Abstract
In hepatic cells, Smad and SnoN proteins converge with p53 to repress transcription of AFP, an oncodevelopmental tumor marker aberrantly reactivated in hepatoma cells. Using p53- and SnoN-depleted hepatoma cell clones, we define a mechanism for repression mediated by this novel transcriptional partnership. We find that p53 anchors activated Smads and the corepressor mSin3A to the AFP distal promoter. Sequential chromatin immunoprecipitation analyses and molecular modeling indicate that p53 and Smad proteins simultaneously occupy overlapping p53 and Smad regulatory elements to establish repression of AFP transcription. In addition to its well-known function in antagonizing transforming growth factor β (TGF-β) responses, we find that SnoN actively participates in AFP repression by positively regulating mSin3A protein levels. We propose that activation of TGF-β signaling restores a dynamic interplay between p53 and TGF-β effectors that cooperate to effectively target mSin3A to tumor marker AFP and reestablish transcription repression.
The intersection of signal transduction pathways creates nodes of regulatory control that allow fine-tuning of target gene regulation and expansion of downstream responses to exogenous signals. A paradigm for these regulatory networks is found in transforming growth factor β (TGF-β) signaling, well-known for its broad influence in development and disease. In this report, we uncover a mechanism by which the TGF-β pathway is harnessed by tumor suppressor p53 to confer repression of gene expression. TGF-β is part of a large superfamily, which includes TGF-βs, activins, bone morphogenetic proteins, nodals, anti-Müllerian hormone, and other structurally related ligands (25). These secreted polypeptides are recognized and bound by cell surface receptors that activate Smad transcription factors and mediate cell growth arrest, apoptosis, or differentiation. In response to ligand binding, the TGF-β1 receptor phosphorylates Smad2 and/or Smad3, which then bind Smad4 to form active heteromeric complexes. These activated complexes translocate to the nucleus where they bind to Smad binding elements (SBEs) of TGF-β-regulated genes (26, 46).
Smad proteins mediate gene activation or repression as a result of promoter-specific interactions with transcription coactivators, such as p300/CBP, or corepressors, such as Ski, SnoN, and TGIF (44). SnoN functions as an important negative regulator in the TGF-β pathway by two potential mechanisms, which are not mutually exclusive. First, binding of SnoN to Smads disrupts the active, DNA-bound heteromeric complex formed between Smad4 and Smad2/3 (22, 47). Second, SnoN binds to transcription corepressors such as N-CoR/SMRT and Sin3A to form a complex with histone deacetylases (HDACs). SnoN, tethered to Smad proteins, may recruit these protein complexes to promoters to repress transcription by mechanisms involving chromatin compaction (22).
Numerous pathways intersect with TGF-β-mediated signal transduction to impose posttranslational modification and/or alter functions of Smad proteins. For example, Ras/mitogen-activated protein kinases phosphorylate and inactivate Smad1, Smad2, and Smad3 (18). Likewise, Cdk2 and Cdk4 cyclin-dependent kinases phosphorylate Smad3 to decrease its function (28, 29). Cross talk also occurs by direct protein-protein interactions between mediators of distinct signaling pathways (27). One example lies in Smad4 interaction with β-catenin and TCF/LEF1, in response to Wnt signaling, to form an activation complex that stimulates Xtwn expression during Xenopus laevis development (35).
Protein-protein interactions may also partly compensate for weak, intrinsic binding affinity of Smad proteins for their target elements. SBEs are rarely found in isolation but rather are buttressed by binding sites for other sequence-specific transcription factors at TGF-β-responsive promoters. For example, the Mix.2 promoter harbors binding sites for both Smads and the winged-helix transcription factor FoxH1, which cooperate in activation (14). The c-myc oncogene is negatively regulated by a TGF-β inhibitory element that contains overlapping binding sites for Smad3/Smad4 and E2F family members E2F4/5, plus retinoblastoma family member p107 (13). These interactions between Smad proteins and their binding partners are thought to enhance TGF-β response at a given promoter (26).
Recently, p53 tumor suppressor was added to the growing list of proteins that collaborate with Smads (5, 41, 45). Smad2/3 and p53 cooperate to activate expression of several TGF-β target genes during Xenopus mesoderm differentiation. This integration is mediated by phosphorylation of p53 by the RTK-Ras-MAPK (RTK stands for receptor tyrosine kinase, and MAPK stands for mitogen-activated protein kinase) pathway, an event that enables binding of p53 to TGF-β-activated Smads (6). Further, p53 is required for full activation of p21 and cell cycle arrest in response to activin signaling in human epithelial cells (5, 41). We discovered that p53 is a requisite, molecular partner of Smads during TGF-β-mediated repression of the hepatic, tumor marker gene AFP encoding alpha-fetoprotein (AFP) (34, 45). AFP is robustly expressed in the developing liver and promotes normal female sexual differentiation and reproductive behavior (3, 10). AFP expression is dramatically shut down at birth but is preferentially reexpressed in liver carcinoma cells and regenerating liver cells. Aberrantly expressed AFP may promote tumor development by enhancing cell growth (31) and triggering apoptosis of antigen-presenting cells (43).
AFP is a classical model of developmental-stage- and tissue-specific expression and is one of few examples of p53-dependent repression by sequence-specific DNA binding (15, 19). The p53 response element of AFP (−850 bp) is intercalated with DNA binding sites for Forkhead protein Foxa1 and Smad proteins to form a Smad binding element and p53 response element (SBE/p53RE). Developmental, postpartum silencing of AFP in hepatocytes correlates with binding of p53, Smad2/4, and SnoN at the SBE/p53RE and repressive histone modifications (45).
The presence of overlapping binding sites for p53 and Smad proteins in the SBE/p53RE led us to investigate the hypothesis that p53 alters or controls Smad and/or SnoN recruitment to this repressor element. Sequential chromatin immunoprecipitation (ChIP) assays of repressed AFP chromatin in normal, differentiated liver and molecular modeling of p53 and Smad proteins bound to the SBE/p53RE support simultaneous binding to the intercalated regulatory elements without steric hindrance. We find that p53 is required to anchor TGF-β-activated Smads and corepressor mSin3A to the AFP SBE/p53RE. SnoN, normally an autoregulated repressor of TGF-β signaling, plays an essential role in AFP repression by positively regulating mSin3A protein levels. We propose that TGF-β and p53 cross talk reestablishes a developmental signature of AFP repression, which relies on the combined action of p53, Smads, and SnoN to culminate in effective recruitment and maintenance of corepressor mSin3A at the AFP promoter.
MATERIALS AND METHODS
Cell lines and plasmids.
