Abstract
Human papillomavirus (HPV) DNA detection and typing are important for diagnosis and management of HPV-associated diseases. One of the most commonly used PCR methods, GP5+/6+, shows weaknesses in amplifying certain types. To circumvent this limitation, we developed and validated broad-spectrum primers targeting the GP5+/6+ region. The addition of eight upstream and two downstream BSGP5+/6+ (BS) primers improved amplification of plasmids of 14 genital HPV types 10- to 1,000-fold versus GP5+/6+ PCR without altering sensitivity for the 10 others. For these 24 types, an analytic sensitivity of ≤1,000 plasmid copies in the presence of 100 ng cellular DNA was obtained. Additionally, we integrated an internal β-globin PCR into both HPV PCR systems, allowing simultaneous DNA quality control without affecting the sensitivity of HPV detection. Furthermore, we describe five additional low-risk HPV probes used in multiplex HPV genotyping (MPG) for simultaneous identification of all 15 high-risk, 3 putative high-risk, and 9 low-risk HPV genotypes. The performance of BSGP5+/6+ multiplexed with β-globin primers was compared to that of standard GP5+/6+ with DNA from 1,112 cervical scrapings. There was 79% overall agreement (kappa = 0.816). BSGP5+/6+ was significantly more sensitive than GP5+/6+ for detection of HPV 30, 39, 42, 44, 51, 52, 53, 68, 73, and 82, detecting 212 additional HPV infections and increasing the proportion of multiple infections from 17.2 to 26.9% in cancer patients. In conclusion, BSGP5+/6+ multiplexed with β-globin PCR provides an improvement in type-specific amplification sensitivity and homogeneity compared to GP5+/6+ and offers simultaneous internal control of DNA quality. BSGP5+/6+-MPG, therefore, is suitable for epidemiologic and also diagnostic applications.
High-risk human papillomavirus (HR HPV) types are causally associated with several malignant diseases, of which cervical cancer has particular significance, being the second most common cancer in women worldwide (11). Until now, more than 100 HPV genotypes are fully characterized based on the isolation of complete genomes, with evidence that a larger number exist (6). There are 48 known mucosal HPV types, which are further divided into three groups based on their epidemiological association with cancer: HR types, including types 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 68, 73, and 82; putative HR (pHR) types, including types 26, 53, and 66; and low-risk (LR) types, including types 6, 11, 40, 42, 43, 44, and 70 (11). HPV genotyping is of particular importance for studying the natural history of HPV infections and the diseases associated therewith, for monitoring vaccine efficacy, and for identifying novel HPV candidates for vaccine development. Further, it is required to diagnose multiple HPV infections.
At present, there are several PCR-based HPV detection methods; one of the most commonly applied uses the GP5+/6+ (GP) primer set (18), which targets conserved sequences within the L1 region of the virus genome flanking highly variable type-specific sequences, allowing detection of a broad range of mucosal HPV types. The HPV genotype can be determined by analysis of the generated PCR product by sequencing, restriction fragment length polymorphism, or hybridization with type-specific probes.
Since the design of GP primers (5), many additional mucosal HPV types have been discovered. It has been shown that the analytic sensitivity of GP PCR for individual genital HPV types differs depending on the number and position of primer mismatches (5). Furthermore, GP PCR detects multiple infections to a lesser extent than other consensus or broad-spectrum PCR systems (9, 13). The selective underamplification of certain HPV types, such as HPV 53, and/or the reduced sensitivity in multiple HPV infections may lead to an underestimation of the prevalence of specific types (1, 2, 13).
Control of template DNA quality is important in the analysis of clinical samples to pinpoint false-negative HPV results. GP PCR is mostly applied to samples prescreened by external β-globin PCR and subsequent PCR product analysis by gel electrophoresis (5, 8). Integration of an internal DNA quality control in a multiplexed fashion has been described for MY09/11 and PGMY09/11 PCR but may decrease the analytic sensitivity for HPV (7). Therefore, an internal DNA control without impairment of the HPV amplification sensitivity is needed not only to reduce assay time and costs but also to provide an internal PCR control for monitoring a failure of the HPV PCR.
Recently, we described a multiplex HPV genotyping assay (MPG) based on GP PCR followed by subsequent detection of the biotinylated products with 22 type-specific oligonucleotide probes covalently coupled to distinct sets of fluorescence-labeled polystyrene beads (Luminex Technology) (17). Meanwhile, MPG allows simultaneous semiquantitative high-throughput analysis for 15 HR, 3 pHR, and 9 LR mucosal HPV genotypes.
To reach a homogeneous analytic sensitivity for all genital HPV types, we designed the novel BSGP5+/6+ (BS) primer set on the basis of mismatch reduction to 48 genital HPV types. Novel β-globin primers were integrated in the BS PCR as well as the standard GP PCR for internal DNA quality control. We compared both PCR-MPG systems with respect to the amplification of 27 genital HPV types in 1,017 cervical samples from a Mongolian population-based study (B. Dondog, G. M. Clifford, S. Vaccarella, T. Waterboer, D. Unurjargal, D. Avirmed, S. Enkhtuya, F. Kommoss, N. Wentzensen, P. J. F. Snijders, C. J. L. M. Meijer, S. Franceschi, and M. Pawlita, submitted for publication) and 95 cervical cancer patients.
MATERIALS AND METHODS
HPV sequence alignment and primer design.
The L1 regions of 48 completely sequenced HPV genotypes (HR HPV types 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 68, 73, 82; pHR HPV types 26, 53, and 66; and LR HPV types 6, 7, 11, 13, 30, 32, 34, 40, 42, 43, 44, 54, 55, 61, 62c, 67, 69, 70, 72, 74, 81, 83, 84, 85, 86c, 87c, CP6108, 90, 97, 102, and 106) were obtained from the National Center for Biotechnology Information nucleotide sequence database (GenBank) and were aligned with T-COFFEE (12). In total, eight forward and two biotinylated reverse primers were designed (Table 1) . The criteria for the design of BS primers were as follows: pools of oligonucleotides were preferred over incorporation of degenerate base sites to avoid synthesis batch variation, and all novel primers were to be identical in length and targeting the same region as GP primers.
TABLE 1.
BSGP5+/6+ primer sequences
| Primer namea | Directionb | Sequencec (5′ to 3′) |
|---|---|---|
| β-Globin | ||
| MS3 | F | AAT ATA TGT GTG CTT ATT TG |
| Bio-MS10 | R | AGA TTA GGG AAA GTA TTA GA |
| HPV | ||
| GP5+ | F | TTT GTT ACT GTG GTA GAT ACT AC |
| BSGP5+-2 | F | TTT GTT ACT GTT GTI GAT ACT AC |
| BSGP5+-3 | F | TTT GTT ACT GTT GTI GAT ACC AC |
| BSGP5+-4 | F | TTT GTT ACT TGT GTI GAT ACT AC |
| BSGP5+-5 | F | TTT TTA ACT GTT GTI GAT ACT AC |
| BSGP5+-6 | F | TTT GTT ACT GTG GTA GAC ACT AC |
| BSGP5+-7 | F | TTT GTT ACA GTI GTA GAC ACT AC |
| BSGP5+-8 | F | TTT GTT ACA GTI GTA GAT ACC AC |
| BSGP5+-9 | F | TTT GTT ACT GTG GTA GAT ACC AC |
| Bio-GP6+ | R | GAA AAA TAA ACT GTA AAT CAT ATT C |
| Bio-BSGP6+-b | R | GAA AAA TAA ATT GTA AAT CAT ACT C |
| Bio-BSGP6+-c | R | GAA AAA TAA ATT GCA ATT CAT ATT C |
Bio-, biotinylated.
