Abstract
The newly proposed variable-number tandem-repeat (VNTR) typing system, which includes a basic 15-locus set and a high-resolution 24-locus set (P. Supply et al., J. Clin. Microbiol. 44:4498-4510, 2006), demonstrated a high power for the discrimination of Mycobacterium tuberculosis isolates collected worldwide. To evaluate its ability to differentiate the Beijing genotype strains from the Beijing area in China, 72 isolates with typical Beijing or Beijing-like spacer oligonucleotide typing profiles were subjected to typing with the VNTR system (24 loci) and typing by restriction fragment polymorphism analysis with IS6110 (IS6110-RFLP). Compared to the “old” 12-locus VNTR typing method, use of the 15- and 24-locus systems had a dramatically improved power to discriminate the Beijing genotype strains. A subtle difference in the Hunter-Gaston discriminatory index (HGI) between the 15-locus and the 24-locus systems resulted from only one locus, Mtub29. However, the VNTR-based clusters could be further differentiated by IS6110-RFLP (HGI by IS6110 RFLP, 0.999), although in one case an IS6110 cluster was subdivided by the 15-locus VNTR system. In this sense, use of the newly proposed 15-locus VNTR system along with the Mtub29 locus can serve as a first-line typing method for the epidemiological study of M. tuberculosis isolates in Beijing, while secondary typing of clustered strains by IS6110-RFLP is still required.
Tuberculosis (TB) remains a big threat to human health, especially in developing countries. China is one of the high-burden countries in the world, with a TB incidence of 101/100,000 population and a mortality rate of 17/100,000 population (23). The emergence of multidrug-resistant Mycobacterium tuberculosis strains and coinfection with M. tuberculosis and human immunodeficiency virus make the situation even worse. In recent years, molecular typing methods have become useful tools for the control of TB and help to indicate possible epidemiological links between TB patients, detect outbreaks and laboratory cross-contamination, and distinguish exogenous reinfection from endogenous reactivation in relapse cases.
In China, the most prevalent Mycobacterium tuberculosis strains belong to the Beijing family, which accounts for more than a quarter of all TB cases worldwide (2, 4). The M. tuberculosis Beijing family is a genetically homogeneous group. These strains are characterized by highly similar multiband patterns by restriction fragment length polymorphism (RFLP) analysis with IS6110 (IS6110-RFLP) and highly similar patterns by spacer oligonucleotide typing (spoligotyping) that show a lack of hybridization signals 1 to 34 and the hybridization of at least three of the spacers from signals 35 to 43 (12, 22). Therefore, spoligotyping is virtually unable to differentiate the members of the Beijing family, whereas IS6110-RFLP remains the “gold standard” method for the typing of these strains, as it has the highest discriminatory power (11-13). Unfortunately, the latter method is cumbersome and time-consuming and requires large quantities of DNA; accordingly, simpler and more discriminatory methods are being searched for and developed.
The development and application of the variable-number tandem-repeat (VNTR) method for the typing of M. tuberculosis became an important methodological achievement toward obtaining a better understanding of the molecular epidemiology of TB (19). The VNTR method is an otherwise well-known approach and technically relies on PCR amplification and calculation of the numbers of repeats on the basis of the size of the amplified product. The 12-locus mycobacterial interspersed repetitive-unit (MIRU)-VNTR scheme has been widely used (1, 8, 17), although it showed a limited ability to discriminate strains of the Beijing family (9, 11, 13, 16, 20). The newly proposed 15- and 24-locus systems were demonstrated to have higher discriminatory powers with a worldwide collection of M. tuberculosis strains (18) and also with members of the Beijing family of strains from Japan (6, 24).
