Abstract
NF-Y is a trimeric transcription factor containing H2A/H2B-like subunits, which specifically binds to the CCAAT box, a common eukaryotic promoter element. To gain insights into NF-Y-dependent transcriptional regulation, we assessed its relationships with positive histone marks by chromatin immunoprecipitation-on-chip and correlative-profiling studies. Unbiased identification of binding sites shows that the majority of genes are bound by NF-Y in the promoter and/or within the coding region. Parallel analysis of H3K9-14ac and H3K4me3 sites indicates that NF-Y loci can be divided in two distinct clusters: (i) a large cohort contains H3K9-14ac and H3K4me3 marks and correlates with expression and (ii) a sizeable group is devoid of these marks and is found on transcriptionally silent genes. Within this class, we find that NF-Y binding is associated with negative histone marks, such as H4K20me3 and H3K27me3. NF-Y removal by a dominant negative NF-YA leads to a decrease in the transcription of expressed genes associated with H3K4me3 and H3K9-14ac, while increasing the levels of many inactive genes. These data indicate that NF-Y is embedded in positive as well as in negative methyl histone marks, serving a dual function in transcriptional regulation, as an activator or as a repressor.
Promoters and enhancers that activate RNA polymerase II (Pol II)-transcribed mRNA genes are formed by a combinatorial puzzle of short sequences recognized by sequence-specific regulators. Among such elements, the CCAAT box is known to be one of the most frequent. This has been illustrated by several unbiased bioinformatic studies of large sets of vertebrate promoters (16, 19, 21, 29, 35, 48, 57). Testing has shown that the CCAAT box significantly contributes to promoter activity (34).
Different entities contain the word CCAAT in their acronyms, but several types of evidence indicate that NF-Y, also termed CBF and HAP2/3/5 for Saccharomyces cerevisiae, is the CCAAT regulator. (i) Highly specific antibodies were used in supershift electrophoretic mobility shift assays and chromatin immunoprecipitations (ChIPs) with a plethora of different promoters (33; D. Dolfini and R. Mantovani, unpublished data). (ii) Specific dominant negative NF-YA vectors were employed in cotransfection and adenovirus infection experiments. (iii) Nucleotides flanking the CCAAT box emerged in the bioinformatic studies cited above, with perfect matches to NF-Y preferences, as assayed by in vitro binding studies (15). It is therefore reasonable to conclude that NF-Y is by far the major protein that regulates this element.
NF-Y is a ubiquitous heteromeric transcription factor (TF) composed of three subunits, NF-YA, NF-YB, and NF-YC, all necessary for DNA binding (34). NF-YB and NF-YC contain conserved histone fold motifs composed of three alpha helices separated by short loop/strand regions (42). NF-YB/NF-YC association is essential for NF-YA binding and sequence-specific DNA interactions. Interestingly, CCAAT boxes are present at a specific location within promoters, typically between −60 and −100 bp from the transcriptional start site (TSS). Within this context, NF-Y is not a powerful activator but rather a promoter organizer that cooperates with the activity of neighboring TFs. The emergence of genome-wide technologies now allows a look at TF binding in a more systematic and unbiased way. Previous studies, performed with CpG island arrays and with an oligonucleotide array representative of a small set of human promoters, left us with an inconclusive picture as to the widespread NF-Y distribution in vivo (12, 51).
Moreover, the simple annotation of TF binding sites is not per se sufficient to obtain satisfactory functional information, since most TFs are known to act in a peculiar chromatin environment, defined by patterns of histone posttranslational modifications. Some of them are associated with accessible, transcriptionally active chromatin, others with repressive heterochromatin, either constitutive or facultative (reviewed in references 4 and 43). In particular, H3K4 trimethylation (H3K4me3) and H3K9-14 acetylations (H3K9-14ac) are associated with an active chromatin environment in regions and promoters that are transcribed or poised for rapid induction by external stimuli (26, 44, 46). Their presence in vivo has been detailed at the single-gene level, and location analysis confirmed their widespread distribution in the proximity of promoters (2, 6, 23, 25, 26, 30, 31, 39, 40, 41). Interestingly, while the presence of these marks precedes gene activation, an increase in their levels is generally noticed in systems of inducible transcription. In NF-Y-dependent endoplasmic reticulum stress promoters, for example, a substantial increase in H3 acetylation and H3K4me3 was seen after induction, while NF-Y binding was detailed before (3, 14). On the other hand, posttranslational modifications such as H3K9, H3K27, and H4K20 methylations are known to be associated with inactive or actively repressed areas of the genome (4, 18, 38, 47). In general, understanding the relationship between histone modifications and TF binding is quite relevant; it is not completely clear, at the moment, whether the binding of certain factors is required for specific modifications to be brought in through recruitment of histone-modifying enzymatic machines or whether the binding of TFs is allowed only by a preexisting nucleosomal environment with an appropriate pattern of histone modifications. To shed light on the relationship between NF-Y and active histone marks, we used high-density tiling arrays of chromosomes 20, 21, and 22 in ChIP-on-chip experiments.
MATERIALS AND METHODS
Cells, infections, and PCR analysis.
HeLa cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum, 1% antibiotics (penicillin and streptomycin), and l-glutamine in 5% CO2. HCT116 p53−/− cells were grown in McCoy's medium. Infection of HCT116p53−/− and HeLa cells with Ad-YAm29, Ad-NF-YA, and Ad-GFP adenoviruses was carried out as described previously (27). Semiquantitative ChIP PCR and reverse transcription-PCR (RT-PCR) analyses were performed according to standard procedures, in the linear range of amplification, essentially as previously described (12).
ChIP, amplicon generation, and ChIP on chip.
ChIPs were performed essentially as described previously (51). Briefly, 5 × 106 cell equivalents of chromatin, <0.5 kb, were immunoprecipitated with 15 μg of anti-YB, anti-H3K4me3 (Abcam), and anti-H3K9-14ac (Upstate) antibodies. Immunoprecipitation-enriched DNAs were used to generate amplicons for hybridization experiments. Parallel ChIPs were run with a Flag control antibody (Sigma), and bona fide NF-Y targets were used to check enrichments before and after the amplification steps. The generation of amplicons from the individual ChIPs was performed by following the protocol of ligation-mediated PCR previously described (9, 54). Design of the oligonucleotides, preparation of the slides, hybridization, and scanning of the fluorescence intensities were performed by Nimblegen. Validations of the results by semiquantitative PCR were performed on independent, nonamplified ChIPs using primers listed in File S1 in the supplemental material. Negative-histone-mark ChIPs were also performed as described previously (51) using anti-H3K9me3 (Abcam), -H3K27me3 (Abcam), and -H4K20me3 (Upstate) antibodies.
ChIP-on-chip data analysis.
The Cy5 and Cy3 raw data obtained from each experiment were normalized by subtracting the median and then adding back the average median of the two channels. The independent median normalization was chosen since channel distributions were not parametric and therefore the mean was not suitable. This approach does not perturb the detection of biologically relevant peaks, while allowing efficient removal of noise due to unbalance between the two channels (not shown). After single-channel normalization, the Cy5/Cy3 ratio was calculated for each probe and then converted in the corresponding Z score. This kind of transformation is useful when seeking to compare the relative standings of items from distributions with different means and/or standard deviations and fitted our requirement of performing comparison between different arrays. Box-and-whisker plots clearly showed that Z-score conversion allowed for a direct comparison of independent data sets derived from biological replicates (not shown). Peak search was then performed essentially as previously reported (9), with the so called “peak first” approach. A given percentile threshold was chosen for all experiments, and all probes with enrichment values greater than the percentile were selected. An NF-Y “peak” was defined as a stretch of at least three adjacent probes (spaced by no more than 150 bp on the genomic sequence) exceeding the percentile threshold. In H3K9-14ac and H3K4me3 experiments, five adjacent probes were considered to define a peak. A hypergeometric P-value-based distribution was then associated with the peaks obtained. Finally, peaks obtained from the different experiments were merged, and a given peak was predicted as biologically relevant if present in at least two out of four experiments for NF-Y or two out of three experiments for active histone marks, with corresponding probability values computed with a binomial distribution.