F, forward; R, reverse.
β-Globin primers are 20 nt, BSGP5+/6+ forward primers are 23 nt, and BSGP5+/6+ backward primers are 25 nt in length. I, inosine.
Seven sets of β-globin primers (MS1-7/MS8-14) with a length of 20 to 22 nucleotides (melting temperature, 45 to 50°C) and allowing amplification of fragments 200 to 240 nucleotides in length were designed with Primer3 software (15), using the β-globin gene (AY260740) as a reference sequence.
Plasmid clones.
Analytic sensitivity of the BS primers was determined for plasmid clones of HPV types 6, 11, 16, 18, 26, 31, 33, 35, 39, 42, 43, 44, 45, 51, 52, 53, 56, 58, 59, 66, 68, 70, 73, and 82 (Table 2). Plasmid preparations were quantified using NanoDrop ND-1000 (NanoDrop Technologies, Wilmington, DE) or a Hitachi U-1100 spectrophotometer (Hitachi Ltd., Tokyo, Japan). Copy numbers were determined on the basis of the molecular weights of the plasmids. Tenfold endpoint dilution series were prepared in 100 ng/μl of human placenta (HP) DNA in a total volume of 30 μl. Two or three replicates of each dilution were assayed independently.
TABLE 2.
HPV type detection limits
| HPV type | Detection limit (plasmid copy number in 100 ng HP DNA)
|
GP/BSa (both with β-globin) | ||
|---|---|---|---|---|
| GP5+/6+ | GP5+/6+ plus β-globin | BSGP5+/6+ plus β-globin | ||
| 33b | 10 | 10 | 1 | |
| 58b | 10 | 10 | 1 | |
| 16c | 100 | 100 | 100 | 1 |
| 66c | 100 | 100 | 100 | 1 |
| 35b | 100 | 10 | 10 | |
| 11c | 100 | 100 | 100 | 1 |
| 18b | 100 | 100 | 1 | |
| 31b | 100 | 100 | 1 | |
| 59b | 100 | 100 | 100 | 1 |
| 51c | 1,000 | 1,000 | 10 | 100 |
| 6c | 1,000 | 1,000 | 100 | 10 |
| 42b | 1,000 | 100 | 10 | |
| 45c | 1,000 | 1,000 | 100 | 10 |
| 52b | 1,000 | 100 | 10 | |
| 70c | 1,000 | 1,000 | 100 | 10 |
| 43b | 1,000 | 1,000 | 1 | |
| 26b | 10,000 | 100 | 100 | |
| 39c | 10,000 | 10,000 | 100 | 100 |
| 56c | 10,000 | 10,000 | 100 | 100 |
| 68c | 10,000 | 10,000 | 100 | 100 |
| 73c | 10,000 | 10,000 | 1,000 | 10 |
| 53b | 100,000 | 100 | 1,000 | |
| 82c | 100,000 | 100,000 | 100 | 1,000 |
| 44c | 1,000,000 | 1,000 | 1,000 | |
Ratio of detection limit for GP5+/6+ to that for BSGP5+/6+, both with β-globin coamplification. 1, equal detection limits; 10, detection limit improved 10-fold by BS.
One dilution series; worst detection limit out of quadruplicate PCR is shown.
Two independent dilution series; worst detection limit out of at least two PCR per dilution series is shown.
For colony PCR, DNA from DH5α bacteria, transformed by high-copy-number plasmids containing the viral genome, replaced the template DNA.
MPG probe design.
Amino-modified oligonucleotide probes for HPV 30 (5′-CAC ACA AAC GTT ATC CAC A-3′), 67 (5′-GGA AAA ATC AGA GGC TAC A-3′), and 69 (5′-CAT CTG CCA CTT TTA AAC C-3′) were newly designed as described previously (17). Probe sequences for HPV 26 (5′-GTA CAT TAT CTG CAG CAT C-3′) and 53 (5′-TGT CTA CAT ATA ATT CAA AGC-3′) are described elsewhere (19). For specificity evaluation, cloned HPV genomes were used (for accession numbers, see Table 3).
TABLE 3.
BSGP5+/6+ and GP5+/6+ sequence alignmentsa
| HPV | Accession no. | Alignment with binding region
|
Alignment with best-fitting primer
|
||
|---|---|---|---|---|---|
| GP5+ | GP6+ | BSGP5+ | BSGP6+ | ||
| 6 | X00203 | ....................C.. | ..G...........A.......... | ....................... | ......................... |
| 7 | NC_001595 | ........A..T........... | ..G.T.................... | .................c..... | ....T.........a.......... |
| 11 | M14119 | ....................C.. | ..G.T.................... | ....................... | ....T.........a.......... |
| 13 | DQ344807 | ...........A..T........ | ....T.........A......... | ...........A........... | ....T.........A.......... |
| 16 | K02718 | ...........T..T........ | ......................... | ....................... | ......................... |
| 18 | X05015 | ....................C.. | ...........G............. | ....................... | ............G............ |
| 26 | NC_001583 | ........CTGT..T.....C.. | ........A.....A.....A.... | ........C...........C.. | ............g.......A.... |
| 30 | X74474 | ...........T..G..C..C.. | ..................G.G.... | .................C..... | ..................G.G.... |
| 31 | J04353 | ....................C.. | ....T.........A.....A.... | ....................... | ....T.........A.....A.... |
| 32 | NC_001586 | ...C.A.....T..G........ | .........A..........A.... | ...C.A................. | .........A..........A.... |
| 33 | M12732 | ....................C.. | .........C........G...... | ....................... | .........C........G...... |
| 34 | NC_001587 | ...T.A.....T........... | ..G.....CC.G......G.G.... | ..............i........ | ........CC.G..a...G.G.... |
| 35 | M74117 | ...........A..T.....A.. | ......................... | ...........A........A.. | ......................... |
| 39 | M62849 | ...C.......T..G..C..... | ..G...........A.....A.... | ...C.............C..... | ....................A.... |
| 40 | X74478 | ........A..T.....C..C.. | ..G.T......G............. | .................C..... | ....T......G..a.......... |
| 42 | M73236 | ...T.A........T........ | .........G.G..A.....A.... | ...........t........... | .........a..........A.... |
| 43 | AJ620205 | ........A...........C.. | .........C.G........A.... | ........A.............. | .........C.G........A.... |
| 44 | U31788 | ...........T........... | ..G.T...C.....A.....G.... | ...........T........... | ....T...C...........G.... |
| 45 | X74479 | ...........A.....C..... | ......................... | ...........A........... | ......................... |
| 51 | M62877 | ...A....CTGT..T........ | ..G.....A..G..A.......... | ...A....C.............. | ..G...................... |
| 52 | X74481 | .....C..A..T..G.....C.. | ....T.........A.......... | .....C..A.............. | ....T.........A.......... |
| 53 | X74482 | .....A.....T..G.....C.. | ........A.....A...G.G.... | .....A................. | ...........g......G.G.... |
| 54 | NC_001676 | ...T.A..A..T........C.. | ....................A.... | ...T.A................. | ....................A.... |
| 55 | U31791 | ...........T........... | ..G.T...C...........G.... | ...........T........... | ....T...C.....a.....G.... |
| 56 | X74483 | ...........A........... | ........A.....A...G...... | ...........A........... | ...........g......G...... |
| 58 | D90400 | ........C.....T.....C.. | ........C.........G...... | ........C..t........... | ........C.........G...... |
| 59 | X77858 | ...T.A..A..T........... | ....T......G........A.... | ........A.....i........ | ....T......G........A.... |
| 61 | U31793 | .....A..C..T..G.....C.. | ..G.T......G..A.......... | .....A..C.............. | ....T......G............. |
| c62 | AY395706 | ..............G........ | ....T......G..A.....A.... | ..............G........ | ....T...a...........A.... |
| 66 | U31794 | ...........T..G........ | ........AC........G.G.... | ....................... | ........AC........G.G.... |
