Abstract
We compared multilocus variable-number tandem-repeat analysis (MLVA) and macrorestriction endonuclease analysis using pulsed-field gel electrophoresis (PFGE) to determine their utility to identify clusters of Clostridium difficile infection (CDI) among 91 isolates of PCR ribotype 027 (NAP1, for North American pulsed-field type 1) from nine hospitals (and 10 general practitioners associated with one institution) in England. We also examined whether mortality in CDI cases was associated with specific MLVA subtypes. PFGE discriminated between ribotype 027 strains at >98% similarity, identifying five pulsovars (I to V) of 1 to 53 isolates. MLVA was markedly more discriminatory, identifying 23 types of 1 to 15 isolates (>71% similarity). PFGE pulsovars I and IV contained 14 and 8 MLVA types, respectively. Twenty-one of twenty-three (91%) of MLVA types were specific to individual PFGE pulsovars. Four CDI clusters were identified in institution A by conventional epidemiological analysis. MLVA typing identified two enlarged and two additional clusters. Thirty of forty-four (68%) patients in institution A with CDI caused by ribotype 027 strains were assigned to seven distinct clusters by a combination of MLVA typing and epidemiological records. Of 33 patients, comprising 14 different MLVA types, nine (27%) died by day 30 (early deaths). Eight of nine (89%) were associated with PFGE type IV C. difficile ribotype 027. Five of nine early deaths were associated with MLVA type 16, which was the dominant type in this cohort (10/33 cases); 4 other distinct MLVA types accounted for the other early deaths. MLVA was far superior to PFGE for analyzing clusters of CDI both within and between institutions. Further study is needed to examine whether subtypes of C. difficile ribotype 027 affect outcome.
Hospital outbreaks of C. difficile infection (CDI) in Canada, starting in 2002 and 2003 and primarily centered on Quebec province, were associated with increased disease severity and mortality (7, 15, 20-22). Detailed analysis by Hubert et al. of these outbreaks showed that two strains predominated, C. difficile PCR ribotypes 027 (often referred to as North American pulsed-field type 1 [NAP1], or group BI by restriction endonuclease analysis [REA]) and 001; these strains were referred to as pulsovars A and B, respectively, as determined by pulsed-field gel electrophoresis (PFGE) (7). C. difficile PCR ribotype 027 and related strains (pulsovar A), all of which had binary toxin genes and a partial tcdC deletion, were twice as likely to cause severe CDI disease as strains that lacked these putative virulence factors (7). C. difficile PCR ribotype 027 is known to have spread throughout Europe and the United States (11, 12, 18). It has also been found in Japan, but more closely resembled the historical fluoroquinolone-susceptible PCR ribotype 027 (9). Since 2005, the United Kingdom has seen outbreaks of CDI caused by C. difficile PCR ribotype 027, which is closely related to the United States/Canadian epidemic strain, and this strain has been identified in more than 90 hospitals in England and Wales (11, 23).
The epidemic spread of C. difficile strains complicates the epidemiological investigation of clusters of CDI cases and nosocomial transmission unless fingerprinting methods can be employed that can discriminate between discrete C. difficile subtypes or clones. Multilocus variable-number tandem-repeat analysis (MLVA) is a highly discriminatory method that can be used to subtype clones of C. difficile (10, 17, 25). A recent international multilaboratory comparison of typing methods for the investigation of C. difficile outbreaks concluded that only REA and MLVA had sufficient discriminatory power to distinguish between strains from different outbreaks (10). We compared MLVA and PFGE to determine their utility to identify clusters of CDI cases among a large collection of C. difficile PCR ribotype 027 isolates and specifically to determine whether we could distinguish clusters between institutions and within individual hospitals. We also examined whether mortality in CDI cases was associated with specific MLVA subtypes.
MATERIALS AND METHODS
A total of 91 C. difficile isolates that had been identified in Leeds as PCR ribotype 027 were examined. These included 81 C. difficile isolates from C. difficile toxin-positive fecal samples that were submitted as part of the routine investigation of diarrhea in nine hospitals that were clustered in four different institutions (each institution was separated by at least 20 miles) in England. The largest group of (consecutive) isolates (n = 53) were recovered from patients in institution A as part of enhanced surveillance, which has been ongoing since November 2005 and included culture and ribotyping of every C. difficile cytotoxin-positive diarrheal fecal sample. These 53 isolates were from 43 patients (34 patients with 1 isolate each, 8 with 2 isolates each, and 1 patient with three isolates). The fecal samples for institutions B, C, and D were referred for ribotyping as part of local investigations of clusters of cases of either increased frequency or severity of CDI. Ten C. difficile isolates were examined that had been recovered from samples submitted from nine different patients by their general practitioners; these general practitioners were situated in the geographical area served by institution A.
PCR ribotyping.