Profiling experiments.
The GEO GSE6022 data set, containing a total of three untreated HeLa cell samples, each being a biological replicate, was used for HeLa profiling experiments. The GEO GSE6207 data set, containing seven untreated samples, was used for HepG2 profiling experiments. The data set from Sato et al. (45), containing three untreated samples, each being a biological replicate (Affimetrix HG-U133-A platform), was used for human embryonic stem (ES) cell profiling experiments. The GEO GSE8884 (Affimetrix HG-U133 plus 2.0 platform) was also used for the latter analysis, yielding essentially the same results despite the larger coverage of the platform (not shown).
Expression signals for microarray data were calculated according to the rma algorithm and used for quality control. Absent (A) and present (P) calls were also calculated according to the open-source version of the MAS5 algorithm and used both for quality control and comparisons. A gene was called P if it was present at least in two out of three of the samples. The same rules were applied for A calls. A gene was called UN if it did not fall in the previous categories. The A, P, and UN groups have no overlaps. The details of the analysis are available upon request.
RESULTS
A widespread distribution of NF-Y binding in CCAAT promoters.
To assay the prevalence of NF-Y binding on CCAAT promoters, we performed ChIP analysis with an anti-NF-YB antibody on chromatins derived from seven different cell lines. We checked 25 randomly picked CCAAT-containing promoters of chromosome 21 genes. Figure 1 indicates that most of the promoters were indeed bound by NF-Y in the majority of cells. However, the degree of enrichment over a control Flag antibody varied substantially and was arbitrarily divided into three levels: high (>30-fold), medium (5- to 30-fold), and low (2- to 5-fold). The expression patterns of these genes were then analyzed according to available Unigene transcriptome data and divided into two broad categories: genes that were ubiquitously expressed and those that had a tissue or cell type preference. With few exceptions, TMEM50B, RUNX1, and DSCR1, the high- and medium-enrichment genes, were all in the ubiquitous category and bound by NF-Y in all cell lines. Four genes, CLDN17, TMPRSS3, AIRE, and ERG, all tissue specific, were negative in all cell lines. Finally, weakly enriched sites were mostly found on tissue-specific genes. We conclude that (i) NF-Y is bound to the majority of CCAAT-containing promoters and (ii) the highest enrichment correlates with a ubiquitous expression profile.
FIG. 1.
NF-Y binding to CCAAT promoters in vivo. Twenty-five randomly picked CCAAT promoters from chromosome 21 were analyzed by ChIP with anti-NF-YB antibody on chromatin derived from seven different cell lines. Enrichment over that for an irrelevant Flag control antibody was assessed by semiquantitative PCR and classified as high (>30-fold), medium (5- to 30-fold), or low (2- to 5-fold). Colored boxes indicate positivity and gray boxes indicate no enrichment in ChIPs. The bottom line refers to percentages of NF-Y-positive sites for each cell line. The expression patterns of the respective genes were analyzed according to available transcriptome data (UniGene build no. 186) and reported on the right. UB (ubiquitous) refers to genes expressed in at least 10 out of 45 body sites of UniGene's EST Profile Viewer, while TS (tissue specific) refers to genes showing expression in less than 10 body sites.
NF-Y ChIP on chip on tiling arrays.
To define in an unbiased way the “landscape” of NF-Y binding in vivo, we performed NF-YB ChIP-on-chip analysis in HeLa cells with a tiling array containing all nonrepetitive sequences of the entire chromosome 21 and large parts of chromosomes 20 and 22. Fifty-mer oligonucleotides are tiled every 50 bp, giving a high degree of resolution and an overall representation of 2.2% of the human genome. To obtain maximal definition of the locations, ChIPs were performed using chromatin shorter than 500 bp, with a mean size of 250 to 300 bp. The enrichments present in the starting ChIPs were assayed against a Flag control antibody and were routinely >100-fold when checked on bona fide NF-Y target genes (see File S2 in the supplemental material). Four NF-YB probes derived from independent ChIPs were Cy5 labeled and hybridized together with the corresponding Cy3 input DNA, used as an internal control. After normalization of single channels, ratios between NF-YB and input probes were calculated and converted to Z scores (see Materials and Methods for details). An NF-Y peak was defined as a set of at least three consecutive probes whose Z scores deviated significantly from the average of normalized data. Typically, selected ratios were above 1.8 to 2.3 depending on the replicate. Only peaks present in at least two out of four replicates were collected for further analysis. Overall, the number of NF-Y binding sites ranged from 757 to 2,120, depending on the stringency applied (P values between 1.6−5 and 1.4−4). The ChIPs performed for Fig. 1 served as a guide for data analysis: none of the NF-Y-negative promoters was scored, while a majority of the positives were retrieved (Table 1), and, with the fourth stringency considered, 14/16 positives were recovered (see File S3 in the supplemental material).
TABLE 1.
NF-Y location analysis: binding sites and TUs
| Stringency (P) | Binding sites
|
TUs (n = 907a)
|
Rate of recovery of prevalidated promoters from HeLa cells (from Fig. 1)
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| Total no. | % (no.) for:
|
% (no.) for:
|
Allb | NF-Y+ | NF-Y− | ||||
| PR | GE | EL | PR | GE | |||||
| 1 (1.06E−05) | 757 | 11.1 (84) | 45.4 (344) | 43.5 (329) | 9.8 (89) | 26.1 (237) | 298 | 9/16 | 0/9 |
| 2 (2.88E−05) | 1,176 | 9.7 (114) | 46.9 (551) | 43.5 (511) | 13.6 (123) | 34.8 (316) | 386 | 11/16 | 0/9 |
| 3 (6.79E−05) | 1,533 | 8.9 (137) | 47.4 (727) | 43.6 (699) | 16.0 (145) | 40.4 (366) | 446 | 11/16 | 0/9 |
| 4 (1.43E−04) | 2,120 | 9.0 (191) | 48.1 (1,020) | 42.9 (909) | 21.6 (196) | 47.4 (430) | 519 | 14/16 | 0/9 |
From RefSeq.
Nonredundant TUs.
Binding sites were then classified and divided into three main categories: (i) “promoters” (PR), describing NF-Y locations residing from −2 kb to +0.5 kb relative to the TSS of a GenBank RefSeq sequence and representing 9 to 11% of the total depending on the stringency; (ii) “genes” (GE), indicating NF-Y locations residing within RefSeq-annotated genes; and (iii) “elsewhere” (EL), referring to locations external to promoters and intergenic regions. The last two categories each accounted for 42 to 48% of the sites.
