Abstract
We conducted a prospective bacteriological survey to investigate antibiotic resistance-related genetic characteristics and the turnover of nasopharyngeal Haemophilus influenzae carriage in healthy children in day-care centers (DCCs). A total of 363 nasopharyngeal mucus samples were collected from children aged 0 to 6 years attending two DCCs in the summer of 2004 (n = 184) and the following winter (n = 179). We obtained 172 H. influenzae isolates and analyzed them by antimicrobial susceptibility testing, PCR for blaTEM-1 and the penicillin-binding protein (PBP) gene, and pulsed-field gel electrophoresis (PFGE). The overall carriage rate was 47.4% (172/363), and 37.2% of the isolates (64/172) were ampicillin (AMP) resistant. All the resistant isolates had a PBP mutation(s), while only three isolates had TEM-1. The carriage rate was significantly higher in the winter than in the summer (56.4% and 38.6%, respectively), owing to the increase in the numbers of AMP-susceptible H. influenzae isolates in the winter. Children aged ≤3 years showed a higher rate of carriage of H. influenzae isolates with an AMP resistance gene(s) than those aged ≥4 years (21.9% and 12.6%, respectively). Forty-two strains with different PFGE patterns were obtained from among the 172 isolates. Only five strains were observed in both seasons. None of the strains isolated in the summer was isolated from the same carrier in the winter. Twenty-seven strains (64.3%) were isolated from two or more children, and 25 of these were each isolated from children belonging to the same DCC. These results indicate the spread of H. influenzae, particularly those with a PBP mutation(s), and the highly vigorous genetic turnover and substantial horizontal transmission of this pathogen in healthy children attending DCCs in Japan.
The nasopharyngeal flora is generally thought to be a major reservoir for bacterial pathogens that cause upper respiratory tract infections (URIs) in children (2, 7). In recent years, the morbidity as a result of recurrent URIs, including acute otitis media, caused by antibiotic-resistant pathogens has been increasing in children, raising a worldwide health problem. Haemophilus influenzae is one of the major bacterial pathogens colonizing the nasopharynx and often causes acute otitis media and sinusitis. H. influenzae is known to acquire antibiotic resistance by two different mechanisms: the production of β-lactamases (TEM-1 or ROB-1 type) (19, 41) and mutations in the penicillin-binding protein (PBP) gene. In Japan, the prevalence of β-lactamase-positive ampicillin (AMP)-resistant (BLPAR) H. influenzae is not high, with such isolates constituting less than 6% of all clinical H. influenzae isolates (11, 13, 34), while recent studies have shown that H. influenzae strains with mutations in the PBP gene, such as β-lactamase-negative AMP-resistant (BLNAR) H. influenzae and β-lactamase-positive amoxicillin-clavulanic acid-resistant (BLPACR) H. influenzae, are increasing (12, 30). Ubukata et al. (37, 38) have discovered several mutations in the ftsI gene, which encodes PBP 3, in clinically isolated H. influenzae strains. Such mutations are known to lower the affinity of PBPs for β-lactams (20, 21).
There has been a growing awareness of increasing morbidity as a result of community-acquired infections, particularly those caused by drug-resistant bacteria. The day-care-center (DCC) attendance of children has been reported to be one of the risk factors for URIs, including acute otitis media (1, 4, 39), and for the nasopharyngeal carriage of bacteria such as H. influenzae and Streptococcus pneumoniae (7, 27, 32). However, the molecular epidemiology of H. influenzae in the nasopharynx in connection with DCC attendance is not fully understood (5, 8, 17, 27, 29, 31, 32, 36, 40, 42). The aim of this study was to investigate antibiotic resistance-related genetic characteristics and the turnover of nasopharyngeal H. influenzae carriage in children attending DCCs.
MATERIALS AND METHODS
Subjects.
Children in two DCCs (DCC-A and DCC-B) in a city in Fukuoka Prefecture in Japan were enrolled in the study. None of the children had been vaccinated against H. influenzae type b. The survey was performed in July 2004 (summer) and February 2005 (winter). The summer survey included 184 children (93 boys, 91 girls) ranging in age from 1 to 6 years (average age, 3.0 years). The winter survey included 179 children (93 boys, 86 girls) ranging in age from 0 to 6 years (average age, 3.7 years). One hundred fifty children participated in both surveys. Written questionnaires about present illness (otitis media, rhinosinusitis, tonsillitis, bronchitis, common cold, and allergic rhinitis) were completed by the children's parents a few days before sampling for bacteria. Informed consent was obtained from the parents, and the study was approved by the Ethics Committee of the University of Occupational and Environmental Health.
Bacterial sampling.
Nasopharyngeal mucus was collected transnasally with a sterile cotton swab (Seed Swab no. 2; Eiken Chemical Co., Ltd., Tokyo, Japan) and subjected to bacterial culture. The samples were placed on chocolate agar plates (Eiken Chemical Co., Ltd.) and incubated under a humidified atmosphere containing 5% CO2 at 35°C overnight. H. influenzae was identified by standard microbiological methods (23). The identification criteria were (i) colony morphology on the culture medium, (ii) negativity by Gram staining, (iii) catalase and oxidase production, and (iv) a requirement for V (hemin) and X (NAD) factors. If colonies of H. influenzae were present on a plate, a major colony was extracted and propagated by the same procedure. The isolates were stored in a stock solution (Microbank; Pro-Lab Diagnostics, Ontario, Canada) at −80°C until the following analyses.
