Abstract
A total of 88 uropathogenic Escherichia coli isolates, including 68 isolates from urine and 20 isolates from blood, were characterized by multilocus sequence typing (MLST). MLST has identified an important genetic lineage of E. coli, designated sequence type 131 (ST-131), represented by 52 of these isolates, 51 of which were resistant to extended-spectrum cephalosporins. ST-131 appears to be a drug-resistant uropathogenic strain of E. coli responsible for causing urinary tract infections and bacteremia and is widely disseminated among both community and hospital patients from different geographical areas in the northwest of England. Application of MLST has helped to define the population biology which may underpin the epidemiology of pathogenic E. coli strains. The portability of MLST allows the accurate monitoring of this antibiotic-resistant uropathogenic strain of E. coli and will enhance surveillance for this important group of organisms.
Urinary tract infection (UTI) is the most common laboratory-confirmed bacterial infection encountered in medical practice in Europe and North America (9). Escherichia coli is a major cause of UTI, and it accounts for approximately 70 to 95% of community-acquired cases and 50% of all hospital-acquired infections (9). These organisms are responsible for significant social and economic costs for both communities and public health resources. It has been estimated that 150 million cases of UTI occur on a global basis per year and cost about 6 billion dollars for national health resources (6).
UTI due to E. coli can progress to bacteremia, which is associated with significant mortality. E. coli is recognized as the one of the two most common causes of bacteremia in England and Wales, and the Health Protection Agency reported 32.5 bacteremia cases per 100,000 subjects in 2005 (7). There is no comprehensive surveillance for community-acquired UTIs in England, and currently, there is only voluntary national surveillance for bacteremia; therefore, it is very difficult to estimate the true incidence of UTI infections and monitor the spread of these organisms (12).
In recent years, there has been an increase in the occurrence cephalosporin-resistant strains of E. coli causing UTI and invasive infections (10). These organisms, which are often also resistant to other widely used antibiotics such as fluoroquinolones and trimethoprim, have particular clinical significance due to the limited therapeutic options that are available. Furthermore, the prevention and control of the spread of uropathogenic E. coli infections are hampered by a poor understanding of the population biology of these pathogens. The potential of particular lineages of antibiotic-resistant or uropathogenic E. coli to disseminate and cause disease is unknown. Improved strain characterization and phylogenetic analysis would improve our understanding of the epidemiology of this pathogen and will allow the development of a rapid assay for monitoring of uropathogenic E. coli.
Multilocus sequence typing (MLST) is a DNA sequence-based method that has been used to study the population biology of pathogenic microorganisms and provides an understanding of the population structure of important medical pathogens including E. coli (http://web.mpiib-berlin.mpg.de), Campylobacter jejuni, Staphylococcus aureus, and Neisseria meningitidis (2, 3, 5). MLST is based upon establishing an allelic profile by sequence analysis of seven housekeeping genes. The resultant allelic profile is summarized by the assignation of a sequence type (ST) via an electronic database. The genetic relatedness between isolates can be compared, and closely related organisms can be grouped as clonal complexes (4). The portability and reproducibility of MLST bring significant advantages to the study of the epidemiology of the emergence of pathogens of major concern to medical microbiology (11).
The aim of the present study was to use MLST for the genetic characterization of uropathogenic E. coli strains and to investigate the population biology of uropathogenic E. coli strains circulating within the northwest of England. Establishing the population structure of emerging pathogens such as E. coli aids in an understanding of the epidemiology and allows the accurate monitoring of the population structure of uropathogenic E. coli strains.
MATERIALS AND METHODS
Bacterial isolates.
A total of 88 routine clinical E. coli isolates from laboratories across the northwest of England were used in this study. Sixty-eight isolates were from urine samples, and 20 isolates were from blood cultures. Isolates were examined for phenotypic resistance to cefpodoxime, as identified using standard British Society for Antimicrobial Chemotherapy disk diffusion methods (1).