Independent ChIP validations were performed on 35 loci derived from the less-stringent criteria: all NF-Y promoter loci tested scored positive in standard ChIPs and so did 11/15 sites among the “genes” cohort (see File S3 in the supplemental material). Thus, based on the prevalidation and validation ChIPs, the highest stringency reported in Table 1 highly underscored the extent of NF-Y binding and the 1.43−4 stringency reflected more closely the actual targets, with 10 to 15% false positives/negatives. Moreover, since the co-occurrence of nearby positive probes is expected to be enriched in location analysis, we monitored our peak finding procedure by randomizing each experimental track; as expected, this procedure dramatically reduced the number of loci identified (see File 4 in the supplemental material), confirming the robustness of our approach. Therefore, we pursued further analysis mainly focusing on this stringency.
We next analyzed NF-Y sites in terms of transcriptional units (TUs), defined as University of California, Santa Cruz, RefSeq-annotated genes (hg17 assembly) with their respective promoters; overall, there are 907 TUs (with no redundant promoter) within the regions considered here. As expected, by comparing the number of NF-Y promoter locations with that of the corresponding TUs (191 versus 196), we could recover, on average, one location per positive promoter. A different picture emerged for NF-Y GE category sites, since the number of NF-Y+ TUs was significantly lower than the overall number of locations: 430 versus 1,020. Considering that some of these locations referred to overlapping or divergent units, this implies that on average two or three NF-Y binding sites were found per positive TU.
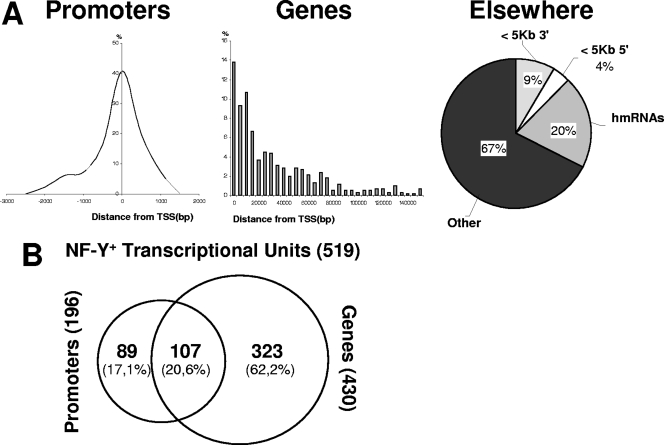
We also analyzed in greater detail the distribution of NF-Y sites (Fig. 2A). Within PR, most sites reside in the core area, as expected. Within GE, the site distribution shows a relatively steep decline as a function of the distance from the TSS, in agreement with a relevant role for NF-Y at the 5′ ends of genes. Interestingly, we found that more than 50% of NF-Y+ promoters possess an additional NF-Y site within the body of the gene (Fig. 2B). Within the EL category, one-third of the locations overlap (20%) or are nearby (9% + 4%) GenBank-annotated mRNAs with no RefSeq definition, which are most likely sites of Pol II activity (Fig. 2A).
FIG. 2.
Categories and locations of NF-Y binding sites. (A) NF-Y promoter sites were plotted according to their positions with respect to the TSS. The frequencies of GE sites versus their distance from the TSS are also represented. EL sites were classified as present within 5 kb to the 5′ or 3′ end of an annotated gene, as present within a GenBank-annotated human mRNA not corresponding to a RefSeq (hmRNAs), or far from annotated genes (other). (B) Venn diagrams showing the overlap between NF-Y TUs containing a site in the promoter and those containing a site within the body of the gene.
Presence of CCAAT boxes in NF-Y locations.
It was of interest to assess whether NF-Y locations contain CCAAT boxes. For this analysis, we considered a short interval of 500 bp centered on the peak, which is most likely a stringent but reliable window considering the length of the original chromatin (see File S2 in the supplemental material). The pentanucleotide was found in 61.6% of total locations, with a higher percentage in PR and lower in GE (Table 2). We also calculated the overall number of CCAAT boxes per location. Based on purely statistical considerations, the expected frequency is around 1 CCAAT box/location; a specific enrichment was clearly evident in PR, since 2 to 2.5 CCAAT boxes per promoter were present (Fig. 3). For the GE and EL locations, a lower enrichment of 1.5 to 1.7 CCAAT boxes per location was scored. Hence, we conclude that CCAAT boxes are present and overrepresented in most NF-Y locations.
TABLE 2.
NF-Y sites containing at least one CCAAT box
| Category | % (no.) of sites containing a CCAAT box (±150 bp) at stringency:
|
|||
|---|---|---|---|---|
| 1 (P = 1.06E−05) | 2 (P = 2.88E−05) | 3 (P = 6.79E−05) | 4 (P = 1.43E−04) | |
| Total | 63.3% (479) | 62.8% (739) | 61.8% (947) | 61.6% (1306) |
| PR | 70.2% (59) | 71.1% (81) | 66.4% (91) | 63.4% (121) |
| GE | 59.6% (205) | 59.3% (327) | 58.5% (425) | 58.6% (598) |
| EL | 66.3% (218) | 64.8% (331) | 64.4% (431) | 64.6% (587) |
FIG. 3.
CCAAT boxes in NF-Y locations. Average numbers of CCAAT boxes present in the different NF-Y location categories were plotted for the four stringencies initially considered. This number was calculated based on the actual mean dimensions of the identified NF-Y binding sites, ranging from 460 to 560 bp depending on the stringency, divided by the theoretical occurrence of the pentanucleotide CCAAT (once per 512 bp).
Correlation between NF-Y, H3K9-14ac, and H3K4me3.
Certain histone modifications are hallmarks of open chromatin clusters, at least in differentiated cells (4, 44). To characterize NF-Y sites in terms of the chromatin environment, we analyzed H3K9-14 acetylations and H3K4 trimethylation on the same tiling platform (Table 3; see File S5 in the supplemental material). A clear proof of the robustness of this approach was gained by comparison of our data with those of Bernstein et al. (6), obtained with HepG2 cells, since an overlap >80% was scored for most features analyzed (for details see File S5A to G in the supplemental material). Briefly, our analysis was consistent with previous results, showing a strong but not absolute correlation (i) between H3K4me3 and H3K9-14ac and (ii) between these marks and gene expression. Moreover, the positions of H3K9-14ac and H3K4me3 sites with respect to TSS were clearly skewed toward the 5′ ends of the genes (see File S6 in the supplemental material) and this was particularly evident for H3K4me3. In the case of H3K4me3-positive promoters, comparison with the data set of Guenther et al., obtained from human ES cells (23), was also reassuring: on average, an overlap of 80% between the two cell lines was scored (see File S5I to K in the supplemental material). Validation of these marks in independent ChIPs confirmed that the vast majority of sites are indeed true positives (see File S3 in the supplemental material). Hence, we felt comfortable in analyzing the NF-Y loci together with H3K4me3 and H3K9-14ac sites derived from our ChIP-on-chip analysis.
TABLE 3.