The Beijing genotype strains constitute a significant proportion of the circulating M. tuberculosis strains within large regions of the world, such as East Asia, Russia, and South Africa (2, 4, 12, 13). The recent global dissemination of these strains is threatening TB control programs worldwide. Nevertheless, the available typing methods lack either discriminatory power or simplicity when they are applied to the Beijing family members (2, 4, 9, 11, 13, 16, 20). Consequently, the aim of the present study was to investigate the power of the use of new combinations of the VNTR loci to differentiate the Beijing family strains from the area of its first description and likely origin, i.e., Beijing, China.
MATERIALS AND METHODS
Mycobacterial isolates.
M. tuberculosis clinical isolates were obtained from the Beijing Chest Hospital from 2002 to 2005. Patient information was collected from the patient's medical history at the same time as isolate collection. Total DNA was isolated from the cultured bacteria by method recommended by Van Embden et al. (21).
Spoligotyping.
All the strains were previously analyzed by spoligotyping (7), as described by Kamerbeek et al. (10). In short, the PCR-amplified biotin-labeled direct-repeat locus is hybridized against an array of immobilized 43 different direct-repeat spacers in a Miniblotter MN45 apparatus (Immunetics, Cambridge, MA). The resulting hybridization signals were revealed by chemiluminescence and were visualized as profiles of discrete dots.
Typing with the VNTR system.
The 24 VNTR loci in the proposed VNTR typing method were used in this study (18). Each locus was amplified by PCR with the primers described elsewhere (3, 16, 18, 19).
PCR mixtures were prepared as follows. The total 25-μl reaction volume included 2 ng template DNA, 2*Taq PCR master mix (Tiangen, Beijing, China), 5% dimethyl sulfoxide, and 10 pmol of primers. Negative controls, consisting of the PCR mixture without template DNA, were run in each amplification procedure. The thermocycler conditions involved 1 cycle at 95°C for 5 min; 40 cycles at 94°C for 50 s, 65°C for 50 s, and 72°C for 1 min; and 1 cycle at 72°C for 5 min. The annealing temperature was changed to 60°C for primers QUB26 and QUB4156. PCR was carried out in a PTC-200 cycler (MJ Research, Watertown, MA).
The PCR products were analyzed by electrophoresis with 1.3% agarose (Besta). Analysis of the sizes of the PCR fragments and assignment of the various VNTR alleles were done with the Image Acquisition and Analysis software of the Alpha Innotech FluoChem system and by comparison with the correspondence table provided by Philip Supply (Institut Pasteur de Lille, Lille, France).
IS6110-RFLP.
DNA fingerprinting with IS6110 as a probe was performed according to the internationally standardized protocol (21). In each Southern blot experiment, reference strain M. tuberculosis 14323 was included as a molecular weight marker. The IS6110-RFLP patterns were analyzed by using Gel Compar (version 4.1) software (Applied Maths, Kortrijk, Belgium). Similarities between RFLP patterns were calculated by using the Dice coefficient, and the dendrogram was produced with the unweighted pair group method with arithmetic averages algorithm. IS6110-RFLP clusters were defined on the basis of fully identical patterns (the same number of IS6110 bands at identical positions).
Statistical analysis.
The allelic diversity (h) at a given VNTR locus was calculated as 1 − ∑xi2 [n/(n − 1)], where xi is the frequency of the ith allele at the locus, and n is the number of isolates in the sample (14).
The level of discriminatory power of each typing method was calculated by using the Hunter-Gaston discriminatory index (HGI) (5), which was calculated by the following equation:
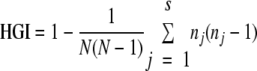 |
(1) |
where N is the total number of strains in the typing sample, s is the total number of different strain types, and nj is the number of strains belonging to the jth type.
The clustering rate was defined as (nc − c)/n, where n is the total number of cases in the sample, c is the number of clusters, and nc is the total number of clustered cases (15).
RESULTS
Study population.
A total of 121 M. tuberculosis strains recovered from pulmonary TB patients from 2002 to 2005 in the Beijing area of China were studied. Among the strains isolated from all the cases, two strains were isolated from the same patients. The other patients were proven to be unlinked on the basis of a standard epidemiological investigation.
