Abstract
An outbreak of infections affecting 311 patients who had undergone different invasive procedures occurred in 2004 and 2005 in the city of Belém, in the northern region of Brazil. Sixty-seven isolates were studied; 58 were from patients who had undergone laparoscopic surgeries, 1 was from a patient with a postinjection abscess, and 8 were from patients who had undergone mesotherapy. All isolates were rapidly growing nonpigmented mycobacteria and presented a pattern by PCR-restriction enzyme analysis of the hsp65 gene with BstEII of bands of 235 and 210 bp and with HaeIII of bands of 200, 70, 60, and 50 bp, which is common to Mycobacterium abscessus type 2, Mycobacterium bolletii, and Mycobacterium massiliense. hsp65 and rpoB gene sequencing of a subset of 20 isolates was used to discriminate between these three species. hsp65 and rpoB sequences chosen at random from 11 of the 58 isolates from surgical patients and the postinjection abscess isolate presented the highest degrees of similarity with the corresponding sequences of M. massiliense. In the same way, the eight mesotherapy isolates were identified as M. bolletii. Molecular typing by pulsed-field gel electrophoresis (PFGE) grouped all 58 surgical isolates, while the mesotherapy isolates presented three different PFGE patterns and the postinjection abscess isolate showed a unique PFGE pattern. In conclusion, molecular techniques for identification and typing were essential for the discrimination of two concomitant outbreaks and one case, the postinjection abscess, not related to either outbreak, all of which were originally attributed to a single strain of M. abscessus.
Rapidly growing mycobacteria (RGM) are widely distributed in the environment, especially in water (rivers, lakes, potable water), and can contaminate reagents and medical equipment. Most RGM infections in humans are caused by species belonging to the Mycobacterium fortuitum, Mycobacterium chelonae-Mycobacterium abscessus, and Mycobacterium smegmatis groups (6).
The M. chelonae-M. abscessus group comprises two genomospecies, M. chelonae and M. abscessus, which have been differentiated on the basis of <70% genomic homology by DNA-DNA hybridization (16, 18). These species have been isolated from sporadic cases of chronic lung disease associated with bronchiectasis and cystic fibrosis, disseminated cutaneous infections, and postsurgical wound infections. They have also been implicated in outbreaks in cardiac, ophthalmologic, and plastic surgeries and pseudo-outbreaks related to contaminated bronchoscopes and contaminated laboratory reagents (6, 12, 20, 25, 26). Mycobacterium immunogenum was included in this group in 2001. It was isolated from metalworking fluids and was associated with cases of hypersensitivity pneumonitis in factory workers. This species has also been detected in cutaneous, catheter-related, articular, and lung infections; in an outbreak related to ophthalmologic surgeries; and in a pseudo-outbreak related to bronchoalveolar lavage procedures (19, 27). Mycobacterium massiliense was validated as a species separate from the M. chelonae-M. abscessus group in 2006 (11). It has been isolated from pulmonary specimens, from a pacemaker pocket infection, and during an outbreak associated with intramuscular injections (4, 15, 22). Mycobacterium bolletii has been isolated from 4 of 59 specimens from a collection of clinical RGM isolates and was recognized as a species separate from the M. chelonae-M. abscessus group in 2006 (1).
The discrimination of species belonging to this group is important for epidemiological studies, the prediction of antibiotic susceptibility, and the recognition of medically important taxa. Few phenotypic tests can distinguish RGM species, and the discriminatory power of lipid analysis by high-performance liquid chromatography is limited due to profile similarities (5). Individual species of the M. chelonae-M. abscessus group present differences in their antimicrobial susceptibilities. The most pronounced differences between M. abscessus and M. chelonae are in their susceptibilities to cefoxitin (with M. abscessus being generally susceptible and M. chelonae being resistant) and tobramycin (with M. abscessus being resistant and M. chelonae being susceptible) (6). M. immunogenum is resistant to both drugs (27). It was reported that M. massiliense could be distinguished by its susceptibility to doxycycline, while other members of this group are resistant to this drug (4). M. massiliense isolates with intermediate susceptibility to doxycycline were also identified (15). M. bolletii was described as a species highly resistant to antimicrobial drugs, including clarithromycin, to which the other members of the group are highly susceptible (1). Five different patterns have been observed by PCR-restriction enzyme analysis of the hsp65 gene (PRA-hsp65) among species belonging to the M. chelonae-M. abscessus group: M. chelonae type 1, M. abscessus type 1 and type 2, and M. immunogenum type 1 and type 2, respectively (8, 19, 27). M. massiliense and M. bolletii present an indistinguishable PRA-hsp65 pattern, described as M. abscessus type 2, precluding the separation of these species by this method. By 16S rRNA sequencing, M. chelonae and M. abscessus differ by only 4 bp and have identical 16S rRNA hypervariable region A sequences; M. immunogenum differs by 8 bp from M. abscessus and by 10 bp from M. chelonae; M. massiliense and M. bolletii show 100% sequence similarity with M. abscessus. Therefore, 16S rRNA sequencing is not adequate for the discrimination of these species. Differentiation is best achieved by analysis of polymorphisms either in the rpoB, hsp65, sodA, and recA genes or in the 16S-23S rRNA internal transcribed sequence (2, 3, 9).