Active histone mark location analysis
| Stringency (P) | H3K9-14ac
|
H3K4me3
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Peaks
|
TUs (n = 907), % (no.) for:
|
Peaks
|
TUs (n = 907), % (no.) for:
|
|||||||||
| Total | % (no.) for:
|
Total | % (no.) for:
|
|||||||||
| PR | GE | EL | Promoters | Genes | PR | GE | EL | Promoters | Genes | |||
| 1 (1.06E−05) | 1,116 | 29.6% (330) | 45.3% (505) | 25.2% (281) | 33.2% (301) | 37.0% (336) | 348 | 71.3% (248) | 14.9% (52) | 13.8% (48) | 30.2% (274) | 6.9% (63) |
| 2 (2.88E−05) | 1,525 | 25.0% (381) | 47.1% (719) | 27.9% (425) | 38.8% (352) | 44.4% (403) | 458 | 65.7% (301) | 18.1% (83) | 16.2% (74) | 35.7% (324) | 9.5% (86) |
| 3 (6.79E−05) | 2,386 | 18.7% (445) | 50.2% (1,198) | 31.1% (743) | 45.2% (410) | 54.9% (498) | 823 | 44.1% (363) | 25.6% (211) | 30.3% (249) | 42.4% (385) | 23.3% (211) |
| 4 (1.43E−04) | 3,148 | 16.6% (521) | 48.2% (1,517) | 35.3% (1,110) | 51.6% (468) | 62.2% (564) | 1,012 | 38.0% (385) | 29.6% (300) | 32.3% (327) | 44.8% (406) | 26.8% (243) |
By combining the three different ChIP-on-chip data sets, we observed that the majority of NF-Y+ promoters were also H3K9-14ac+ or H3K4me3+ (68% and 61%, respectively [Fig. 4A, left]); a good proportion, 57%, contained both marks. Correlative P values were highly significant. Within NF-Y+ genes, the percentage of H3K9-14ac+ GE sites was higher, at 85%, while the percentage of H3K4me3+ GE sites was lower, at 42%, and, consequently, the percentage of NF-Y+ genes double positive for histone marks was also around 40% (Fig. 4A, right). However, the correlation between NF-Y and active histone marks within this category was stronger, with a P value of E−27 for NF-Y+ H3K9-14ac+ H3K4me3+ genes, indicating that essentially all of NF-Y+/H3K4me3+ sites were also acetylated. Somewhat surprisingly, we noticed that a sizeable set of NF-Y+ TUs were neither acetylated nor trimethylated, both within promoters (27%) and within the bodies of the genes (18%). This cluster was unexpected, at least in these proportions, and was further analyzed below.
FIG. 4.
Correlation between NF-Y binding and active histone marks. (A) Percentages of NF-Y+ TUs that scored positive for either H3K9-K14ac or H3K4me3 or for both histone marks. (B) Percentages of H3K9-K14ac- or H3K4me3-positive TUs and of H3K9-K14ac/H3K4me3 double positives that shared at least one NF-Y location. (C) Position of NF-Y binding sites and histone modification islands within a representative cluster of five genes.
When we reversed the analysis, asking how many H3K9-14ac+ and/or H3K4me3+ TUs were bound by NF-Y, we found that 28% of H3K9-14ac+ and 30% of H3K4me3+ promoters were also NF-Y+ (Fig. 4B, left); this is quite significant, considering the promoters that are not active in HeLa cells (see below) and considering that the overall percentage of NF-Y+ promoters was 21% (Table 1). Within the GE category the percentage of H3K9-14ac+ TUs positive for NF-Y was 64% and the percentages of double positives were as high as 79% (Fig. 4B, right). A representative cluster of five genes is shown in Fig. 4C, with two divergent promoters positive for NF-Y binding and for active histone marks, and one, DONSON, devoid of the former. Altogether, these data indicate a significant correlation between NF-Y binding and these histone modifications, either within promoters or within the bodies of the genes, and highlight a smaller population of NF-Y-positive locations that are neither H3K9-14 acetylated nor H3K4 trimethylated.
Correlation between NF-Y, H3K9-14ac, H3K4me3, and gene expression.
Next, we interrogated the list of NF-Y+ TUs with respect to both histone marks and expression using available HeLa profiling data sets obtained on the Affymetrix U133 2.0 Plus platform. Overall, 59% of the 907 TUs analyzed here were in the P set and 39% in the A set. As expected, the majority, 75%, of NF-Y+ promoters followed P calls (Fig. 5A). NF-Y+ genes showed modest skewing toward expression, with P calls at 67% (Fig. 5B). We then integrated the active histone mark data into this analysis. Within promoters, the NF-Y+ H3K9-14ac+ and NF-Y+/H3K4me3+ clusters showed clear skewing toward P calls, and even more did the triple-positive NF-Y+/H3K9-14ac+/H3K4me3+ clusters; only a residual 10% of TUs were scored as A in the latter class (Fig. 5A). On the other hand, the NF-Y+/H3K9-14ac−/H3K4me3− promoters were strongly enriched in A calls (Fig. 5A), indicating (i) that NF-Y is bound at these promoters in the absence of active histone marks and (ii) that this cohort of genes is not expressed. A somewhat similar situation emerged for NF-Y+ genes: enrichment of NF-Y+/H3K9-14ac+/H3K4me3+ clusters toward expression was less evident, but once again the majority of NF-Y+/H3K9-14ac−/H3K4me3− genes were found in A calls (Fig. 5B). To gain insights into this dual behavior, NF-Y-positive TUs were subjected to functional enrichment analysis after dividing them into those containing at least one active histone mark and those devoid of both modifications. Pathways expected to be positively affected by NF-Y (cell cycle, cell signaling) were present in the NF-Y+/H3K9-14ac+/H3K4me3+ cohort of genes, but no single function was overwhelmingly enriched; this is consistent with the notion that NF-Y is a broad transcriptional regulator, with specific roles in certain cellular functions (see File S7A in the supplemental material). A different picture emerged with NF-Y+ TUs devoid of active marks, since functions expected to be repressed in HeLa cells, sensory perception and immune response, were enriched (see File S7B in the supplemental material). A graphic representation of an NF-Y+/H3K9-14ac−/H3K4me3− locus, SUHW1, is shown in Fig. 5C, and the complete list of NF-Y+/H3K9-14ac−/H3K4me3− TUs is reported in File S8 in the supplemental material. Altogether, these data indicate that NF-Y binding is associated with two different chromatin states and opposite functional outcomes: in the presence of the two active histone marks, NF-Y is bound to expressed loci; binding within areas devoid of these modifications is coupled to inactive genes.
FIG. 5.
Correlation between NF-Y binding, active histone marks, and gene expression. (A) NF-Y+ promoters were scored for the presence (P) or absence (A) in the GSE6022 data set after the intersection with histone mark experiments. (B) Same as panel A, except that NF-Y+ genes were analyzed. (C) An NF-Y locus within the promoter of a representative nonexpressed gene, devoid of active histone marks, is shown.
NF-Y associates with chromatin with negative histone marks.
We further investigated the cluster of nonexpressed NF-Y+/H3K9-14ac−/H3K4me3− TUs. To assay the possibility that negative histone marks were present in these areas, we performed ChIP experiments with antibodies directed against H3K9me3, H3K27me3, and H4K20me3, which have been associated, in various terms, with inactive or partially inactive chromatin environments (8, 19). As shown in Fig. 6, with one exception, these loci were confirmed to be NF-Y positive, most of them with a low or medium enrichment level compared to genes targeted by NF-Y and transcribed, such as the SON gene (Fig. 6, top).
FIG. 6.
ChIP with negative histone marks in nonexpressed NF-Y+ loci. Twelve NF-Y+/H3K9-K14ac−/H3K4me3− sites, randomly selected from the list of targets not expressed in HeLa cells (see File S7 in the supplemental material) were analyzed by ChIP with anti-NF-YB, anti-H3K27me3, anti-H3K9me3, and anti-H4K20me3 antibodies. Enrichment over that for an irrelevant Flag control antibody was assessed by semiquantitative PCR in duplicate experiments; enrichments greater than twofold were considered significant. The chromosome 11 satellite centromeric region and the promoter of the transcribed NF-Y target SON were used as internal controls.