Murine hepatoma cells (Hepa 1-6) and human embryonic kidney cells (293) were cultured in Dulbecco's modified essential medium (Gibco) supplemented with 10% fetal bovine serum and 1% antibiotics. AFP LacZ has been previously described (19). Myc-Smad4 and -Smad2 constructs were kindly provided by Michael Haymann. Wild-type (WT) and mutant p53 constructs were kind gifts of J. Manfredi. p53 and SnoN short hairpin RNA (shRNA) constructs have been previously described (45). The p53 rescue plasmid was generated from a p53-pBABE construct encoding WT murine p53 (generous gift of Guillermina Lozano). Site-directed mutagenesis was performed to introduce three missense mutations using a Stratagene site-directed mutagenesis kit per the manufacturer's specifications. The primers used for this purpose were as follows: p53-pBABE-rescue, 5′CACTACAAGTACATGTGCAACAGCTCCTGCATGGGG and 3′CCCCATGCAGGAGCTGTTGCACATGTACTTGTAGTG.
Transfections and reporter assays.
Hepa 1-6 and 293 cells were transfected with the indicated constructs using Lipofectamine 2000 (Invitrogen Inc.) per the manufacturer's instructions and as previously described (19, 45). Samples were analyzed per the manufacturer's instructions (Dual-Luciferase assay; Promega). All transfections were normalized using a Renilla luciferase construct, pRL-TK (23).
TGF-β treatment, RNA isolation, RT-PCR, and primers used.
Hepa 1-6 cells were exposed to TGF-β1 ligand (R&D Systems) at a concentration of 3 ng/ml or to vehicle control (4 mM HCl containing 0.1% bovine serum albumin [BSA]) for the indicated time points. RNA was isolated at the indicated time points using TRIzol reagent (Invitrogen Inc.) per the manufacturer's instructions. Synthesis of cDNA was performed using 1 μg of RNA per the manufacturer's instructions using a reverse transcription-PCR (RT-PCR) kit (Invitrogen Inc.). PCRs were set up in 96-well plates and read in an Applied Biosystems 7500 fast real-time PCR instrument and analyzed using the 7500 fast system sequence detection software, version 1.3.1 (ΔΔCT relative quantification method [where CT is threshold cycle]). AFP primers spanning an exon-intron junction were used for accurately measuring pre-mRNA levels. Primers used for the assays are as follows: for AFP mRNA, 5′CAGGCAACAACCATTATTAAGC and 3′TTCCTTGGCAACACTCCTC; for AFP pre-mRNA, 5′CCTTTACCCAGTTTGTTCCGG and 3′CGCTTGCTCTGCTTCAACAGT; for actin mRNA, 5′AGGGAAATCGTGCGTGAC and 3′CTCGTTGCCAATAGTGATGAC; for p53 mRNA, 5′AACCGCCGACCTATCCTTACCATC and 3′AGGCCCCACTTT CTTGACCATTG; for SnoN mRNA, 5′CGCACAGATCCCCTGACAA and 3′GTGCCACTTGGCTGACTCAA.
Solid-phase protein pull downs.
TGF-β- or vehicle-treated nuclear lysates from p53 KD (p53 knockdown) Hepa 1-6 cells transfected with either WT p53 or its DNA binding mutant, 143A, were prepared as described previously (45). Approximately 500 μg of individual lysates was diluted to 500 μl with dilution buffer (60 mM NaCl, 20 mM HEPES [pH 7.8], 5 mM MgCl2, 0.2 mM EDTA, 10% glycerol, 1 mM dithiothreitol, 2% BSA, and 0.1% NP-40) and precleared with 150 ng of biotinylated control oligonucleotide (−1007) and 20 μl of streptavidin-agarose (Millipore) for 1 hour at 4°C. The cleared lysates were collected and further incubated with 500 ng SBE/p53E or control oligonucleotide (−1007) overnight at 4°C. Protein-oligonucleotide complexes were collected by incubation with 50 μl of streptavidin-agarose for 2 h, followed by three or four washes with wash buffer (dilution buffer lacking BSA). The immunoprecipitated complexes were resuspended in 1× sodium dodecyl sulfate (SDS) buffer for analysis by immunoblotting.
The sequences of biotinylated oligonucleotides are as follows: SBE/p53RE, 5′-Bio-GATCCTTAGCAAACATGTCTGGACCTCTAGAC, and −1007, 5′-Bio-GATCCAATATCCTCTTGAC, where Bio is biotin.
Coimmunoprecipitation assays.
Whole-cell lysates from 293 cells transfected individually with Myc- and hemagglutinin (HA)-tagged proteins were prepared, and amounts equal to total lysate protein of 500 μg were mixed along with ethidium bromide at a final concentration of 100 μg/ml (33). Coimmunoprecipitations were performed in the absence of TGF-β treatment, as exogenously expressed Smad proteins enter the nucleus and bypass the requirement for ligand stimulation (17, 42). Proteins bound to Myc antibody-agarose beads (Bethyl Laboratories) were washed with wash buffer containing 25 mM Tris, 500 mM NaCl, 1 mM EDTA, 0.25% NP-40, and 1 mM phenylmethylsulfonyl fluoride three times for 10 min at 4°C prior to being directly dissolved in SDS buffer and processed for Western blot analysis.
Immunoblotting and antibodies used.
Immunoblotting of whole-cell lysates and nuclear/cytoplasmic fractions were performed using standard SDS-polyacrylamide gel electrophoresis methodology as previously described (45). The primary antibodies used are as follows: p53 (Ab1 [Oncogene] and p53DO1 [Santa Cruz]), SnoN (SC-9141; Santa Cruz), phospho-Smad2 (catalog no. 07-392; Upstate), Smad4 (catalog no. 06-693 [Upstate] and SC-7966 [Santa Cruz]), AFP (SC C-19; Santa Cruz), mSin3A (SC AK11; Santa Cruz), actin (A5316; Sigma), anti-HA (12CA5; Roche), and anti-Myc (9E10; Santa Cruz). All antibodies were used at a dilution of 1:1,000 with the following exceptions: p53 Ab1, actin, and anti-HA were used at a dilution of 1:500, 1:10,000, and 1:5,000, respectively.
ChIPs.
The ChIP assay was performed as previously described (45) with the following modifications. The purified DNA was used as template in real-time PCRs. Primers against AFP SBE/p53RE and the albumin enhancer were designed with Primer Express 2.0 (Applied Biosystems). Ten-microliter PCRs containing 1× fast TaqMan mix (Applied Biosystems), 4 μl of the ChIP-enriched DNA, and 100 nM primers were set up in 96-well plates and read in an Applied Biosystems 7500 fast real-time PCR instrument and analyzed using the 7500 fast system sequence detection software, version 1.3.1 (ΔΔCT relative quantification method). Input DNA values were used to normalize the values from ChIP samples.
