Abstract
The common respiratory viruses, including influenza A, influenza B, and newly emerging severe acute respiratory syndrome (SARS) viruses, may cause similar clinical symptoms. Therefore, differential diagnosis of these virus pathogens is frequently required for single clinical samples. In addition, there is an urgent need for noninfectious and stable RNA standards and controls for multivirus detection. In this study, reverse transcription-PCR (RT-PCR) targeting of the RNAs of influenza A and influenza B viruses and SARS coronavirus was performed, and the resulting products were spliced into a fragment which was packaged into armored RNA for use as a noninfectious, quantifiable synthetic substitute. Furthermore, in the present study we developed a multiplex real-time RT-PCR assay in which the armored RNA was used as an external positive control and the three RNA viruses could be detected simultaneously in a single reaction mix. The detection limit of the multiplex real-time PCR was 10 copies/μl of armored RNA.
Respiratory infections are causes of upper and lower respiratory tract diseases, mainly causing croup, bronchiolitis, and pneumonia (11). Influenza virus infection is a highly contagious respiratory disease that can spread easily by droplets or nasal secretions and is responsible for considerable morbidity and mortality each year (18). Due to virus evolution and migration, the transmission of respiratory viruses between animals and humans has been a very severe matter. In particular, the highly pathogenic influenza virus and the severe acute respiratory syndrome coronavirus (SARS-CoV) have posed critical threats to public health. SARS-CoV infection is a transmissible, febrile, and newly emerging respiratory disease. Reverse transcription-PCR (RT-PCR) has been found to be sensitive, specific, and rapid for detection of influenza and SARS viruses (8, 18). External positive controls (EPC) are important components of nucleic acid-based assays. The lack of safe and stable EPC could become a serious problem in cases of SARS and influenza, where cross-infection could sometimes happen during sample exchange between laboratories (4).
Armored RNA (AR) is an engineered MS2 phage assembly that encapsulates an RNA fragment of a target gene after expression in Escherichia coli. AR is a noninfectious RNA virus surrogate and can be designed for use as a calibration standard or external control for RT-PCR or real-time RT-PCR. AR is resistant to RNase digestion and shows high stability in plasma, nasopharyngeal secretions, feces, and water (3, 9, 12, 15, 19). Due to the extreme lability of RNA, AR controls have been used widely (1, 3-7, 9, 12, 14, 16, 19). At present, a >1,200-nucleotide (nt) heterologous RNA can be accommodated and modified into AR (14).
Most current AR package the RNA of a single respiratory virus (4, 5, 14). Since the common respiratory viruses may cause similar clinical signs and symptoms, differential diagnosis of pathogens is therefore required. To reduce costs and simplify multivirus detection procedures, AR derived from three respiratory viruses, namely, influenza A virus, influenza B virus, and SARS-CoV, were designed, and the resulting single AR species might be used as a positive control for detecting the viruses. Furthermore, we developed a multiplex real-time RT-PCR assay in which the AR was used as an EPC and the three RNA viruses could be detected in a single reaction tube simultaneously.
MATERIALS AND METHODS
Primers and probes.
Primers and probes were designed to amplify highly conserved regions of the matrix protein (M) gene of influenza A virus (Am fragment) and the M gene of influenza B virus (Bm fragment). Primers and probes were optimized with Beacon Designer, version 2.0. The primers and probe for SARS-CoV were synthesized to amplify open reading frame (ORF) 1b of the RNA polymerase gene, as previously described (8). The primers and probes are showed in Table 1. The reporter dye FAM (5′-6-carboxyfluorescein), HEX (5′-6-carboxy-2′,4,4′,5′,7,7′-hexachlorofluorescein), or ROX (6-carboxyrhodamine) was conjugated at the 5′ end of each probe, and the quencher dye 6-carboxytetramethylrhodamine (TAMRA) was conjugated at the 3′ end. The primers used for splicing by overlap extension (SOE) by PCR and the other primers used in this study are also listed in Table 1.
TABLE 1.