Interestingly, all 12 tested sites scored positive for H4K20me3, as did 7 out of 12 for H3K27me3; among these SUHW1, encoding the human homologue of Drosophila Suppressor of Hairy-wing insulator binding protein, and the vitamin D receptor-activated CYP24A1; only three sites, APOBEC3, FTCD, and C21orf81, were H3K9me3+. Reassuringly, with just one exception, ChIPs confirmed negativity for the active histone mark H3K4me3 (not shown). These results are consistent with the idea that NF-Y can be associated in vivo with areas of the genome containing negative histone marks.
NF-Y is an activator as well as a repressor.
The binding of a TF to a target DNA sequence does not necessarily imply direct transcriptional regulation. To assay the role of NF-Y binding in gene expression, we infected HeLa and HCT116 cells with adenoviral vectors coding for an NF-YA dominant negative protein (Ad-YA-m29) containing a mutation in the DNA binding domain; the mutant still associates with the histone fold NF-YB/NF-YC dimer but renders the trimer incapable of CCAAT association (see reference 27 and references therein). In parallel, we infected cells with wild-type Ad-NF-YA and Ad-GFP viruses. Under these conditions, expression of CCAAT-containing, NF-Y-dependent promoters was previously shown to be crippled (51). Figure 7 shows RT-PCR analysis of expressed NF-Y targets associated with positive histone marks (top); dominant negative YA treatment led to decrease in expression of the genes analyzed, while little effects were observed in the control wild-type Ad-GFP or Ad-NF-YA infections. The mRNA levels of control, NF-Y-independent genes such as GAPDH were unaffected. Similar results were obtained with the HCT116 cells (Fig. 7, top right). In parallel, we also analyzed NF-Y loci not expressed and associated with negative histone marks; some of these genes, notably CYP24A1 and SERPIND, were clearly and specifically up-regulated by the YA-m29 treatment, both in HeLa and in HCT116 cells. Note that the former had high levels of H3K27me3 and low H4K20me3; SERPIND showed the opposite pattern. SUHW1, which was low on both these marks, was very modestly induced by YA-m29. Interestingly, all these genes contain prototypical CCAAT boxes in their promoters. We conclude that the presence of an active NF-Y trimer is required both for the expression of active genes and for repression of some of the NF-Y targets associated with negative histone marks.
FIG. 7.
Effects of NF-Y removal on gene expression. HeLa cells (left) were infected in parallel with control Ad-GFP, wild-type Ad-NF-YA, and the dominant negative Ad-YAm29 adenovirus. RT-PCR analyses of infected cells were performed in the linear range of amplification for the indicated genes. HCT116 cells (right) were analyzed under the same conditions. Western blot analysis confirmed equivalent overexpression of wild-type NF-YA and of YAm29 proteins (not shown).
DISCUSSION
In this report, we correlate the in vivo binding of NF-Y with the presence of H3-K9-14 acetylations and H3-K4 trimethylation using high-density arrays covering chromosome 21 and large parts of chromosomes 20 and 22. We came to the following relevant conclusions. A majority of genes analyzed here are targeted by NF-Y, either in the promoter or in the 5′ end region of the gene. Functionally, two types of NF-Y loci exist: one has active histone marks and correlates with transcription, and the other is found in sites that are transcriptionally silent and loaded with negative histone marks. NF-Y serves as an activator of the former and as a repressor of the latter.
NF-Y location analysis.
Several bioinformatic studies have found CCAAT boxes in >60% of human promoters (19, 35, 48, 49); specific pathways, such as those involving cell cycle- and endoplasmic reticulum stress-regulated genes, were found to be quite enriched for CCAAT-containing genes (16, 50, 57). Interestingly, analysis of microRNA control sequences identifies NF-Y as one of the few TFs involved in the regulation of all of them (29).
The first issue tackled here is how frequent are NF-Y binding sites. The answer to this depends, to some extent, upon the stringency of the analysis. Monitoring 25 promoters (Fig. 1) and validations of additional loci helped us fine-tune our analysis, so that we “lose” only 10 to 15% of the positive locations, keeping the false-positive sites at the same level. The two extremes range from 757 to 2,120 sites; the former is an absolute minimum that considers only very-high-affinity sites, the latter is a wider look at low-affinity sites or at sites present only in a subcategory of cells, as ChIPs were made from a heterogeneous population. Extrapolation of these numbers to the whole genome brings NF-Y sites to 35,000 to 80,000, which is high compared to extrapolations performed for other TFs such as E2F1 (20,000 to 30,000 binding sites [9]). However, it should be noted that there are at least 200,000 NF-YB molecules within the nuclei of various cell lines (R. Mantovani, unpublished data). These data are consistent with the idea that NF-Y is involved in a wide range of RNA production procedures, both positive and negative; indeed, the TUs that contained at least one NF-Y site are a majority, 519 out of 907.
The second issue tackled by this work concerns the presence of the CCAAT consensus in the identified NF-Y sites. Previous location analysis experiments performed with similar platforms reported a low rate of consensus sites near locations of TFs (9, 11, 17), while others suggested significant variation from the in vitro-derived consensus (56). It was found, using stringent criteria, that >60% of NF-Y binding sites contain a CCAAT consensus, particularly promoter sites. Furthermore, 1.5 to 2.5 CCAAT boxes per locus are found; thus, the presence of one or more CCAAT boxes is in general important for NF-Y binding, in agreement with biochemical in vitro work (15). Note that the total number of CCAAT-containing promoters in the cluster analyzed here is 463 out of 907 (51%), which is lower than the 60 to 67% derived from other studies (48). Nonetheless, around 35% of the identified NF-Y loci are apparently devoid of CCAAT boxes. There are explanations for this finding. (i) Variation of a single nucleotide in the CCAAT pentanucleotide was reported to be compatible with NF-Y binding, especially when other functionally important overlapping sites are involved (20, 37). A degenerate CCAAT box would be missed in our stringent analysis. (ii) The ChIP-on-chip experiments were performed with an anti-NF-YB antibody; since NF-YB and NF-YC subunits are in excess with respect to NF-YA (Mantovani, unpublished), some CCAAT-less sites devoid of the sequence-specific NF-YA might have emerged.
The last point raised by our analysis is related to the genomic location of the identified NF-Y binding sites. The CCAAT box has long been considered almost exclusively as a promoter element crucial for Pol II recruitment (24). In keeping with expectations (33), the vast majority of NF-Y PR sites, 70%, are positioned near the TSS, and there is clear enrichment of sites near the TSS also within the GE cohort (Fig. 2). However, NF-Y locations appear to be more scattered, and roughly 50% of NF-Y-positive promoters contain an extra site within the body of the gene (Fig. 2). It is possible that their role in many such cases is related to promoter-enhancer connections, while other GE locations could be alternative promoters of the same gene or sites of divergent RNA production. Moreover, within the EL cohort, a large number of sites are located in mRNA-expressing areas, sites of Pol II activity. This accounts for 20% of the 909 EL peaks (Table 1 and Fig. 2A). If we apply to these Pol II/NF-Y+ units the same ratio of 2 sites/unit, as measured in bona fide NF-Y RefSeq GE sites, at least 100 additional Pol II units would be added. Extrapolation to the whole genome gives 4,500 NF-Y+ units outside of the annotated RefSeq genes. The remaining EL peaks could either be enhancer sequences located at a distance from transcribed regions or TUs whose RNAs have not been annotated yet.
NF-Y, active histone marks, and transcription.