On the basis of spoligotyping, 103 (85.1%) of 121 isolates showed a profile typical of that for the Beijing genotype, which consisted of signals 35 to 43, and 8 strains had abridged Beijing-like spoligotyping profiles (7). We treated these 103 strains as Beijing family strains. Of these, 72 strains had a sufficient quantity of DNA for further genotyping by both the VNTR method and IS6110-RFLP.
Genotyping.
IS6110 fingerprinting subdivided the 72 Beijing genotype isolates into 66 unique types and three clusters (Fig. 1). The copy number of IS6110 was high among these strains and varied from 10 to 23.
FIG. 1.
IS6110-RFLP-based dendrogram and 24-locus MIRU-VNTR profiles of the 72 Beijing genotype strains. The boxes indicate clustered isolates.
The proposed 24-locus scheme (18) was used for the typing of the selected 72 Beijing genotype strains by the VNTR method. The use of the old 12-locus combination (19) generated 19 unique strains and 53 clustered strains (HGI, 0.788), with a clustering rate of 59.7% (Table 1). A significantly increased resolution with 55 unique and 17 grouped strains was observed by application of the 24-locus system (HGI, 0.992), with the clustering rate decreased to 15.3%. The use of the basic 15-locus subset did not remarkably affect the discriminatory power (HGI, 0.990), with a clustering rate of 18.1% that resulted from addition of two clustered isolates. Finally, use of the old 12-locus scheme plus three exact tandem repeats (ETRs) only slightly improved the resolution, resulting in 29 unique profiles and 43 shared types (HGI, 0.849; clustering rate, 48.6%).
TABLE 1.
Clustering results and discriminatory power of the genotyping methods
| Method | No. of unique isolates | No. of clustered isolates | No. of clusters | No. of distinct types | Clustering rate (%) | HGI |
|---|---|---|---|---|---|---|
| Spoligotyping | 2 | 70 | 1 | 3 | 95.8 | 0.055 |
| VNTR system with 12 old loci | 19 | 53 | 10 | 29 | 59.7 | 0.788 |
| VNTR system with 12 old loci + 3 ETRs | 29 | 43 | 8 | 37 | 48.6 | 0.849 |
| VNTR system with 12 old loci + spoligotyping | 31 | 41 | 10 | 41 | 43.1 | 0.813 |
| VNTR system with 15 basic loci | 53 | 19 | 6 | 59 | 18.1 | 0.990 |
| VNTR system with 15 basic loci + spoligotyping | 54 | 18 | 6 | 60 | 16.7 | 0.991 |
| VNTR system with 15 basic loci + Mtub29 | 55 | 17 | 6 | 61 | 15.3 | 0.992 |
| VNTR system with 24 loci | 55 | 17 | 6 | 61 | 15.3 | 0.992 |
| VNTR system with 24 loci + spoligotyping | 56 | 16 | 6 | 62 | 13.9 | 0.993 |
| IS6110-RFLP | 66 | 6 | 3 | 69 | 4.2 | 0.999 |
The allelic diversity differed significantly among the VNTR loci (Table 2). The highest allelic diversity among the 72 Beijing genotype strains was observed for the QUB-11b locus (0.651), and the lowest (null) allelic diversity was found for the monomorphic MIRU2 and MIRU24 loci. The nine supplemental loci had a relatively lower allelic diversity, with the highest score of 0.119 achieved for the Mtub29 and MIRU39 loci.
TABLE 2.