The TaqMan primers used for the assay are as follows. For AFP SBE/p53RE, the TaqMan primers were 5′CTACATATGAAGCCTTAGCAAACATGT and 3′ACTCAGACGTTGGCGTGTCA, and the Mgb probe was 6FAM-CCTCTAGACACACAGACT-MGB where 6FAM stands for 6-carboxyfluorescein. For the albumin gene ALB, the TaqMan primers were 5′TGCTGATACCAGGGAATGTTTGT and 3′AAACTGGCCAAGGCAAACAC, and the Mgb probe was 6FAM-CTTAAATACCATCATTCCGG-MGB.
Sequential ChIPs.
Sequential ChIPs were performed exactly as described by Cui et al. (8) except that 12.5 μl of p53 FL393 (SC-6243) antibody was used for immunoprecipitation.
Generation of p53 and SnoN knockdown stables.
The construct used to target p53 and SnoN for RNA interference (RNAi)-mediated decay has been previously described (45). Hepa 1-6 cells were cotransfected with 1 μg of this p53 or SnoN shRNA expression plasmid as well as 100 ng of an expression plasmid containing a zeocin-resistant gene expression cassette (pcDNA3.1/Zeo, Invitrogen) followed by G4-18 antibiotic selection at a concentration of 1 mg/ml of medium for cell clones that had stably integrated the plasmid.
Rescue of AFP repression.
Hepa 1-6 cells were cotransfected with 2 μg of green fluorescent protein (GFP) vector (pEGFP) (catalog no. 6077-1; Clontech) and 300 ng of p53 rescue plasmid or vector control for 24 h, following which the cells were treated with TGF-β at a concentration of 3 ng/ml for 4 h. GFP-positive cells were enriched using fluorescence-activated cell sorting using the BD Aria cell sorter (BD, San Jose, CA). GFP-positive cells were collected in 500 μl Dulbecco's modified essential medium supplemented with 10% serum and 1% antibiotic and pelleted at 4°C for 10 min at 2,000 × g. The cell pellet was resuspended in 100 μl TRIzol and processed for cDNA analysis as described above.
Molecular modeling.
To construct a model of p53 and Smad proteins binding to the upstream, 5′ p53 regulatory element of AFP (p53 dimer consensus binding site), the p53 DNA (1TUP) and the Smad3-MH1 DNA (1QZJ) pdb coordinates were read into the modeling program O (16), and the model was constructed as follows. First, the two p53 DNA binding domains that are nonspecifically bound to the DNA (chain C) or not bound to DNA (chain A) in the 1TUP coordinates were eliminated, leaving only the specific p53-DNA complex (protein chain B and the DNA duplex). The DNA was extended to include the Smad site. The Smad3-MH1 domain was then docked onto the Smad DNA site. The model was then checked for steric clash using the program CNS (4).
RESULTS
Molecular model for simultaneous interaction of p53 and Smad proteins at the SBE/p53RE.
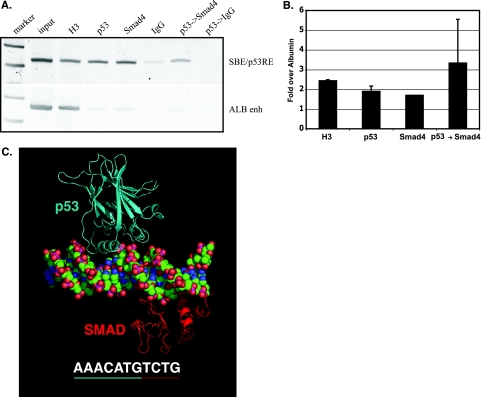
One potential mechanism underlying a role for p53 in recruiting Smad proteins is simultaneous occupancy of p53 and Smad proteins at their intercalated binding sites (SBE/p53RE) within the AFP distal promoter. To determine whether this was feasible, we used two approaches: sequential ChIP (or re-ChIP) of p53 and Smad proteins in adult mouse liver, where postnatal AFP silencing occurs, and molecular modeling of structurally defined domains of p53 and Smad proteins bound to the SBE/p53RE. Previous, locus-specific ChIP studies found correlation between p53, Smad2/4, and SnoN binding to the SBE/p53RE and developmental repression of AFP (34). Sequential ChIP analyses of liver tissue from 2-month-old mice showed that eluted, p53-bound chromatin fragments were specifically enriched by a second round of immunoprecipitation with a Smad4 antibody, demonstrating that Smad4 and p53 coexist on the AFP SBE/p53RE (Fig. 1A and B). The efficiency of immunoprecipitation by Smad4 and p53 antibodies cannot be directly compared to each other; therefore, we compared protein-chromatin interactions at the repressed AFP locus (SBE/p53RE region) and the actively expressing ALB gene (ALB enhancer region). Levels of nucleosomal occupancy, measured by histone H3 levels, are greatly increased at the repressed AFP chromatin, compared to the active ALB locus, supporting an integral role of chromatin accessibility in establishing regulation of transcription.
FIG. 1.
p53 and Smads simultaneously occupy an intercalated SBE/p53RE. (A and B) ChIP analysis of normal, differentiated liver tissue from mice correlates with repression of AFP expression. (A) PCR products. Input DNA (diluted 1/10) and DNA enriched by ChIP (histone H3 [diluted 1/10], immunoglobulin G [IgG] control, p53, and Smad4 antibodies) and sequential ChIP (p53→Smad4, primary antibody enrichment followed by secondary antibody enrichment) of p53, Smad4, and both p53 and Smad4 proteins bound to the AFP SBE/p53RE and negative, distal control albumin enhancer (ALB enh) regions. (B) Quantified ChIP and sequential ChIP of p53 and Smad4 interaction with chromatin, as in panel A. The graph shows changes in enrichment of indicated proteins bound at AFP SBE/p53RE compared to that of the ALB enhancer region. All values, generated by real-time PCR analysis, were normalized to control IgG precipitations and represent means plus standard deviations (error bars) of triplicate determinations. (C) Molecular model of simultaneous binding of p53 and Smad to the AFP SBE/p53RE. DNA sequence shown in its entirety is the upstream, p53 regulatory element (p53 dimer binding site) with the Smad binding sequence underlined in red, centered at −850 of AFP. The molecular model reveals no clash between p53 and the Smad3 MH1 domains even without energy minimization, indicating that the two proteins can bind the site simultaneously.
The structural feasibility of simultaneous occupation of the SBE/p53RE by p53 and Smad proteins was modeled by utilizing crystal structures of the p53 core domain bound to DNA and the Smad3-MH1 domain-DNA complex. The upstream p53 and Smad consensus sites within the composite SBE/p53RE were chosen for modeling, as they are essential for p53 binding in vitro and AFP repression in vivo (8, 11, 19, 45, 48). The resulting model reveals no steric hindrance or interactions between the p53 core and Smad3-MH1 domains (Fig. 1C). Even accounting for regions or domains of the proteins not included in this model, e.g., MH2 for Smads, N and C termini for p53, this finding suggests that the DNA binding domains of p53 and Smad proteins simultaneously occupy the AFP SBE/p53RE and that any interactions between the two proteins are mediated by regions outside the DNA binding domains.