Nucleic acid sequences of primers and probes
| Primer or probe | Sequence (5′→3′) | Target fragmenta | Note on underlined sequence |
|---|---|---|---|
| MS2F | ATGGATCCTCCTGCTCAACTTCCTGTCG | MS2g1, MS2g2, and pac site | BamHI restriction site |
| MS2R | GCAAGCTTTCTTCGACATGGGTAATCCT | MS2g1, MS2g2, and pac site | HindIII restriction site |
| BN IoutS2-SOE | AAGGCAAAGCAGATGAATTACCAAGTCAATGGTTAC | BNI 1 | Overlap sequence identical to Bm fragment |
| BN IoutAS-SOE | GTCAAGCTTCATAACCAGTCGGTACAGCTAC | BNI 1 | HindIII restriction site |
| AM119F-SOE | CTCAAGCTTAGGCACTCATGGAATGGCTAAAG | Am | HindIII restriction site |
| AM248R-SOE | TAAGAAAGTGCCGCATTTTGGACAAAGCGTCTAC | Am | Overlap sequence complementary to Bm fragment |
| BM11F-SOE | TGTCCAAAATGCGGCACTTTCTTAAAATGTCGCT | Bm | Overlap sequence identical to Am fragment |
| BM91R-SOE | TGGTAATTCATCTGCTTTGCCTTCTCCATCT | Bm | Overlap sequence complementary with SARS BNI 1 fragment |
| BN IoutS2 | ATGAATTACCAAGTCAATGGTTAC | BNI 1 | |
| BN IoutAS | CATAACCAGTCGGTACAGCTAC | BNI 1 | |
| BN IinS | GAAGCTATTCGTCACGTTCG | BNI 1 | |
| BN IinAS | CTGTAGAAAATCCTAGCTGGAG | BNI 1 | |
| Am119F | AGGCWCTCATGGARTGGCTAAAGb | Am | |
| Am248R | GCATTYTGGACAAAKCGTCTACc | Am | |
| pAm221 | HEX-AGTCCTCGCTCACTGGGCACGGT-TAMRA | Am | |
| Bm11F | GGCACTTTCTTAAAATGTCGCT | Bm | |
| Bm91R | CTGCTTTGCCTTCTCCATCT | Bm | |
| pBm36 | FAM-TGGAGACACAATTGCCTACCTGCTTTCA-TAMRA | Bm | |
| SARS1 | TTATCACCCGCGAAGAAGCT | BNI 1 | |
| SARS2 | CTCTAGTTGCATGACAGCCCTC | BNI 1 | |
| pSARS | ROX-TCGTGCGTGGATTGGCTTTGATGT-TAMRA | BNI 1 | |
| FluA(GJ)-F | GACCRATCCTGTCACCTCTGAC | Am | |
| FluA(GJ)-R | GGGCATTYTGGACAAAKCGTCTACGc | Am | |
| FluA(GJ)-Pb | FAM-TGCAGTCCTCGCTCACTGGGCACG-TAMRA | Am |
MS2g1, MS2g2, and pac site, fragment encoding maturase and coat protein and carrying the packaging site of MS2 bacteriophage; BNI 1, fragment encoding open reading frame 1b of the RNA polymerase gene of SARS-CoV; Am, fragment of the influenza A virus matrix protein gene; Bm, fragment of the influenza B virus matrix protein gene.
W = A or T and R = A or G.
Y = T or C and K = G or T.
RNAs of influenza A and B viruses and SARS-CoV.
An influenza A3 virus strain (A/Hangzhou/189/2005) (H3N2) and an influenza B virus strain (B/Hangzhou/42/2005) stored in our laboratory were used to extract RNAs for construction of AR. The RNA of SARS-CoV stored in our laboratory was collected from an oral swab specimen from a SARS patient during the 2003 SARS outbreak. RNAs were extracted with a QIAamp viral RNA extraction kit (Qiagen, Inc., Germany) according to the manufacturer's instructions.
Synthesis of Am-Bm-BNI 1 fragment by SOE-PCR.
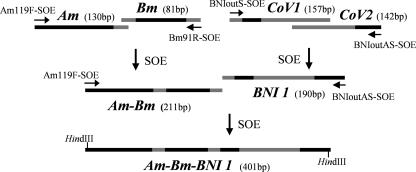
For synthesis of the Am-Bm-BNI 1 fragment by SOE-PCR (13), a 401-bp nucleic acid sequence was designed to include PCR targets for three respiratory viruses (Fig. 1).
FIG. 1.