Isolates were chosen to represent geographical spread over an area including Barrow-in-Furness, Central Manchester, Lancaster, Preston, South Manchester, and Stockport, with the greatest distance between the furthest locations being 99 miles. The isolates were obtained from six hospital laboratories between November 2004 and October 2005 and included those from patients and from general practice (community specimens).
Serotyping.
Serotyping was performed on a representative collection of 43 E. coli isolates at the Escherichia, Shigella, Yersinia, and Vibrio Reference Unit, Laboratory of Enteric Pathogens, Centre for Infections, Health Protection Agency, Colindale, United Kingdom.
DNA isolation.
DNA isolation was performed by using a PrepMan Ultra sample preparation reagent (Applied Biosystems) according to the manufacturer's instructions.
MLST.
This study used the MLST scheme for E. coli developed by M. Achtamn and others (http://web.mpiib-berlin.mpg.de). The scheme uses the following seven housekeeping genes: adk (adenylate kinase), fumC (fumarate hydratase), gyrB (DNA gyrase), icd (isocitrate dehydrogenase), mdh (malate dehydrogenase), purA (adenylosuccinate synthetase), and recA (ATP/GTP binding motif). The primer sequences are available at http://web.mpiib-berlin.mpg.de. Amplification was carried out in 50-μl reaction mixture volumes containing 1 μl of DNA, 5 μl of each primer (10 pmol/μl), 5 μl of 10× PCR buffer (Qiagen, West Sussex, United Kingdom), 10 μl of 1 mM deoxynucleoside triphosphates (Roche, Hertfordshire, United Kingdom), and 0.25 units of Taq DNA polymerase (Qiagen, United Kingdom). The reaction conditions were an initial denaturation step at 94°C for 2 min, followed by 35 cycles of the following conditions: denaturation at 94°C for 1 min, annealing temperature for each primer set for 1 min, and extension at 72°C for 1 min, with a final extension step at 72°C for 5 min. The presence of the correct size PCR product was confirmed by agarose gel electrophoresis.
PCR products were purified using a Whatman (Middlesex, United Kingdom) PCR Cleanup Unifilter according to instructions provided by the manufacturer.
Sequencing.
Sequencing reactions were carried out by using CEQ Dye terminator cycle sequencing with a Quick Start kit (Beckman Coulter, Buckinghamshire, United Kingdom). CEQ Dye terminator cycle sequencing reactions were carried out in 10 μl of one-quarter-strength reaction mixture volumes containing 0.5 μl of purified PCR product, 0.5 μl of primer (10 pmol/μl), 1 μl of halfCEQ buffer (Genetix, Hampshire, United Kingdom), 2 μl of DTCS Quick Start Master mix (Beckman Coulter, Buckinghamshire, United Kingdom), and 6 μl of sterile water. The reaction conditions were 40 cycles of 96°C for 20 s and 60°C for 4 min. Details of sequencing primers are listed in Table 1. Ethanol precipitation was used for the postsequencing cleanup. The reaction products were separated and detected with a CEQ 8000 genetic analysis system. Sequences were determined on each DNA strand and were assembled from the chromatograms with Sequencher software (Gene Codes Corporation, MI). The allelic profile was summarized by the assignation of an ST via an electronic database (http://web.mpiib-berlin.mpg.de.).
TABLE 1.
Details of sequencing primers used for analysis of amplification products during MLST
| Gene | Primera | Primer sequence (5′-3′) |
|---|---|---|
| adk | adk seq F | GCAATGCGTATCATTCTGCT |
| adk seq R | CAGATCAGCGCGAACTTCAG | |
| fumC | fumC seq F | CCACCTCACTGATTCATGCG |
| fumC seq R | CGGTGCACAGGTAATGACTG | |
| gyrB | gyrB seq F | CGGGTCACTGTAAAGAAATTATCG |
| gyrB seq R | GTCCATGTAGGCGTTCAGGG | |
| icd | icd seq F | TACATTGAAGGTGATGGAATCG |
| icd seq R | GTCTTTAAACGCTCCTTCGG | |
| mdh | mdh seq F | TCTGAGCCATATCCCTACTG |
| mdh seq R | CGATAGATTTACGCTCTTCCA | |
| purA | purA seq F | CTGCTGTCTGAAGCATGTCC |
| purA seq R | CAGTTTAGTCAGGCAGAAGC | |
| recA | recA seq F | AGCGTGAAGGTAAAACCTGTG |
| recA seq R | ACCTTTGTAGCTGTACCACG |
F, forward; R, reverse; seq, sequencing.