In vitro system to address mechanisms of p53 and TGF-β partnership.
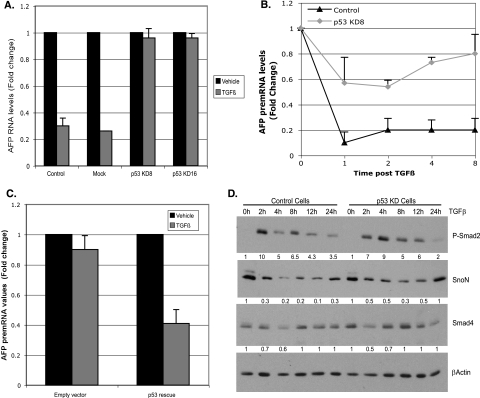
To uncover the molecular mechanisms of TGF-β/p53 interplay, we turned to exogenous addition of the TGF-β1 ligand and depletion of p53 from cultured hepatoma cells that express AFP as a tumor marker. Mouse hepatoma (Hepa 1-6) cells respond to TGF-β1 ligand addition by repression of endogenous AFP expression in correlation with p53, SnoN, phosphorylated Smad2 (P-Smad2), and Smad4 recruitment to the SBE/p53RE (45). We generated stable clones of Hepa 1-6 cells depleted of p53 by shRNA-mediated depletion of p53 expression to determine the p53 dependence of recruitment of transcription factors and corepressors to chromatin over the time course of TGF-β1 treatment. Two zeocin-resistant clones, which stably integrated an shRNA plasmid targeted to p53 RNA, had marked depletion of p53 expression: knockdown (KD) clones 8 and 16 showed reductions of 5- and 10-fold in the levels of p53 RNA and protein, respectively, compared to the levels in parental or luciferase shRNA control cells (see Fig. S1 in the supplemental material). After 24 h of exposure to TGF-β1 ligand, p53 KD cells exhibited little or no decrease in stable, endogenous AFP mRNA levels, which were repressed three- to fourfold in control cells (Fig. 2A).
FIG. 2.
Stable depletion of p53 compromises AFP repression in response to TGF-β. (A) Quantitative RT-PCR analysis of AFP RNA. Hepa 1-6 parental cells (control), mock selected cells, and two independent p53 knockdown (KD) clones (p53 KD clone 8 [KD8] and KD16) were treated with TGF-β for 24 h and harvested for RNA analysis. AFP RNA levels in untreated cells were set at 1. All the values, normalized to actin RNA levels represent averages plus standard deviations (SD) (error bars) from three experiments. Black and gray bars indicate vehicle- and TGF-β-treated samples, respectively. p53 RNA levels were regularly monitored (data not shown) to ensure maintenance of p53 depletion. In subsequent analyses, p53 KD clone 8 was used, since these cells maintained p53 knockdown more effectively over passage of the cells (data not shown). (B) Graph showing the changes in AFP pre-mRNA levels compared to the time zero value over the indicated time course of TGF-β treatment in control and p53 KD Hepa 1-6 cells. Values represent averages plus SD (error bars) from three or four experiments. (C) Rescue of AFP repression by introduction of p53 in KD cells. AFP repression was monitored by measuring AFP pre-mRNA levels under the conditions described above for panel B. All values were normalized to actin and represent means plus SD (error bars) for triplicate determinations. (D) Stable depletion of p53 does not significantly alter the global levels of TGF-β effectors. Western blot analysis performed on nuclear extracts of control and p53 KD8 Hepa 1-6 cells for the indicated proteins over the time course of TGF-β treatment. Actin was used to control for loading and to normalize quantified protein levels. The amount of change (n-fold) in normalized protein expression is shown below the lanes in the immunoblots. The expression levels of each protein at the 0-h time point were set at 1.
TGF-β elicits rapid changes in gene expression, typically within a few hours and often measurably within 1 hour (25). To determine whether this is true of AFP repression, AFP pre-mRNA levels were measured over the time course of TGF-β treatment. Real-time RT-PCR analysis showed that control Hepa 1-6 cells exhibited almost a 10-fold reduction in the level of AFP pre-mRNA within 1 h of TGF-β treatment. After 1 h, the response leveled off to an approximately fivefold reduction for up to 8 h (Fig. 2B, control). By comparison, p53 KD cells exhibited a blunted response to TGF-β treatment at all time points (Fig. 2B, p53 KD), where the magnitude of repression was reduced three- to sixfold compared to control cells. A low level of repression (20% to 40%) can be detected at all time points, supporting a model where p53 alone is insufficient to repress AFP expression in the presence of TGF-β but is essential for the full response.
To assess whether the results obtained with the p53-targeted shRNA plasmid were specifically due to depletion of p53 or due to off-target effects, we performed a p53 complementation assay with a p53 rescue construct that expresses p53 RNA refractory to RNAi-mediated degradation (see Fig. S2 in the supplemental material). The p53 rescue construct robustly expressed p53 RNA, exhibiting an eightfold increase compared to cells that were transfected only with a GFP-expressing construct. Expression of the p53 rescue construct led to a decrease in AFP pre-mRNA levels by 2.5-fold in response to TGF-β (Fig. 2C). These results indicate that the impaired repression of AFP in the p53 KD cells is primarily the direct result of depleted levels of p53.
Effect of p53 on expression of TGF-β effectors in response to TGF-β.
We tested whether p53 directly regulates expression of TGF-β effectors, which would in turn alter response to TGF-β treatment in p53 KD cells. We determined the levels of Smad4, phosphorylated Smad2, and SnoN proteins in nuclear extracts of TGF-β-treated control and p53 KD cells by immunoblot analysis (Fig. 2D). In control cells, P-Smad2 levels were robustly induced as early as 2 h after administration of TGF-β, whereas Smad4 levels were relatively unchanged (Fig. 2D, control). SnoN protein levels decreased shortly after treatment and showed partial recovery within 8 h to the levels observed in the absence of ligand. A similar pattern of P-Smad2, Smad4, and SnoN expression was observed in p53 KD cells (Fig. 2D), indicating that p53 does not directly regulate the genes encoding these TGF-β effectors. Collectively, these results indicate that depletion of p53 does not significantly alter nuclear levels of Smad4, Smad2, and SnoN.
p53-dependent recruitment of P-Smad2 and Smad4.