| 67 | D21208 | ...........T.....C..... | ...........G........A.... | ...........T........... | ...........G........A.... |
| 68 (ME180) | AJ831568 | ...C.......T..G.....C.. | ...........G..A.....A.... | ...C................... | ........a...........A.... |
| 68 | X67161 | ...C....C..T..G.....A.. | ....................A.... | ...C....C...........A.. | ....................A.... |
| 69 | AB027020 | .........TGT........... | .........A..........A.... | ..............i........ | ........A...........A.... |
| 70 | U22461 | ...A..........G..C..... | ..............A.....A.... | ...A..........G........ | ..............A.....A.... |
| 72 | X94164 | .....G..A..T........... | ....T......G........A.... | .....G..A..T........... | ....T......G........A.... |
| 73 | NC_006165 | ...T.A.....T........... | ..G.T.............G...... | ..............i........ | ....T.........a...G...... |
| 74 | AF436130 | ........A..T..G.....C.. | ....T......G..A.......... | ........A.............. | ....T...a................ |
| 81 | AJ620209 | ........A.....G........ | ...........G...........C. | ........A..t........... | ...........G...........C. |
| 82 | AB027021 | ...A.....TGT..T..C..... | ........A..G..A.....A.... | ..A..............C..... | ....................A.... |
| 83 | AF151983 | ........A..T........... | ........C......G....A..G. | .................c..... | ........C......G....A..G. |
| 84 | AF293960 | .....C..G...........C.. | ...........G........A..C. | .....C..G.............. | ...........G........A..C. |
| c85 | AF131950 | ...A.A...........C..A.. | ..............A.....A.... | ...A.A..............A.. | ..............A.....A.... |
| c86 | AF349909 | ..............C..C..C.. | ...........G..A.......... | ..............C..C..... | ........a................ |
| c87 | AJ400628 | .....A..G..T..T........ | ......................... | .....A..G.............. | ......................... |
| CP6108 | U12478 | ....................C.. | ........CC..........A..C. | ....................C.. | ........CC..........A..C. |
| 90 | NC_004104 | .....A........T........ | ....T......G...........C. | .....A........T........ | ....T......G...........C. |
| 97 | DQ080080 | ..............G..C..A.. | ..G...................... | ..............G.....A.. | ..............a.......... |
| 102 | DQ080083 | .....A..A..T........... | ..............A.....C.... | .....A...........c..... | ..............A.....C.... |
| 106 | DQ080082 | .....C..............C.. | ....T......G..A.......... | .....C..............C.. | ....T...a................ |
Nucleotide homology is indicated with dots, and mismatches in the HPV sequence are in uppercase. For alignments with the respective best fitting BSGP5+/6+ primer, new mismatches are shown in lowercase.
A second universal probe (5′-GMC AYR CAG ARG AAT ATG A-3′) (M = A/C; Y = C/T; R = A/G) was designed as described elsewhere (17) to accommodate mismatches found between the first universal probe and HPV 34, 42, 53, 61, 72, 73, 81, and 83.
The β-globin probe (5′-CTT CTT TTA ATA TAC TTT TTT GTT-3′) was designed for the MS3/MS10 PCR product using the reference sequence AY260740 and following the criteria for HPV type-specific probe design (17).
DNA isolation from cervical scrapings.
For DNA isolation, 2.0 ml (from a total of 20 ml) of cervical scrapings collected with a cytobrush (Cervex-Brush, Rovers Medical Devices B.V., Oss, The Netherlands) in PreserveCyt solution (Cytyc Corp., Boxborough, MA) were purified with a Roche High Pure PCR template preparation kit according to the manufacturer's instructions. DNA was eluted in 0.2 ml of elution buffer (10 mM Tris, pH 8.5) and stored at −20°C until further use. Scrapes were kept 2 months at 4°C before DNA extraction.
Oligonucleotide synthesis.
All primers and probes were purchased from MWG-Biotech AG, Ebersberg, Germany.
GP5+/6+ PCR.
GP PCR was performed as previously described (5) with some modifications. Briefly, 10 μl of DNA extracted from cervical scrapings or 1 μl of HPV plasmid dilution was amplified in 50 μl containing 50 mM KCl, 0.8 g/liter Nonidet P40, 10 mM Tris HCl (pH 8.8) (10× PCR buffer; MBI Fermentas GmbH, St. Leon Roth, Germany), 200 μM of each deoxynucleoside triphosphate, 3.5 mM MgCl2 (Biozym Scientific GmbH, Hessisch Oldendorf, Germany), 1 U of DNA AmpliTaq polymerase (Roche Applied Biosystems, Mannheim, Germany) and 500 nM each of the GP5+ and 5′-biotinylated GP6+ primers. In the case of the integrated β-globin-GP5+/6+ PCR, 100 nM each of the β-globin primers MS3 and 5′biotinylated MS10 were added to the PCR mixture. A 4-min denaturation step at 94°C was followed by 40 cycles of amplification with a PCR thermocycler (Gene Amp PCR system 2400; Perkin-Elmer, Wellesley, MA) or a Mastercycler (Eppendorf). Each cycle included a denaturation step at 94°C for 20 s, an annealing step at 38°C for 30 s, and an elongation step at 71°C for 80 s. The final elongation step was prolonged for 4 min further. Ramping rates for the Mastercycler were adjusted as described recently (18): 1.8°C/s from 94°C to 38°C, 2.0°C/s from 38°C to 71°C, and 2,8°C/s from 71°C to 94°C. Each PCR experiment included samples with reference plasmids as positive controls and several samples lacking template DNA as contamination controls.
BSGP5+/6+ PCR.
For the initial BS PCR, eight additional forward (BSGP5+-2 to -9) and two additional 5′-biotinylated reverse (BSGP6+-b and -c) primers were added to the GP PCR (Table 1). Two hundred nanomolar of each forward primer (including GP5+), 400 nM of each reverse primer (including GP6+), and 300 nM each of the β-globin primers MS3 and 5′-biotinylated MS10 were used. Otherwise, the PCR buffers, reagents, and temperature profiles were identical to those described above.
Coupling of oligonucleotide probes.
5′-Amino-modified C-12-linked oligonucleotide probes were coupled to carboxylated beads (xMAP; Luminex Corp., Austin, TX) by a carbodiimide-based coupling procedure as described elsewhere (17).
MPG.
Following PCR amplification, 10 μl of each reaction was analyzed by MPG as described elsewhere (17) with some modifications. Instead of casein, 0.02% Tween 20 was added to the wash buffer, and casein was also omitted from the 2.0 M tetramethylammonium chloride staining buffer.
Briefly, GP PCR products were generated, denatured, and hybridized to the bead-coupled probes in 96-well plates, allowing PCR products from 96 samples to be processed in parallel. After transfer into wash plates with filter bottoms, unhybridized DNA was removed. Subsequently, biotinylated PCR products were stained by streptavidin-R-phycoerythrin conjugate. After further washing steps, beads were analyzed in a Luminex reader (Luminex Corp.), which contains two lasers to identify the bead set by the internal bead color and to quantify the reporter fluorescence on the bead. The result was expressed as the median fluorescence intensity (MFI) of at least 100 beads per set.