This report describes the molecular identification and typing of isolates obtained during an outbreak of infections after invasive procedures in Belém, Brazil. To our knowledge, this is the first report of infections caused by the recently described species M. massiliense and M. bolletii in Brazil.
MATERIALS AND METHODS
Mycobacterial isolates.
From February 2004 to June 2005, 311 patients who had undergone different invasive procedures, performed in 16 private hospitals and clinics in the city of Belém, state of Pará, in the northern region of Brazil, presented with signs and symptoms of localized infection. One-hundred seventy specimens were collected at the Instituto Evandro Chagas by needle aspiration of secretions, by surgical biopsy, or from the open drainages of abscesses and were cultivated at 37°C on Löwenstein-Jensen solid medium for up to 4 weeks. Sixty-seven isolates were included in this study. Fifty-eight isolates were obtained from patients who had undergone surgery; eight were from patients who had undergone mesotherapy, a procedure comprising multiple subcutaneous injections of pharmaceutical and/or homeopathic medications for cosmetic purposes; and one was from an abscess after the intramuscular injection of a contraceptive.
Species identification.
All isolates were identified by analysis of their characteristics on culture (the time for growth and pigment production) and by PRA-hsp65. In brief, a 441-bp fragment of the hsp65 gene, Telenti's fragment, was amplified with primers Tb11 (ACCAACGATGGTGTGTCCAT) and Tb12 (CTTGTCGAACCGCATACCCT), as described before (17, 23). The PCR mixture was used as a negative control. The amplicons were digested with BstEII (Promega) and HaeIII (Invitrogen), and the restriction fragments were separated by electrophoresis in 3% Seakem LE (BioWhittaker Molecular Applications) agarose gels stained with ethidium bromide. Fifty- and 25-bp DNA ladders (Invitrogen) were used as molecular standards.
As the PRA-hsp65 pattern obtained was common to the three different species, a subset of isolates was submitted to DNA sequencing to discriminate between these species. Two DNA targets were sequenced: a 667-bp fragment of the hsp65 gene containing Telenti's fragment was amplified with primers hsp667forward (GGCCAAGACAATTGCGTACG) and hsp667-reverse (GGAGCTGACCAGCAGGATG) (21), and a 752-bp fragment of rpoB region V was amplified with primers MycoF (GGCAAGGTCACCCCGAAGGG) and MycoR (AGCGGCTGCTGGGTGATCATC) (2). The PCR mixture was used as negative control. To obtain consensus hsp65 and rpoB sequences, the amplicons from isolate B5, chosen at random from among the 58 isolates from surgical patients, were cloned before sequencing by using the TOPO TA cloning kit for sequencing (Invitrogen), according to the manufacturer's recommendations. Recombinant plasmids were purified from the isolated colonies with the Wizard Plus SV Minipreps DNA purification system (Promega) and were sequenced with BigDye Terminator cycle sequencing reagents and primers M13F and M13R (Invitrogen). Cycle sequencing was performed with a Perkin-Elmer 9600 GeneAmp PCR system programmed for 25 cycles at 96°C for 20 s, 50°C for 10 s, and 60°C for 4 min. The sequencing products were cleaned with CentriSep spin columns, according to the manufacturer's instructions (Princeton Separations, Applied Biosystems), and were then analyzed on a ABI Prism 3100 sequencer (Perkin-Elmer, Applied Biosystems).
The sequences were assembled from the ABI chromatograms by using the PHRED, PHRAP, and CONSED programs (13). The quality of the assembled sequences (the phred scores) ranged from 0 to 1.45 per 10 kb, as estimated with the CONSED program. Consensus sequences corresponding to 401 bp of the hsp65 gene (the 441-bp Telenti's fragment without the sequences of the primers at both ends) and to 711 bp of the rpoB gene (752 bp from rpoB region V without the sequences of the primers at both ends) were identified by similarity analysis with the sequences in the GenBank database by use of the BLAST program (Basic Local Alignment Search Tool; http://www.ncbi.nlm.nih.gov/BLAST).