PCR ribotyping was performed on all C. difficile isolates in Leeds as described previously (24), with modifications. Briefly, bacteria were harvested from 48-h-old cultures on modified Brazier's cycloserine-cefoxitin-egg yolk agar (CCEYL) (BioConnections, Bardsey, United Kingdom), with the omission of egg yolk and supplemented with 5 mg/ml lysozyme. Each isolate was resuspended in 5% (wt/vol) Chelex-100 solution (Bio-Rad, Hertfordshire, United Kingdom), boiled for 10 min, and centrifuged (13,000 rpm, 2 min). Ten microliters of supernatant was added to a 90-μl PCR mixture containing 50 pmol of primer mix 5′-CTGGGGTGAAGTCGTAACAAGG-3′ and 5′-GCGCCCTTTGTAGCTTGACC-3′, 2 U of Taq polymerase, and 2.25 mM MgCl2 (Sigma, Poole, United Kingdom). PCR mixtures were subjected to a thermal cycling program consisting of 35 cycles at 94°C for 1 min, 55°C for 1 min, and 72°C for 2 min. Amplimer was concentrated to an approximate final volume of 25 μl by heating at 75°C before electrophoresis (150 mA) in 3% MetaSeive agarose (Flowgen, Ashby de la Zouch, United Kingdom). A 100-bp molecular standard (Fermentas, York, United Kingdom) was used for data normalization. Agarose gels were soaked in ethidium bromide solution, and DNA profiles were visualized and documented using the GeneGenius bioimaging system (Syngene, Cambridge, United Kingdom).
Macrorestriction endonuclease analysis using PFGE.
Macrorestriction endonuclease analysis using PFGE was performed in Leeds, United Kingdom, on all C. difficile isolates as described previously (3). Briefly, isolates were cultured anaerobically in prereduced Schaedler's anaerobic broth (Oxoid, Basingstoke, United Kingdom) overnight at 37°C. Cells were harvested from 5 ml broth culture by centrifugation, and the resultant pellets were washed twice in 5 ml sterile phosphate-buffered saline and resuspended in 100 μl lysis buffer (10 mM Tris, 0.5 mM EDTA, 0.8% N-lauryl sarcosine, 5 mg/ml lysozyme). This suspension was cast into 1% (wt/vol) agarose plugs and immersed in lysis buffer for 1 h at 37°C and then 1 ml ESP buffer (0.5 mM EDTA, 1% N-lauryl sarcosine, 10 mg/ml proteinase K) for 22 h at 50°C (with ESP buffer refreshed after 16 h). The plugs were washed four times in TE buffer (10 mM Tris, 1 mM EDTA). DNA was digested with 20 U of SmaI restriction enzyme for 5 h at 30°C. The digestion products were separated in a 1% PFGE-grade agarose gel using CHEF II PulseMaster PFGE apparatus (Bio-Rad, Hertfordshire, United Kingdom). Digestion products were exposed to a field strength of 6 V/cm, with linear ramping from 5 s to 55 s, over 21 h. A lambda DNA concatemer (Bio-Rad) was used for data normalization. Agarose gels were soaked in ethidium bromide solution, and DNA profiles were visualized and documented using the GeneGenius bioimaging system (Syngene, Cambridge, United Kingdom).
MLVA fingerprinting.
All C. difficile isolates were typed by MLVA in Leiden, The Netherlands, using automated fragment analysis and multicolored capillary electrophoresis, as described previously (25). MLVA typing was carried out with the investigators blinded to the origin of the isolates. Briefly, seven regions with short tandem repeats spread over the genome, designated as C. difficile MLVA markers A6, B7, C6, E7, F3, G8, and H9, were used. For marker G8, a new reverse primer was developed and used in this study: 5′ ACCAAAAATTTCTAACCCAAC 3′. Three separate duplex PCRs (MLVA A6-H9, B7-F3, and C6-E7) and one singleplex PCR (MLVA G8) were developed. The repeats were amplified using a single PCR protocol. The forward primer of each PCR was labeled at the 5′ end with either carboxyfluorescein (FAM), hexachlorofluorescein (HEX), 2′-chloro-7′-phenyl-1,4-dichloro-6-carboxyfluorescein (VIC), or 2′-chloro-5′-fluoro-7′,8′-fused phenyl-1,4-dichloro-6-carboxyfluorescein (NED). PCR fragments were analyzed using multicolored capillary electrophoresis on an ABI3100 genetic analyzer, with a ROX500 marker as an internal marker for each sample. The size of each marker was determined by Genescan software (Applied Biosystems).
Analysis of PFGE and MLVA profiles.