Cutoff definition and statistics.
For each probe, MFI values in reactions with no PCR product added to the hybridization mixture were considered background values. Net MFI values were computed by subtraction of 1.1 times the median background value. For all probes, this cutoff value was above the mean background plus three times the standard deviation. Reactions with net MFI values above 5 were defined as positive reactions. Correlation between the two PCR methods was assessed using kappa statistics. The coefficient of variation (CV) was computed to describe assay reproducibility. Fisher's exact test was used to compare dichotomous variables between two groups. P values of <0.05 were considered statistically significant.
RESULTS
Development of BS primers.
The analytic sensitivity for detection of individual HPV types by GP PCR differed up to 1,000-fold (Table 2). To obtain a more homogeneous analytic sensitivity, additional PCR primers were designed based on the L1 sequence of 48 HPV types (Table 3). The BS primer set consists of a pool of oligonucleotides which bind in the same region as the GP primers, thereby minimizing the number of mismatches to individual HPV types (Table 3). In total, we added eight upstream and two 5′-biotinylated downstream primers to the standard GP primer pair to form the BS primer set.
To assess the efficacy of the novel BS PCR in comparison to GP, plasmids containing HPV L1 sequences from 27 different genotypes were analyzed by both methods. PCR products of the expected size of about 150 bp were obtained from all plasmids, as determined by gel electrophoresis and subsequent genotyping by MPG (data not shown).
The analytic sensitivity of the GP and BS primer sets was compared after integration of the β-globin primers (see next paragraph). PCR was performed at least in duplicate using serial 10-fold dilutions of plasmids containing genomic DNA from 24 HPV types in 100 ng of HP DNA (Table 2). PCR products were analyzed by MPG. The BS primer set detected all 24 HPV genotypes with a sensitivity between 10 and 1,000 copies, while the analytic sensitivity of GP PCR varied between 10 and 1,000,000 copies. For HPV 11, 16, 18, 31, 33, 43, 58, 59, and 66, both PCR primer sets demonstrated the same sensitivity. For HPV 6, 35, 42, 45, 52, 70, and 73 the BS primer set was 10-fold, for HPV 26, 39, 56, 68, and 51 100-fold, and for HPV 44, 53, and 82 1,000-fold more sensitive than GP. These differences were independent of β-globin coamplification (see below).
Development of novel β-globin primers.
No internal DNA quality control has been described for the GP PCR. Instead, external β-globin PCR serves to control for DNA integrity, but it cannot control for the PCR efficacy in the HPV PCR itself. To overcome this limitation, novel β-globin primers were integrated into the BS PCR as well as the standard GP PCR. The CO3/5 primer set (5) and seven newly designed primer pairs (MS1-7/MS8-14) were included in various combinations in GP or BS PCR, and 10-fold dilutions of plasmids containing HPV 16, 18, 31, or 33 DNA were amplified in a background of 100 ng of human cellular DNA. β-Globin and HPV PCR products were analyzed by agarose gel electrophoresis (data not shown). The CO3/5 primers failed to coamplify the β-globin sequence, while three combinations of newly designed primers succeeded. These primer sets were further tested with DNA from SiHa and CasKi cells and from clinical specimens. Only the MS3/MS10 primers generated PCR products visible in the gel (data not shown).
Integration of β-globin primers in the HPV PCR.
Next, we determined the β-globin primer concentration that maximized β-globin signals after integration in the BS and GP PCR without concurrent competition of HPV PCR.
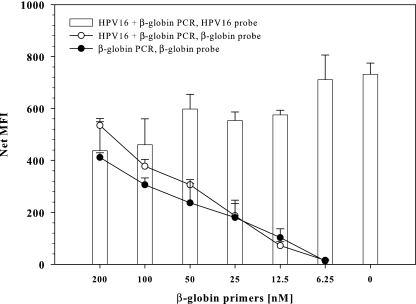
First, different amounts of MS3/MS10 β-globin primers were tested in the presence of HPV 16 (5,000 copies) and 100 ng of cellular DNA. β-Globin primer concentrations of 100 nM in the GP PCR (Fig. 1) and 300 nM in the BS PCR yielded similar β-globin Luminex signals (data not shown). Titration of cellular DNA resulted in a detection limit for β-globin of less than 10 ng (approximately 1,700 genome equivalents) by GP and BS PCR (data not shown).
FIG. 1.
Dependence of HPV amplification on β-globin coamplification. Twofold dilutions of β-globin primers with stable amounts of GP primers (500 nM) were tested on 100 ng of HP DNA spiked with (empty symbols) or without (filled symbols) 5,000 copies of HPV 16. PCR was performed in duplicate; mean net MFI values and standard deviations are indicated. HPV 16 signals are shown in bars, β-globin signals are shown in circles. No HPV 16 was detected in β-globin singleplex PCR.
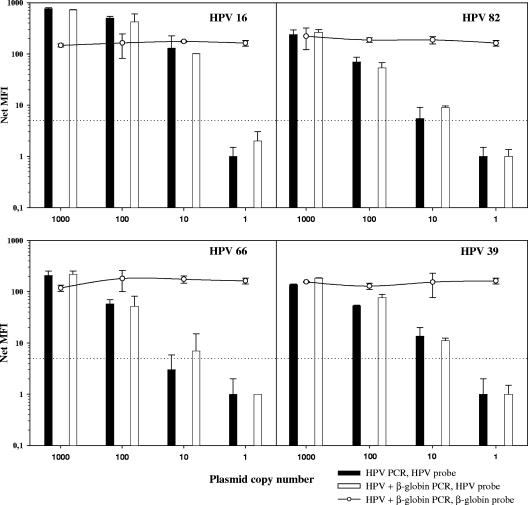
Second, serial dilutions of HPV in 100 ng of cellular DNA were amplified by PCR with and without β-globin coamplification. HPV detection limits were independent of β-globin coamplification in both GP and BS PCR (Fig. 2; Table 2).
FIG. 2.
Influence of HPV and β-globin coamplification on HPV amplification sensitivity. HPV 16, 39, 66, and 82 plasmids diluted in 100 ng HP DNA were amplified in duplicate by BSGP5+/6+ PCR with (empty symbols) and without (filled symbols) β-globin primers. HPV 16 signals are shown by bars, and β-globin signals are shown by circles; the cutoff is indicated by the dotted line.
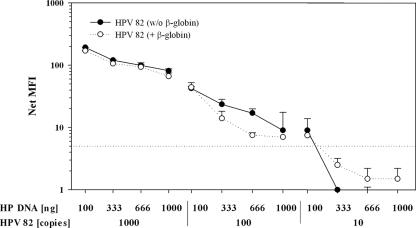
To study the effect of increasing amounts of cellular DNA on the HPV detection limit, serial dilutions of HPV 82 DNA (1,000 to 10 copies) in a background of 100 to 1,000 ng of HP DNA were examined by BS PCR with and without β-globin coamplification (Fig. 3). Amplification of HPV 82 in 1,000 ng of HP DNA resulted in two- to fourfold-weaker signals than a background of 100 ng of cellular DNA. The HPV 82 detection limit worsened from 10 to 100 copies in the presence of 333 to 1,000 ng of HP DNA. Compared to BS PCR without β-globin primers, the coamplification of β-globin did not change the detection limit of HPV 82 and showed consistent HPV signal strengths when 100 to 1,000 ng of background HP DNA was used. In summary, the ability of the BS PCR to detect HPV 82 was reciprocally correlated with the amount of background HP DNA and independent of β-globin coamplification.
FIG. 3.