Antimicrobial susceptibility.
The AMP MICs were determined by the broth microdilution method, in accordance with the guidelines of the Japanese Society of Chemotherapy (15), by using an Eiken Dry Plate (cation-adjusted Mueller-Hinton broth with NAD [15 mg/liter], yeast extract [5 g/liter], and lysed horse blood [20 ml/liter]) (16). Briefly, each well of the plate was inoculated with a 100-μl inoculum containing approximately 5 ×104 CFU, and the plate was incubated at 35°C for 20 h. The MIC was read as the lowest concentration of an antimicrobial agent at which there was no visible growth. Breakpoints for susceptibility were provided according to the guidelines of the Clinical and Laboratory Standards Institute (formerly the National Committee for Clinical Laboratory Standards) (25). β-Lactamase production was detected with nitrocefin disks (cefinase; Becton, Dickinson and Company, Sparks, MD).
PCR analysis.
Four sets of primers (Wakunaga Pharmaceutical Co. Ltd., Osaka, Japan) were used. P6 primers were used to amplify the p6 gene, which encodes the P6 membrane protein specific for H. influenzae (26). The primers can generally distinguish H. influenzae from Haemophilus haemolyticus but cannot completely rule out the presence of some nonhemolytic H. haemolyticus strains (22). TEM-1 primers were used to amplify a part of the blaTEM-1 gene (33). PBP3-S primers were used to identify an Asn-526→Lys-526 substitution in the ftsI gene (11, 12, 38). PBP3-BLN primers were used to identify a Ser-385→Thr-385 substitution in the ftsI gene (11, 12, 38). A single colony of H. influenzae on the chocolate agar plate was extracted and suspended in 30 μl of lysis solution (1 M Tris [pH 8.9] containing 4.5% Nonidet P-40, 4.5% Tween 20, and 10 mg/ml proteinase K) in a microcentrifuge tube. The tube was placed in a thermal cycler (PCR thermal cycler MP; Takara Bio Inc., Shiga, Japan) and heated at 60°C for 10 min and at 94°C for 5 min to lyse the bacterial cells. Two microliters of the bacterial lysate was then dispensed into each of three tubes: The tubes contained two sets of primers for P6 and TEM-1, primers for PBP3-S, and primers for PBP3-BLN, respectively, dissolved in 30 μl of a reaction mixture consisting of 3 μl of 10× PCR buffer, 3 μl of a deoxynucleoside triphosphate mixture at a concentration of 2 mM, 1.2 U of Tth DNA polymerase (Toyobo, Co., Ltd., Osaka, Japan), and 4 pmol of both the sense and the reverse primers. The PCR cycling conditions were 30 cycles at 94°C for 15 s, 53°C for 15 s, and 72°C for 15 s. The amplified DNA fragments were analyzed by gel electrophoresis with 3% agarose.
On the basis of the PCR results, the H. influenzae strains tested were classified into six groups (11, 12, 38): β-lactamase-negative AMP-susceptible (BLNAS) strains, which lack all the resistance genes; BLPAR strains, which have the blaTEM-1 gene; low-BLNAR strains, which show low-level resistance associated with the substitution of Lys-526 in ftsI; BLNAR strains, which show resistance associated with the substitution of Lys-526 and Thr-385 in ftsI; BLPACR I strains, which have the blaTEM-1 gene and ftsI with the same substitution as the low-BLNAR strains; and BLPACR II strains, which have the blaTEM-1 gene and ftsI with the same substitution as the BLNAR strains.
PFGE.
Pulsed-field gel electrophoresis (PFGE) analysis was carried out by a modification of the method of Yano et al. (42). The bacteria were suspended in 50 mM EDTA (pH 8.0) at a concentration of a no. 1 McFarland standard. The pellet was mixed with an equal volume of melted 1.2% chromosomal-grade agarose (Bio-Rad Laboratories, Richmond, CA). The mixture was poured in a mold and chilled, and the resultant plugs were treated with 0.5 ml of lysis solution I (6 mM Tris-HCl [pH 8.0], 0.1 M EDTA, 0.5% Brij 58, 0.2% sodium deoxycholate, 0.5% Sarkosyl, 0.8 mg/ml of lysozyme [Wako Pure Chemical Industries, Ltd., Japan], and 10 μg/ml of RNase [Nippon Gene, Co., Ltd., Tokyo, Japan]) at 37°C for 12 h and treated with 0.5 ml of lysis solution II (0.5 M EDTA [pH 8.0], 1% Sarkosyl, and 0.25 mg/ml of proteinase K [Wako Pure Chemical Industries, Ltd.]) at 50°C for 24 h. For restriction endonuclease digestion, the plugs were incubated with 30 U of SmaI (Takara Bio Inc.) in restriction enzyme buffer at 30°C for 16 h.