Allelic diversity of each VNTR locus in the M. tuberculosis Beijing genotype strains from Beijing, China
| VNTR locus combinations | VNTR locusa | VNTR alias | No. of alleles | No. of repeats (range) | Allelic diversity |
|---|---|---|---|---|---|
| Basic 15 | 2163b | QUB-11b | 5 | 3-7 | 0.651 |
| 1955 | Mtub21 | 5 | 1-6 | 0.556 | |
| 4052 | QUB-26 | 8 | 3-10 | 0.518 | |
| 4156 | QUB-4156 | 5 | 1-5 | 0.395 | |
| 2996 | MIRU26 | 6 | 3-9 | 0.353 | |
| 424 | Mtub04 | 4 | 2-5 | 0.306 | |
| 2165 | ETR-A | 3 | 2-4 | 0.232 | |
| 802 | MIRU40 | 4 | 1-4 | 0.194 | |
| 3690 | Mtub39 | 5 | 1-6 | 0.171 | |
| 3192 | MIRU31, ETR-E | 3 | 4-6 | 0.169 | |
| 960 | MIRU10 | 3 | 1-3 | 0.144 | |
| 580 | MIRU4, ETR-D | 4 | 0-3′ | 0.120 | |
| 577 | ETR-C | 4 | 2-6 | 0.094 | |
| 1644 | MIRU16 | 3 | 2-4 | 0.068 | |
| 2401 | Mtub30 | 4 | 2-5 | 0.068 | |
| Supplemental 9 | 2347 | Mtub29 | 3 | 2-4 | 0.119 |
| 4348 | MIRU39 | 3 | 2-4 | 0.119 | |
| 3007 | MIRU27, QUB-5 | 2 | 2-3 | 0.014 | |
| 2461 | ETR-B | 2 | 1-2 | 0.014 | |
| 3171 | Mtub34 | 2 | 3-4 | 0.014 | |
| 2531 | MIRU23 | 2 | 5-6 | 0.014 | |
| 2059 | MIRU20 | 2 | 1-2 | 0.014 | |
| 154 | MIRU2 | 1 | 2 | 0 | |
| 2687 | MIRU24 | 1 | 1 | 0 |
The loci within different VNTR locus combinations are listed in descending order of allelic diversity. The 12 old VNTR loci are underlined.
DISCUSSION
Molecular typing methods are generally and justly assumed to be useful tools for the resolution of various issues related to the classical epidemiology of human pathogens, including M. tuberculosis. Their application in the field and discriminatory abilities may be challenged by a disequilibrium of the local population structures of a pathogen, manifested as the predominance of a particular, homogeneous clone(s). This situation applies to the M. tuberculosis population in China. The Beijing genotype was identified for the first time in strains isolated in the Beijing area of China, which resulted in the name of the genotype (22). Interestingly, in the first study of the Beijing genotype, it was identified in 89.4% of the M. tuberculosis strains collected from 1992 to 1994 (22). Our recent study identified this genotype in 91.9% of the strains from Beijing studied (7). Therefore, it appears that the 10 years of the National TB Control Program has not altered the predominance of the Beijing genotype strains in the local population of M. tuberculosis, although it has decreased the incidence of TB. Furthermore, clones of the Beijing family of strains are spread worldwide and the causes of epidemics in many areas (2). Consequently, a method of effective and easy typing for the characterization and differentiation of strains of this family is urgently needed.
The sample used in the present study included 72 M. tuberculosis strains from the Beijing area of China previously shown to belong to the Beijing genotype on the basis of spoligotyping (7). Thus, spoligotyping could not effectively distinguish them. These strains were further subjected to the newly proposed VNTR (24 loci [18] and IS6110-RFLP typing methods. The ability to differentiate Beijing genotype strains varied among the different typing methods. IS6110-RFLP (69 types; HGI, 0.999) was superior to the 24-locus VNTR system (61 types; HGI, 0.992) and the basic 15-locus VNTR system (59 types; HGI, 0.990), whereas all methods were highly superior to spoligotyping (3 types; HGI, 0.055).