Synthesis of Am-Bm-BNI 1 fragment by SOE-PCR. The bars in gray are overlap sequences identical with the target fragments. Primers used for SOE are showed by arrows. Am, fragment of the influenza A virus matrix protein gene; Bm, fragment of the influenza B virus matrix protein gene; CoV1, fragment of SARS-CoV amplified by primers BN IoutS2 and BN IinAS; CoV2, fragment of SARS-CoV amplified by primers BN IinS and BN IoutAS; BNI 1, spliced fragment derived from CoV1 and CoV2; Am-Bm, spliced fragment derived from Am and Bm; Am-Bm-BNI 1, spliced fragment derived from Am, Bm, and BNI 1.
Naked SARS-CoV RNA was used as a template, and RT-PCR was performed using the primers BN IoutS2 and BN IoutAS. Seminested PCR was then performed using the primers BN IoutS2/BN IinAS and BN IinS/BN IoutAS, respectively. The lengths of the products were 157 bp and 142 bp, respectively. The two fragments were purified by gel electrophoresis and were spliced using SOE-PCR, and a 10-ng aliquot of each of the PCR products was used as template DNA in a 20-μl reaction mixture comprised of 1× buffer, MgCl2 (2 mM), deoxynucleoside triphosphates (0.2 mM), and ExTaq DNA polymerase (25 U/ml). The PCR profile was as follows: an initial denaturation step at 94°C for 3 min; 10 cycles of denaturation at 94°C for 10 s, annealing at 66°C for 10 s, and extension at 72°C for 30 s; after spiking of primers BN IoutS2 and BN IoutAS into the reaction mixture, 10 cycles of denaturation at 94°C for 10 s, annealing at 66°C for 10 s, and extension at 72°C for 30 s, with the annealing temperature reduced 1°C per cycle; 32 cycles of denaturation at 94°C for 10 s, annealing at 56°C for 10 s, and extension at 72°C for 20 s; and 10 cycles of denaturation at 94°C for 10 s and extension at 72°C for 20 s. A 190-bp amplicon (BNI 1) was separated by agarose gel electrophoresis.
RT-PCR was then applied to the naked RNAs of an influenza virus H3N2 strain (A/Hangzhou/189/2005) and an influenza B virus strain (B/Hangzhou/42/2005), using the primers AM119F-SOE/AM248R-SOE and BM11F-SOE/BM91R-SOE, respectively. Both the 130-bp and 81-bp RT-PCR products were purified and spliced using SOE-PCR. The reaction conditions were as described above. A 211-bp DNA fragment (Am-Bm) was separated by agarose gel electrophoresis. We then spliced the purified BNI 1 and Am-Bm fragments by SOE-PCR. A 401-bp DNA fragment (Am-Bm-BNI 1) was achieved (Fig. 1).
Preparation of AR.
A DNA fragment encoding maturase and coat protein and carrying the packaging (pac) site of MS2 bacteriophage was synthesized from plasmid pMS27 (BCCM, Ghent, Belgium) by PCR using the primers MS2F and MS2R (9), and the exact position of the fragment was at positions 21 to 1770 (GenBank accession no. NC 001417). The resulting PCR product was cloned into the BamHI and HindIII sites of the expression vector pET30b (Novagen, Inc.). The recombinant plasmid was named pAR-1. The Am-Bm-BNI 1 fragment was inserted into the HindIII site downstream of the pac site on pAR-1, which formed the new recombinant plasmid pAR-2. The plasmid pAR-2 was then transformed into Escherichia coli strain BL21(DE3). Protein expression was induced with 1 mM isopropyl-l-thio-d-galactopyranoside (IPTG) at 37°C for 16 h. The AR packaging the RNA of the Am-Bm-BNI 1 fragment was named AR-2. Cells were collected by centrifugation and lysed by ultrasonic disruption. After a brief centrifugation step (12,000 × g for 10 min), the supernatant was incubated with excess DNase I and/or RNase A at 37°C overnight. Aliquots of the product were checked by 1% agarose gel electrophoresis with ethidium bromide staining.
Purification of AR.
AR was purified as an RNA-protein complex which could be electroeluted from an agarose gel with a GeBAflex tube (Gene Bio-Application Ltd., Israel). The sonication supernatant containing AR-2 particles was incubated at 37°C for 16 h with superfluous RNase A and DNase I (Sabc, Luoyang, China), followed by incubation with 10 U DNase (Ambion, Inc., Austin, TX) for 15 min at 37°C to remove any traces of DNA. After fractionation by gel electrophoresis, the slice of gel containing the desired RNA-protein complex was transferred to a GeBAflex tube (membrane cutoff, molecular weight of 12,000 to 14,000). An electric current was run for approximately 20 to 30 min until the RNA-protein complex was eluted from the gel slice. After the solution containing the purified AR was incubated at 85°C for 5 min, real-time RT-PCR and real-time PCR were performed to check the purity.