The hsp65 and rpoB amplicons of 19 additional isolates were submitted to direct sequencing: 10 isolates (isolates B51, B49, B37, B31, B25, B58, B52, B43, B50, and B30) randomly chosen from among the 58 available isolates from surgical patients operated on in different hospitals, 8 isolates from mesotherapy patients (isolates B59 to B66), and 1 isolate (isolate B67) from the postinjection abscess. As these sequences were obtained from noncloned PCR products, the data output from the sequencer had to be processed, and a pipeline was developed for this purpose by using the Egene platform (10). The trace files were initially submitted to analysis with the PHRED program for base calling and quality assessment. Then, the primers sequences were masked by use of the cross_match program (P. Green, 1997, http://www.phrap.org/phredphrapconsed.html). After masking of the primer sequences, a trimming process was performed to eliminate masked and low-quality (phred value, 15) regions from the 3′ and the 5′ ends of the sequence. The surviving sequences were subjected to a size filter (minimum sizes, 350 bp for hsp65 and 670 bp for rpoB). Next, the resulting sequences with less than 90% of the bases with phred values above 15 across the minimum size were filtered. These sequences were aligned by using the BioEdit program (version 7.0.5.3) (14) and the CLUSTALW algorithm.
Molecular typing.
Pulsed-field gel electrophoresis (PFGE) was performed as described by Coleman and Spain (7), with modifications. Colonies were cultivated in 30 ml of Mueller-Hinton broth supplemented with 0.1% Tween 80 at 37°C on a rotatory shaker until an optical density of 0.64 at 650 nm was achieved. The cells were pelleted and frozen at −80°C for 1 h. After they were thawed, the cell pellets were resuspended in a solution of 100 mM NaCl, 10 mM Tris, pH 8.0, 50 mM EDTA, and 0.1% Tween 80. The suspension was mixed with an equal volume of 1% low-melting-point preparative-grade agarose (Bio-Rad Laboratories) in 125 mM EDTA prewarmed to 55°C, and the mixture was cast into plug molds. The plugs were treated with 2 ml of 10 mg/ml lysozyme in a solution of 100 mM NaCl, 10 mM Tris, pH 8.0, and 50 mM EDTA at 37°C overnight and were then incubated at 4°C for 1 h in 0.5 M EDTA plus 1% Sarkosyl. Proteinase K (BioAmerica, Brazil) was added at a final concentration of 2 mg/ml, and the plugs were incubated at 55°C for 24 h and then at 4°C for 1 h. The plugs were washed with 1× Tris-EDTA (TE) and incubated in 1× TE containing 0.12 mg/ml phenylmethylsulfonyl fluoride (Sigma) for 1 h at 55°C. The plugs were washed once in 1× TE and once in 0.1% Triton for 90 min and digested with 30 U of DraI (Promega) at 37°C overnight or were stored in 0.5 M EDTA at 4°C until they were used. After digestion, the plugs were washed in 0.05 M EDTA, loaded into 1% pulsed-field-certified agarose gels (Bio-Rad) prepared in 0.5× TBE buffer (45 mM Tris-HCl, 45 mM boric acid, 1 mM EDTA), and subjected to electrophoresis in 0.5× TBE buffer. PFGE was carried out in a CHEF-DR III system (Bio-Rad) at 14°C for 24 h at 6 V/cm, with a switch time of 1.6 to 21.3 s. A bacteriophage lambda ladder PFG marker (New England BioLabs) was used as a molecular standard.
The PFGE gel images were analyzed with the BioNumerics program (version 4.5; AppliedMaths, Sint-Martens-Latem, Belgium). The PFGE patterns of type strains M. abscessus ATCC 19977 and M. chelonae ATCC 35752 were included in the analysis. The band-based Dice unweighted pair group method with arithmetic mean method was used to prepare dendrograms of the PFGE profiles, based on 2% optimization and position tolerance. The PFGE interpretative criteria were those described by Tenover et al. (24).
Nucleotide sequence accession numbers.
The consensus sequences obtained from isolate B5 were deposited in GenBank under accession numbers EU117205 (hsp65) and EU117207 (rpoB). The hsp65 and rpoB gene sequences from isolates B67, B60, B62, and B66, which represented all sequevars detected in this work, were deposited in GenBank under accession numbers EU220417 to EU220424.
RESULTS
Outbreak description.
From February 2004 to June 2005, 311 patients who underwent different types of surgeries involving the use of laparoscopes or cannulas or other invasive procedures, such as mesotherapy and intramuscular injection (Table 1), presented with signs and symptoms of localized infection. The clinical manifestations included local hyperemia; vesicles; abscess formation with either an acute inflammatory reaction and festering or a chronic evolution; nodules; ulceration at the portal of entry of laparoscopes; fistulization with serous, bloody, or purulent secretions; difficulty in cicatrization; low fever; and a lack of a response to conventional treatment for common skin bacterial pathogens. Generalized infections or deaths were not reported.