It is clear that H3K4me3 acetylation and H3K9-14 acetylation are hallmarks of active genes (6, 40, 43). Our data for HeLa cells confirm previous results for HepG2, obtained using similar criteria of analysis (6). First of all we find an excellent correlation between active mark islands and gene expression in HeLa cells (see File S5 in the supplemental material). Second, as the overlap between expressed and not expressed genes in HeLa and HepG2 cells is quite high, 85%, we found a remarkable degree of coincidence between histone mark sites in the two cell lines, with deviations only for H3K4me3 within genes, an effect due to cell type-specific patterns and/or to our underestimation of such a category (see File S3 in the supplemental material). A similar picture emerged also by comparison with the data set of Guenther et al. (23), obtained with human ES cells (see File S5I to K in the supplemental material). As expected, skewing toward expression was less evident for H3K4me3-positive promoters in ES cells.
NF-Y binding has been so far mostly associated with two possible transcriptional states: an actively transcribing gene and a poised promoter, with a prebound NF-Y ready to help recruit whichever TF is specifically responsible for full activation, following a stimulus. Indeed, the majority of NF-Y-bound units contain H3K9-14ac and H3K4me3 sites and are expressed. Note that active genes are expected to be readily discernible in Affymetrix profiling data, while the levels of the inducible ones might not, thus underscoring this already significant correlation.
It has long been known that the binding of TFs and cofactors to promoters is a hallmark of expression, by signaling to the Pol II transcription machinery the positional coordinates (52). It would seem logical, therefore, to postulate that positive histone marks are positioned according to a code determined by the sequence-specific TFs. Reassuringly, several studies have confirmed that TF binding indeed correlates with the presence of “active” marks and with gene expression. In a thorough study performed with quantitative PCR on the correlation between >30 histone modifications and MYC sites, Guccione et al. (22) concluded that the presence of H3K4me2/3, H3K9-14ac, and H3K79me2 is a prerequisite for MYC binding. Interestingly, elimination of MYC had little effect on the levels of these modifications, unlike what was found for H4 acetylations, which were decreased (22). Clearly, this is not the case for NF-Y, since our unbiased analysis identified NF-Y regions devoid of these active histone marks.
The most surprising result is the identification of the discrete cohort of NF-Y+ TUs in which H3K9-14ac and H3K4me3 islands are absent and specifically enriched in A calls (almost 80%); this indicates that NF-Y is not always associated with active/inducible transcription. This cluster comprises mostly tissue-specific and developmentally regulated genes (see File S7 in the supplemental material for a complete list) and is associated with repressive histone marks such as H4K20me3 and H3K27me3. While hints at a negative role for NF-Y in transcription had previously been reported (5, 27, 32, 53), we are intrigued by the extent of this phenomenon, which involves 20 to 25% of the total binding sites. Significantly, we find that removal of the NF-Y trimer leads to activation of these loci. Among the derepressed, CYP24A1, but not SERPIND, is H3K27me3+. It is known that this modification is associated with Polycomb (47); hints to a possible mechanism of repression through deposition of this negative mark come from recent experiments with Caenorhabditis elegans, in which ceNF-Y function has been genetically linked to Polycomb through direct interaction with the ESC/E(Z) component (13).
The structural resemblance of NF-YB-NF-YC to H2A-H2B should be remembered when considering the bifunctional behavior of NF-Y. The first sign of an opening chromatin cluster, and one specifically required for H3K4me3, is monoubiquitination of H2B on K123 (28); we noted that lysines are present in the corresponding region of the H2B-like NF-YB, and indeed NF-YB is monoubiquitinated (G. Donati and R. Mantovani, data not shown). On the other hand, H2A is monoubiquitinated by Polycomb components, and the functional significance of this modification is opposite to that of H2B-ubiquitin, leading to repression of transcription (55). Thus, H2A-ubiquitin could serve as a signal to recruit H3K27 methyltransferase. Given the dual dominant NF-Y function uncovered here, we are tempted to speculate that a code of posttranslational modifications may exist for the histone-like NF-Y.
The bivalent behavior of NF-Y and its independence from a specific pattern of activating versus repressive histone modifications might have important consequences for transcriptional regulation at bivalent loci that have been recently mapped in both ES and lineage-committed cells. At these sites, in fact, the activating H3K4me3 and the repressive H3K27me3 are present together on areas of the genome “poised” for alternative developmental fates (1, 7, 36). Interestingly, the role of NF-Y in stem cells has been highlighted by two recent studies: (i) NF-YA was shown to be required for maintenance of hematopoietic stem cells (58) and (ii) the CCAAT box was found to be specifically enriched in conserved regions of genes highly expressed in mouse and human ES cells, with NF-Y being required both for regulation of these elements and for cell survival (21). Moreover, a switch in the two major isoforms of NF-YA was noticed upon differentiation, with the “short” form being highest in stem cells and decreasing in embryonic bodies. Note that this isoform is specifically required for stemness in the hematopoietic system (58). These results, considered together with the widespread bifunctional role of NF-Y and its independence from specific methyl marks shown here in committed HeLa cells, suggest that NF-Y might be associated with bivalent sites in ES cells, possibly even regulating their positioning. Because of its structure and histone interactions (10), NF-Y would be ideal to maintain nucleosome-free areas, accommodate accessibility of nearby TFs, and recruit modifying complexes, positive or negative. This hypothesis and the cause-and-effect relationships between the positioning of histone marks and NF-Y binding will now be investigated with appropriate genetic experiments.
Supplementary Material
Acknowledgments
A.M.V. was supported by a FIRB Giovani Ricercatori contract. R.M. was supported by grants from PRIN-Miur, FIRB, CARIPLO, and AIRC.