Analytic sensitivity of BSGP5+/6+ PCR for HPV 82 in different amounts of cellular DNA. Dilution series of HPV 82 plasmids in 100 to 1,000 ng of HP DNA were amplified in the presence and absence of β-globin primers. The cutoff is indicated by the dotted line.
Extension of MPG probe spectrum.
To increase the total number of HPV types detected by the bead-based MPG system from 22 to 27, additional type-specific probes for HPV 26, 30, 53, 67, and 69 were integrated and validated using HPV plasmid-derived PCR products as described elsewhere (17). The universal probe uni1, designed to detect HPV types not tested for by MPG, was complemented by a second universal probe, uni2. uni2 targets the same region of the GP PCR product as uni1 and overlaps the GP6+ primer sequence by eight nucleotides. To reduce the number of mismatches present between uni1 and some HPV types, such as 34, 42, 53, 61, 72, 73, 81, and 83, degenerated nucleotides were included at four positions. Of 44 HPV sequences examined, 15 showed no mismatches and the other 29 showed only one mismatch with either of the universal probes. The universal probes recognized all 27 mucosal HPV types amplified with GP and BS PCR (data not shown).
For multiplex detection of the HPV (150 nucleotides) and β-globin PCR products (208 nucleotides), a specific β-globin probe was designed and integrated in MPG (Fig. 1 to 4).
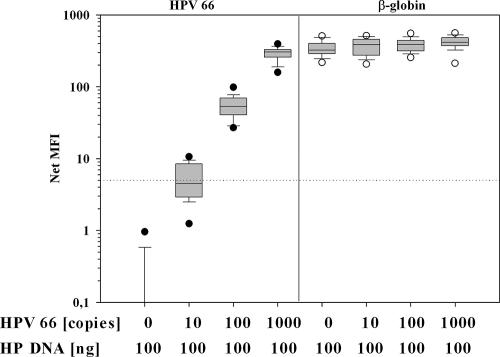
FIG. 4.
Reproducibility of BSGP5+/6+ PCR with β-globin coamplification. Tenfold dilution series of plasmids containing genomic HPV 66, diluted in 100 ng of HP DNA, were subjected 32 times to BS PCR with β-globin coamplification. Coamplified HPV 66 and β-globin PCR products were analyzed by MPG. The lower and upper edges of the boxes are the 25th and 75th percentiles, respectively. Median values are shown by the lines in the boxes, the whiskers represent the 5th and 95th percentiles, outliers are indicated by circles, and the cutoff is indicated by the dotted line.
Reproducibility and dose response of BSGP5+/6+ PCR.
We analyzed reproducibility of the BS PCR with integrated β-globin. Within 2 weeks, the PCR was repeated 32 times using 1 μl of the same 10-fold dilution series of plasmid containing 10, 100, and 1,000 copies of HPV 66, and the PCR products were analyzed on different days by MPG. The assay relies on end-point PCR detection that has a limited ability to achieve quantitative results. However, the high reproducibility of MPG (17) and the semiquantitative data read-out allowed us to discriminate a 10-fold HPV copy number difference in all 32 experiments (Fig. 4). Signals obtained with the HPV 66 probe exhibited a decreasing CV with increasing copy number, ranging from 53.4% (10 copies; median MFI, 4.5) over 34.7% (100 copies; median MFI, 53.4) to 22.9% (1,000 copies; median MFI, 302.7). In 14 of 32 PCRs (43.8%), 10 copies of HPV 66 were positive. The median CV for β-globin signals was 22.1% (range, 20.8 to 25.2%).
Semiquantitative HPV detection, which is independent of β-globin coamplification, was possible for all HPV types and is exemplarily depicted for HPV 16, 39, 66, and 82 (Fig. 2). In addition, quantification was also feasible when three HPV plasmid DNA dilution series were mixed, with 10- to 1,000-fold differences in concentration. The resulting MFI values for each HPV type closely reflected the relative concentrations of targets included in the PCR (data not shown). Altogether, these data demonstrated the interassay consistency of MPG and its ability to quantify several HPV types, especially in multiple infections.
Evaluation of BSGP5+/6+ PCR with clinical samples.
We compared the performance of BS plus integrated β-globin PCR against that of GP without β-globin PCR on DNA from exfoliated cervical cells from 1,017 women of the general population in Mongolia (Dondog et al., submitted) and 95 Mongolian cervical cancer patients. To minimize variation, both PCRs were performed in parallel and the products were analyzed on the same plate by MPG. Of the 1,112 samples, 27 were excluded because they were negative for β-globin and HPV amplification by BS; three of the excluded samples strongly tested HPV 16 or 31 positive by GP PCR, while the other 24 were HPV negative.
HPV prevalence in the remaining 1,085 clinical samples is presented in Tables 4 and 5. Of all typing results (1,085 samples and 27 HPV types analyzed), 639 (2.2%) were concordantly positive, 28,378 (96.9%) were concordantly negative, and 278 (0.9%), discussed in detail in the next paragraph, were discordant. These numbers yielded a kappa value of 0.816 (95% confidence interval, 0.797 to 0.836). Identical typing results were obtained for 858 (79.1%) of the 1,085 clinical samples. Despite β-globin coamplification, the overall HPV prevalence was higher (46.0%) by BS than GP (41.4%).
TABLE 4.
Detection of HPV genotypes in 1,085 clinical samples by BSGP5+/6+ or GP5+/6+ PCR followed by MPG
| HPV type | No. of positive reactions with indicated PCR and signal strength (net MFI)a
|
Rprevalence BS/GPb | Rsignal BS/GPc | |||||
|---|---|---|---|---|---|---|---|---|
| BS only
|
GP only
|
BS and GP | BS and/or GP | |||||
| >15 | <15 | >15 | <15 | |||||
| 6 | 11 | 11 | 1.00 | 0.93 | ||||
| 11 | 1 | 2 | 1 | 1 | 16 | 21 | 1.06 | 0.90 |
| 16 | 12 | 25 | 2 | 12 | 130 | 181 | 1.16 | 1.31 |
| 18 | 3 | 3 | 36 | 42 | 1.00 | 0.90 | ||
| 26 | 8 | 8 | 1.00 | 1.86 | ||||
| 30 | 9 | 1 | 13 | 23 | 1.77 | 33.81 | ||
| 31 | 6 | 7 | 36 | 49 | 0.73 | 0.46 | ||
| 33 | 1 | 2 | 1 | 29 | 33 | 0.94 | 0.51 | |
| 35 | 1 | 3 | 1 | 2 | 23 | 30 | 1.04 | 0.91 |
| 39 | 7 | 4 | 12 | 23 | 1.92 | 2.37 | ||
| 42 | 6 | 7 | 27 | 40 | 1.48 | 4.45 | ||
| 43 | 1 | 8 | 9 | 1.13 | 0.73 | |||
| 44 | 11 | 11 | ||||||
| 45 | 1 | 1 | 2 | 23 | 27 | 1.00 | 1.04 | |
| 51 | 9 | 9 | 1 | 38 | 57 | 1.44 | 3.46 | |
| 52 | 8 | 4 | 1 | 28 | 41 | 1.38 | 4.93 | |
| 53 | 24 | 10 | 4 | 38 | 9.50 | 15.12 | ||
| 56 | 1 | 2 | 4 | 23 | 30 | 0.96 | 1.05 | |
| 58 | 4 | 1 | 29 | 34 | 1.10 | 1.77 | ||
| 59 | 1 | 2 | 2 | 3 | 30 | 38 | 0.94 | 0.75 |
| 66 | 8 | 36 | 44 | 0.82 | 0.53 | |||
| 67 | 13 | 13 | 1.00 | 0.61 | ||||
| 68 | 11 | 4 | 2 | 17 | 8.50 | 29.30 | ||
| 70 | 2 | 1 | 2 | 22 | 27 | 0.96 | 0.89 | |
| 73 | 6 | 1 | 2 | 24 | 33 | 1.19 | 1.76 | |
| 82 | 10 | 8 | 1 | 18 | 37 | 1.89 | 6.03 | |
| Total | 118 | 94 | 16 | 50 | 639 | 918 | ||
Number of positive reactions for given HPV type using a net MFI cutoff of >5. For example, from a total of 42 HPV 18-positive samples, 36 were detected by both methods and three additional reactions each with values below a net MFI of 15 were each detected by BS and GP PCR. HPV 69 showed no positive reaction.