Molecular typing results were analyzed using BioNumerics software (Applied Maths, Kortrijk, Belgium). For PFGE, dendrograms were constructed by the unweighted-pair group method with arithmetic mean clustering (UPGMA), using the Dice correlation coefficient (data not shown). A threshold of 98% similarity was used to assign pulsovars. For MLVA, the repeat numbers were analyzed using UPMGA with the multistate categorical similarity coefficient. All markers were given equal weight, irrespective of the number of repeats. Subsequently, if two strains had an equal number of repeats in five of seven markers (i.e., 71% similar) they were considered to be closely related. This threshold of similarity was based on the previous results of 027 typing studies (17, 25).
Epidemiological and follow-up data.
Case clusters in time and place were identified by examination of microbiology, infection control, and hospital databases and individual patient medical records. Specifically, if CDI patients overlapped during their hospital stay (i.e., on the same specific hospital ward) with at least one other patient, they were classified as being part of a cluster. MLVA-defined clusters met these criteria, but additionally shared the same MLVA type. In the light of MLVA data implicating additional hitherto nonclustered cases, the records of the dates of patient stay and their locations were rechecked, including verification of the accuracy of location databases; this was carried out by review of individual ward/unit records. There was no fixed time period used prospectively to define a cluster as the length of stays of individual patients varied. Mortality data were obtained for 33/44 patients from institution A by review of patient medical records and death certificates. Early deaths were defined as those occurring within 30 days of the diagnosis of CDI.
RESULTS
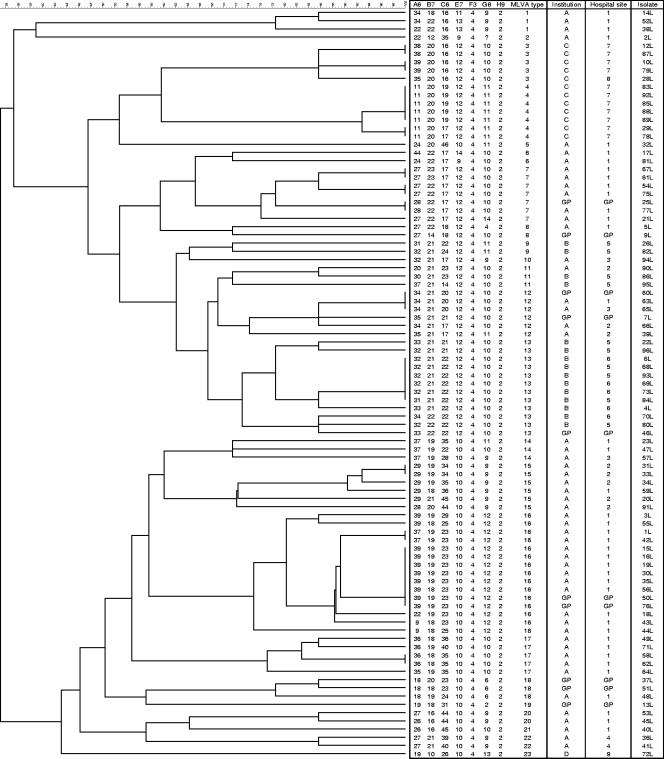
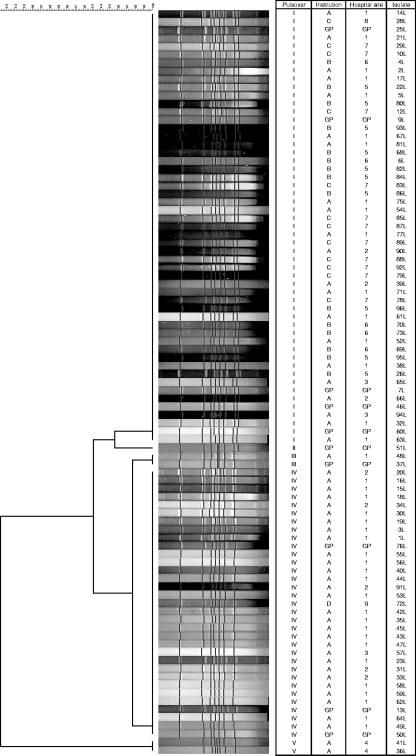
Ninety-one C. difficile isolates were examined from a total of 80 patients with a median age of 73 years (range, 21 to 99 years). Table 1 shows the distribution of C. difficile isolates, MLVA types (Fig. 1), and PFGE pulsovars (Fig. 2) according to hospital and institution. PFGE discriminated between C. difficile ribotype 027 strains at >98% similarity. On this basis, PFGE identified five pulsovars (I to V) of 53, 1, 2, 33, and 2 isolates, respectively (Fig. 2). Using a threshold of >71% similarity, MLVA was markedly more discriminatory than PFGE, identifying 23 types of 1 to 15 isolates (Table 1). PFGE pulsovars I and IV contained 14 and 8 MLVA types, respectively. Six C. difficile MLVA types were represented by a single C. difficile isolate. The great majority (21/23 [91%]) of MLVA types were specific to individual PFGE pulsovars. Thus, only MLVA types 17 and 18 were associated with isolates in more than one PFGE pulsovar (I [n = 1] and IV [n = 4]; and II [n = 1] and III [n = 2], respectively).