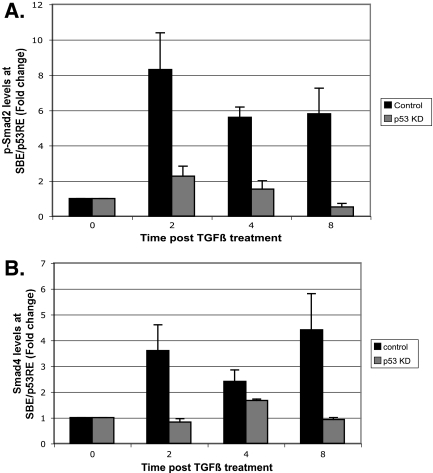
To determine whether p53 promotes recruitment of TGF-β effector molecules to chromatin, we performed ChIP analysis of control and p53 KD cells, incubated with or without TGF-β1 ligand. Treatment with TGF-β greatly increased binding of P-Smad2 and Smad4 at the SBE/p53RE, as early as 2 h after the addition of ligand (Fig. 3A and B, control). Rapid recruitment of a Smad heteromeric complex is consistent with reports demonstrating that Smads are rapidly activated and translocated into the nucleus after TGF-β treatment (38). In contrast, p53 KD cells exhibited little or reduced recruitment of Smad proteins (Fig. 3A and B), indicating that recruitment of Smad4 and P-Smad2, in response to TGF-β signaling, depends on p53. Determinations of absolute, quantified levels of Smad4 and P-Smad2 bound to AFP chromatin in the absence of TGF-β treatment showed that these effectors resided at the SBE/p53E at levels near background (data not shown).
FIG. 3.
p53 anchors Smads to the AFP SBE/p53RE. (A and B) Quantified ChIP analysis of phosphorylated Smad2 (p-Smad2) (A) and Smad4 (B) binding to SBE/p53RE. Control Hepa 1-6 cells and p53 KD cells were treated with TGF-β and harvested at the indicated time points for pSmad2 and Smad4 ChIP analyses. The graphs show the levels of the indicated proteins recruited to SBE/p53RE in control and p53 KD clone 8 cells over the indicated time course. All data were normalized to input measurements and represent averages plus standard deviations (error bars) of triplicate determinations. Control immunoglobulin G immunoprecipitations were performed in parallel to quantify antibody specificity (not shown).
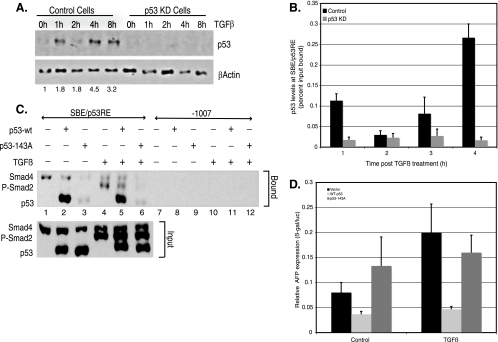
In addition to eliciting recruitment of Smad proteins, TGF-β signaling may trigger increased binding of p53 itself to the SBE/p53RE. If this were so, it would provide a simple explanation for how TGF-β elicits repression of transcription together with p53, as we showed that p53 is essential for this process (Fig. 2). We examined whether TGF-β treatment had any effect on the global levels of p53. Immunoblot analysis of TGF-β-treated nuclear lysates revealed that while p53 levels increased only marginally at the earlier time points, there was a substantial increase observed by 8 h of treatment (Fig. 4A). Inconsistent with its global expression levels, ChIP analysis revealed that p53 is bound to the SBE/p53RE region of AFP, even in the absence of TGF-β (Fig. 4B). Furthermore, the amount of p53 bound to the AFP regulatory region actually decreased in level (albeit modestly) at the earliest time of analysis after TGF-β treatment (2 h in Fig. 4B, control cells). While TGF-β did increase p53 binding at 8 h (Fig. 4B, control cells), repression of AFP transcription was well established by this time of incubation (Fig. 2B). As expected, p53 KD cells had no measurable binding of p53 to the AFP SBE/p53RE (Fig. 4B). The discovery that p53 is bound to AFP chromatin in the absence of TGF-β treatment implies that p53 binding, while necessary, is not sufficient to confer transcriptional repression. Interaction of p53 with P-Smad2 and Smad4 may be required, as recruitment of Smad proteins to the AFP regulatory region is dependent on both TGF-β and p53 (Fig. 3). In line with this prediction, Smad3 and p53 have been shown to interact directly via the N terminus of p53 in both Xenopus embryos and mammalian cells (6). These observations would predict that TGF-β signaling prompts the formation of a Smad-p53 complex that is targeted to gene promoters.
FIG. 4.
DNA binding of p53 at the SBE/p53RE is essential for AFP repression by TGF-β. (A) Immunoblot analysis of p53 protein levels in response to TGF-β treatment in control and p53 KD cells, performed as described in the legend to Fig. 2D. (B) p53 is poised at the AFP SBE/p53RE prior to TGF-β treatment. The absolute levels of p53 bound to SBE/p53RE in control and p53 KD Hepa 1-6 cells treated with TGF-β over the indicated time course, as determined by ChIP analysis of p53, are shown. All data were normalized to input and represent averages plus standard errors of the means (error bars) from four or five experiments. (C) Interaction of Smad proteins at the SBE/p53RE depends on p53-DNA binding. The panel represents Western blot analyses for endogenous P-Smad2, Smad4, and exogenous p53. Lanes 2 and 5 show protein complexes from p53 KD Hepa 1-6 cells transfected with wild-type p53 (p53-wt), untreated (−) or treated with TGF-β (3 ng/ml) (+), were precipitated using biotinylated oligonucleotides containing the SBE/p53RE consensus. Lanes 3 and 6 show the same experimental conditions as described for lanes 2 and 5 except that the cells were transfected with a DNA binding mutant of p53 (p53-143A) (+). For a negative control, the protein complexes were precipitated using biotinylated oligonucleotides containing distal promoter elements of AFP (−1007 to −977). (D) p53 KD Hepa 1-6 hepatoma cells were transfected with the AFP/lacZ reporter construct (1 μg/plate) along with the indicated expression vectors (WT p53 or p53-143A, 50 ng/plate each). Each plate was also cotransfected with the Renilla luciferase (500 ng/plate) to standardize and control for transfection efficiency. Eight hours posttransfection, the cells were treated with TGF-β or its vehicle (control) for 24 h. Expression levels relative to baseline are indicated for AFP/lacZ (black). Relative AFP expression is shown as the ratio of β-galactosidase to luciferase expressed (β-gal/luc).
Binding of p53 to DNA is essential for TGF-β-mediated AFP repression.