TABLE 1.
Type of surgery or invasive procedure performed for patients from this study
| Procedure | No. of patients | % of all patients |
|---|---|---|
| Cholecystectomy | 182 | 58.52 |
| Hiatal hernia | 22 | 7.07 |
| Diagnostic laparoscopy | 15 | 4.82 |
| Bariatric surgery | 11 | 3.54 |
| Other surgical proceduresa | 42 | 13.5 |
| Mesotherapy | 14 | 4.5 |
| NRb | 25 | 8.04 |
| Total | 311 | 100 |
Other surgical procedures included mammaplasty, liposuction, ooforectomy, miomectomy, appendectomy, salpingectomy, cystectomy, and kidney stone extraction.
NR, not reported.
All surgeries were performed in 16 private hospitals in the city of Belém and mesotherapy was performed in one private clinic in Belém. Sixty-seven percent of the cases were concentrated in three hospitals, and in 17 cases, the institutions where the procedures were performed were not reported. One patient received a contraceptive injection in a different city and developed a local abscess, which was initially attributed to a Proteus sp. Treatment for this abscess, including antibiotics and surgical debridement, was performed in Belém, and an additional specimen yielded a positive culture for RGM.
A total of 225 (72.35%) patients were female and 86 (27.65%) were male, and for 1 (0.31%) patient this information was unknown. The patients’ ages ranged from 10 to 89 years (mean, 45 years; standard deviation, 13.87 years).
Identification results.
Sixty-seven isolates, which were obtained at the Instituto Evandro Chagas, were identified as rapidly growing nonpigmented mycobacteria and presented a pattern by PRA-hsp65 with BstEII with bands of 235 of 210 bp and with HaeIII with bands of 200, 70, 60, and 50 bp, which is common to M. abscessus type 2, M. bolletii, and M. massiliense.
To discriminate between these three species, a subset of 20 isolates, including 11 isolates chosen at random from among the 58 isolates from surgical patients, all 8 isolates from mesotherapy patients, and the single isolate from the postinjection abscess, was submitted to DNA sequencing of the hsp65 and rpoB genes. Consensus hsp65 and rpoB sequences were initially obtained by cloning the amplicons from isolate B5, which was chosen at random for this purpose. BLASTn analysis of the hsp65 and rpoB consensus sequences showed the highest indexes of similarity with the corresponding sequences from M. massiliense (GenBank accession numbers AY596465 and AY593981) and M. bolletii (GenBank accession numbers AY859675 and AY859692) (Table 2).
TABLE 2.
BLASTn analysis of the consensus hsp65 (401 bp) and rpoB (711 bp) sequences from isolate B5
| Gene region | GenBank accession no. | Strain | No. of identical nucleotides/total no. | Index of similarity (%) to isolate B5 |
|---|---|---|---|---|
| hsp65 | AY596465 | M. massiliense CIP 108297 | 401/401 | 100 |
| AY859675 | M. bolletii CIP 108541 | 398/401 | 99.25 | |
| AY458075 | M. abscessus CIP 104536 | 396/401 | 98.75 | |
| AY458074 | M. chelonae CIP 104535 | 371/401 | 92.5 | |
| DQ288262 | M. immunogenum F1112 | 370/401 | 92.26 | |
| AY458081 | M. immunogenum CIP 106684 | 367/401 | 91.52 | |
| rpoB | AY593981 | M. massiliense CIP 108297 | 706/711 | 99.29 |
| AY859692 | M. bolletii CIP 108541 | 700/711 | 98.45 | |
| DQ288264 | M. immunogenum F1112 | 633/651 | 97.23 | |
| AY262739 | M. immunogenum CIP 106684 | 691/711 | 97.18 | |
| AY147164 | M. abscessus CIP 104536 | 686/711 | 96.48 | |
| AY147163 | M. chelonae CIP 104535 | 679/711 | 95.49 |
The other sequences were obtained by direct sequencing of the PCR products. CLUSTALW analysis of the hsp65 sequences revealed that the sequences from the surgical isolates and the postinjection abscess isolate presented 100% similarity to the sequence of M. massiliense CIP 108297 deposited in the GenBank database and differed from the sequence of M. bolletii CIP 108541 at three positions (99.25% similarity). Six of the eight sequences from the mesotherapy isolates were 100% similar to the sequence from the M. bolletii type strain (GenBank accession number AY859675); two isolates (isolates B59 and B60) presented a single substitution at position 253 (99.75% similarity).