TABLE 1.
Distribution of C. difficile isolates, MLVA types, and PFGE pulsovars according to hospital and institution
| Institution | Hospital | No. of patients | No. of C. difficile isolates | PFGE pulsovar(s) (no. of isolates) | MLVA type(s) (no. of isolates) |
|---|---|---|---|---|---|
| A | 1 | 32 | 40 | I (16), III (1), IV (23) | 1 (3), 2 (1), 5 (1), 6 (2), 7 (6), 8 (1), 12 (2), 14 (2), 16 (13), 17 (5), 18 (1), 20 (2), 21 (1) |
| 2 | 8 | 8 | I (3), IV (5) | 11 (1), 12 (1), 15 (6) | |
| 3 | 3 | 3 | I (2), IV (1) | 10 (1), 12 (1), 14 (1) | |
| 4 | 1 | 2 | V (2) | 22 (2) | |
| B | 5 | 10 | 10 | I (10) | 9 (2), 11 (2), 13 (6) |
| 6 | 5 | 5 | I (5) | 13 (5) | |
| C | 7 | 11 | 11 | I (11) | 3 (4), 4 (7) |
| 8 | 1 | 1 | I (1) | 3 (1) | |
| D | 9 | 1 | 1 | IV (1) | 23 (1) |
| General practices | NAa | 10 | 9 | I (5), II (1), III (1), IV (3) | 7 (1), 8 (1), 12 (2), 13 (1), 16 (2), 18 (2), 19 (1) |
NA, not applicable.
FIG. 1.
Dendrogram for 91 C. difficile 027 isolates to indicate relatedness according to seven MLVA markers. Strains 20L and 34L have PCR-ribotype profiles that are very similar to those for ribotype 036, but are epidemiologically related to and cluster closely with other 027 isolates studied here. Further investigation of these isolates is ongoing. GP, general practitioner.
FIG. 2.
Dendrogram for 91 C. difficile 027 isolates to indicate relatedness according to macrorestriction endonuclease profile using PFGE. Strains 20L and 34L have PCR-ribotype profiles that are very similar to those for ribotype 036, but are epidemiologically related to and cluster closely with other 027 isolates studied here. Further investigation of these isolates is ongoing. GP, general practitioner.
Disregarding C. difficile isolates from GP samples (as they do not necessarily relate to specific hospitals), we found that 17/22 (77%) MLVA types clustered exclusively to a single hospital (Table 1). Furthermore, 21/22 (95%) MLVA types clustered exclusively to an institution. The one exception was C. difficile MLVA type 11, which was isolated from one and two patients in institutions A and B, respectively. We are unaware of any epidemiological link between these cases.
Clusters of C. difficile infection in time and place.
We provisionally identified five clusters of C. difficile infection in time and place in institution A by prospective surveillance (i.e., before the results of MLVA typing were available). MLVA typing grouped 3/3, 3/4, 2/3, 2/3, and 10/11 of these cases in the gut surgery (MLVA type 15), neurosurgery (MLVA type 15), hematology (MLVA type 7), renal medicine (MLVA type 17), and elderly medicine (MLVA type 16) specialities, respectively. We reanalyzed the epidemiological data for cases of C. difficile infection in institution A in the light of the MLVA typing results. MLVA typing results confirmed the provisionally identified case clusters and additionally showed hitherto unrecognized epidemiological links (in time and place) between other cases. Thus, taking into account the results of MLVA typing we identified two enlarged and two additional clusters of cases of C. difficile infection as follows: hematology (n = 5), renal medicine (n = 4), respiratory medicine (n = 2; MLVA type 20), and general surgery (n = 2; MLVA type I), compared with those identified prospectively. Individual clusters in institution A occurred over a maximum of 12 weeks. Overall, 30 out of 44 patients in institution A with infection caused by C. difficile ribotype 027 strains could be connected by a combination of MLVA typing and epidemiological records. These 30 cases comprised a total of seven distinct clusters (ranging from 2 to 11 cases).
Two of the (epidemiologically unlinked) clusters, in gut surgery (hospital 1) and neurosurgery (hospital 2), were both caused by C. difficile MLVA type 15. C. difficile MLVA type 14 was found in three patients distributed across two hospitals in institution A. There was no epidemiological link between these cases. Interestingly, C. difficile MLVA type 14 was the first PCR ribotype 027 strain identified in this institution. The patient was a transfer from a hospital in an area of the United Kingdom that had endemic C. difficile ribotype 027. Similarly, C. difficile MLVA type 12 was found in four patients distributed across three hospitals in institution A. There was no epidemiological link between these cases. Of nine patients from whom more than one C. difficile isolate was fingerprinted, seven had the same MLVA type; the respective patient isolates were recovered up to 2 months apart. Two in-patients had C. difficile isolates with different MLVA types recovered (from cytotoxin-positive samples) 2 weeks and 6 months apart; tandem repeat data for each isolate pair differed in three of seven loci. Both of these patients had multiple ward stays/transfers in the periods around when the distinct MLVA type C. difficile isolates were recovered.