Footnotes
Published ahead of print on 22 January 2008.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Azuara, V., P. Perry, S. Sauer, M. Spivakov, H. F. Jorgensen, R. M. John, M. Gouti, M. Casanova, G. Warnes, M. Merkenschlager, and A. G. Fisher. 2006. Chromatin signatures of pluripotent cell lines. Nat. Cell Biol. 8532-538. [DOI] [PubMed] [Google Scholar]
- 2.Barski, A., S. Cuddapah, K. Cui, T. Y. Roh, D. E. Schones, Z. Wang, G. Wei, I. Chepelev, and K. Zhao. 2007. High-resolution profiling of histone methylations in the human genome. Cell 129823-837. [DOI] [PubMed] [Google Scholar]
- 3.Baumeister, P., S. Luo, W. C. Skarnes, G. Sui, E. Seto, Y. Shi, and A. S. Lee. 2005. Endoplasmic reticulum stress induction of the Grp78/BiP promoter: activating mechanisms mediated by YY1 and its interactive chromatin modifiers. Mol. Cell. Biol. 254529-4540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berger, S. L. 2007. The complex language of chromatin regulation during transcription. Nature 447407-412. [DOI] [PubMed] [Google Scholar]
- 5.Bernadt, C. T., T. Nowling, M. S. Wiebe, and A. Rizzino. 2005. NF-Y behaves as a bifunctional transcription factor that can stimulate or repress the FGF-4 promoter in an enhancer-dependent manner. Gene Expr. 12193-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bernstein, B. E., M. Kamal, K. Lindblad-Toh, S. Bekiranov, D. K. Bailey, D. J. Huebert, S. McMahon, E. K. Karlsson, E. J. Kulbokas III, T. R. Gingeras, S. L. Schreiber, and E. S. Lander. 2005. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell 120169-181. [DOI] [PubMed] [Google Scholar]
- 7.Bernstein, B. E., T. S. Mikkelsen, X. Xie, M. Kamal, D. J. Huebert, J. Cuff, B. Fry, A. Meissner, M. Wernig, K. Plath, R. Jaenisch, A. Wagschal, R. Feil, S. L. Schreiber, and E. S. Lander. 2006. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125315-326. [DOI] [PubMed] [Google Scholar]
- 8.Bernstein, B. E., A. Meissner, and E. S. Lander. 2007. The mammalian epigenome. Cell 128669-681. [DOI] [PubMed] [Google Scholar]
- 9.Bieda, M., X. Xu, M. Singer, R. Green, and P. J. Farnham. 2006. Unbiased location analysis of E2F1 binding sites suggests a widespread role for E2F1 in the human genome. Genome Res. 16595-605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Caretti, G., M. C. Motta, and R. Mantovani. 1999. NF-Y associates H3-H4 tetramers and octamers by multiple mechanisms. Mol. Cell. Biol. 198591-8603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carroll, J. S., X. S. Liu, A. S. Brodsky, W. Li, C. A. Meyer, A. J. Szary, J. Eeckhoute, W. Shao, E. V. Hestermann, T. R. Geistlinger, E. A. Fox, P. A. Silver, and M. Brown. 2005. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell 12233-43. [DOI] [PubMed] [Google Scholar]
- 12.Ceribelli, M., M. Alcalay, M. A. Vigano, and R. Mantovani. 2006. Repression of new p53 targets revealed by ChIP on chip experiments. Cell Cycle 51102-1110. [DOI] [PubMed] [Google Scholar]
- 13.Deng, H., Y. Sun, Y. Zhang, X. Luo, W. Hou, L. Yan, Y. Chen, E. Tian, J. Han, and H. Zhang. 2007. Transcription factor NFY globally represses the expression of the C. elegans Hox gene Abdominal-B homolog egl-5. Dev. Biol. 308583-592. [DOI] [PubMed] [Google Scholar]
- 14.Donati, G., C. Imbriano, and R. Mantovani. 2006. Dynamic recruitment of transcription factors and epigenetic changes on the ER stress response gene promoters. Nucleic Acids Res. 343116-3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dorn, A., J. Bollekens, A. Staub, C. Benoist, and D. Mathis. 1987. A multiplicity of CCAAT binding proteins. Cell 50863-872. [DOI] [PubMed] [Google Scholar]
- 16.Elkon, R., C. Linhart, R. Sharan, R. Shamir, and Y. Shiloh. 2003. Genome-wide in silico identification of transcriptional regulators controlling the cell cycle in human cells. Genome Res. 13773-780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Euskirchen, G., T. E. Royce, P. Bertone, R. Martone, J. L. Rinn, F. K. Nelson, F. Sayward, N. M. Luscombe, P. Miller, M. Gerstein, S. Weissman, and M. Snyder. 2004. CREB binds to multiple loci on human chromosome 22. Mol. Cell. Biol. 243804-3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fang, J., Q. Feng, C. S. Ketel, H. Wang, R. Cao, L. Xia, H. Erdjument-Bromage, P. Tempst, J. A. Simon, and Y. Zhang. 2002. Purification and functional characterization of SET8, a nucleosomal histone H4-lysine 20-specific methyltransferase. Curr. Biol. 121086-1099. [DOI] [PubMed] [Google Scholar]
- 19.FitzGerald, P. C., A. Shlyakhtenko, A. A. Mir, and C. Vinson. 2004. Clustering of DNA sequences in human promoters. Genome Res. 141562-1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gilthorpe, J., M. Vandromme, T. Brend, A. Gutman, D. Summerbell, N. Totty, and P. W. Rigby. 2002. Spatially specific expression of Hoxb4 is dependent on the ubiquitous transcription factor NFY. Development 1293887-3899. [DOI] [PubMed] [Google Scholar]
- 21.Grskovic, M., C. Chaivorapol, A. Gaspar-Maia, H. Li, and M. Ramalho-Santos. 2007. Systematic identification of cis-regulatory sequences active in mouse and human embryonic stem cells. PLoS Genet. 3e145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guccione, E., F. Martinato, G. Finocchiaro, L. Luzi, L. Tizzoni, V. Dall' Olio, G. Zardo, C. Nervi, L. Bernard, and B. Amati. 2006. Myc-binding-site recognition in the human genome is determined by chromatin context. Nat. Cell Biol. 8764-770. [DOI] [PubMed] [Google Scholar]
- 23.Guenther, M. G., S. S. Levine, L. A. Boyer, R. Jaenisch, and R. A. Young. 2007. A chromatin landmark and transcription initiation at most promoters in human cells. Cell 13077-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kabe, Y., J. Yamada, H. Uga, Y. Yamaguchi, T. Wada, and H. Handa. 2005. NF-Y is essential for the recruitment of RNA polymerase II and inducible transcription of several CCAAT box-containing genes. Mol. Cell. Biol. 25512-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim, T. H., L. O. Barrera, M. Zheng, C. Qu, M. A. Singer, T. A. Richmond, Y. Wu, R. D. Green, and B. Ren. 2005. A high-resolution map of active promoters in the human genome. Nature 436876-880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kurdistani, S. K., S. Tavazoie, and M. Grunstein. 2004. Mapping global histone acetylation patterns to gene expression. Cell 117721-733. [DOI] [PubMed] [Google Scholar]
- 27.Imbriano, C., A. Gurtner, F. Cocchiarella, S. Di Agostino, V. Basile, M. Gostissa, M. Dobbelstein, G. Del Sal, G. Piaggio, and R. Mantovani. 2005. Direct p53 transcriptional repression: in vivo analysis of CCAAT-containing G2/M promoters. Mol. Cell. Biol. 253737-3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laribee, R. N., S. M. Fuchs, and B. D. Strahl. 2007. H2B ubiquitylation in transcriptional control: a FACT-finding mission. Genes Dev. 21737-743. [DOI] [PubMed] [Google Scholar]