The rpoB sequences from the surgical isolates analyzed were identical. Their sequences presented five mismatches compared to the sequence of M. massiliense type strain CIP 108297 deposited in the GenBank database (99.29% similarity) and 11 mismatches compared to the sequence of M. bolletii CIP 108541 (98.45% similarity). The rpoB sequence of the postinjection abscess isolate presented 99.1% similarity with the sequence of M. massiliense CIP 108297 and 98% similarity with that of M. bolletii CIP 108541. It also differed from the rpoB sequence of the surgical isolates at five positions (99.25% similarity).
The mesotherapy isolates comprised three rpoB sequevars. The rpoB sequences from two isolates (isolates B59 and B60) presented 100% similarity to the corresponding sequence of M. bolletii CIP 108541 and 98% similarity to the sequence of M. massiliense CIP 108297. Compared to the rpoB sequence of M. bolletii CIP 108541, the rpoB sequences of three isolates (isolates B61, B62, and B65) had two substitutions (99.56% similarity) and those of the remaining isolates (isolates B63, B64, and B66) had one substitution (99.85% similarity). Compared to the rpoB sequence of M. massiliense CIP 108297, these two sequevars presented 97.66% and 97.93% similarities, respectively.
In conclusion, the isolates from the surgical patients and from the postinjection abscess were identified by the highest similarity of sequences as M. massiliense, while the mesotherapy isolates were classified as M. bolletii.
Molecular typing results.
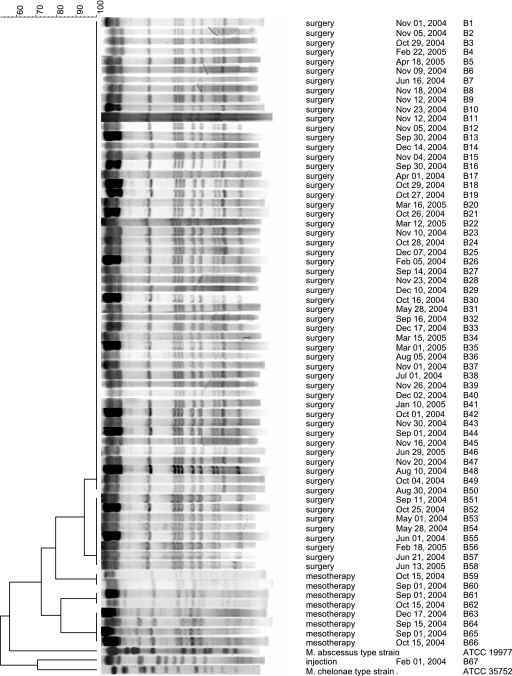
All isolates generated interpretable patterns by PFGE. Except for eight isolates that lacked one band, the isolates from surgical patients presented indistinguishable patterns (Fig. 1). According to the criteria proposed by Tenover et al. (24), these isolates were considered highly related and to belong to the same strain. The M. massiliense isolate from the postinjection abscess presented a clearly distinct PFGE pattern, showing 46.7% similarity with the patterns from the surgical isolates.
FIG. 1.
Dendrogram of PFGE patterns of isolates from this study. Indistinguishable patterns grouped the isolates from surgical patients; however, the patterns of eight isolates lacked one restriction band. Three clusters of indistinguishable patterns were detected in the mesotherapy isolates, but none of the patterns matched the patterns of the surgical isolates. The columns on the right indicate the type of procedure, the procedure date, and the isolate number. Comparison was performed with the BioNumerics (version 4.5) program and by analysis by the Dice unweighted pair group method with arithmetic mean method, based on 2% optimization and position tolerance. The PFGE patterns of M. abscessus ATCC 19977 and M. chelonae ATCC 35752 were included in the analysis.
The mesotherapy isolates were distributed in three clusters, each of which showed indistinguishable PFGE patterns. The clusters comprised four isolates (isolates B63 to B66), two isolates (isolates B59 and B60), and two isolates (isolates B61 and B62), respectively. These PFGE patterns differed from the patterns of the surgical isolates and the postinjection abscess isolate (Fig. 1). The overall similarity of the PFGE patterns of the mesotherapy isolates was 68.7%.
DISCUSSION
From 1999 to 2003, between 4 and 12 nontuberculous mycobacterial infections were diagnosed each year at the Mycobacteria Laboratory of the Instituto Evandro Chagas in Belém, Brazil, and no more than one isolate per year presented the PRA-hsp65 pattern reported here. Most isolates were obtained from respiratory specimens. Unexpectedly, from October 2004 on, several isolates that presented this PRA-hsp65 pattern were isolated from specimens from patients who had undergone invasive procedures. An epidemiological investigation was initiated, and in parallel, the isolates were identified and typed by the use of molecular methods.