C. difficile 027 isolates from general practitioner samples.
The 10 C. difficile isolates that were recovered from general practitioner patient fecal samples came from nine patients (both isolates from the patient with two isolates were MLVA type 18 and were isolated 6 weeks apart). Of these nine patients, eight had been hospitalized in institution A within the previous 3 months. Case cluster analysis showed that six of these eight patients were related in time and space to other patients that shared the same MLVA types. The two remaining patients had distinct MLVA types that were not seen in any other patients in institution A. There was a solitary patient who also had a unique C. difficile MLVA type (type 19) isolated from a cytotoxin-positive sample that was submitted by the general practitioners after a 3-week history of antibiotic-associated diarrhea. The patient had not been admitted to institution A previously, and there was no general practitioner record of recent hospitalization at any other institution.
Mortality and C. difficile MLVA/PFGE type.
Of 33 patients, comprising 14 different MLVA types, that were found in institution A, 9 (27%) died by day 30 (early deaths). Eight of nine (89%) cases were associated with PFGE pulsovar IV C. difficile ribotype 027. Five of nine early deaths were associated with MLVA type 16, which was the dominant type in this cohort (10/33 cases); 4 other distinct MLVA types accounted for the other early deaths. We were unable to analyze patient deaths according to MLVA type in the other institutions because of a lack of follow-up data for these cases.
DISCUSSION
Several epidemic C. difficile strains have been recognized, including C. difficile PCR ribotypes 001, 027, and 017 (1-3, 7, 15, 18, 24). Investigation of the spread of such strains within individual hospitals is poorly understood, not least because the different fingerprinting methods commonly used for C. difficile have generally lacked discriminatory power. For example, Hubert and colleagues recently found that although PFGE of 478 C. difficile isolates yielded 61 PFGE profiles, three pulsovars accounted for 57%, 10%, and 8% of these: i.e., 75% of the entire collection (7). The predominant pulsovar (A) was identical to that of strain NAP1, but the distribution and spread of clones within this epidemic pulsovar type, which was significantly associated with poor outcome, was not described.
C. difficile PCR ribotype 027 has spread dramatically between and within institutions in multiple countries, including those with traditionally excellent infection control capacity to minimize nosocomial transmission of pathogens (7, 9, 11, 12, 15, 18). This strain has 18- and 1-bp deletions in tdcC, a gene that negatively regulates toxin production. These deletions, particularly the latter that results in a frameshift, are thought to cause premature cessation of tdcC transcription, resulting in increased transcription of the toxin genes due to the lack of down-regulation (16). We have shown in the human gut model that the duration of germination and toxin production by C. difficile ribotype 027 is significantly longer (5). The significance of the binary toxin produced by C. difficile ribotype 027 strains remains uncertain. However, a toxin A-negative, toxin B-negative, binary toxin-positive strain induced fluid secretion in rabbit ileal loops but was not pathogenic (no diarrhea or death) in a hamster model (6). C. difficile ribotype 027 (and 001) sporulates more frequently than other strains, which may contribute to survival and spread (4, 26). These potential virulence determinants may account for the increased diarrheal symptoms that are seen in some patients with C. difficile infection caused by ribotype 027, which in turn may promote spread and cross-infection within health care institutions. Institution A in the present study continues to experience sporadic cases, with occasional clusters, of CDI caused by C. difficile ribotype 027. Using a combination of conventional infection control measures, antibiotic restriction, and intensive surveillance, we have to date kept the prevalence of this ribotype to ∼5% over a 2-year period.
In an earlier study, we found that MLVA could discriminate between all the reference strains that were examined, which included 31 serogroups, 25 toxinotypes, and 7 subtypes of PCR ribotype 001 (25). A group of 28 isolates of PCR ribotype 027 collected from outbreaks in The Netherlands were separated into 13 clusters. Additionally, 29 toxin A-negative, toxin B-positive isolates belonging to PCR ribotype 017 were separated into country-specific clusters. Marsh et al., using dissimilar MLVA targets and a different analysis method, also found that MLVA was potentially useful for tracking nosocomial C. difficile infections (17). These findings have been extended in our present study where we examined a considerably larger collection of PCR ribotype 027 isolates.