Statistical analysis.
The association between sequence types and serogroup was independently assessed by chi-square tests using Statistical Package for the Social Sciences software, version 14.0. P values of <0.05 were the criteria chosen for statistical significance. Data were subjected to analysis using the package eBURST V3, which assesses the relationship within the clonal complexes but does not reconstruct evolutionary pathways between clonal complexes. The entire E. coli MLST database (1,506 isolates and 695 STs) (as of 25 September 2007) and 88 E. coli isolates from this study were entered into eBURST. The eBURST V3 program is available at http://eburst.mlst.net.
RESULTS
Characterization of uropathogenic E. coli isolates by MLST.
MLST analysis of the 88 cephalosporin-resistant organisms studied identified 22 different STs and 10 clonal complexes for 68 urinary tract (uropathogenic) E. coli isolates and 20 E. coli isolates from blood cultures (Table 2). The predominant ST was ST-131, which was comprised of 52 isolates (59%), followed by ST-73 and ST-95, each with six isolates (6.8%). Of 22 different STs, 14 STs occurred only once in the data set, and two of the STs (ST-457 and ST-458) were novel to this study.
TABLE 2.
MLST of 88 E. coli isolates from urine and blood samples
| Clonal complex | ST | No. of isolates
|
|
|---|---|---|---|
| Urine | Blood | ||
| 14 | 14 | 1 | 0 |
| 23 | 23 | 1 | 0 |
| 350 | 57 | 1 | 0 |
| 59 | 59 | 0 | 1 |
| 69 | 69 | 1 | 1 |
| 73 | 73 | 4 | 2 |
| 568 | 80 | 0 | 1 |
| 23 | 88 | 1 | 0 |
| 95 | 95 | 4 | 2 |
| Unassigned | 127 | 1 | 0 |
| Unassigned | 131 | 43 | 9 |
| Unassigned | 141 | 0 | 1 |
| Unassigned | 144 | 1 | 0 |
| 155 | 155 | 2 | 0 |
| Unassigned | 372 | 1 | 0 |
| Unassigned | 391 | 2 | 0 |
| 394 | 394 | 1 | 0 |
| 23 | 410 | 2 | 0 |
| 95 | 416 | 0 | 2 |
| 95 | 421 | 0 | 1 |
| Unassigned | 457 | 1 | 0 |
| 73 | 458 | 1 | 0 |
Resistance to expanded-spectrum cephalosporins.
Resistance to cefpodoxime was seen in 65 (74%) of the isolates examined. Most notable was the finding that 51 of the 52 ST-131 isolates were resistant to cefpodoxime. Of the organisms recovered from urine, 12 (18%) (ST-14, ST-69, ST-73, ST-95, ST-127, ST-131, ST-144, and ST-372) were susceptible to cefpodoxime, as were 11 isolates (55%) (ST-59, ST-69, ST-80, ST-95, ST-141, ST-416, and ST-421) from blood.
Relationship between ST and samples.
The isolates from urine were more diverse than those from blood, and they belonged to 17 different STs. Of the 68 urine isolates, the majority (63% [n = 43]) belonged to ST-131. Four urine isolates belonged to ST-73 and ST-95 (Table 2), and the remaining isolates belonged to one of 14 STs. Of the 20 bacteremia isolates, 45% (n = 9) belonged to ST-131, and 55% (n = 11) of the isolates belonged to a variety of STs (Table 2). Fifty-two urine and 14 blood isolates shared the same STs (ST-69, ST-73, ST-95, and ST-131). A specific ST, ST-131, was overrepresented in urine and blood samples (63% of urine isolates and 45% of blood isolates).