This PRA-hsp65 pattern was initially described as characteristic of that of M. abscessus (8) and was later observed in two recently described species from the M. chelonae-M. abscessus group, M. massiliense and M. bolletii (1, 4). Discrimination of these closely related species is best achieved by DNA sequencing. By consideration of the 401-bp fragment from the hsp65 gene, the sequences of M. massiliense (GenBank accession number AY596465) and M. bolletii (GenBank accession number AY859675) differ at three nucleotides (99.25% similarity); however, this polymorphism does not affect the PRA-hsp65 pattern. Region V of the rpoB gene was shown to be more polymorphic (1, 2). In general, the RGM interspecies sequence divergence is >3%, and isolates presenting less than 1.7% sequence divergence are considered to belong to the same species (2). M. massiliense CIP 108297 and M. bolletii CIP 108541 have 98% similarity in the rpoB region V sequences and are presently considered to be two separate species.
As the differentiation of these species by hsp65 and rpoB sequencing relies on few polymorphisms, the quality of the sequences obtained is of utmost importance. For this reason, a strategy was devised in which one isolate (isolate B5), chosen from among the 58 isolates from surgical patients which presented indistinguishable PFGE patterns, had hsp65 and rpoB amplicons cloned to obtain reliable consensus sequences. The highest similarities of the consensus sequences of isolate B5 were observed with the respective sequences of M. massiliense (Table 2). The hsp65 and rpoB sequences of 10 additional isolates from surgical patients, obtained by direct sequencing of the PCR products, were indistinguishable from the hsp65 and rpoB sequences of isolate B5.
It is important to point out that even though the highest sequence similarity for rpoB (99.29%) was observed with the corresponding sequence from M. massiliense, the similarity to the rpoB sequence from M. bolletii was also high (98.45%), corresponding to divergences of <1.7% in both cases. According to the criteria proposed by Adekambi et al. (2), these isolates could be identified as either M. massiliense or M. bolletii. The rpoB sequences from surgical isolates showed 100% similarity to two recently deposited rpoB sequences (GenBank accession numbers EU090065 and EU090066) from isolates identified as M. massiliense in a recent publication (22). Taking all of this into account, M. massiliense was considered the conclusive identification for the isolates from the surgical patients described here.
The hsp65 and rpoB sequences of the postinjection abscess isolate also showed the highest similarities to the corresponding sequences of M. massiliense. Compared to the sequences of the surgical isolates, the hsp65 sequence of the postinjection abscess isolate was identical and the rpoB sequence of the postinjection abscess isolate presented five mismatches, suggesting that this isolate belonged to a different strain of M. massiliense and indicating that this patient was not epidemiologically related to the surgical cases. Molecular typing results confirmed that this isolate presented a distinct PFGE pattern, strongly suggesting that this abscess represented an isolated infection not related to the outbreak.
M. massiliense was reported to be susceptible to doxycycline, which could distinguish it from resistant M. abscessus and M. bolletii isolates (4). M. massiliense isolates with intermediate susceptibility to doxycycline have also been described (15). In the present study, nine M. massiliense isolates from surgical patients (isolates B51, B41, B31, B5, B22, B28, B43, B48, and B19) were shown to be resistant to doxycycline (MICs > 64 μg/ml) (data not shown) These results agree with those reported by Simmon et al. (22), indicating that this species does not have a unique pattern of susceptibility to this drug. Moreover, the doxycycline resistance and the identity of the rpoB region V sequences of Brazilian and U.S. isolates (22) could suggest the existence of a variant clone of M. massiliense in the Americas.
A different result was obtained with isolates from mesotherapy patients. The hsp65 and rpoB sequences presented the highest similarities with the corresponding sequences from M. bolletii CIP 108541, and this is probably the correct identification for these isolates. Isolates from mesotherapy patients also presented three distinct PFGE patterns that were clearly separated from the patterns observed for the surgical isolates. Therefore, even though the mesotherapy infections were concomitant with the surgical cases, they are most likely unrelated to the outbreak of laparoscopic surgeries and may have had different sources.
The sources of the infections for the surgical cases have not been identified here. The patients were operated on by different surgeons, who used their own laparoscopic equipment, which was disinfected by immersion in 2% glutaraldehyde between surgeries and which was used in different hospitals. Glutaraldehyde, a frequently employed biocide, can be used for the high-level disinfection of some medical equipment after careful cleaning for the elimination of organic substances. Without adequate cleaning, surgical or medical equipment can accumulate organic material, generating biofilms and preventing the action of disinfectants. Our hypothesis is that inconsistencies in equipment cleaning, glutaraldehyde concentrations, or contact times may have contributed to the selection of the M. massiliense strain described here, which contaminated the surgical equipment. The use of laparoscopic equipment in several hospitals disseminated the strain to the different outbreak settings. Cases of infection after laparoscopic surgeries subsided after the cleaning and sterilization procedures were reviewed. No RGM infections were reported in patients who underwent different surgical procedures, in which laparoscopic equipment or cannulas and disinfection with glutaraldehyde were not employed, during the same period and in the same hospitals.