The concentration of AR particles was determined using an extinction coefficient of 8 mg/ml of MS2 bacteriophage per absorbance unit at 260 nm and a molecular weight of 3.0 × 106 (9).
The stability of the AR was evaluated by spiking AR-2 (∼104 copies) into RNase-rich oral swab fluid (20 μl). The oral swab fluid was Hank's buffer into which an oral swab was spiked. Oral swab fluids were stored at 37°C for 24 h and 48 h, at room temperature (20°C to 23°C) for 48 h and 14 days, and at 4°C for 14 and 30 days. Freeze-thaw action was applied to the oral swab fluids 3, 6, and 12 times. At the appointed times, samples were collected and kept at −30°C. Finally, their RNAs were extracted using a viral RNA extraction kit and tested using real-time RT-PCR.
Multiplex real-time RT-PCR detection of Am, Bm, and BNI 1.
Viral RNA was extracted with a QIAamp viral RNA extraction kit (Qiagen, Inc., Germany), whereas AR-2 was released from its phage coat by incubation at 85°C for 5 min or by extraction with a viral RNA extraction kit. The reaction conditions of the multiplex real-time RT-PCR were optimized in a matrix. The multiplex real-time RT-PCR was performed using a real-time RT-PCR kit (Takara, Dalian, China) with a 20-μl volume containing 1× one-step RT-PCR buffer, ExTaq HS (2U), Moloney murine leukemia virus RNase (40 U), RNase inhibitor (16 U), three sets of primers (0.4 μmol/liter [each]), and three sets of probes (0.24 μmol/liter [each]). The PCR thermal profile included an initial cDNA step of 15 min at 42°C, denaturation at 95°C for 2 min, and 45 cycles of 95°C for 5 s and 60°C for 30 s. Amplification, detection, and data analysis were performed with a Rotor Gene real-time detection system (RG-3000). The single primer-probe reaction was compared with the multiplex primer-probe reaction to determine any significant differences in the cycle threshold (CT) values. AR (∼104 copies) was amplified in both single and multiplex reactions (n = 3 replicates). To demonstrate the detection range of the multiplex real-time RT-PCR, 10-fold serial dilutions containing 106 to 100 copies/μl of AR particles were assayed. FAM-, HEX-, and ROX-labeled probes were used for the detection of the Bm, Am, and BNI 1 fragments, respectively. Fluorescence signals were measured at 60°C.
Multiplex real-time RT-PCR used for clinical samples.
From September 2005 to January 2007, 290 respiratory samples (throat swabs) were received from the Center for Disease Control and Prevention (CDC) in Hangzhou County or District. From each sample, an aliquot was stored at −70°C for multiplex real-time RT-PCR analysis. The specificity of the multiplex real-time RT-PCR was evaluated by detection of influenza A viruses in clinical samples and comparison with a commercial Biose-A-qRT-PCR kit (Biose, Hangzhou, China) and single-reaction real-time RT-PCR, whose primers [FluA(GJ)-F and FluA(GJ)-R] and probe [FluA(GJ)-Pb] were recommended by the Chinese CDC. The 80 respiratory samples received were detected by both the multiplex real-time RT-PCR assay and the commercial Biose-A-qRT-PCR kit. The other 97 respiratory samples received were detected in both the multiplex real-time RT-PCR assay and the single-reaction real-time RT-PCR (recommended by the Chinese CDC).
RESULTS
Synthesis of Am-Bm-BNI 1 fragment.
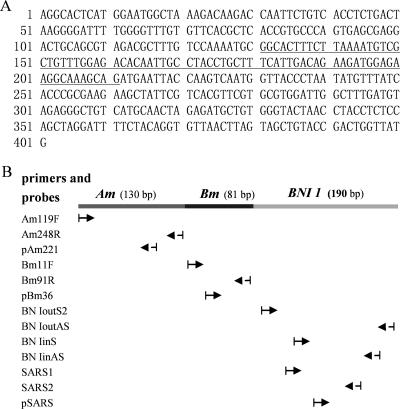
The Am-Bm-BNI 1 fragment was assembled successfully using SOE-PCR (Fig. 1). The sequence is listed in Fig. 2A. The targets included the forward and reverse primer sites as well as probe binding sites for each of the three RT-PCR assays (Fig. 2B). The target sequence for influenza A virus was 130 bp in length, the influenza B virus target was 81 bp, and the SARS-CoV target was 190 bp.