Ratio of additional detection for BSGP5+/6+ versus GP5+/6+. For example, HPV 53 was detected 38 times by BS versus 4 times by GP, resulting in 9.5 times more frequent detection.
Mean of ratio of net MFI intensity for BSGP5+/6+ versus GP5+/6+ for double-positive reactions. For example, for HPV 68, there were two double-positive reactions (for one of the double-positive reactions, the values for BS and GP were 326 and 6, respectively; for the other double-positive reaction, the values were 386 and 75, respectively), with pairwise ratios of 53.5 and 5.1, yielding a mean ratio of 29.3.
TABLE 5.
Number of HPV types found in 1,085 clinical samples with BSGP5+/6+ and GP5+/6+ PCR, respectively
| PCR type | No. of samples with indicated no. of HPV typesa
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | |
| BS | 586 | 307 | 108 | 43 | 21 | 10 | 6 | 3 | 1 |
| GP | 636 | 293 | 103 | 32 | 7 | 2 | 12 | ||
Number of samples obtained using a cutoff net MFI of >5. 0, HPV negative; 1 to 8, single to octuple infection.
Compared to GP, BS failed to detect a total of 66, mostly borderline, infections but identified 212 additional infections. Table 4 shows the difference in the detection rates for the individual HPV types. HPV 69 was not detected in any sample. Differences clustered in three groups: (i) some HPV types were systematically better detected by one method or the other (see next paragraph); (ii) others (HPV 11, 16, 18, 33, 35, 43, 45, 56, 59, and 70) showed small differences with a few additional weakly positive reactions (net MFI below 15) by either of the two PCR methods; (iii) no difference was found for HPV 6, 26, and 67. Systematically, BS showed 1.2- to 9.5-times-higher detection rates (Rprevalence BS/GP) for HPV types 30, 39, 42, 44, 51, 52, 53, 68, 73, and 82 (Table 4). At least 50% of these additionally detected cases (Table 4) showed strong signals (net MFI of >15). Among the additional reactions, strong BS signals (above one-third of the maximal signal [signalmax] for this HPV type) were also missed by GP PCR. In single as well as multiple infections, BS primers additionally detected HPV 82 with signals above one-third of signalmax, HPV 30 and 68 with signals above half signalmax, and HPV 44 and 53 with full signalmax. LR HPV 44 was exclusively detected by BS primers. On the other hand, GP primers demonstrated better detection of HPV types 31 and 66, showing 1.4 and 1.2 times more frequent detection, respectively. Out of 13 HPV 31 infections additionally detected by GP, 7 showed borderline signals (net MFI below 15), and the remaining 6 (mean net MFI of 48) were all found in multiple infections with at least one concomitant strong infection by HPV 16, 51, 52, 53, 58, or 59. All HPV 66 infections detected solely by GP primers showed borderline net MFI values below 15.
Between 15% (HPV 73) and 73% (HPV 44) of the HPV infections detected by BS only were single infections. However, the majority of the additionally detected infections was found in multiple infections, resulting in 38.5% multiple infections (192 of 499 total HPV-positive samples) by BS versus 34.7% (156 of 449) by GP (Table 5). Among the general population (n = 992) the proportion of multiple infections was 16.8% by BS versus 14.1% by GP. In cancer patients (n = 93), multiple infections were significantly more frequently found using BS (26.9%, P = 0.007), while the frequency found using GP was not significantly different from that in the general population (17.2%, P = 0.084).
Amplification analysis of HPV types.
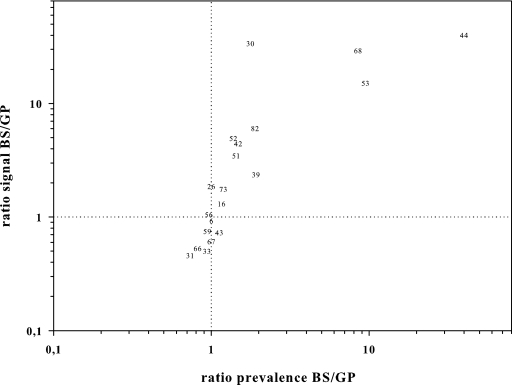
To further characterize the different affinities of both primer sets to individual HPV types, we took advantage of the quantitative read-out of MPG. For the reactions positive (net MFI of >5) with both primer sets, we computed the mean of the ratio: BS signal over GP signal (Rsignal BS/GP). This ratio allowed us to identify HPV types in two distinct patterns: P1 had an Rsignal BS/GP between 0.5 and 2.0 (positive signals differing less than twofold between PCRs), and P2 had an Rsignal BS/GP above 2.0 (>2-fold-higher signals with BS). According to these criteria, P1 was found for HPV 6, 11, 16, 18, 26, 31, 33, 35, 43, 45, 56, 58, 59, 66, 67, 70, and 73, and P2 for HPV 30, 39, 42, 44, 51, 52, 53, 68, and 82 (Table 4). No Rsignal BS/GP could be computed for HPV 44, grouped in P2, because it was found exclusively by BS. The Rsignal BS/GP closely correlated with the number of additional HPV reactions found by BS or GP (Fig. 5).
FIG. 5.
Relative HPV detection ability and signal strength in positive reactions with BSGP5+/6+ and GP5+/6+. The ratio of additional HPV detections by BSGP5+/6+ to those by GP5+/6+ (x axis) is plotted against the ratio of hybridization signals with BSGP5+/6+ versus GP5+/6+ for reactions positive by both methods (y axis) in a logarithmical scale. Missing HPV types overlap with other types shown; e.g., HPV 6 clusters with 11, 18, 35, and 70, HPV 56 with 45, and HPV 73 with 58.
Improvement of HPV 31 sensitivity with BSGP5+/6+ PCR.
To examine the reason for the apparent lower sensitivity of the BS primers for HPV 31, we increased the concentrations of the standard GP5+ and GP6+ primers in the BS PCR from 200 nM to 500 nM and 400 nM to 600 nM, respectively, while all other primer concentrations were maintained. All 13 clinical samples with multiple infections that were previously HPV 31 negative by BS PCR were reanalyzed, and 7 of 13 previously missed HPV 31 reactions were detected. Amplification of HPV 66 was also improved, resulting in the detection of three out of eight previously BS PCR-negative infections. Detection of other HPV types was not altered (data not shown).
DISCUSSION
PCR methods using general or consensus primers are highly convenient. Because only one primer pair is employed to amplify DNA of a large variety of HPV genotypes in a single reaction, this PCR is easy to standardize. Under conditions tolerating mismatches, GP can amplify at least 37 mucosal HPV types (19). As a result, GP is one of the most used PCR systems for large epidemiological studies (18). However, since the design of these primers (5), additional mucosal HPV types have been discovered, and still more are presumed to exist. GP showed differences in its ability to amplify mucosal HPV types depending on the number and position of primer mismatches (5, 13). We consistently reproduced these results with endpoint dilution series of 24 cloned HPV types. Our data indicated that the number of mismatches is reciprocally correlated with PCR sensitivity; however, the location of mismatches also plays an important role.