DNA fragments were electrophoresed in a 1% agarose gel at 6 V/cm and 14°C for 22 h with a 3- to 60-s pulse time with CHEF Mapper and CHEF Mapper XA pulsed-field electrophoresis systems (Bio-Rad Laboratories). A bacteriophage lambda ladder (Bio-Rad Laboratories) was used as a size standard. The PFGE patterns were visually compared and evaluated according to the criteria developed by Tenover et al. (35). Isolates that showed indistinguishable PFGE patterns were considered the same strain (42).
Statistics.
Statistical analyses were performed with StatView-J software (version 5.0; SAS Institute Inc., Cary, NC). The chi-square test or Fisher's exact test was used to evaluate the statistical significance of the differences. P values of <0.05 were considered statistically significant.
RESULTS
The overall percentages of children who were under treatment for otitis media, rhinosinusitis, tonsillitis, bronchitis, common cold, and allergic rhinitis were 1.4% (5/363), 4.7% (17/363), 0.6% (2/363), 0.3% (1/363), 0.8% (3/363), and 2.5% (9/363), respectively. No significant difference in morbidity was observed between seasons or ages.
There were 172 H. influenzae isolates from 363 samples during the two seasons, and all possessed the p6 gene. Three isolates were positive for β-lactamase, as detected with nitrocefin disks, and they all had the TEM-1-type β-lactamase gene. Among the 172 isolates, the percentages of isolates of each resistance class were as follows: 62.8% (108/172) for BLNAS strains, 10.5% (18/172) for low-BLNAR strains, 25.0% (43/172) for BLNAR strains, and 1.7% (3/172) for BLPACR II strains. Neither BLPAR nor BLPACR I strains were detected.
Table 1 summarizes the genetic differences of the H. influenzae strains isolated between seasons and age groups. The overall carriage rate was 47.4% (172/363), and the rate of carriage of H. influenzae strains in which any of the AMP resistance genes were detected by PCR was 17.6% (64/363). TEM-1 was detected in only 0.8% of the samples (3/363). The rate of nasopharyngeal carriage of H. influenzae was significantly increased in winter than in summer (56.4% and 38.6%, respectively; P < 0.01): the rate of carriage of H. influenzae without an AMP resistance gene(s) was higher in winter than in summer (39.7% and 20.1%, respectively; P < 0.05), whereas the rate of carriage of H. influenzae with an AMP resistance gene(s) was not statistically different between the seasons. On the other hand, children aged ≤3 years showed a higher rate of carriage of H. influenzae with an AMP resistance gene(s) than those aged ≥4 years (21.9% and 12.6%, respectively; P < 0.05), but the rate of carriage of H. influenzae without an AMP resistance gene(s) was not statistically different between the age groups.
TABLE 1.
Genetic differences of the H. influenzae strains isolated by season and age
| Isolate | No. of isolates (% carriage)
|
||||||
|---|---|---|---|---|---|---|---|
| Season
|
Age
|
Overall (n = 363) | |||||
| Summer (na = 184) | Winter (n = 179) | P value | ≤3 yr (n = 196) | ≥4 yr (n = 167) | P value | ||
| All H. influenzae isolates | 71 (38.6) | 101 (56.4) | <0.01 | 98 (50.0) | 74 (44.3) | NSb | 172 (47.4) |
| H. influenzae without AMP resistance gene(s) | 37 (20.1) | 71 (39.7) | <0.05 | 55 (28.1) | 53 (31.7) | NS | 108 (29.8) |
| H. influenzae with AMP resistance gene(s) | 34 (18.5) | 30 (16.8) | NS | 43 (21.9) | 21 (12.6) | <0.05 | 64 (17.6) |
| Asn-526→Lys-526 substitution | 34 (18.5) | 30 (16.8) | NS | 43 (21.9) | 21 (12.6) | <0.05 | 64 (17.6) |
| Ser-385→Thr-385 substitution | 23 (12.5) | 23 (12.8) | NS | 33 (16.8) | 13 (7.8) | <0.05 | 46 (12.7) |
| TEM-1 | 3 (1.6) | 0 (0) | NS | 3 (1.5) | 0 (0) | NS | 3 (0.8) |
n, total number of samples.
NS, not significant.
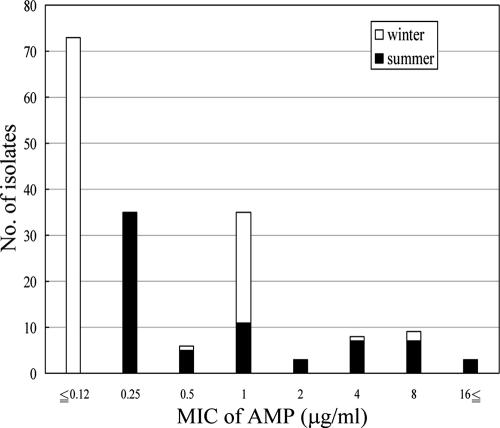
Figure 1 shows a histogram of the AMP MICs. Overall, 86.6% of the isolates (149/172) were AMP susceptible (MICs ≤ 1 μg/ml) and 11.6% of the isolates (20/172) were AMP resistant (MICs ≥ 4 μg/ml). Among the isolates collected in the summer, 71.8% (51/71) were AMP susceptible and 23.9% (17/71) were AMP resistant. Among the isolates collected in the winter, the proportion of AMP-susceptible isolates increased from that in the summer to 97.0% (98/101), while only 3.0% of isolates (3/101) were AMP resistant.