We previously showed that repression of AFP transcription by p53 displays a strict requirement for DNA binding to the SBE/p53RE (19). It was therefore surprising to note that binding of p53 to AFP chromatin decreased modestly at the early time points of Smad recruitment and AFP repression (Fig. 2 and 4). Although p53 recruitment subsequently increased in parallel with maximal recruitment of Smad2 and Smad4, it is formally possible that Smad proteins are recruited independently of p53 at the SBE/p53RE due to their affinity for Smad binding elements. We therefore tested whether p53 DNA binding was essential for TGF-β-mediated AFP repression by performing two sets of experiments. We examined binding of Smads to the AFP promoter in the presence of WT p53 and, in parallel, a mutant form of p53, compromised in its ability to bind DNA (Fig. 4C). Nuclear extracts of p53 KD cells, which were transfected with either a plasmid expressing WT or mutant p53 (p53-143A), were incubated with biotinylated oligonucleotides spanning the SBE/p53RE of AFP (residues −860 to −830) and harvested with streptavidin-coupled agarose beads. The SBE/p53RE affinity-purified protein complexes were analyzed by immunoblotting by serial additions of Smad4, P-Smad2, and p53 antibodies. WT p53 displayed high affinity for the SBE/p53RE oligomers in both TGF-β-treated and untreated nuclear extracts (Fig. 4C, lanes 2 and 5), but DNA binding mutant p53-143A failed to bind with any robustness in either condition (lanes 3 and 6). Surprisingly, binding of Smad4 was detected even in the absence of p53 in both untreated and treated conditions (lanes 1, 2, 4, and 5). Similarly, P-Smad2 interaction at the SBE/p53RE, while dependent on TGF-β signaling, occurred even in the absence of p53 (lanes 4 and 5). p53-independent interactions of Smad proteins in this assay are likely to occur due to the presence of Foxa1 protein bound at this element (data not shown; also see Discussion). However, in extracts of cells that express p53-143A, which does not bind DNA, P-Smad2 and Smad4 fail to bind the SBE/p53RE oligomers (lanes 3 and 6), and TGF-β treatment had no effect. The interaction of freely soluble p53 and Smad proteins is supported by previous work of Cordenonsi and colleagues (5) and coimmunoprecipitation of p53-Smad protein complexes from extracts of Hepa 1-6 cells (see Fig. S3 in the supplemental material). Mutant p53-143A cannot bind the SBE/p53RE or tether p53-Smad protein complexes to DNA, acting in a dominant-negative manner for Smad-DNA interaction. Taken together, these results suggest that soluble p53-Smad protein complexes form independently of DNA in vitro and are recruited and anchored to the SBE/p53RE in a WT p53-dependent manner.
We complemented the in vitro binding assays with a functional assessment of TGF-β-mediated repression of AFP by either WT p53 or p53-143A expression in p53 KD Hepa 1-6 cells (Fig. 4D). The AFP reporter construct harbors an SBE/p53RE at −850 within 3.8 kb of upstream AFP regulatory sequences, fused to a bacterial β-galactosidase gene (AFP/lacZ) (19). Introduction of WT p53 alone (Fig. 4D, WT p53, control) repressed AFP reporter expression twofold, whereas p53-143A (Fig. 4D, p53-143A, control) conferred minor activation. The addition of TGF-β ligand, together with expression of p53, caused synergistic repression of approximately fivefold (Fig. 4D, WT p53, TGF-β). This repression was dramatically reduced in the presence of p53-143A (Fig. 4D, p53-143A, TGF-β). The changes do not reflect differences in the expression of p53, as determined by immunoblot analysis of p53 protein (Fig. 4C, input; also data not shown). Interestingly, the addition of TGF-β ligand activated the AFP reporter construct in the absence of p53, further supporting binding of Smad proteins to the SBE/p53RE, observed in vitro (Fig. 4C). Collectively, these experiments highlight the importance of DNA binding by p53 in mediating AFP repression in the presence of active Smad complexes.
SnoN plays an essential role in TGF-β-mediated AFP repression.
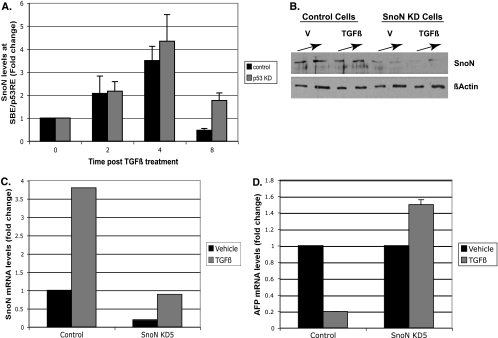
We previously showed that SnoN is required for AFP repression and is recruited to the AFP promoter in response to TGF-β (45). We predicted that SnoN recruitment would be impaired in p53 KD cells, since these cells are compromised for Smad binding at the SBE/p53RE (Fig. 3). Previous studies supported a model of Smad-dependent recruitment of SnoN to TGF-β-regulated genes, as SnoN does not directly bind DNA (22). Loss of SnoN binding would potentially explain lack of AFP repression, since SnoN associates with corepressors and HDAC proteins (22). However, to our surprise, SnoN recruitment in p53 KD cells was nearly equal to levels seen in control cells (Fig. 5A). It is important to note that SnoN, although bound to chromatin, is not sufficient to repress AFP expression in p53 KD cells (Fig. 2B), which lack p53 and Smad2/4 interaction at the SBE/p53RE (Fig. 3 and 4).
FIG. 5.
SnoN is essential for TGF-β-mediated AFP repression. (A) SnoN recruitment to SBE/p53RE is independent of p53. Quantified ChIP analysis for SnoN protein bound to SBE/p53RE was performed with a SnoN-specific antibody as described in the legend to Fig. 3. (A and B) Stable depletion of SnoN compromises AFP repression in response to TGF-β. (B) Analysis of SnoN protein levels. Control and SnoN KD Hepa 1-6 cells were treated with vehicle (V) or TGF-β and harvested for nuclear extract preparation. Standard Western blot analysis was performed to determine the levels of SnoN protein depletion. Actin levels were used to control for loading errors. (C) Quantitative RT-PCR analysis of SnoN RNA was performed as described in the legend to Fig. S1 in the supplemental material to ensure maintenance of SnoN KD under the experimental conditions shown in panel D. (D) Quantitative RT-PCR analysis of AFP RNA was performed as described in the legend to Fig. 2A for the indicated cell types. Black and gray bars indicate vehicle- and TGF-β-treated cells, respectively.
In order to clarify a role for SnoN in AFP repression, we generated SnoN KD Hepa 1-6 cells (Fig. 5B and C). SnoN KD cells, which stably express an shRNA plasmid targeted to SnoN RNA, had marked depletion of SnoN protein (Fig. 5B) and RNA (Fig. 5C) levels compared to parental control cells. Similar to p53 depletion, continuous depletion of SnoN abrogated AFP repression in response to TGF-β (Fig. 5D). Thus, SnoN is clearly required for cross talk of p53 and TGF-β and, although bound to AFP chromatin in p53 KD cells, needs p53 to promote AFP repression.
p53 and SnoN are both needed to recruit the mSin3A corepressor.