IS6110-RFLP is considered the gold standard method for the typing of M. tuberculosis strains, especially strains with high IS6110 copy numbers. In the present study, all 72 M. tuberculosis isolates showed high-IS6110-copy-number (10 to 23) fingerprints. IS6110-RFLP analysis revealed three clusters, with two isolates in each cluster (Fig. 1). The strains in cluster 2 were isolated from the same patients over an interval of 2 weeks, and these strains also had the same spoligotyping profile with typical 9-locus signals and identical 24-locus VNTR patterns. The isolates grouped in clusters 1 and 3 had no epidemiological links, while the isolates in cluster 3 were further subdivided on the basis of one repeat difference in ETR-A within a basic 15-locus combination. At the same time, five of six VNTR-defined clusters were subdivided by RFLP typing. Taken together, these data confirmed recent reports that typing with the VNTR system alone, even when the types are determined on the basis of the use of more loci, can overestimate the recent transmission of the M. tuberculosis Beijing genotype strains (11, 24). The explanation may be that IS6110-RFLP profiles are evolving faster than VNTRs in these multiband strains.
The allelic diversity of the VNTR loci varied significantly at each locus. Among the 24 loci investigated in this study, QUB-11b (h = 0.651), Mtub21 (h = 0.556), and QUB-26 (h = 0.518) were the relatively more discriminatory loci; but the MIRU2 (h = 0) and MIRU24 (h = 0) loci did not differentiate the Beijing genotype strains. All the loci of the supplemental nine-locus set showed low levels of polymorphism (h, less than 0.119) among the Beijing genotype strains, as was also shown in previous studies (6, 24), indicating the close genetic relationship between the members of this specific family.
Different VNTR typing sets showed various efficiencies in differentiating Beijing genotype strains (Table 1). Until recently, the most widely used 12-locus set had a limited discriminatory power (HGI, 0.788), with a clustering rate 59.7%, as reported previously (8, 13, 20). Its discriminatory ability was not improved even by combination with three additional ETR loci or spoligotyping. Application of the newly proposed 15-locus basic VNTR typing system and the 24-locus high-resolution VNTR typing system dramatically increased the discriminatory power, with HGIs of 0.990 and 0.992, respectively. A very subtle difference in HGIs was due to the Mtub29 locus included in the 24-locus system, whereas the supplemental 8 loci of the 24-locus system did not contribute to further differentiation of the Beijing genotype strains. This confirmed the findings described in previous reports that the supplemental nine loci are not polymorphic in Beijing genotype strains (9, 11, 16, 20). However, the strains clustered by typing by use of the VNTR system could still be further subdivided by typing by IS6110-RFLP, which is consistent with other evaluations of this new 24-locus system in Japan, a country where the Beijing genotype also constitutes a predominant clone (6, 24).
In conclusion, the population of M. tuberculosis in Beijing, China, continues to be dominated by strains of the Beijing family. The basic 15-locus VNTR system plus locus Mtub29 were shown to achieve a good discrimination of these strains, so we suggest that this 16-locus VNTR scheme be used as the first-line screening method for the routine epidemiological investigation of M. tuberculosis isolates in the Beijing area. However, secondary subtyping, either by IS6110-RFLP or by use of the hypervariable VNTR loci (6), should be performed for clustered isolates, although a standardized approach remains to be developed for these isolates.
Acknowledgments
We are grateful to Philip Supply for kindly providing his VNTR manual.
This study was supported by a grant (no. 30471841) from the National Natural Science Foundation of China.
Footnotes
Published ahead of print on 16 January 2008.