FIG. 1.
Histogram of AMP MICs.
The characteristics of the isolated H. influenzae strains with different PFGE patterns are listed in Table 2. Forty-two different PFGE patterns and, thus, 42 different strains were obtained from among the 172 isolates and were designated strains S1 to S42. Of the 42 strains, 37 strains consisted of isolates in a single PCR-based class. Three isolates classified as BLPACR II strains by the PCR results were identified as the same strain (strain S42). Every strain except strains S24 and S28 showed a narrow range of AMP MICs. Only five strains (strains S17, S18, S19, S24, and S31) were observed in both seasons. None of the strains isolated in the summer was isolated from the same carrier in the winter. Twenty-seven strains (64.3%) were isolated from two or more children, and 25 of these were each isolated from children belonging to the same DCC. Twelve strains (28.6%) were isolated from five or more children, and 10 of these were isolated from children belonging to the same DCC. Such findings suggesting horizontal transmission were not dependent on the season, the age of the child, or genetic mutations in the PBP gene.
TABLE 2.
Characteristics of H. influenzae strains with different PFGE patterns
| PFGE pattern | Strain type by PCR | AMP MIC range (μg/ml) | No. of carriers
|
|||
|---|---|---|---|---|---|---|
| Age
|
DCC-A/DCC-B
|
|||||
| ≤3 yr | ≥4 yr | Summer | Winter | |||
| S1 | BLNAS | ≤0.12 | 2 | 1 | 3/0 | 0/0 |
| S2 | BLNAS | ≤0.12 | 7 | 0 | 0/0 | 7/0 |
| S3 | BLNAS | ≤0.12 | 1 | 2 | 0/0 | 3/0 |
| S4 | BLNAS | ≤0.12 | 1 | 3 | 0/0 | 4/0 |
| S5 | BLNAS | ≤0.12 | 0 | 1 | 0/0 | 1/0 |
| S6 | BLNAS | ≤0.12 | 1 | 4 | 0/0 | 5/0 |
| S7 | BLNAS | ≤0.12 | 0 | 1 | 0/0 | 1/0 |
| S8 | BLNAS | ≤0.12 | 1 | 5 | 0/0 | 0/6 |
| S9 | BLNAS | ≤0.12 | 7 | 10 | 0/0 | 0/17 |
| S10 | BLNAS | ≤0.12 | 0 | 5 | 0/0 | 0/5 |
| S11 | BLNAS | ≤0.12 | 1 | 1 | 0/0 | 0/2 |
| S12 | BLNAS | ≤0.12 | 2 | 6 | 0/0 | 0/8 |
| S13 | BLNAS | ≤0.12 | 1 | 3 | 0/0 | 0/4 |
| S14 | Low-BLNAR | ≤0.12 | 1 | 0 | 0/0 | 0/1 |
| S15 | BLNAR | ≤0.12 | 0 | 1 | 0/0 | 1/0 |
| S16 | BLNAR | ≤0.12 | 0 | 1 | 0/0 | 0/1 |
| S17 | BLNAS | ≤0.12-0.25 | 5 | 2 | 3/1 | 3/0 |
| S18 | BLNAS | ≤0.12-0.25 | 2 | 5 | 0/4 | 3/0 |
| S19 | BLNAS, BLNAR | ≤0.12-0.25 | 1 | 3 | 0/3 | 1/0 |
| S20 | BLNAS | 0.25 | 3 | 1 | 4/0 | 0/0 |
| S21 | BLNAS | 0.25 | 1 | 0 | 0/1 | 0/0 |
| S22 | BLNAS | 0.25-0.5 | 12 | 1 | 13/0 | 0/0 |
| S23 | BLNAS | 0.25-0.5 | 4 | 0 | 4/0 | 0/0 |
| S24 | BLNAS | 0.25-4 | 2 | 0 | 0/1 | 0/1 |
| S25 | Low-BLNAR | 0.5 | 1 | 0 | 1/0 | 0/0 |
| S26 | BLNAR | 0.5 | 1 | 0 | 0/0 | 0/1 |
| S27 | BLNAS, BLNAR | 0.5-1 | 2 | 0 | 2/0 | 0/0 |
| S28 | Low-BLNAR, BLNAR | 0.5-8 | 2 | 0 | 2/0 | 0/0 |
| S29 | Low-BLNAR | 1 | 1 | 4 | 0/5 | 0/0 |
| S30 | Low-BLNAR | 1 | 0 | 2 | 0/2 | 0/0 |
| S31 | Low-BLNAR, BLNAR | 1 | 16 | 6 | 2/0 | 9/11 |
| S32 | Low-BLNAR, BLNAR | 1 | 3 | 1 | 0/0 | 0/4 |
| S33 | BLNAR | 1 | 0 | 1 | 1/0 | 0/0 |
| S34 | BLNAR | 2-4 | 8 | 1 | 0/9 | 0/0 |
| S35 | BLNAR | 4 | 1 | 0 | 1/0 | 0/0 |
| S36 | Low-BLNAR | 8 | 1 | 0 | 0/0 | 0/1 |
| S37 | BLNAR | 8 | 1 | 0 | 1/0 | 0/0 |
| S38 | BLNAR | 8 | 1 | 0 | 1/0 | 0/0 |
| S39 | BLNAR | 8 | 1 | 2 | 0/3 | 0/0 |
| S40 | BLNAR | 8 | 1 | 0 | 0/1 | 0/0 |
| S41 | BLNAR | 8 | 0 | 1 | 0/0 | 1/0 |
| S42 | BLPACR II | ≤16 | 3 | 0 | 0/3 | 0/0 |
DISCUSSION
The overall rate of nasopharyngeal carriage of H. influenzae in the present study was 47.4%, roughly equal to that reported by Masuda et al. (53.2%) in Japanese children in a similar situation (18). The rates of H. influenzae carriage in Europe and North America range from 11.7% to 95.0% among children attending DCCs, living in orphanages, or attending schools in various situations (3, 5, 8, 17, 18, 27, 28, 31, 32, 36). Such a wide dispersion may be explained by the fact that the carriage rate is dependent on multiple factors, such as age, season, the size of the facility, antibiotic treatment, morbidity from acute URIs, the sampling technique, and the type of infant feeding (9).