Our novel primer set demonstrated an increased sensitivity for at least 10 HPV types, despite the coamplification of β-globin by the MS3/MS10 primer pair. The overall amplification of mucosal HPV types under standardized experimental conditions was more homogeneous, reaching detection limits for all 24 HPV types analyzed between 10 and 1,000 copy numbers. The improved sensitivity for HPV types 30, 39, 42, 44, 51, 52, 53, 68, 73, and 82 was confirmed with clinical samples and correlated with the prediction based on the mismatch analysis with target HPV regions. In addition, BS primers have shown an improved detection of HPV 61 and 84 (I. Sabol, M. Salakova, J. Smahelova, M. Pawlita, M. Schmitt, N. M. Gas̆perov, M. Grce, and R. Tachezy, submitted for publication).
The increased sensitivity of BS is entirely due to primer differences, since the same detection system (MPG) was used for both methods. The BS primer pool not only reduced mismatches to HPV types that were not sensitively detected by GP but also reduced competition in multiple HPV infections. However, increased primer concentrations in BS can generate larger amounts of PCR products, which may result in weak cross-reactivities with related probes. Cross-reactivities were determined by colony PCR for all 27 HPV types. These PCR products, originating from millions of HPV copies, revealed a single cross-reactivity; i.e., very strong HPV 51 amplification showed weak hybridization (less than 5% of HPV 51 signal) with the HPV 82 probe (data not shown). Such HPV 82 signals were excluded when HPV positivity was scored.
HPV 31 detection by BS PCR was reduced in clinical samples with low viral loads or multiple infections. Hitherto, we could not find a plausible explanation for this observation. However, increased concentrations of GP5+ and GP6+ primers in the BS mixture resulted in stronger HPV 31 amplification, permitting the detection of 7 out of 13 previously missed HPV 31 infections. Improved amplification was also observed for HPV 66 infections, while the detection of HPV types preferentially amplified by the initial BS PCR protocol remained unchanged. As a consequence, the modified protocol will be used in future studies.
The quantitative read-out of MPG provided a second measure to assess both PCRs’ abilities to amplify distinct HPV types, including types with a low prevalence in the study. Overall, Rsignal BS/GP values were largely consistent with findings in the clinical samples, confirming the reduced amplification of HPV 31 and 66 and the improved amplification of HPV 30, 39, 42, 44, 51, 52, 53, 68, 73, and 82 by BS (Table 4; Fig. 5).
Likewise, the quantitative data obtained from MPG can be used to better characterize multiple HPV infections. The resulting intensities of MFI signals for each HPV type in a multiple infection faithfully reflected the relative concentrations of targets included in the PCR. This was demonstrated in dilution series of various cloned HPV types. As a result, the dominant HPV type(s) present in an infection, i.e., the type(s) with the highest viral loads, could be easily identified. It has been shown that 20 to 30% of HPV-positive women harbor multiple types that were acquired simultaneously or successively (10). It remains controversial whether an infection with multiple HPV DNA types is a risk factor for HPV persistence and for cervical lesions (14, 16). However, a reliable and unbiased profiling of individual HPV types in multiple HPV infections will be important in evaluating the efficacy of HPV vaccine implementation. In the present study, we demonstrated the improved detection of multiple HPV infection by the novel BS PCR in comparison to GP PCR. A significantly higher prevalence of multiple infections was found in cancer samples than in samples from the general population only by using the BS PCR system. This can be explained by the presence of very high viral loads of the “driving” HR HPV type in cervical cancer. As a consequence, PCR reagents, i.e., GP primers, are preferentially consumed by this type, thereby weakening the amplification of other coinfecting types. Thus, addition of several different BS primers minimizes the risk of amplification bias by a dominant HPV infection, as is in particular the case for cervical cancer samples. Therefore, BS primers may be the better tool for future studies analyzing multiple HPV infections as a risk factor for developing cervical cancer.
More studies are required to show whether the additional detection of some HPV types by the novel BS primers will be of value for predicting the risk of having or developing cervical lesions. However, we believe that the detection of high copy numbers of “rare” HPV types, currently being missed by GP PCR, may have an impact on the management of HPV-related diseases. As such, GP PCR-reverse line blot-based epidemiological studies, summarized in recently published meta-analyses (3, 4), found HPV 39, 51, 66, 68, and 82 in either single or multiple infections in less than 2.9% of high-grade squamous intraepithelial lesions and less than 0.6% of squamous cell carcinoma of the cervix. The distribution of HPV 26 and 53 was not described. The use of MPG in combination with the novel BS PCR, however, suggests that in squamous cell carcinoma of the cervix, all these types may be present more frequently than described so far (unpublished data).
Internal DNA quality controls (e.g., β-globin) possess the advantage of simultaneous HPV and genomic DNA amplification; however, they may also decrease the analytic sensitivity of the HPV PCR (7). This decrease can be explained by highly efficient β-globin amplification, which competes for DNA polymerase and deoxynucleoside triphosphates and exhibits higher sensitivity than the HPV primers. As a consequence, most HPV PCRs, such as GP, lack an internal DNA quality control and require sample prescreening by an external β-globin PCR and subsequent gel electrophoresis (5, 8). However, the use of internal DNA quality controls without concurrent impairment of the HPV PCR is desirable. In addition to gaining time and reducing study costs and sample consumption, an internal DNA quality control can also monitor a failure of the HPV PCR. To this end, we designed a novel β-globin primer pair, MS3/MS10, and showed that its integration had no influence on the sensitivity of the conventional GP PCR or the novel BS PCR. This was achieved by adjusting the β-globin primer concentration to reach an analytic sensitivity of approximately 1,700 genome equivalents, which was purposely lower than for any of the HPV types. Thus, clinical samples containing large amounts of HPV copy numbers showed no or suppressed β-globin signals, while strong HPV values were measured. Nevertheless, this effect is highly desirable, minimizing the risk of false-negative HPV results due to a dominant β-globin coamplification. In contrast, HPV PCR sensitivity decreased with an increasing amount of cellular background DNA, independent of simultaneous β-globin coamplification and the amount of β-globin PCR product obtained.
The analytic sensitivity of a HPV genotyping system is composed of the amplification sensitivity and the detection limit of the read-out method. The selective overamplification of certain HPV types, as well as limiting sensitivity of read-out techniques, however, can skew the reported frequency of mucosal HPV types. Moreover, methodological bias leads to an underestimation of the prevalence of distinct types in cervical specimens (1). Improved HPV genotyping is needed (i) to detect a higher variety of HPV types, (ii) to reduce the number of false-negative and false-positive results, (iii) to better characterize multiple infections, and (iv) to save time and cost. New HPV types have continuously been discovered, and a yet-larger number is expected to exist. Therefore, HPV amplification and genotyping methods that allow specific, sensitive, unbiased, and robust detection of HPV DNA and yet are flexible enough for the integration of additional HPV types are needed. Hitherto, genotyping of GP5+/6+ PCR products has been based on type-specific hybridization with oligonucleotide probes immobilized to membranes (RLB) (19) or to fluorescent beads (MPG) (17). The latter easily allows extension of the probe number to cover all mucosal HPV types with known sequences in the amplified L1 region. The automated read-out of MPG provides semiquantitative analysis of PCR products, avoids mistakes during data entry, and allows genotyping of up to 1,000 PCR products per day.