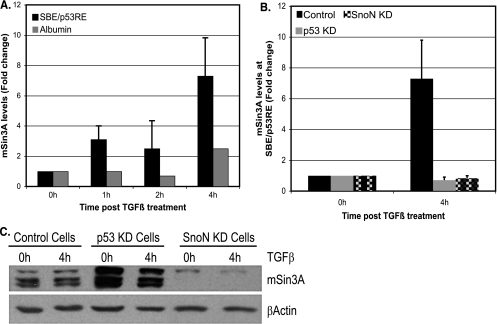
Current models of p53-mediated repression of transcription generally invoke mSin3A protein as a predominant corepressor in complex with HDAC protein(s) (15, 32). HDAC proteins provide enzymatic activity and promote transcription repression by deacetylating histones (39). Previously, we showed that developmentally repressed AFP displays hallmarks of repressed chromatin and recruitment of mSin3A/HDAC1 repressor complexes (34, 45). A connection between SnoN and mSin3A is provided in studies showing that SnoN binds to mSin3A and related corepressor complexes (22, 36). We predicted that SnoN and/or p53 were likely important for mSin3A recruitment to the AFP SBE/p53RE. To test this directly, ChIP assays for mSin3A in control, p53 KD, and SnoN KD cells were performed. TGF-β stimulation engendered a robust recruitment of mSin3A to the SBE/p53RE (Fig. 6A) over 4 h of TGF-β treatment, whereas recruitment was severely impaired in both p53 and SnoN KD cells (Fig. 6B).
FIG. 6.
p53 recruits mSin3A to the AFP SBE/p53RE, while SnoN acts to maintain mSin3A protein levels. (A and B) ChIP analysis of mSin3A interaction with chromatin in control cells (A) and p53-SnoN KD cells (B). (A) Graph showing levels of mSin3A recruitment to AFP SBE/p53RE at the indicated times after TGF-β treatment. PCR amplifications were performed with albumin primers as a distal control of specificity. (B) Graph showing the levels of mSin3A recruited to SBE/p53RE in control cells, p53 KD cells, and SnoN KD cells 4 h after TGF-β treatment. (C) SnoN is required to maintain mSin3A protein levels in Hepa 1-6 cells. Western blot analysis for mSin3A levels in nuclear extracts prepared from control, p53 KD, and SnoN KD cells. Actin levels serve to normalize loading errors.
Loss of mSin3A recruitment could potentially result from loss of global levels of mSin3A protein, although no regulatory role for p53 or SnoN in mSin3A expression has previously been reported. Analysis of mSin3A levels in control, p53 KD, and SnoN KD Hepa 1-6 cells revealed that mSin3A protein levels were drastically reduced in SnoN KD cells, whereas mSin3A levels were maintained in p53 KD cells and control Hepa 1-6 cells (Fig. 6C). Two independent SnoN KD clones, both of which were compromised for AFP repression, displayed similar reductions in mSin3A protein levels compared to control cells (data not shown). Since TGF-β treatment results in initial degradation of SnoN protein (Fig. 2D), we tested whether mSin3A levels decreased in parallel as well, which would further support a role for SnoN in regulating mSin3A protein stability. As seen in Fig. S4A in the supplemental material, the levels of SnoN and mSin3A proteins mirror each other, after TGF-β treatment, in a cycle of degradation and resynthesis that peaks approximately 4 h after ligand exposure. This time point coincides with maximal recruitment of these factors at the SBE/p53RE (Fig. 5A and 6A).
SnoN-mediated regulation of mSin3A likely occurs at the level of protein stability or translation, as mSin3A mRNA levels are not altered in p53 KD and SnoN KD cells (see Fig. S4B in the supplemental material). Future studies will determine this, as well as the contribution of SnoN regulation of corepressor mSin3A in the oncogenic function of SnoN. Collectively, our results show that mSin3A is an essential component in p53/TGF-β-mediated repression of AFP transcription. Chromatin-bound p53 recruits mSin3A and maintains Smad protein interactions with the SBE/p53RE. Surprisingly, the essential role of SnoN is in maintaining mSin3A protein levels.
DISCUSSION
AFP SBE/p53RE, a novel p53/TGF-β response element.
Utilizing an experimentally accessible, TGF-β- and p53-responsive hepatoma cell line as a model system, we addressed key features of p53 as a partner for transcription repression conferred by TGF-β. We find that p53 is essential for temporal recruitment and anchoring of an activated Smad heteromeric complex and mSin3A corepressor to the AFP distal promoter. Our finding that p53 is required for stable association of Smad proteins with chromatin furthers our molecular understanding of the cooperativity between Smad2/3 and p53, first shown in activation of multiple TGF-β target genes during differentiation of Xenopus mesoderm and cell cycle arrest in human epithelial cells (41).
In addition to p53 and Smads, a Forkhead family member, Foxa1, binds to the SBE/p53RE and, in contrast to DNA-bound p53/Smads, activates AFP expression (7, 19). Intriguingly, this activation also likely involves Smad4/2 proteins, as we observe a correlation between robust AFP expression in vivo and Foxa1/Smad occupancy, without p53, at the SBE/p53RE (34, 45). Forkhead transcription factors are known to bind to the MH2 domain of Smad2/3 and are required for binding of Smad protein complexes to specific target promoters (14). It is therefore not surprising that we detected Smad4/2 binding to the SBE/p53RE in vitro in the absence of p53, since Foxa1 is also bound to its consensus site within this element (data not shown). When p53-mediated repression is blocked by RNAi-mediated depletion of endogenous p53 and/or expression of a dominant-negative p53, addition of TGF-β1 activated AFP expression. On the basis of these observations, we hypothesize that the SBE/p53RE, a complex DNA binding element with Foxa1/p53 tetramer/Smad dimer binding sites intercalated within 30 bp, acts an interface for Smad-mediated activation as well as repression of AFP gene expression.
p53-Smad protein complex.
In the absence of TGF-β, low levels of p53 bind to the SBE/p53RE repressor of AFP but do not induce repression of transcription. This finding is consistent with previous reports of p53 bound to a p53RE as a latent, inactive protein in the absence of stress stimuli (2, 12). Mdm2 protein, a major regulator of p53 protein stability and degradation (21), is reportedly associated with inactive, chromatin-bound p53. Stress-induced signaling leads to the dissociation of Mdm2 to facilitate posttranslational modification of DNA-bound p53 or to Mdm2-mediated ubiquitylation and degradation of p53 to allow binding de novo of activated p53 (30). Both mechanisms of p53 activation may be at work here. The N terminus of p53, a target of several posttranslational modifications in response to specific stimuli (1), is required for p53 and Smad interaction in a complex, as shown by Piccolo and colleagues (6). Our ChIP assays show that p53 interactions decrease slightly in response to TGF-β, before a considerable increase in parallel with soluble p53 protein levels.
SnoN-dependent regulation of mSin3A levels.