We found that MLVA was far superior to PFGE (23 versus 5 subtypes) for analyzing clusters of CDI cases both within a single institution and between hospitals. Interestingly, the two dominant PFGE pulsovars that we identified (I and IV, which accounted for 95% of the isolates examined) were indistinguishable from isolates that we obtained from the United States and a major United Kingdom outbreak, respectively (18). We found that MLVA identified clustering of C. difficile ribotype 027 subtypes within hospitals and institutions. In contrast, distinct subtypes were usually seen in geographically discrete institutions. We provisionally identified five clusters of C. difficile infection in time and place in institution A using conventional prospective surveillance. MLVA typing results confirmed these findings. Crucially, however, MLVA also showed that two of these five initial clusters were larger than originally suspected and identified two additional clusters of cases of C. difficile infection. We emphasize that considerable time and resources may be expended when searching for possible epidemiological links between nosocomial CDI cases. Use of a highly discriminatory fingerprinting method such as MLVA has the potential to ease this process and at the same time increase the sensitivity and specificity of the detection of linked cases.
MLVA analysis of C. difficile ribotype 027 isolates recovered from community patients showed that the majority of patients (6/8) were associated with other hospitalized patients; all but one patient had been hospitalized within the previous 3 months. Two patients recently discharged from hospital had distinct MLVA types that were not seen in any other patients. One community patient, from whom we isolated a unique C. difficile MLVA type, had antibiotic-associated diarrhea but no record of recent hospitalization. Thus, this case may represent a case of community-acquired CDI caused by ribotype 027.
For all 91 type 027 isolates in this study, markers F3 and H9 contained four and two repeats, respectively. Marker E7 showed 10 to 12 repeats for 84 of 89 (94.4%), marker G8 contained 9 to 12 repeats in 81 (91.0%) of 89 samples, and 18 to 22 repeats for marker B7 in 81 (91.0%) of 89 samples. This was comparable to the results in the study by Killgore et al. (10). Type 027 isolates showed the same number of repeats for markers F3 and H9. Marker E7 had 10 repeats for 16 of 22 (72.7%) samples. Of interest is how subtypes emerge within a clone. We previously reported that multiple subcultures of a C. difficile 027 strain (n = 30) resulted in a difference of only one repeat unit between strains for markers A6 and C6 (25). Based on these stability tests, it was concluded that a difference of 1 or 2 repeat units between strains should not be interpreted as indicative of separate types or subtypes, which is also in complete concordance with the study of the stability of C. difficile MLVA loci by Marsh et al. (17). Using our epidemiological data to analyze isolates, we conclude that differences of up to two repeat units (i.e., >71% similarity) indicate closely related strains (i.e., members of the same MLVA type).
Killgore et al. recently examined the discriminatory power of seven DNA fingerprinting techniques when applied to 42 prevalent C. difficile strains collected in four countries, including 22 C. difficile ribotype 027 isolates (referred to as allelic profile B by the authors) (10). Four methods, MLVA, REA, slpA sequence typing, and PFGE, recognized subtypes under primary types, while the other four methods, United Kingdom and United States PCR-ribotyping, multilocus sequence typing, and amplified fragment length polymorphism, did not identify subtypes. Only REA and MLVA had sufficient power to distinguish strains from different outbreaks and to discriminate between North America and European CD 027 isolates. MLVA and REA subtyped CD 027 strains into 11 and 8 different types, respectively. Pasqualotto et al. recently reported that there may be size differences of up to 6 bases between sequencing data and sizing by capillary electrophoresis and as many as 3-base differences between different capillary electrophoresis instruments (19). This could limit the interlaboratory reproducibility of MLVA data, unless a correction factor or standardized equipment is used. Alternatively, sample products could be sequenced to check for such variability.
We found that 27% of patients for whom follow-up data were available died within 30 days of the diagnosis of CDI. The most common PFGE pulsovar/MLVA-type C. difficile isolate associated with these deaths was the predominant strain causing infection in these patients. Furthermore, a total of five distinct MLVA types of C. difficile were the cause of infection in these fatal cases. Thus, we could not show that increased likelihood of death was associated with a specific MLVA-type or PFGE pulsovar, in contrast to the finding by Hubert et al. (7). It is likely that the risk of death reflects pathogen virulence, the presence of risk factors such as multiple antibiotic exposure, and host susceptibility, for example, related to humoral immune response to C. difficile toxin A and host interleukin-8 AA genotype (8, 13, 14). Further study is needed to examine the contribution of C. difficile strains and, specifically, subtypes of C. difficile ribotype 027, to outcome.
In summary, we recommend that MLVA be used to characterize and improve the understanding of the transmission of epidemic strains within and between health care institutions. More research is needed to understand the clonal spread and long-term epidemiology of epidemic C. difficile strains. Furthermore, MLVA has the potential to improve our understanding of the interplay between community and health care-associated C. difficile strains.