In conclusion, this study describes the development and validation of a novel BS PCR for the sensitive detection of HPV in single as well as multiple infections. Our data suggest that especially for pHR HPV type 53 and HR HPV types 39, 51, 52, 68, 73, and 82, use of BSGP5+/6+ PCR in clinical practice will lead to more reliable estimates of infection rates with these types. Further studies will clarify whether the use of BSGP5+/6+ PCR in combination with MPG has a positive impact on the prediction of developing cervical lesions caused by these types. We believe that by using the BSGP5+/6+ PCR with internal DNA quality control in conjunction with MPG, researchers will obtain a powerful, highly flexible and sensitive tool for a more homogeneous amplification and genotyping of HPV that could be used in large epidemiological studies, in particular in vaccinated populations.
Acknowledgments
HPV 6, 11, 16, 18, 26, 30, 51, 53, 69, and 73 plasmid DNA was kindly provided by E.-M. de Villiers (DKFZ, Heidelberg, Germany), HPV 33, 39, 42, 45, 66, and 70 plasmid DNA by G. Orth (Institut Pasteur, Paris, France), HPV 31, 35, 43, 44, and 56 plasmid DNA by A. Lörincz (Digene Corp., Gaithersburg, MD), HPV 58, 59, 67, and 82 plasmid DNA by T. Matsukura (National Institute of Infectious Diseases, Tokyo, Japan), HPV 52 plasmid DNA by W. Lancaster (Wayne State University, Detroit, MI), and ME180 (HPV 68) plasmid DNA by E. Schwarz (DKFZ, Heidelberg, Germany). We are grateful to Lutz Gissmann for critical reading of the manuscript.
Footnotes
Published ahead of print on 16 January 2008.
REFERENCES
- 1.Baay, M. F., W. G. Quint, J. Koudstaal, H. Hollema, J. M. Duk, M. P. Burger, E. Stolz, and P. Herbrink. 1996. Comprehensive study of several general and type-specific primer pairs for detection of human papillomavirus DNA by PCR in paraffin-embedded cervical carcinomas. J. Clin. Microbiol. 34745-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chan, P. K., T. H. Cheung, A. O. Tam, K. W. Lo, S. F. Yim, M. M. Yu, K. F. To, Y. F. Wong, J. L. Cheung, D. P. Chan, M. Hui, and M. Ip. 2006. Biases in human papillomavirus genotype prevalence assessment associated with commonly used consensus primers. Int. J. Cancer 118243-245. [DOI] [PubMed] [Google Scholar]
- 3.Clifford, G. M., J. S. Smith, T. Aguado, and S. Franceschi. 2003. Comparison of HPV type distribution in high-grade cervical lesions and cervical cancer: a meta-analysis. Br. J. Cancer 89101-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clifford, G. M., J. S. Smith, M. Plummer, N. Munoz, and S. Franceschi. 2003. Human papillomavirus types in invasive cervical cancer worldwide: a meta-analysis. Br. J. Cancer 8863-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Roda Husman, A. M., J. M. Walboomers, A. J. van den Brule, C. J. Meijer, and P. J. Snijders. 1995. The use of general primers GP5 and GP6 elongated at their 3′ ends with adjacent highly conserved sequences improves human papillomavirus detection by PCR. J. Gen. Virol. 761057-1062. [DOI] [PubMed] [Google Scholar]
- 6.de Villiers, E. M., C. Fauquet, T. R. Broker, H. U. Bernard, and H. zur Hausen. 2004. Classification of papillomaviruses. Virology 32417-27. [DOI] [PubMed] [Google Scholar]
- 7.Gravitt, P. E., C. L. Peyton, T. Q. Alessi, C. M. Wheeler, F. Coutlee, A. Hildesheim, M. H. Schiffman, D. R. Scott, and R. J. Apple. 2000. Improved amplification of genital human papillomaviruses. J. Clin. Microbiol. 38357-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jacobs, M. V., J. M. Walboomers, P. J. Snijders, F. J. Voorhorst, R. H. Verheijen, N. Fransen-Daalmeijer, and C. J. Meijer. 2000. Distribution of 37 mucosotropic HPV types in women with cytologically normal cervical smears: the age-related patterns for high-risk and low-risk types. Int. J. Cancer 87221-227. [PubMed] [Google Scholar]
- 9.Kleter, B., L. J. van Doorn, L. Schrauwen, A. Molijn, S. Sastrowijoto, J. ter Schegget, J. Lindeman, B. ter Harmsel, M. Burger, and W. Quint. 1999. Development and clinical evaluation of a highly sensitive PCR-reverse hybridization line probe assay for detection and identification of anogenital human papillomavirus. J. Clin. Microbiol. 372508-2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mendez, F., N. Munoz, H. Posso, M. Molano, V. Moreno, A. J. van den Brule, M. Ronderos, C. Meijer, and A. Munoz. 2005. Cervical coinfection with human papillomavirus (HPV) types and possible implications for the prevention of cervical cancer by HPV vaccines. J. Infect. Dis. 1921158-1165. [DOI] [PubMed] [Google Scholar]
- 11.Munoz, N., F. X. Bosch, S. de Sanjose, R. Herrero, X. Castellsague, K. V. Shah, P. J. Snijders, and C. J. Meijer. 2003. Epidemiologic classification of human papillomavirus types associated with cervical cancer. N. Engl. J. Med. 348518-527. [DOI] [PubMed] [Google Scholar]
- 12.Notredame, C., D. G. Higgins, and J. Heringa. 2000. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 302205-217. [DOI] [PubMed] [Google Scholar]
- 13.Qu, W., G. Jiang, Y. Cruz, C. J. Chang, G. Y. Ho, R. S. Klein, and R. D. Burk. 1997. PCR detection of human papillomavirus: comparison between MY09/MY11 and GP5+/GP6+ primer systems. J. Clin. Microbiol. 351304-1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rousseau, M. C., J. S. Pereira, J. C. Prado, L. L. Villa, T. E. Rohan, and E. L. Franco. 2001. Cervical coinfection with human papillomavirus (HPV) types as a predictor of acquisition and persistence of HPV infection. J. Infect. Dis. 1841508-1517. [DOI] [PubMed] [Google Scholar]
- 15.Rozen, S., and H. Skaletsky. 2000. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132365-386. [DOI] [PubMed] [Google Scholar]
- 16.Schiffman, M. H., H. M. Bauer, R. N. Hoover, A. G. Glass, D. M. Cadell, B. B. Rush, D. R. Scott, M. E. Sherman, R. J. Kurman, S. Wacholder, et al. 1993. Epidemiologic evidence showing that human papillomavirus infection causes most cervical intraepithelial neoplasia. J. Natl. Cancer Inst. 85958-964. [DOI] [PubMed] [Google Scholar]
- 17.Schmitt, M., I. G. Bravo, P. J. Snijders, L. Gissmann, M. Pawlita, and T. Waterboer. 2006. Bead-based multiplex genotyping of human papillomaviruses. J. Clin. Microbiol. 44504-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Snijders, P. J., A. J. van den Brule, M. V. Jacobs, R. P. Pol, and C. J. Meijer. 2005. HPV DNA detection and typing in cervical scrapes. Methods Mol. Med. 119101-114. [DOI] [PubMed] [Google Scholar]
- 19.van den Brule, A. J., R. Pol, N. Fransen-Daalmeijer, L. M. Schouls, C. J. Meijer, and P. J. Snijders. 2002. GP5+/6+ PCR followed by reverse line blot analysis enables rapid and high-throughput identification of human papillomavirus genotypes. J. Clin. Microbiol. 40779-787. [DOI] [PMC free article] [PubMed] [Google Scholar]