Previous reports on seasonal differences in the rate of H. influenzae carriage have been controversial: Some authors have documented an increased rate in the spring (32), while some others have found no significant differences between seasons (5, 17). Our results showed that the carriage rate was significantly higher in the winter than in the summer, as listed in Table 1. There are several potential reasons for this trend: cold weather increases the opportunity for indoor activities, leading to close interpersonal contact. Insufficient room ventilation would facilitate the airborne transmission of pathogens. Viral URIs, which predominantly occur in winter, may also impair the local host defense and help H. influenzae adhere to the nasopharyngeal mucosa.
The present study also indicated that the increase in the H. influenzae carriage rate in the winter was due to the increase in the proportions of antibiotic-susceptible H. influenzae isolates; by contrast, the rate of carriage of antibiotic-resistant H. influenzae did not differ between the two seasons. Antibiotic use is known to be a major risk factor for the occurrence of resistant bacteria. Several studies have revealed the potential impact of antibiotic use on the increase in the rate of nasopharyngeal carriage of resistant H. influenzae isolates in children (3, 6, 28, 29, 40). In our study, the percentage of children who were under treatment for common infantile inflammatory diseases did not differ between the summer and the winter. This finding suggests that the percentage of children who were given antibiotics also did not differ between the two seasons, corresponding to the constant rate of carriage of resistant strains.
The present study demonstrated that there was no significant difference in the rate of H. influenzae carriage between the age groups; however, the rate of carriage of antibiotic-resistant H. influenzae isolates was higher in younger children than in older children. It has been known that children aged 1 to 2 years are more likely than older children to contract repeated ear infections (10) and that an early age at which attendance at a DCC begins increases the risk of recurrent otitis media (24). This may suggest that younger children have more of a chance of receiving antibiotic treatment, which would potentially induce resistance genes.
The majority of the resistant isolates in our study were low-BLNAR/BLNAR strains, whereas only three isolates (1.7% of all H. infuenzae isolates) were β-lactamase positive. In other countries, BLPAR strains are more prevalent, while BLNAR strains are present at low levels; the rates of carriage of BLPAR and BLNAR strains have been reported to be from 3.7 to 55.3% and 0.9 to 2.8%, respectively, in children attending DCCs or living in orphanages (3, 5, 28, 32). In contrast, converse observations have been documented in Japan (11, 13, 18, 34). Hasegawa et al. (11) found that 44.9% of H. influenzae clinical isolates were resistant and that low-BLNAR/BLNAR strains accounted for 39.6% of clinical isolates (88.2% of resistant strains). Recent surveys in Japan also revealed that the percentage of AMP-resistant isolates among clinically isolated H. influenzae strains ranges from 4.8% to 15.7% (11, 13, 37). The present results showed similar percentages of low- BLNAR/BLNAR strains (35.5%) and AMP-resistant H. influenzae strains (11.6%), suggesting the spread of these resistant strains in healthy children attending DCCs in Japan. Such peculiar characteristics of antibiotic-resistant H. influenzae strains, i.e., the small percentage of β-lactamase-positive strains and the predominance of low-BLNAR/BLNAR strains, may have been caused by the extensive use of cephem antibiotics throughout Japan (11, 12).