REFERENCES
- 1.Banu, S., S. V. Gordon, S. Palmer, M. R. Islam, S. Ahmed, K. M. Alam, S. T. Cole, and R. Brosch. 2004. Genotypic analysis of Mycobacterium tuberculosis in Bangladesh and prevalence of the Beijing strain. J. Clin. Microbiol. 42674-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bifani, P. J., B. Mathema, N. E. Kurepina, and B. N. Kreiswirth. 2002. Global dissemination of the Mycobacterium tuberculosis W-Beijing family strains. Trends Microbiol. 1045-52. [DOI] [PubMed] [Google Scholar]
- 3.Frothingham, R., and W. A. Meeker-O'Connell. 1998. Genetic diversity in the Mycobacterium tuberculosis complex based on variable numbers of tandem DNA repeats. Microbiology 1441189-1196. [DOI] [PubMed] [Google Scholar]
- 4.Glynn, J. R., J. Whiteley, P. J. Bifani, K. Kremer, and D. van Soolingen. 2002. Worldwide occurrence of Beijing/W strains of Mycobacterium tuberculosis: a systematic review. Emerg. Infect. Dis. 8843-849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hunter, P. R., and M. A. Gaston. 1998. Numerical index of the discriminatory ability of typing systems: an application of Simpsons's index of diversity. J. Clin. Microbiol. 262465-2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iwamoto, T., S. Yoshida, K. Suzuki, M. Tomita, R. Fujiyama, N. Tanaka, Y. Kawakami, and M. Ito. 2007. Hypervariable loci that enhance the discriminatory ability of newly proposed 15-loci and 24-loci variable-number tandem repeat typing method on Mycobacterium tuberculosis strains predominated by the Beijing family. FEMS Microbiol. Lett. 272282-283. [DOI] [PubMed] [Google Scholar]
- 7.Jiao, W. W., I. Mokrousov, G. Z. Sun, M. Li, J. W. Liu, O. Narvskaya, and A. D. Shen. 2007. Molecular characteristics of rifampin and isoniazid resistant Mycobacterium tuberculosis strains from Beijing, China. Chin. Med. J. (Engl.). 120814-819. [PubMed] [Google Scholar]
- 8.Kam, K. M., C. W. Yip, L. W. Tse, K. L. Wong, T. K. Lam, K. Kremer, B. K. Au, and D. van Soolingen. 2005. Utility of mycobacterial interspersed repetitive unit typing for differentiating multidrug-resistant Mycobacterium tuberculosis isolates of the Beijing family. J. Clin. Microbiol. 43306-313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kam, K. M., C. W. Yip, K. L. Leung, K. L. Wong, W. M. Ko, and W. S. Wong. 2006. Optimization of variable number tandem repeat typing set for differentiating Mycobacterium tuberculosis strains in the Beijing family. FEMS Microbiol. Lett. 256258-265. [DOI] [PubMed] [Google Scholar]
- 10.Kamerbeek, J., L. Schouls, A. Kolk, M. van Agterveld, D. van Soolingen, S. Kuijper, A. Bunschoten, H. Molhuizen, R. Shaw, M. Goyal, and J. D. A. van Embden. 1997. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J. Clin. Microbiol. 35907-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kremer, K., B. K. Au, P. C. Yip, R. Skuce, P. Supply, K. M. Kam, and D. van Soolingen. 2005. Use of variable-number tandem-repeat typing to differentiate Mycobacterium tuberculosis Beijing family isolates from Hong Kong and comparison with IS6110 restriction fragment length polymorphism typing and spoligotyping. J. Clin. Microbiol. 43314-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kremer, K., J. R. Glynn, T. Lillebaek, S. Niemann, N. E. Kurepina, B. N. Kreiswirth, P. J. Bifani, and D. van Soolingen. 2004. Definition of the Beijing/W lineage of Mycobacterium tuberculosis on the basis of genetic markers. J. Clin. Microbiol. 424040-4049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mokrousov, I., H. M. Ly, T. Otten, N. N. Lan, B. Vyshnevskyi, S. Hoffner, and O. Narvskaya. 