Acknowledgments
As possible conflicts of interest, M.H.W. has received honoraria for consultancy work, financial support to attend meetings, and research funding from Astra-Zeneca, Bayer, Cerexa, Genzyme, Nabriva, Pfizer, Targanta, Vicuron, and Wyeth.
Footnotes
Published ahead of print on 23 January 2008.
REFERENCES
- 1.Brazier, J. S., B. Patel, and A. Pearson. 2007. Distribution of Clostridium difficile PCR ribotype 027 in British hospitals. Eur. Surveill. 12(4):E070426.2. http://www.eurosurveillance.org/ew/2007/070426.asp#2. [DOI] [PubMed] [Google Scholar]
- 2.Drudy, D., N. Harnedy, S. Fanning, M. Hannan, and L. Kyne. 2007. Emergence and control of fluoroquinolone-resistant, toxin A-negative, toxin B-positive Clostridium difficile. Infect. Control Hosp. Epidemiol. 28932-940. [DOI] [PubMed] [Google Scholar]
- 3.Fawley, W. N., P. Parnell, P. Verity, J. Freeman, and M. H. Wilcox. 2005. Molecular epidemiology of endemic Clostridium difficile infection and the significance of subtypes within the United Kingdom epidemic strain (PCR ribotype 1). J. Clin. Microbiol. 432685-2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fawley, W. N., S. Underwood, J. Freeman, S. D. Baines, K. Saxton, K. Stephenson, R. C. Owens, Jr., and M. H. Wilcox. 2007. Efficacy of hospital cleaning agents and germicides against epidemic Clostridium difficile strains. Infect. Control Hosp. Epidemiol. 28920-925. [DOI] [PubMed] [Google Scholar]
- 5.Freeman, J., S. D. Baines, K. Saxton, and M. H. Wilcox. 2007. Effect of metronidazole on growth and toxin production by epidemic Clostridium difficile PCR ribotypes 001 and 027 in a human gut model. J. Antimicrob. Chemother. 6083-91. [DOI] [PubMed] [Google Scholar]
- 6.Geric, B., R. J. Carman, M. Rupnik, C. W. Genheimer, S. P. Sambol, D. M. Lyerly, D. N. Gerding, and S. Johnson. 2006. Binary toxin-producing, large clostridial toxin-negative Clostridium difficile strains are enterotoxic but do not cause disease in hamsters. J. Infect. Dis. 1931143-1151. [DOI] [PubMed] [Google Scholar]
- 7.Hubert, B., V. G. Loo, A. M. Bourgault, L. Poirier, A. Dascal, E. Fortin, M. Dionne, and M. Lorange. 2007. Portrait of the geographic dissemination of the Clostridium difficile North American pulsed-field type 1 strain and the epidemiology of C. difficile-associated disease in Quebec. Clin. Infect. Dis. 44238-244. [DOI] [PubMed] [Google Scholar]
- 8.Jiang, Z. D., K. W. Garey, M. Price, G. Graham, P. Okhuysen, T. Dao-Tran, M. Larocco, and H. L. Dupont. 2007. Association of interleukin-8 polymorphism and immunoglobulin G anti-toxin A in patients with Clostridium difficile-associated diarrhea. Clin. Gastroenterol. Hepatol. 5964-968. [DOI] [PubMed] [Google Scholar]
- 9.Kato, H., Y. Ito, R. J. van den Berg, E. J. Kuijper, and Y. Arakawa. 2007. First isolation of Clostridium difficile 027 in Japan. Eur. Surveill. 12(1)E070111.3. http://www.eurosurveillance.org/ew/2007/070111.asp#3. [DOI] [PubMed] [Google Scholar]
- 10.Killgore, G., A. Thompson, S. Johnson, J. Brazier, E. Kuijper, J. Pepin, P. Savelkoul, B. Nicholson, R. J. van den Berg, H. Kato, C. Woods, D. Gerding, and C. McDonald. 26 November 2007. Comparison of seven techniques for typing international epidemic strains of Clostridium difficile: REA, PFGE, PCR-ribotyping, MLST, MLVA, AFLP, and slpAST. J. Clin. Microbiol. doi: 10.1128/JCM.01484-07. [DOI] [PMC free article] [PubMed]
- 11.Kuijper, E., B. Coignard, J. Brazier, C. Suetens, D. Drudy, C. Wiuff, H. Pituch, P. Reichert, F. Schneider, A. F. Widmer, K. E. Olsen, F. Allerberger, D. W. Notermans, F. Barbut, M. Delmée, M. Wilcox, A. Pearson, B. C. Patel, D. J. Brown, R. Frei, T. Åkerlund, I. R. Poxton, and P. Tüll. 2007. Update of Clostridium difficile-associated disease due to PCR ribotype 027 in Europe. Eur. Surveill. 12(6). http://www.eurosurveillance.org/em/v12n06/1206-221.asp. [DOI] [PubMed] [Google Scholar]
- 12.Kuijper, E. J., B. Coignard, and P. Tull. 2006. Emergence of Clostridium difficile-associated disease in North America and Europe. Clin. Microbiol. Infect. 12(Suppl. 6)2-18. [DOI] [PubMed] [Google Scholar]