Because PFGE analysis does not necessarily reflect all genetic mutations, isolates indistinguishable by PFGE may not be genetically identical. Molecular research with H. influenzae strains has revealed that strains with an indistinguishable PFGE pattern may have heterogeneous mutations in the PBP gene (14). However, the present study demonstrated a considerable correlation between PFGE patterns and PCR results: of the 42 strains, 37 strains consisted of isolates in a single PCR-based class. Although there were five strains that included isolates in the different PCR-based classes, three of them comprised closely related strains: low-BLNAR and BLNAR strains. Moreover, almost every strain showed a narrow range of AMP MICs. These observations indicate that molecular typing by PFGE is a useful tool for the epidemiological investigation of H. influenzae.
We observed a highly vigorous genetic turnover of H. influenzae strains in the nasopharynx: none of the children carried the same strain in both the summer and the winter, and only five strains were detected in both seasons. Several previous studies have used PFGE to investigate the molecular epidemiology, dynamics, and variable genetic heterogeneity of H. influenzae colonizing the upper respiratory tracts of children (5, 8, 27, 31, 32, 42). Peerbooms et al. (27) documented that 35.7% of children attending DCCs were H. influenzae positive in the first sampling, about half of the carriers became negative in the second sampling performed 1 month later, and only 7.0% of the remaining half carried an isolate with the same genotype as that in the first sampling. Sulikowska et al. (32) conducted bacterial sampling from the nasopharynges of children in a DCC or an orphanage in the winter and in the following spring and found that of the 22 pairs of H. influenzae isolates, 19 pairs were heterogeneous by PFGE analysis.
We also revealed that about two-thirds of the strains colonized two or more children, who mostly belonged to the same DCC. This finding suggests the substantial occurrence of the horizontal transmission of H. influenzae among healthy children in DCCs.
In conclusion, we investigated the characteristics of nasopharyngeal H. influenzae carriage in children attending DCCs by means of antimicrobial susceptibility testing, PCR, and PFGE in the summer of 2004 and in the following winter. Nearly half of the children were H. influenzae carriers, and 37.2% of the isolates were resistant, most of which were low-BLNAR and BLNAR. We also observed a highly vigorous genetic turnover and the substantial occurrence of the horizontal transmission of H. influenzae among these children. Hygienic and prophylactic countermeasures against this health problem need to be taken without delay.
Acknowledgments
We thank Kimiko Ubukata of the Laboratory of Infectious Agents Surveillance, Kitasato Institute for Life Sciences, Kitasato University, Tokyo, Japan, for her valuable technical advice.
This research was partially supported by the Ministry of Education, Science, Sports and Culture through a Grant-in-Aid for Young Scientists (B) (grant 16791032, 2004).
Footnotes
Published ahead of print on 9 January 2008.
REFERENCES
- 1.Alho, O. P., E. Laaro, and H. Oja. 1996. Public health impact of various risk factors for acute otitis media in northern Finland. Am. J. Epidemol. 1431149-1156. [DOI] [PubMed] [Google Scholar]
- 2.Aniansson, G., B. Alm, B. Andersson, P. Larsson, O. Nylen, H. Peterson, P. Rigner, M. Svanborg, and C. Svanborg. 1992. Nasopharyngeal colonization during the first year of life. J. Infect. Dis. 165(Suppl. 1)S38-S42. [DOI] [PubMed] [Google Scholar]
- 3.Barbosa-Cesnik, C., R. S. Farjo, M. Patel, J. Gilsdorf, S. I. McCoy, M. M. Pettigrew, C. Marrs, and B. Foxman. 2006. Predictors for Haemophilus influenzae colonization, antibiotic resistance and for sharing an identical isolate among children attending 16 licensed day-care centers in Michigan. Pediatr. Infect. Dis. J. 25219-223. [DOI] [PubMed] [Google Scholar]
- 4.Collet, J. P., T. Ducruent, D. Floret, J. Cogan-Collet, D. Honneger, and J. P. Boissel. 1991. Daycare attendance and risk of first infectious disease. Eur. J. Pediatr. 150214-216. [DOI] [PubMed] [Google Scholar]
- 5.Dabernat, H., M. A. Plisson-Saune, C. Delmas, M. Seguy, G. Faucon, R. Pelissier, H. Carsenti, C. Pradier, M. Roussel-Delvallez, J. Leroy, M. J. Dupont, F. De Bels, and P. Dellamonica. 2003. Haemophilus influenzae carriage in children attending French day care centers: a molecular epidemiological study. J. Clin. Microbiol. 411664-1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dabernat, H., P. Geslin, F. Megraud, P. Begue, J. Boulesteix, C. Dubreuil, F. de La Roque, A. Trinh, and A. Scheimberg. 2001. Effects of cefixime or co-amoxiclav treatment on nasopharyngeal carriage of Streptococcus pneumoniae and Haemophilus influenzae in children with acute otitis media. J. Antimicrob. Chemother. 41253-258. [DOI] [PubMed] [Google Scholar]