FIG. 2.
(A) Sequence of Am-Bm-BNI 1 RNA packaged within AR-2. The underlined sequence is the Bm fragment. (B) Positions of primers and probes located on the Am-Bm-BNI 1 fragment.
Preparation of AR.
AR-2 particles were detected as a fluorescent band migrating at about 900 to 1,000 bp compared to the DNA markers run on the same gel. AR-2 was resistant to RNase digestion. Treatment of AR-2 with DNase or RNase did not affect its intensity of staining or electrophoretic mobility. AR-2 was also stable under many storage conditions according to analysis of the CT values of the samples. AR-2 incubated at 4°C in oral swab fluid was stable over a 30-day challenge. AR-2 incubated at 37°C for 48 h and at room temperature for 14 days did not show any signal loss. The amount of AR-2 frozen at −30°C and thawed at room temperature 12 times had no visible loss. The stability of AR indicated that the packaged RNA might withstand extreme environmental conditions. Extraction of RNA from AR-2 by incubation at 85°C for 5 min had a higher efficiency than did extraction with a QIAamp viral RNA extraction kit. Approximately 1.0 × 1012 AR-2 particles could be purified from 1 ml of culture.
Purification of AR-2.
AR-2 could be electroeluted from an agarose gel with a GeBAflex tube. After the sample was purified as an RNA-protein complex, both PCR and RT-PCR were applied to the purified AR solution. Real-time RT-PCR was positive, and real-time PCR was negative. The results showed that E. coli genomic DNA and plasmid DNA had been removed and that only AR-2 particles were retained in the harvested sample.
Multiplex real-time RT-PCR detection of Am, Bm, and BNI 1.
Tenfold serial dilutions containing 106 to 100 copies/μl of AR-2 particles were assayed in the multiplex real-time RT-PCR format. The detection limit of the multiplex real-time PCR assay was 101 copies/μl of AR-2, and the three genes had the same detection limit (Fig. 3). The single primer-probe reaction was compared with the multiplex primer-probe reaction to determine the differences in CT values. The average CT values for the two reaction formats did not show a significant difference. AR-2 (∼104 copies) was used as template RNA, and the average CT values for the Am, Bm, and BNI 1 fragments in single primer-probe reactions were 25.92, 26.46, and 26.95, respectively. In contrast, the average CT values for the multiplex format were 26.07, 26.34, and 27.15, respectively.
FIG. 3.
Amplification of AR-2 as the EPC in multiplex real-time RT-PCR. The AR-2 template ranged from 1.0 × 106 to 1.0 × 101 copies/μl (from left to right). Water was used as a negative control. (A) Real-time RT-PCR curves of Bm fragment in the FAM channel. (B) Real-time RT-PCR curves of Am fragment in the HEX channel. (C) Real-time RT-PCR curves of SARS-CoV BNI 1 fragment in the ROX channel.
Multiplex real-time RT-PCR detection of respiratory samples.
Using AR-2 as the EPC, the multiplex real-time RT-PCR was applied to respiratory samples. Among 290 respiratory samples, 90 samples were detected as influenza A virus positive, 78 samples were detected as influenza B virus positive, and no BNI 1 fragment-positive samples were monitored. Among 80 respiratory samples, the positive rate of influenza A virus detection in the multiplex real-time RT-PCR format was 37.5% (30/80 samples), and the positive rate determined by a commercial Biose-A-qRT-PCR kit was 38.75% (31/80 samples). The positive coincidence rate for the two methods was 96.8%. Among the other 97 respiratory samples tested, the positive rate of influenza A virus detection in the multiplex real-time RT-PCR format was 61.8% (60/97 samples), and that for the single real-time RT-PCR format for influenza A virus (recommended by the Chinese CDC) was 58.8% (57/97 samples). The positive coincidence rate for the two methods was 95.3%.
DISCUSSION
Most current AR are packaged with the RNA of a single respiratory virus. A 359-nt SARS-CoV fragment was assembled with viral coat proteins into a pseudoviral particle for use as an RNase-resistant RNA control (4). A 228-nt matrix fragment of avian influenza virus was packaged into AR (5). Furthermore, a 1,200-nt RNA containing gene fragments of hepatitis C virus, human immunodeficiency virus type 1, SARS-CoV1, and SARS-CoV2 was packaged into AR (14). Influenza and SARS are both highly contagious respiratory diseases and can be transmitted easily by droplets. Influenza viruses and SARS-CoV frequently need to be detected in clinical respiratory samples.