2005. Origin and primary dispersal of the Mycobacterium tuberculosis Beijing genotype: clues from human phylogeography. Genome Res. 151357-1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Selander, R. K., D. A. Caugant, H. Ochman, J. M. Musser, M. N. Gilmour, and T. S. Whittam. 1986. Methods of multilocus enzyme electrophoresis for bacterial population genetics and systematics. Appl. Environ. Microbiol. 51873-884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Small, P. M., P. C. Hopewell, S. P. Singh, A. Paz, J. Parsonnet, D. C. Ruston, G. F. Schecter, C. L. Daley, and G. K. Schoolnik. 1994. The epidemiology of tuberculosis in San Francisco. A population-based study using conventional and molecular methods. N. Engl. J. Med. 3301703-1709. [DOI] [PubMed] [Google Scholar]
- 16.Smittipat, N., P. Billamas, M. Palittapongarnpim, A. Thong-On, M. M. Temu, P. Thanakijcharoen, O. Karnkawinpong, and P. Palittapongarnpim. 2005. Polymorphism of variable-number tandem repeats at multiple loci in Mycobacterium tuberculosis. J. Clin. Microbiol. 435034-5043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun, Y. J., R. Bellamy, A. S. Lee, S. T. Ng, S. Ravindran, S. Y. Wong, C. Locht, P. Supply, and N. I. Paton. 2004. Use of mycobacterial interspersed repetitive unit-variable-number tandem repeat typing to examine genetic diversity of Mycobacterium tuberculosis in Singapore. J. Clin. Microbiol. 421986-1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Supply, P., C. Allix, S. Lesjean, M. Cardoso-Oelemann, S. Rusch-Gerdes, E. Willery, E. Savine, P. de Haas, H. van Deutekom, S. Roring, P. Bifani, N. Kurepina, B. Kreiswirth, C. Sola, N. Rastogi, V. Vatin, M. C. Gutierrez, M. Fauville, S. Niemann, R. Skuce, K. Kremer, C. Locht, and D. van Soolingen. 2006. Proposal for standardization of optimized mycobacterial interspersed repetitive unit-variable-number tandem repeat typing of Mycobacterium tuberculosis. J. Clin. Microbiol. 444498-4510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Supply, P., E. Mazars, S. Lesjean, V. Vincent, B. Gicquel, and C. Locht. 2000. Variable human minisatellite-like regions in the Mycobacterium tuberculosis genome. Mol. Microbiol. 36762-771. [DOI] [PubMed] [Google Scholar]
- 20.Surikova, O. V., D. S. Voitech, G. Kuzmicheva, S. I. Tatkov, I. V. Mokrousov, O. V. Narvskaya, M. A. Rot, D. van Soolingen, and M. L. Filipenko. 2005. Efficient differentiation of Mycobacterium tuberculosis strains of the W-Beijing family from Russia using highly polymorphic VNTR loci. Eur. J. Epidemiol. 20963-974. [DOI] [PubMed] [Google Scholar]
- 21.van Embden, J. D. A., M. D. Cave, J. T. Crawford, J. W. Dale, K. D. Eisenach, B. Gicquel, P. Hermans, C. Martin, R. McAdam, T. M. Shinnik, and P. M. Small. 1993. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J. Clin. Microbiol. 31406-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Soolingen, D., L. Qian, P. E. W. de Haas, J. T. Douglas, H. Traore, F. Portaels, Z. Quing, D. Enkhasaikan, P. Nymadawa, and J. D. A. van Embden. 1995. Predominance of a single genotype of Mycobacterium tuberculosis in countries of East Asia. J. Clin. Microbiol. 333234-3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.World Health Organization. 2006. Global tuberculosis control: surveillance, planning, financing. WHO report WHO/HTM/TB/2006.362. World Health Organization, Geneva, Switzerland.
- 24.Yokoyama, E., K. Kishida, M. Uchimura, and S. Ichinohe. 2007. Improved differentiation of Mycobacterium tuberculosis strains, including many Beijing genotype strains, using a new combination of variable number of tandem repeats loci. Infect. Genet. Evol. 7499-508. [DOI] [PubMed] [Google Scholar]