- 29.Lee, J., Z. Li, R. Brower-Sinning, and B. John. 2007. Regulatory circuit of human microRNA biogenesis. PLoS Comput. Biol. 3e67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liang, G., J. C. Lin, V. Wei, C. Yoo, J. C. Cheng, C. T. Nguyen, D. J. Weisenberger, G. Egger, D. Takai, F. A. Gonzales, and P. A. Jones. 2004. Distinct localization of histone H3 acetylation and H3-K4 methylation to the transcription start sites in the human genome. Proc. Natl. Acad. Sci. 1017357-7362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu, C. L., T. Kaplan, M. Kim, S. Buratowski, S. L. Schreiber, N. Friedman, and O. J. Rando. 2005. Single-nucleosome mapping of histone modifications in S. cerevisiae. PLoS Biol. 3e328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Manni, I., G. Mazzero, A. Gurtner, R. Mantovani, U. Haugwitz, K. Krause, K. Engeland, A. Sacchi, S. Soddu, and G. Piaggio. 2001. NF-Y mediates the transcriptional inhibition of the cyclin B1, cyclin B2, and Cdc25C promoters upon induced G2 arrest. J. Biol. Chem. 2765570-5576. [DOI] [PubMed] [Google Scholar]
- 33.Mantovani, R. 1998. A survey of 178 NF-Y binding CCAAT boxes. Nucleic Acids Res. 261135-1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mantovani, R. 1999. The molecular biology of the CCAAT-binding factor NF-Y. Gene 23915-27. [DOI] [PubMed] [Google Scholar]
- 35.Marino-Ramirez, L., J. L. Spouge, G. C. Kanga, and D. Landsman. 2004. Statistical analysis of over-represented words in human promoter sequences. Nucleic Acids Res. 32949-958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mikkelsen, T. S., M. Ku, D. B. Jaffe, B. Issac, E. Lieberman, G. Giannoukos, P. Alvarez, W. Brockman, T. K. Kim, R. P. Koche, W. Lee, E. Mendenhall, A. O'Donovan, A. Presser, C. Russ, X. Xie, A. Meissner, M. Wernig, R. Jaenisch, C. Nusbaum, E. S. Lander, and B. E. Bernstein. 2007. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448553-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Milos, P. M., and K. S. Zaret. 1992. A ubiquitous factor is required for C/EBP-related proteins to form stable transcription complexes on an albumin promoter segment in vitro. Genes Dev. 6991-1004. [DOI] [PubMed] [Google Scholar]
- 38.Nishioka, K., J. C. Rice, K. Sarma, H. Erdjument-Bromage, J. Werner, Y. Wang, S. Chuikov, P. Valenzuela, P. Tempst, R. Steward, J. T. Lis, C. D. Allis, and D. Reinberg. 2002. PR-Set7 is a nucleosome-specific methyltransferase that modifies lysine 20 of histone H4 and is associated with silent chromatin. Mol. Cell 91201-1213. [DOI] [PubMed] [Google Scholar]
- 39.Pokholok, D. K., C. T. Harbison, S. Levine, M. Cole, N. M. Hannett, T. I. Lee, G. W. Bell, K. Walker, P. A. Rolfe, E. Herbolsheimer, J. Zeitlinger, F. Lewitter, D. K. Gifford, and R. A. Young. 2005. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell 122517-527. [DOI] [PubMed] [Google Scholar]
- 40.Roh, T. Y., S. Cuddapah, and K. Zhao. 2005. Active chromatin domains are defined by acetylation islands revealed by genomewide mapping, Genes Dev. 19542-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roh, T. Y., S. Cuddapah, K. Cui, and K. Zhao. 2006. The genomic landscape of histone modifications in human T cells. Proc. Natl. Acad. Sci. USA 10315782-15787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Romier, C., F. Cocchiarella, R. Mantovani, and D. Moras. 2003. The crystal structure of the NF-YB/NF-YC heterodimer gives insight into transcription regulation and DNA binding and bending by transcription factor NF-Y. J. Biol. Chem. 2781336-1345. [DOI] [PubMed] [Google Scholar]
- 43.Ruthenburg, A. J., C. D. Allis, and J. Wysocka. 2007. Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. Mol. Cell 2515-30. [DOI] [PubMed] [Google Scholar]
- 44.Santos-Rosa, H., R. Schneider, A. J. Bannister, J. Sherriff, B. E. Bernstein, N. C. Emre, S. L. Schreiber, J. Mellor, and T. Kouzarides. 2002. Active genes are tri-methylated at K4 of histone H3. Nature 419407-411. [DOI] [PubMed] [Google Scholar]
- 45.Sato, N., I. M. Sanjuan, M. Heke, M. Uchida, F. Naef, and A. H. Brivanlou. 2003. Molecular signature of human embryonic stem cells and its comparison with the mouse. Dev. Biol. 260404-413. [DOI] [PubMed] [Google Scholar]
- 46.Schneider, R., A. J. Bannister, F. A. Myers, A. W. Thorne, C. Crane-Robinson, and T. Kouzarides. 2004. Histone H3 lysine 4 methylation patterns in higher eukaryotic genes. Nat. Cell Biol. 673-77. [DOI] [PubMed] [Google Scholar]
- 47.Squazzo, S. L., H. O'Geen, V. M. Komashko, S. R. Krig, V. X. Jin, S. W. Jang, R. Margueron, D. Reinberg, R. Green, and P. J. Farnham. 2006. Suz12 binds to silenced regions of the genome in a cell-type-specific manner. Genome Res. 16890-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Suzuki, Y., T. Tsunoda, J. Sese, H. Taira, J. Mizushima-Sugano, H. Hata, T. Ota, T. Isogai, T. Tanaka, Y. Nakamura, A. Suyama, Y. Sakaki, S. Morishita, K. Okubo, and S. Sugano. 2001. Identification and characterization of the potential promoter regions of 1031 kinds of human genes. Genome Res. 11677-684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Suzuki, Y., R. Yamashita, M. Shirota, Y. Sakakibara, J. Chiba, J. Mizushima-Sugano, A. E. Kel, T. Arakawa, P. Carninci, J. Kawai, Y. Hayashizaki, T. Takagi, K. Nakai, and S. Sugano. 2004. Large-scale collection and characterization of promoters of human and mouse genes. In Silico Biol. 4429-444. [PubMed] [Google Scholar]
- 50.Tabach, Y., M. Milyavsky, I. Shats, R. Brosh, O. Zuk, A. Yitzhaky, R. Mantovani, E. Domany, V. Rotter, and Y. Pilpel. 2005. The promoters of human cell cycle genes integrate signals from two tumor suppressive pathways during cellular transformation. Mol. Syst. Biol. 1E1-E15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Testa, A., G. Donati, P. Yan, F. Romani, T. H. Huang, M. A. Viganò, and R. Mantovani. 2005. ChIP on chip experiments uncover a widespread distribution of NF-Y binding CCAAT sites outside of core promoters. J. Biol. Chem. 28013606-13615. [DOI] [PubMed] [Google Scholar]
- 52.Thomas, M. C., and C. M. Chiang. 2006. The general transcription machinery and general cofactors. Crit. Rev. Biochem. Mol. Biol. 41105-178. [DOI] [PubMed] [Google Scholar]
- 53.Uramoto, H., D. Wetterskog, A. Hackzell, Y. Matsumoto, and K. Funa. 2004. p73 competes with co-activators and recruits histone deacetylase to NF-Y in the repression of PDGF beta-receptor. J. Cell Sci. 1175323-5331. [DOI] [PubMed] [Google Scholar]
- 54.Viganò, M. A., J. Lamartine, B. Testoni, D. Merico, D. Alotto, C. Castagnoli, A. Robert, E. Candi, G. Melino, X. Gidrol, and R. Mantovani. 2006. New p63 targets in keratinocytes identified by a genome-wide approach. EMBO J. 255105-5116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang, H., L. Wang, H. Erdjument-Bromage, M. Vidal, P. Tempst, R. S. Jones, and Y. Zhang. 2004. Role of histone H2A ubiquitination in Polycomb silencing. Nature 431873-878. [DOI] [PubMed] [Google Scholar]
- 56.Wei, C. L., Q. Wu, V. B. Vega, K. P. Chiu, P. Ng, T. Zhang, A. Shahab, H. C. Yong, Y. Fu, Z. Weng, J. Liu, X. D. Zhao, J. L. Chew, Y. L. Lee, V. A. Kuznetsov, W. K. Sung, L. D. Miller, B. Lim, E. T. Liu, Q. Yu, H. H. Ng, and Y. Ruan. 2006. A global map of p53 transcription-factor binding sites in the human genome. Cell 124207-219. [DOI] [PubMed] [Google Scholar]
- 57.Zhu, Z., J. Shendure, and G. M. Church. 2005. Discovering functional transcription-factor combinations in the human cell cycle. Genome Res. 15848-855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhu, J., Y. Zhang, G. J. Joe, R. Pompetti, and S. G. Emerson. 2005. NF-Ya activates multiple hematopoietic stem cell (HSC) regulatory genes and promotes HSC self-renewal. Proc. Natl. Acad. Sci. USA 10211728-11733. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.