In this study, we spliced DNA fragments of three respiratory viruses by using SOE-PCR. This resulted in the design and production of AR-2 for influenza A virus, influenza B virus, and SARS-CoV. The multitarget approach for AR preparation is practical and reduces the labor and cost of quality control of multiplex RNA virus assays. Influenza A viruses have many serotypes. Sixteen subtypes of hemagglutinin (HA), H1 to H16, and nine subtypes of neuraminidase (NA), N1 to N9, have been identified among influenza A viruses, with minimal serological cross-reactivity between subtypes (10, 17). This brings great difficulty to the diagnosis of influenza virus infections. In this study, we designed specific primers and probes to amplify a highly conserved region of the matrix gene, which subsequently was packaged into AR-2. Using these primers and probes, almost all influenza viruses with different serum types could be detected in a real-time RT-PCR format, so they were suitable for screening of influenza A and influenza B viruses at the molecular level. In the detection of SARS-CoV, most published RT-PCR assays target ORF1b of the RNA polymerase gene (2, 8). Therefore, a 190-nt fragment (BNI 1) was packaged into AR-2, and it included the primer and probe areas used in nested PCR and real-time PCR and covered partial sequences in the region of ORF1b of the SARS-CoV genome. For detection of small amounts of SARS-CoV RNA in a sample, nested RT-PCR can be combined with real-time PCR, and modified nested real-time RT-PCR had a higher sensitivity than did routine real-time RT-PCR (20).
The current AR purification method involves the combined use of gradient ultracentrifugation, column chromatography, and Co2+ affinity chromatography (4, 9, 15). In this study, AR was purified as an RNA-protein complex and electroeluted from an agarose gel with a GeBAflex tube. Compared to purification of AR using traditional CsCl fractionation, the method we used was efficient and easy to operate.
Multiplex PCR for clinical diagnosis has a significant advantage, as it permits simultaneous amplification of several viruses in a single reaction mixture, facilitating cost-effective diagnosis. However, there is a lack of multiplex real-time RT-PCR assays to detect influenza A and B viruses and SARS-CoV. This study developed a multiplex real-time RT-PCR assay for detecting the pathogenic respiratory RNA viruses influenza A virus, influenza B virus, and SARS-CoV. The multiplex real-time RT-PCR assay had a high sensitivity, and the detection limit of each channel (FAM, HEX, or ROX) was 101 copies/μl of AR-2. We used AR-2 as a standard and positive control for multiplex real-time RT-PCR. As a virus surrogate, AR-2 could provide a means of evaluating all steps of the assay.
The advantages of AR controls include safety, stability, and physical characteristics that mimic the natural virus. AR-2 was able to be a good substitute for the three target viruses within the respective real-time RT-PCR assays.
Acknowledgments
This work was supported by the Hangzhou Municipal Bureau of Health under grant no. 2005A009.
We are grateful to Qi Wu for his technical support.
Footnotes
Published ahead of print on 26 December 2007.