Relationship between ST and community and hospital isolates.
The 88 isolates investigated were comprised of 39 E. coli isolates that were from community-acquired infections and 25 E. coli isolates that came from hospital cases (24 E. coli isolates were excluded due to a lack of clinical/epidemiological information). The isolates from community specimens were more diverse than those from hospital specimens, belonging to 12 different STs (ST-23, ST-57, ST-73, ST-88, ST-95, ST-131, ST-141, ST-155, ST-391, ST-394, ST-410, and ST-458), whereas hospital isolates belonged to five different STs (ST-69, ST-131, ST-410, ST-421, and ST-457). In both settings, ST-131 predominated and was responsible for 64% of community infections and 84% of hospital infections.
Relationship between ST and serogroups.
A representative subset of 43 E. coli isolates were serotyped: the majority (48.8% [n = 21]) belonged to serogroup O25, and they were all ST-131 isolates (Table 3). Three isolates were unidentifiable by serotyping. There is an association between a particular ST and serogroup; for example, serogroup O25 and ST-131 were significantly associated (P < 0.01 by chi-square test). However, some isolates with the same serogroup did not have the same STs (Table 3).
TABLE 3.
MLST and serogroups of 43 E. coli isolates
| ST | Serogroup | No. of isolates |
|---|---|---|
| 14 | O18 | 1 |
| 59 | O1 | 1 |
| 69 | O11 | 1 |
| 69 | O15 | 1 |
| 73 | O6 | 2 |
| 73 | O18 | 1 |
| 80 | O75 | 1 |
| 88 | O8 | 1 |
| 95 | O1 | 2 |
| 95 | O unidentifiable | 1 |
| 127 | O6 | 1 |
| 131 | O25 | 21 |
| 131 | O unidentifiable | 1 |
| 141 | O2 | 1 |
| 144 | O16 | 1 |
| 372 | O75 | 1 |
| 391 | O1 | 2 |
| 410 | O unidentifiable | 1 |
| 416 | O18 | 1 |
| 421 | O rough | 1 |
Population biology of uropathogenic E. coli.
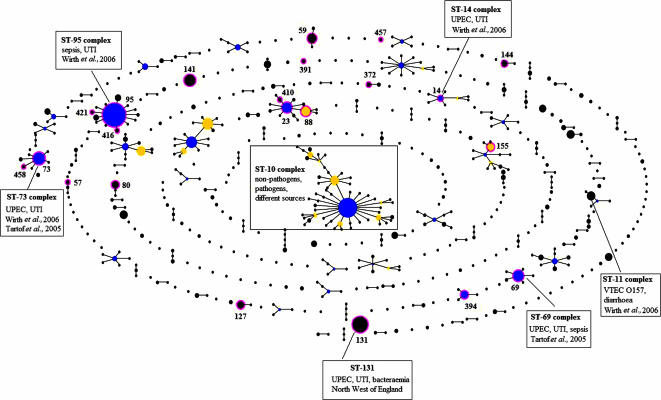
Analysis with eBURST provided an overview of the E. coli population: 88 E. coli isolates from this study were compared with 1,506 isolates from the E. coli MLST database that were isolated from different sources such as humans, animals, food, and the environment. The data set includes strains representing different groups of pathogenic E. coli, such as verocytotoxin-producing E. coli (VTEC), including strains of serogroup O157; enteropathogenic E. coli; enterotoxigenic E. coli; and enteroinvasive E. coli (http://web.mpiib-berlin.mpg.de.) (14). The relationships among members of clonal complexes were represented by the eBURST diagram (Fig. 1). There are 54 clonal complexes and 695 STs within the E. coli population, and some groups of pathogenic E. coli strains are associated with particular STs or ST complexes. For example, all O157:H7 VTEC strains are in ST-11 (http://web.mpiib-berlin.mpg.de). Previous studies reported that some uropathogenic E. coli strains belong to ST-14, ST-69, ST-73, and ST-95 complexes (Fig. 1) (13, 14). A search of the E. coli MLST database revealed four previously submitted isolates belonging to ST-131: one was isolated from a case of cystitis, one was from a patient with septicemia, and the remaining two isolates were from a dog and an unknown source.
