were not tested for Ureaplasma until several days after the onset of specific treatments, and although selective media were used, the cultures were overgrown by other bacteria. Therefore, evidence for Ureaplasma in the urogenital tract of the patient could not be supplied.
The amplification of Ureaplasma DNA and the growth of U. urealyticum from the CSF clearly demonstrate the etiologic role of this unusual bacterium in the nosocomial meningitis of our patient. The strength of using a molecular method to detect microorganisms in culture-negative patient samples lies in both the universal spectrum of pathogens targeted and the high sensitivity of the method itself. In our case, the impact of the result of the PCR for universal bacteria on the anti-infection strategy was crucial for the survival of the patient.
The isolated U. urealyticum strain designated CM239 showed resistance to ciprofloxacin and intermediate susceptibility to ofloxacin in vitro. Recent studies have addressed the question of the genetic background of quinolone-resistant Ureaplasma strains in the quinolone resistance-determining regions of the DNA gyrase- and topoisomerase IV-coding genes (gyrA, gyrB, parC, and parE) (2, 3, 12, 13), which are well known as hot spots of mutation in other bacteria. The D95E (GyrA) mutation of our strain has already been described for 1 (3), 11 (13), 8 (2), and 2 (12) quinolone-resistant isolates, respectively, of U. urealyticum. Interestingly, as in isolate CM239, this mutation was combined with A123T and A134T (both ParC) alterations in all 9 strains reported from France (2, 3) but in none of the 13 strains from China (12, 13). In summary, the genotype of Ureaplasma isolate CM239 has been linked to quinolone resistance in earlier studies and is in accordance with the elevated MICs of quinolone detected by using the mycoplasma IST kit. Of the Ureaplasma strains we isolated at our hospital from 1995 to 2006, 71% were ofloxacin susceptible (n = 395) and as few as 12% were ciprofloxacin susceptible (n = 95). The quinolone resistance of our patient may have been selected for by prior medication with quinolone, although he was exposed to ciprofloxacin only once during his hospital stay 2 months earlier (300 mg q8h for 11 days).
In the situation of a life-threatening infection, we decided to change therapy from moxifloxacin to chloramphenicol as soon as results of the in vitro antibiotic susceptibility test became available. Chloramphenicol, but not moxifloxacin, seemed to reach a high enough concentration in the CSF to eliminate Ureaplasma infection and resolve the inflammatory response which caused the central respiratory insufficiency (6). Hence, one has to keep in mind that chloramphenicol reaches high levels of active compound in the CSF, and despite its adverse side effects, it may still be a valuable choice for second- or third-line therapy for certain infections, especially severe meningitis (1). Interestingly, although it has not so far been reported that U. urealyticum secretes known activators of Toll-like receptor-induced cytokines such as IL-6, we found significantly increased levels of this and other cytokines.
U. urealyticum is well known as a cause of meningitis in newborns or premature infants (11), but it can be a causative agent of meningitis in adults as well, which may be overlooked as a consequence of its unusual growth requirements. Additionally, the obvious value of using PCR to detect any bacterial DNA in specimens that were previously culture negative cannot be overestimated, and this important procedure should be applied whenever the timely diagnosis of infectious diseases of unknown origin in critically ill persons is mandatory.
Nucleotide sequence accession numbers.
The sequences of the gyrA, gyrB, and parC quinolone resistance-determining regions of the U. urealyticum isolate CM239 have been deposited in the GenBank database under accession numbers EU27940 to EU27942.
Footnotes
Published ahead of print on 3 January 2008.
REFERENCES
- 1.Balbi, H. J. 2004. Chloramphenicol: a review. Pediatr. Rev. 25284-288. [DOI] [PubMed] [Google Scholar]
- 2.Bébéar, C. M., H. Renaudin, A. Charron, M. Clerc, S. Pereyre, and C. Bébéar. 2003. DNA gyrase and topoisomerase IV mutations in clinical isolates of Ureaplasma spp. and Mycoplasma hominis resistant to fluoroquinolones. Antimicrob. Agents Chemother. 473323-3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bebear, C. M., H. Renaudin, A. Charron, D. Gruson, M. Lefrancois, and C. Bebear. 2000. In vitro activity of trovafloxacin compared to those of five antimicrobials against mycoplasmas including Mycoplasma hominis and Ureaplasma urealyticum fluoroquinolone-resistant isolates that have been genetically characterized. Antimicrob. Agents Chemother. 442557-2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chapin, K. C., and T.-L. Lauderdale. 2003. Reagents, stains, and media: bacteriology, p. 365-366. In P. M. Murray, E. J. Baron, J. H. Jorgensen, M. A. Pfaller, and R. H. Yolken (ed.), Manual of clinical microbiology. ASM Press, Washington, DC.
- 5.Kong, F., Z. Ma, G. James, S. Gordon, and G. L. Gilbert. 2000. Species identification and subtyping of Ureaplasma parvum and Ureaplasma urealyticum using PCR-based assays. J. Clin. Microbiol. 381175-1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Matlow, A., C. Th'ng, D. Kovach, P. Quinn, M. Dunn, and E. Wang. 1998. Susceptibilities of neonatal respiratory isolates of Ureaplasma urealyticum to antimicrobial agents. Antimicrob. Agents Chemother. 421290-1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ponticelli, C., and M. R. Campise. 2005. Neurological complications in kidney transplant recipients. J. Nephrol. 18521-528. [PubMed] [Google Scholar]
- 8.Relman, D. A., T. M. Schmidt, R. P. MacDermott, and S. Falkow. 1992. Identification of the uncultured bacillus of Whipple's disease. N. Engl. J. Med. 327293-301. [DOI] [PubMed] [Google Scholar]
- 9.Robertson, J. A., G. W. Stemke, J. W. Davis, Jr., R. Harasawa, D. Thirkell, F. Kong, M. C. Shepard, and D. K. Ford. 2002. Proposal of Ureaplasma parvum sp. nov. and emended description of Ureaplasma urealyticum (Shepard et al. 1974) Robertson et al. 2001. Int. J. Syst. Evol. Microbiol. 52587-597. [DOI] [PubMed] [Google Scholar]
- 10.Tunkel, A. R., B. J. Hartman, S. L. Kaplan, B. A. Kaufman, K. L. Roos, W. M. Scheld, and R. J. Whitley. 2004. Practice guidelines for the management of bacterial meningitis. Clin. Infect. Dis. 391267-1284. [DOI] [PubMed] [Google Scholar]
- 11.Waites, K. B., B. Katz, and R. L. Schelonka. 2005. Mycoplasmas and ureaplasmas as neonatal pathogens. Clin. Microbiol. Rev. 18757-789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xie, X., and J. Zhang. 2006. Trends in the rates of resistance of Ureaplasma urealyticum to antibiotics and identification of the mutation site in the quinolone resistance-determining region in Chinese patients. FEMS Microbiol. Lett. 259181-186. [DOI] [PubMed] [Google Scholar]
- 13.Zhang, W., Y. Wu, W. Yin, and M. Yu. 2002. Study of isolation of fluoroquinolone-resistant Ureaplasma urealyticum and identification of mutant sites. Chin. Med. J. (Engl.) 1151573-1575. [PubMed] [Google Scholar]