REFERENCES
- 1.Beld, M., R. Minnaar, J. Weel, C. Sol, M. Damen, H. van der Avoort, P. Wertheim-van Dillen, A. van Breda, and R. Boom. 2004. Highly sensitive assay for detection of enterovirus in clinical specimens by reverse transcription-PCR with an armored RNA internal control. J. Clin. Microbiol. 423059-3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bressler, A. M., and F. S. Nolte. 2004. Preclinical evaluation of two real-time, reverse transcription-PCR assays for detection of severe acute respiratory syndrome coronavirus. J. Clin. Microbiol. 42987-991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown, D., and B. L. Pasloske. 2001. Ribonuclease-resistant RNA controls and standards. Methods Enzymol. 341648-654. [DOI] [PubMed] [Google Scholar]
- 4.Cheng, Y., J. Niu, Y. Zhang, J. Huang, and Q. Li. 2006. Preparation of His-tagged armored RNA phage particles as a control for real-time reverse transcription-PCR detection of severe acute respiratory syndrome coronavirus. J. Clin. Microbiol. 443557-3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Das, A., E. Spackman, D. Senne, J. Pedersen, and D. L. Suarez. 2006. Development of an internal positive control for rapid diagnosis of avian influenza virus infections by real-time reverse transcription-PCR with lyophilized reagents. J. Clin. Microbiol. 443065-3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Donia, D., M. Divizia, and A. Pana. 2005. Use of armored RNA as a standard to construct a calibration curve for real-time RT-PCR. J. Virol. Methods 126157-163. [DOI] [PubMed] [Google Scholar]
- 7.Drosten, C., E. Seifried, and W. K. Roth. 2001. TaqMan 5′-nuclease human immunodeficiency virus type 1 PCR assay with phage-packaged competitive internal control for high-throughput blood donor screening. J. Clin. Microbiol. 394302-4308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Drosten, C., S. Günther, W. Preiser, S. Werf, H. R. Brodt, S. Becker, H. Rabenau, M. Panning, L. Kolesnikova, R. A. Fouchier, A. Berger, A. M. Burguière, J. Cinatl, M. Eickmann, N. Escriou, K. Grywna, S. Kramme, J. C. Manuguerra, S. Müller, V. Rickerts, M. Stürmer, S. Vieth, H. D. Klenk, A. D. Osterhaus, H. Schmitz, and H. W. Doerr. 2003. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 3481967-1976. [DOI] [PubMed] [Google Scholar]
- 9.DuBois, D. B., M. M. Winkler, and B. L. Pasloske. October 1997. U.S. patent 5,677,124.
- 10.Fouchier, R. A., V. Munster, A. Wallensten, T. M. Bestebroer, S. Herfst, D. Smith, G. F. Rimmelzwaan, B. Olsen, and A. D. Osterhaus. 2005. Characterization of a novel influenza A virus hemagglutinin subtype (H16) obtained from black-headed gulls. J. Virol. 792814-2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hall, C. B. 2001. Respiratory syncytial virus and parainfluenza virus. N. Engl. J. Med. 3441917-1928. [DOI] [PubMed] [Google Scholar]
- 12.Hietala, S. K., and B. M. Crossley. 2006. Armored RNA as virus surrogate in a real-time reverse transcriptase PCR assay proficiency panel. J. Clin. Microbiol. 4467-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Horton, R. M., H. D. Hunt, S. N. Ho, J. K. Pullen, and L. R. Pease. 1989. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene 7761-68. [DOI] [PubMed] [Google Scholar]
- 14.Huang, Q., Y. Cheng, Q. Guo, and Q. Li. 2006. Preparation of a chimeric armored RNA as a versatile calibrator for multiple virus assays. Clin. Chem. 521446-1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pasloske, B. L., C. R. Walkerpeach, R. D. Obermoeller, M. Winkler, and D. B. DuBois. 1998. Armored RNA technology for production of ribonuclease-resistant viral RNA controls and standards. J. Clin. Microbiol. 363590-3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pasloske, B. L., D. B. DuBois, D. M. Brown, and M. M. Winkler. June 2002. Methods of quantifying viral load in an animal with a ribonuclease resistant RNA preparation. U.S. patent 6,399,307.
- 17.Swayne, D. E., and D. L. Suarez. 2000. Highly pathogenic avian influenza. Rev. Sci. Technol. 19463-482. [DOI] [PubMed] [Google Scholar]
- 18.Templeton, K. E., S. A. Scheltinga, M. F. Beersma, A. C. Kroes, and E. C. Claas. 2004. Rapid and sensitive method using multiplex real-time PCR for diagnosis of infections by influenza A and influenza B viruses, respiratory syncytial virus, and parainfluenza viruses 1, 2, 3, and 4. J. Clin. Microbiol. 421564-1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.WalkerPeach, C. R., M. Winkler, D. B. DuBois, and B. L. Pasloske. 1999. Ribonuclease-resistant RNA controls (armored RNA) for reverse transcription-PCR, branched DNA, and genotyping assays for hepatitis C virus. Clin. Chem. 452079-2085. [PubMed] [Google Scholar]
- 20.Ye, R., J. C. Pan, Z. C. Huang, H. Wang, H. Q. Wang, D. F. Wei, K. Xu, H. G. Wen, and K. K. Chen. 2005. Detection of severe acute respiratory syndrome probable patients' virus RNA in Hangzhou by using a two loci and a modified nested real-time reverse transcription polymerase chain reaction. Chin. J. Prev. Med. 39129-132. [PubMed] [Google Scholar]