FIG. 1.
Population snapshots of E. coli clusters of related STs and individual unrelated STs within the entire E. coli MLST database. STs in circles are those identified in this study. This diagram does not show the genetic distance between unrelated STs and clonal complexes. The primary founder of each clonal complex is at a central position in the diagram. Two STs separated by one node represent a single-locus variation between two isolates (a single-locus variant). UPEC, uropathogenic E. coli.
DISCUSSION
The population biology of uropathogenic E. coli is poorly understood. It is generally accepted that the capacity of E. coli to cause disease among humans varies according to the presence and type of virulence factors within each lineage. Lineages that possess factors which promote adherence to the uroepithelium, for example, are more likely to result in a UTI than strains lacking such a factor. The existence of a predominant sequence type, ST-131, among antibiotic-resistant clinical isolates of E. coli suggests that this genetic lineage may have acquired virulence determinants that enable it to overcome host defense mechanisms and therefore cause UTIs. While this would need to be confirmed by pathogenicity studies, it is it is biologically plausible that such uropathogenic strains would be more likely to be exposed to antibiotic treatment courses than less pathogenic stains, resulting in the coselection of uropathogenicity and antibiotic resistance determinants over time.
Uropathogenic E. coli strains usually belonged to a limited number of O serogroups, mainly O1, O2, O6, O18, and O75 (8). However, in this study, ST-131 was significantly associated with serogroup O25. The strong association between ST-131 and serogroup O25 indicates that this group of E. coli isolates could now be considered to be a new emerging strain associated with community and hospital cases of UTIs.
The population structure of uropathogenic E. coli needs to be elucidated in order to understand the spread of the different clones and to investigate the dissemination of this pathogen to humans. ST-131 was the predominant ST in the northwest region of England, and it accounted for 59% of UTIs and bacteremia. The data strongly suggest that E. coli ST-131 is a strain of uropathogenic E. coli responsible for causing UTIs. This study has demonstrated that ST-131 has become widely disseminated in hospital and community patients in the northwest of England. In addition, the high prevalence of ST-131 isolates in blood samples indicates that this lineage is particularly pathogenic to humans and warrants a full investigation with respect to geographic distribution, phylogeny, and carriage of antibiotic resistance determinants in this lineage. The potential emergence of a cephalosporin-resistant, uropathogenic E. coli ST-131 strain presents a serious threat to public health due to the limited options available to clinicians treating patients infected with this strain (10).
The use of MLST to investigate the population structure and the relationship of different groups of pathogenic E. coli strains revealed that VTEC O157 and the majority of representatives of other pathogenic groups of E. coli (E. coli associated with diarrheal illness) were genetically distinct from uropathogenic E. coli strains (14). MLST has identified a number of important genetic lineages that are related to particular pathogenic groups such as VTEC O157 that all belong to the ST-11 complex (14). In addition, four major ST complexes that are associated with uropathogenic E. coli have been identified; these are ST-14, ST-69, ST-73, and ST-95 complexes (13, 14). The ST-69 complex is commonly found in the United States but was relatively uncommon in this study. A previous study by Tartof et al. showed that there are several different disseminated uropathogenic E. coli lineages present in different countries (13). The portability and reproducibility of MLST (2, 11) will provide useful and important information about E. coli genetic lineages responsible for UTI. In this study, ST-131 isolates appeared to be associated with UTI and life-threatening bacteremia, which highlights the need for robust molecular epidemiological data and risk factor analysis to allow a more accurate understanding of the biology of this organism so that the future spread of this ST in England and the rest of the world can be monitored. Currently, there is no comprehensive surveillance system for UTI in the community in England and Wales (12); however, MLST can provide new information for enhanced surveillance for the potential emergence of uropathogenic E. coli isolates.