- 7.Faden, H., L. Duffy, R. Wasielewski, J. Wolf, D. Krystofik, and Y. Tung. 1997. Relationship between nasopharyngeal colonization and the development of otitis media in children. J. Infect. Dis. 1751440-1445. [DOI] [PubMed] [Google Scholar]
- 8.Farjo, R. S., B. Foxman, M. J. Patel, L. Zhang, M. M. Pettigrew, S. I. Mccoy, C. F. Maars, and J. R. Gilsdorf. 2004. Diversity and sharing of Haemophilus influenzae strains colonizing healthy children attending day-care centers. Pediatr. Infect. Dis. J. 2341-46. [DOI] [PubMed] [Google Scholar]
- 9.Garcia-Rodriguez, J. A., and M. J. F. Martinez. 2002. Dynamics of nasopharyngeal colonization by potential respiratory pathogens. J. Antimicrob. Chemother. 50(Suppl. S2)59-73. [DOI] [PubMed] [Google Scholar]
- 10.Hardy, A. M., and M. G. Fowler. 1993. Child care arrangements and repeated ear infections in younger children. Am. J. Public Health 831321-1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hasegawa, K., K. Yamamoto, N. Chiba, R. Kobayashi, K. Nagai, M. R. Jacobs, P. C. Appelbaum, K. Sunakawa, and K. Ubukata. 2003. Diversity of ampicillin-resistance genes in Haemophilus influenzae in Japan and the United States. Microb. Drug Resist. 939-46. [DOI] [PubMed] [Google Scholar]
- 12.Hasegawa, K., N. Chiba, R. Kobayashi, S. Murayama, S. Iwata, K. Sunakawa, and K. Ubukata. 2004. Rapidly increasing prevalence of β-lactamase-nonproducing, ampicillin-resistant Haemophilus influenzae type b in patients with meningitis. Antimicrob. Agents Chemother. 481509-1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hotomi, M., K. F. Sakai, D. S. Billal, J. Shimada, M. Suzumoto, and N. Yamanaka. 2006. Antimicrobial resistance in Haemophilus influenzae isolated from the nasopharynx among Japanese children with acute otitis media. Acta Otolaryngol. 126130-137. [DOI] [PubMed] [Google Scholar]
- 14.Hotomi, M., N. Yamanaka, D. S. Billal, A. Sakai, K. Yamauchi, M. Suzumoto, S. Takei, N. Yasui, S. Moriyama, and K. Kuki. 2004. Genotyping of Streptococcus pneumoniae and Haemophilus influenzae isolated from paired middle ear fluid and nasopharynx by pulsed-field gel electrophoresis. ORL J. Otorhinolaryngol. Relat. Spec. 6233-240. [DOI] [PubMed] [Google Scholar]
- 15.Japanese Society of Chemotherapy. 1990. Standard method of Japanese Society of Chemotherapy for MIC determination by the broth microdilution method. Chemotherapy 38103-105. (In Japanese.) [Google Scholar]
- 16.Manome, I., M. Ikedo, Y. Saito, K. Ishii, and M. Kaku. 2003. Evaluation of a novel automated chemiluminescent assay system for antimicrobial susceptibility testing. J. Clin. Microbiol. 41279-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marchissio, P., S. Gironi, S. Esposito, G. C. Schito, S. Mannelli, N. Principi, and the Ascanius Project Collaborative Group. 2001. Seasonal variations in nasopharyngeal carriage of respiratory pathogens in healthy Italian children attending day-care centers or schools. J. Med. Microbiol. 501095-1099. [DOI] [PubMed] [Google Scholar]
- 18.Masuda, K., R. Masuda, J. Nishi, K. Tokuda, M. Yoshinaga, and K. Miyata. 2002. Incidences of nasopharyngeal colonization of respiratory bacterial pathogens in Japanese children attending day-care centers. Pediatr. Int. 44376-380. [DOI] [PubMed] [Google Scholar]
- 19.Medeiros, A. A., R. Levesque, and G. A. Jacoby. 1986. An animal source for the ROB-1 β-lactamase of Haemophilus influenzae type B. Antimicrob. Agents Chemother. 29212-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mendelman, P. M., D. O. Chaffin, J. M. Musser, R. DeGroot, D. A. Serfass, and R. K. Selander. 1987. Genetic and phenotypic diversity among ampicillin-resistant, non-β-lactamase-producing, nontypeable Haemophilus influenzae isolates. Infect. Immun. 552585-2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mendelman, P. M., D. O. Chaffin, T. L. Stull, C. E. Rubens, K. D. Mack, and A. L. Smith. 1984. Characterization of non-β-lactamase-mediated ampicillin resistance in Haemophilus influenzae. Antimicrob. Agents Chemother. 26235-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murphy, T. F., A. L. Brauer, S. Sethi, M. Kilian, X. Cai, and A. J. Lesse. 2007. Haemophilus haemolyticus: a human respiratory tract commensal to be distinguished from Haemophilus influenzae. J. Infect. Dis. 19581-89. [DOI] [PubMed] [Google Scholar]
- 23.Murray, P. R., E. J. Baron, M. A. Pfaller, F. C. Tenover, and R. H. Yolken (ed.). 1999. Manual of clinical microbiology, 7th ed. American Society for Microbiology, Washington, DC.
- 24.Nafstad, P., J. A. Hagen, L. Oie, P. Magnus, and J. J. K. Jaakkola. 1999. Day care centers and respiratory health. Pediatrics 103753-758. [DOI] [PubMed] [Google Scholar]
- 25.National Committee for Clinical Laboratory Standards. 2000. Approved standard M7-A5, M100-S10. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. National Committee for Clinical Laboratory Standards, Wayne, PA.