- 13.Kyne, L., M. Warny, A. Qamar, and C. P. Kelly. 2000. Asymptomatic carriage of Clostridium difficile and serum levels of IgG antibody against toxin A. N. Engl. J. Med. 342390-397. [DOI] [PubMed] [Google Scholar]
- 14.Kyne, L., M. Warny, A. Qamar, and C. P. Kelly. 2001. Association between antibody response to toxin A and protection against recurrent Clostridium difficile infection. Lancet 357189-193. [DOI] [PubMed] [Google Scholar]
- 15.Loo, V. G., L. Poirier, M. A. Miller, M. Oughton, M. D. Libman, S. Michaud, A. M. Bourgault, T. Nguyen, C. Frenette, M. Kelly, A. Vibien, P. Brassard, S. Fenn, K. Dewar, T. J. Hudson, R. Horn, P. Rene, Y. Monczak, and A. Dascal. 2005. A predominantly clonal multi-institutional outbreak of Clostridium difficile-associated diarrhea with high morbidity and mortality. N. Engl. J. Med. 3532442-2449. [DOI] [PubMed] [Google Scholar]
- 16.MacCannell, D. R., T. J. Louie, D. B. Gregson, M. Laverdiere, A. C. Labbe, F. Laing, and S. Henwick. 2006. Molecular analysis of Clostridium difficile PCR ribotype 027 isolates from eastern and western Canada. J. Clin. Microbiol. 442147-2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marsh, J. W., M. M. O'Leary, K. Shutt, A. W. Pasculle, S. Johnson, D. N. Gerding, C. A. Muto, and L. H. Harrison. 2006. Multilocus variable-number tandem-repeat analysis for investigation of Clostridium difficile transmission in hospitals. J. Clin. Microbiol. 442558-2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McDonald, L. C., G. E. Killgore, A. Thompson, R. C. Owens, Jr., S. V. Kazakova, S. P. Sambol, S. Johnson, and D. N. Gerding. 2005. An epidemic, toxin gene-variant strain of Clostridium difficile. N. Engl. J. Med. 3532433-2441. [DOI] [PubMed] [Google Scholar]
- 19.Pasqualotto, A. C., D. W. Denning, and M. J. Anderson. 2007. A cautionary tale: lack of consistency in allele sizes between two laboratories for published multilocus microsatellite typing system. J. Clin. Microbiol. 45522-528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pepin, J., M. E. Alary, L. Valiquette, E. Raiche, J. Ruel, K. Fulop, D. Godin, and C. Bourassa. 2005. Increasing risk of relapse after treatment of Clostridium difficile colitis in Quebec, Canada. Clin. Infect. Dis. 401591-1597. [DOI] [PubMed] [Google Scholar]
- 21.Pepin, J., L. Valiquette, M. E. Alary, P. Villemure, A. Pelletier, K. Forget, K. Pepin, and D. Chouinard. 2004. Clostridium difficile-associated diarrhea in a region of Quebec from 1991 to 2003: a changing pattern of disease severity. Can. Med. Assoc. J. 171466-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pepin, J., L. Valiquette, and B. Cossette. 2005. Mortality attributable to nosocomial Clostridium difficile-associated disease during an epidemic caused by a hypervirulent strain in Quebec. Can. Med. Assoc. J. 1731037-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smith, A. 2005. Outbreak of Clostridium difficile infection in an English hospital linked to hypertoxin-producing strains in Canada and the US. Eur. Surveill. 10(6)E050630.2. http://www.eurosurveillance.org/ew/2005/050630.asp#2. [DOI] [PubMed] [Google Scholar]
- 24.Stubbs, S. L., J. S. Brazier, G. L. O'Neill, and B. I. Duerden. 1999. PCR targeted to the 16S-23S rRNA gene intergenic spacer region of Clostridium difficile and construction of a library consisting of 116 different PCR ribotypes. J. Clin. Microbiol. 37461-463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van den Berg, R. J., I. Schaap, K. E. Templeton, C. H. W. Klaassen, and E. J. Kuijper. 2007. Typing and subtyping of Clostridium difficile isolates by using multiple-locus variable-number tandem-repeat analysis. J. Clin. Microbiol. 451024-1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wilcox, M. H., and W. N. Fawley. 2000. Hospital disinfectants and spore formation by Clostridium difficile. Lancet 3561324. [DOI] [PubMed] [Google Scholar]