Acknowledgments
We thank Manchester Royal Infirmary, Royal Preston Hospital, Stepping Hill Hospital, and Wythenshawe Hospital microbiology laboratories for submitting isolates to the study. This publication made use of the Escherichia coli MLST website (http://web.mpiib-berlin.mpg.de.) and the eBURST V3 website (http://eburst.mlst.net), which are sited at the Max Planck Institute für Infektionsbiologie and the Department of Infectious Disease Epidemiology at Imperial College London, respectively.
S.H.L. is supported by a Ph.D. studentship from the Health Protection Agency.
Footnotes
Published ahead of print on 23 January 2008.
REFERENCES
- 1.Andrews, J. M. 2005. BSAC standardized disc susceptibility testing method (version 4). J. Antimicrob. Chemother. 5660-76. [DOI] [PubMed] [Google Scholar]
- 2.Dingle, K. E., F. M. Colles, D. R. Wareing, R. Ure, A. J. Fox, F. E. Bolton, H. J. Bootsma, R. J. Willems, R. Urwin, and M. C. Maiden. 2001. Multilocus sequence typing system for Campylobacter jejuni. J. Clin. Microbiol. 3914-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Enright, M. C., N. P. Day, C. E. Davies, S. J. Peacock, and B. G. Spratt. 2000. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 381008-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Enright, M. C., and B. G. Spratt. 1999. Multilocus sequence typing. Trends Microbiol. 7482-487. [DOI] [PubMed] [Google Scholar]
- 5.Feavers, I. M., S. J. Gray, R. Urwin, J. E. Russell, J. A. Bygraves, E. B. Kaczmarski, and M. C. Maiden. 1999. Multilocus sequence typing and antigen gene sequencing in the investigation of a meningococcal disease outbreak. J. Clin. Microbiol. 373883-3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harding, G. K. M., and A. R. Ronald. 1994. The management of urinary tract infections: what have we learned in the past decade? Int. J. Antimicrob. Agents 483-88. [DOI] [PubMed] [Google Scholar]
- 7.Health Protection Agency. 2006. Trends in antimicrobial resistance in England and Wales, 2004-2005. Health Protection Agency, London, United Kingdom.
- 8.Johnson, J. R. 1991. Virulence factors in Escherichia coli urinary tract infection. Clin. Microbiol. Rev. 480-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kucheria, R., P. Dasgupta, S. H. Sacks, M. S. Khan, and N. S. Sheerin. 2005. Urinary tract infections: new insights into a common problem. Postgrad. Med. J. 8183-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paterson, D., and R. Bonomo. 2005. Extended-spectrum β-lactamases: a clinical update. Clin. Microbiol. Rev. 18657-686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maiden, M. C. J., J. A. Bygraves, E. Feil, G. Morelli, J. E. Russell, R. Urwin, Q. Zhang, J. Zhou, K. Zurth, D. A. Caugant, I. M. Feavers, M. Achtman, and B. G. Spratt. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. USA 953140-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mayor, S. 2005. UK agency recommends better monitoring of multidrug resistant E. coli. BMJ 331594. [Google Scholar]
- 13.Tartof, S. Y., O. D. Solberg, A. R. Manges, and L. W. Riley. 2005. Analysis of a uropathogenic Escherichia coli clonal group by multilocus sequence typing. J. Clin. Microbiol. 435860-5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wirth, T., D. Falush, R. Lan, F. Colles, P. Mensa, L. H. Wieler, H. Karch, P. R. Reeves, M. C. Maiden, H. Ochman, and M. Achtman. 2006. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol. Microbiol. 601136-1151. [DOI] [PMC free article] [PubMed] [Google Scholar]