The prevalence of this particular M. massiliense strain could have originated from its atypical survival ability in different environments, resistance to biocides, and/or enhanced biofilm formation. It is also possible that this particular strain may be unusually resistant to the disinfectant used, and experiments to test this hypothesis are under way.
In conclusion, this outbreak, which was initially attributed to M. abscessus, in fact consisted of two concomitant but different outbreaks. The main outbreak grouped 298 surgical patients who were operated on in several hospitals and was probably caused by a single strain of M. massiliense. The other outbreak clustered 14 mesotherapy patients who were treated in a private clinic and involved multiple strains of M. bolletii. The postinjection abscess case was not related to either outbreak.
Acknowledgments
Rosângela Siqueira de Oliveira is acknowledged for performing the doxycycline susceptibility tests, and Silvia Yukie Kawashita is acknowledged for helping with sequence analysis.
This work was supported by the Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP; process no. 06/01533-9) and the Conselho Nacional de Desenvolvimento Científico e Tecnológico (process no. 480594-06-3 universal). C.V.-N. was the recipient of a fellowship from FAPESP (proc. no. 04/07394-5).
Footnotes
Published ahead of print on 3 January 2008.
REFERENCES
- 1.Adekambi, T., P. Berger, D. Raoult, and M. Drancourt. 2006. rpoB gene sequence-based characterization of emerging non-tuberculous mycobacteria with descriptions of Mycobacterium bolletii sp. nov., Mycobacterium phocaicum sp. nov. and Mycobacterium aubagnense sp. nov. Int. J. Syst. Evol. Microbiol. 56133-143. [DOI] [PubMed] [Google Scholar]
- 2.Adekambi, T., P. Colson, and M. Drancourt. 2003. rpoB-based identification of nonpigmented and late-pigmenting rapidly growing mycobacteria. J. Clin. Microbiol. 415699-5708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adekambi, T., and M. Drancourt. 2004. Dissection of phylogenetic relationships among 19 rapidly growing Mycobacterium species by 16S rRNA, hsp65, sodA, recA and rpoB gene sequencing. Int. J. Syst. Evol. Microbiol. 542095-2105. [DOI] [PubMed] [Google Scholar]
- 4.Adekambi, T., M. Reynaud-Gaubert, G. Greub, M. J. Gevaudan, B. La Scola, D. Raoult, and M. Drancourt. 2004. Amoebal coculture of “Mycobacterium massiliense” sp. nov. from the sputum of a patient with hemoptoic pneumonia. J. Clin. Microbiol. 425493-5501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown-Elliott, B. A., D. E. Griffith, and R. J. Wallace, Jr. 2002. Diagnosis of nontuberculous mycobacterial infections. Clin. Lab. Med. 22911-925, vi. [DOI] [PubMed] [Google Scholar]
- 6.Brown-Elliott, B. A., and R. J. Wallace, Jr. 2002. Clinical and taxonomic status of pathogenic nonpigmented or late-pigmenting rapidly growing mycobacteria. Clin. Microbiol. Rev. 15716-746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coleman, N. V., and J. C. Spain. 2003. Distribution of the coenzyme M pathway of epoxide metabolism among ethene- and vinyl chloride-degrading Mycobacterium strains. Appl. Environ. Microbiol. 696041-6046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Devallois, A., K. S. Goh, and N. Rastogi. 1997. Rapid identification of mycobacteria to species level by PCR-restriction fragment length polymorphism analysis of the hsp65 gene and proposition of an algorithm to differentiate 34 mycobacterial species. J. Clin. Microbiol. 352969-2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Devulder, G., M. Perouse de Montclos, and J. P. Flandrois. 2005. A multigene approach to phylogenetic analysis using the genus Mycobacterium as a model. Int. J. Syst. Evol. Microbiol. 55293-302. [DOI] [PubMed] [Google Scholar]
- 10.Durham, A. M., A. Y. Kashiwabara, F. T. Matsunaga, P. H. Ahagon, F. Rainone, L. Varuzza, and A. Gruber. 2005. EGene: a configurable pipeline generation system for automated sequence analysis. Bioinformatics 212812-2813. [DOI] [PubMed] [Google Scholar]
- 11.Euzéby, J. P. 2007, posting date. List of bacterial names with standing in nomenclature. http://www.bacterio.cict.fr/m/mycobacterium.html. Societé de Bactériologie Systématique et Vétérinaire, Toulouse, France.