- 26.Nelson, M. B., M. A. Apicella, T. F. Murphy, H. Vankeulen, L. D. Spotila, and D. Rekosh. 1988. Cloning and sequencing of Haemophilus influenzae outer membrane protein P6. Infect. Immun. 56128-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peerbooms, P. G. H., M. N. Engelen, D. A. J. Stokman, B. H. B. van Benthem, M. L. van Weert, S. M. Bruisten, A. van Belkum, and R. A. Coutinho. 2002. Nasopharyngeal carriage of potential bacterial pathogens related to day care attendance, with special reference to the molecular epidemiology of Haemophilus influenzae. J. Clin. Microbiol. 402832-2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Principi, N., P. Marchisio, G. C. Schito, and S. Mannelli. 1999. Risk factors for carriage of respiratory pathogens in the nasopharynx of healthy children. Pediatr. Infect. Dis. J. 18517-523. [DOI] [PubMed] [Google Scholar]
- 29.Samuelson, A., A. Freijd, J. Jonasson, and A. A. Lindberg. 1995. Turnover of nonencapsulated Haemophilus influenzae in the nasopharynges of otitis-prone children. J. Clin. Microbiol. 332027-2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seki, H., Y. Kasahara, K. Ohta, Y. Saikawa, R. Sumita, A. Yachie, S. Fujita, and S. Koizumi. 1999. Increasing prevalence of ampicillin- resistant, non-beta-lactamase-producing strains of Haemophilus influenzae in children in Japan. Chemotherapy 4515-21. [DOI] [PubMed] [Google Scholar]
- 31.St. Sauver, J., C. F. Marrs, B. Foxman, P. Somsel, R. Madera, and J. R. Gilsdorf. 2000. Risk factors for otitis media and carriage of multiple strains of Haemophilus influenzae and Streptococcus pneumoniae. Emerg. Infect. Dis. 6622-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sulikowska, A., P. Grzesiowski, E. Sadowy, J. Fiett, and W. Hryniewicz. 2004. Characteristics of Streptococcus pneumoniae, Haemophilus influenzae, and Moraxella catarrhalis isolated from the nasopharynges of asymptomatic children and molecular analysis of S. pneumoniae and H. influenzae strain replacement in the nasopharynx. J. Clin. Microbiol. 423942-3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sutcliffe, J. G. 1978. Nucleotide sequence of the ampicillin resistance gene of Escherichia coli plasmid pBR322. Proc. Natl. Acad. Sci. USA 753737-3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Suzuki, K., T. Nishimura, and S. Baba. 2003. Current status of bacterial resistance in the otolaryngology field: results from the second nationwide survey in Japan. J. Infect. Chemother. 946-52. [DOI] [PubMed] [Google Scholar]
- 35.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 332233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Trottier, S., K. Stenberg, and C. Svanborg-Eden. 1989. Turnover of nontypable Haemophilus influenzae in the nasopharynges of healthy children. J. Clin. Microbiol. 272175-2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ubukata, K., N. Chiba, K. Hasegawa, Y. Shibasaki, K. Sunakawa, M. Nonoyama, S. Iwata, and M. Konno. 2002. Differentiation of β-lactamase-negative ampicillin-resistant Haemophilus influenzae from other H. influenzae strains by a disc method. J. Infect. Chemother. 850-58. [DOI] [PubMed] [Google Scholar]
- 38.Ubukata, K., Y. Shibasaki, K. Yamamoto, N. Chiba, K. Hasegawa, Y. Takeuchi, K. Sunakawa, M. Inoue, and M. Konno. 2001. Association of amino acid substitutions in penicillin-binding protein 3 with β-lactam resistance in β-lactamase-negative ampicillin-resistant Haemophilus influenzae. Antimicrob. Agents Chemother. 451693-1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Uhari, M., K. Mantysaari, and M. Niemela. 1996. A meta-analytic review of risk factors for acute otitis media. Clin. Infect. Dis. 221079-1083. [DOI] [PubMed] [Google Scholar]
- 40.Varon, E., C. Levy, F. De La Rocque, M. Boucherat, D. Deforche, I. Podglajen, M. Navel, and R. Cohen. 2000. Impact of antimicrobial therapy on nasopharyngeal carriage of Streptococcus pneumoniae, Haemophilus influenzae, and Branhamella catarrhalis in children with respiratory tract infections. Clin. Infect. Dis. 31477-481. [DOI] [PubMed] [Google Scholar]
- 41.Vega, R., H. L. Sadoff, and M. J. Patterso. 1976. Mechanism of ampicillin resistance in Haemophilus influenzae type b. Antimicrob. Agents Chemother. 9164-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yano, H., M. Suetake, A. Kuga, K. Irinoda, R. Okamoto, T. Kobayashi, and M. Inoue. 1999. Pulsed-field gel electrophoresis analysis of nasopharyngeal flora in children attending a day care center. J. Clin. Microbiol. 38625-629. [DOI] [PMC free article] [PubMed] [Google Scholar]