- 12.Freitas, D., L. Alvarenga, J. Sampaio, M. Mannis, E. Sato, L. Sousa, L. Vieira, M. C. Yu, M. C. Martins, A. Hoffling-Lima, and R. Belfort, Jr. 2003. An outbreak of Mycobacterium chelonae infection after LASIK. Ophthalmology 110276-285. [DOI] [PubMed] [Google Scholar]
- 13.Gordon, D., C. Abajian, and P. Green. 1998. Consed: a graphical tool for sequence finishing. Genome Res. 8195-202. [DOI] [PubMed] [Google Scholar]
- 14.Hall, T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 4195-98. [Google Scholar]
- 15.Kim, H. Y., Y. J. Yun, C. G. Park, D. H. Lee, Y. K. Cho, B. J. Park, S. I. Joo, E. C. Kim, Y. J. Hur, B. J. Kim, and Y. H. Kook. 2007. Outbreak of Mycobacterium massiliense infection associated with intramuscular injections. J. Clin. Microbiol. 453127-3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kusunoki, S., and T. Ezaki. 1992. Proposal of Mycobacterium peregrinum sp. nov., nom. rev., and elevation of Mycobacterium chelonae subsp. abscessus (Kubica et al.) to species status: Mycobacterium abscessus comb. nov. Int. J. Syst. Bacteriol. 42240-245. [DOI] [PubMed] [Google Scholar]
- 17.Leao, S. C., A. Martin, G. I. Mejia, J. C. Palomino, J. Robledo, M. A. S. Telles, and F. Portaels. 2004. Practical handbook for the phenotypic and genotypic identification of mycobacteria. Vanden Broelle, Bruges, Belgium.
- 18.Lévy-Frébault, V., F. Grimont, P. A. D. Grimont, and H. L. David. 1986. Deoxyribonucleic acid relatedness study of the Mycobacterium fortuitum-Mycobacterium chelonae complex. Int. J. Syst. Bacteriol. 36458-460. [Google Scholar]
- 19.Sampaio, J. L., D. N. Junior, D. de Freitas, A. L. Hofling-Lima, K. Miyashiro, F. L. Alberto, and S. C. Leao. 2006. An outbreak of keratitis caused by Mycobacterium immunogenum. J. Clin. Microbiol. 443201-3207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sampaio, J. L., C. Viana-Niero, D. de Freitas, A. L. Hofling-Lima, and S. C. Leao. 2006. Enterobacterial repetitive intergenic consensus PCR is a useful tool for typing Mycobacterium chelonae and Mycobacterium abscessus isolates. Diagn. Microbiol. Infect. Dis. 55107-118. [DOI] [PubMed] [Google Scholar]
- 21.Selvaraju, S. B., I. U. Khan, and J. S. Yadav. 2005. A new method for species identification and differentiation of Mycobacterium chelonae complex based on amplified hsp65 restriction analysis (AHSPRA). Mol. Cell. Probes 1993-99. [DOI] [PubMed] [Google Scholar]
- 22.Simmon, K. E., J. I. Pounder, J. N. Greene, F. Walsh, C. M. Anderson, S. Cohen, and C. A. Petti. 2007. Identification of an emerging pathogen, Mycobacterium massiliense, by rpoB sequencing of clinical isolates collected in the United States. J. Clin. Microbiol. 451978-1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Telenti, A., F. Marchesi, M. Balz, F. Bally, E. C. Bottger, and T. Bodmer. 1993. Rapid identification of mycobacteria to the species level by polymerase chain reaction and restriction enzyme analysis. J. Clin. Microbiol. 31175-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 332233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tiwari, T. S., B. Ray, K. C. Jost, Jr., M. K. Rathod, Y. Zhang, B. A. Brown-Elliott, K. Hendricks, and R. J. Wallace, Jr. 2003. Forty years of disinfectant failure: outbreak of postinjection Mycobacterium abscessus infection caused by contamination of benzalkonium chloride. Clin. Infect. Dis. 36954-962. [DOI] [PubMed] [Google Scholar]
- 26.Wallace, R. J., Jr. 1994. Recent changes in taxonomy and disease manifestations of the rapidly growing mycobacteria. Eur. J. Clin. Microbiol. Infect. Dis. 13953-960. [DOI] [PubMed] [Google Scholar]
- 27.Wilson, R. W., V. A. Steingrube, E. C. Bottger, B. Springer, B. A. Brown-Elliott, V. Vincent, K. C. Jost, Jr., Y. Zhang, M. J. Garcia, S. H. Chiu, G. O. Onyi, H. Rossmoore, D. R. Nash, and R. J. Wallace, Jr. 2001. Mycobacterium immunogenum sp. nov., a novel species related to Mycobacterium abscessus and associated with clinical disease, pseudo-outbreaks and contaminated metalworking fluids: an international cooperative study on mycobacterial taxonomy. Int. J. Syst. Evol. Microbiol. 511751-1764. [DOI] [PubMed] [Google Scholar]