To the best of our knowledge, the discovery that SnoN regulates mSin3A protein levels is unprecedented, occurring at either the level of transcription, translation, protein stability, or subcellular localization. Transcription is unlikely to play a role, as we saw no change in mSin3A mRNA levels after SnoN depletion. Subcellular mislocalization of mSin3A does not occur due to SnoN depletion because loss of mSin3A protein is seen in both nuclear fractions (Fig. 6C) and cytoplasmic fractions (data not shown) of SnoN KD cells. Although it is unlikely, SnoN protein may associate with mSin3A mRNA transcripts and regulate their translation. On the basis of our observations and published work demonstrating that SnoN physically interacts with mSin3A (22), we hypothesize that SnoN acts to stabilize mSin3A protein levels. SnoN itself is under the control of ubiquitin-mediated degradation in response to TGF-β signaling, an event proposed to control the timing and duration of Smad activity (40). It is therefore interesting to note that mSin3A protein levels mirror this response at early time points of TGF-β stimulation, suggesting that SnoN, mSin3A, and perhaps HDACs associated with this complex are subject to regulation by TGF-β. This regulation would be critical in determining activation versus repression mediated by Smad-p53 complexes, which can associate with either corepressor and coactivator proteins.
The functional consequences of a regulatory connection between SnoN and mSin3A are manifold. mSin3A is essential for embryonic development and positively regulates cellular proliferation and survival, in part by controlling p21 gene expression and deacetylation of p53 (9). The corepressor SnoN is aberrantly overexpressed in multiple tumor-derived cells, including Hepa 1-6 (24, 45, 49), which may lead to increased mSinA protein. Such an event could directly alter the activity of p53, leading to abrogation of its tumor suppressor functions and, as a consequence, increased proliferation and survival.
Although oncogenic SnoN is typically thought to antagonize TGF-β-mediated gene activation by disrupting Smad complexes, our work and recently published studies of others suggest that SnoN retains the ability to be harnessed by the TGF-β pathway for active participation in gene regulation together with Smad proteins (20, 45). When TGF-β ligand is added, overexpressed SnoN protein is degraded in a process mediated by the E3-ubiquitin ligase Arkadia (20). Only when existing SnoN is depleted and then synthesized de novo, can the machinery of TGF-β signaling proceed with normal, Smad-mediated regulation of gene expression. Control of mSin3A protein levels may also be integral to this regulation. Our temporal studies of chromatin binding support this view and further suggest that degradation of SnoN, while necessary, is not sufficient to promote an ordered progression of transcription factor interactions with chromatin and regulation of expression.
Understanding whether aberrant reactivation of AFP is due to dysfunction in p53, TGF-β signaling, or both has prognostic and therapeutic value. AFP is reactivated as a tumor marker in 70% to 85% of cases of hepatocellular carcinoma and is widely used to stage aggressiveness and growth of the tumor. Progression, but not initiation, of hepatocellular carcinoma is frequently associated with mutations in p53 (37). Our studies suggest that when p53 is not mutated, reactivation of TGF-β signaling is sufficient to reinstate differentiation-associated regulation of transcription at the level of tumor marker AFP expression in hepatoma cells.
Supplementary Material
Acknowledgments
This work was supported in part by National Institutes of Health training grant T32-HD07325 to D.S.W., National Institutes of Health grant GM53683 to M.C.B., and a National Cancer Institute Cancer Center Support Grant to the University of Texas M. D. Anderson Cancer Center.
We gratefully acknowledge J. Manfredi, L. Resnick-Silverman, G. Lozano, S. Dent, and members of the Barton and Dent laboratories for materials and/or helpful discussions. D.S.W. especially thanks M. Wilkinson for advice and support.
Footnotes
Published ahead of print on 22 January 2008.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Appella, E., and C. W. Anderson. 2000. Signaling to p53: breaking the posttranslational modification code. Pathol. Biol. (Paris) 48227-245. [PubMed] [Google Scholar]
- 2.Arva, N. C., T. R. Gopen, K. E. Talbott, L. E. Campbell, A. Chicas, D. E. White, G. L. Bond, A. J. Levine, and J. Bargonetti. 2005. A chromatin-associated and transcriptionally inactive p53-Mdm2 complex occurs in mdm2 SNP309 homozygous cells. J. Biol. Chem. 28026776-26787. [DOI] [PubMed] [Google Scholar]
- 3.Bakker, J., C. De Mees, Q. Douhard, J. Balthazart, P. Gabant, J. Szpirer, and C. Szpirer. 2006. Alpha-fetoprotein protects the developing female mouse brain from masculinization and defeminization by estrogens. Nat. Neurosci. 9220-226. [DOI] [PubMed] [Google Scholar]
- 4.Brunger, A. T., P. D. Adams, G. M. Clore, W. L. DeLano, P. Gros, R. W. Grosse-Kunstleve, J. S. Jiang, J. Kuszewski, M. Nilges, N. S. Pannu, R. J. Read, L. M. Rice, T. Simonson, and G. L. Warren. 1998. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. Sect. D 54905-921. [DOI] [PubMed] [Google Scholar]
- 5.Cordenonsi, M., S. Dupont, S. Maretto, A. Insinga, C. Imbriano, and S. Piccolo. 2003. Links between tumor suppressors: p53 is required for TGF-beta gene responses by cooperating with Smads. Cell 113301-314. [DOI] [PubMed] [Google Scholar]
- 6.Cordenonsi, M., M. Montagner, M. Adorno, L. Zacchigna, G. Martello, A. Mamidi, S. Soligo, S. Dupont, and S. Piccolo. 2007. Integration of TGF-beta and Ras/MAPK signaling through p53 phosphorylation. Science 315840-843. [DOI] [PubMed] [Google Scholar]
- 7.Crowe, A. J., L. Sang, K. K. Li, K. C. Lee, B. T. Spear, and M. C. Barton. 1999. Hepatocyte nuclear factor 3 relieves chromatin-mediated repression of the α-fetoprotein gene. J. Biol. Chem. 27425113-25120. [DOI] [PubMed] [Google Scholar]
- 8.Cui, R., T. T. Nguyen, J. H. Taube, S. A. Stratton, M. H. Feuerman, and M. C. Barton. 2005. Family members p53 and p73 act together in chromatin modification and direct repression of α-fetoprotein transcription. J. Biol. Chem. 28039152-39160. [DOI] [PubMed] [Google Scholar]
- 9.Dannenberg, J. H., G. David, S. Zhong, J. van der Torre, W. H. Wong, and R. A. Depinho. 2005. mSin3A corepressor regulates diverse transcriptional networks governing normal and neoplastic growth and survival. Genes Dev. 191581-1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Mees, C., J. F. Laes, J. Bakker, J. Smitz, B. Hennuy, P. Van Vooren, P. Gabant, J. Szpirer, and C. Szpirer. 2006. Alpha-fetoprotein controls female fertility and prenatal development of the gonadotropin-releasing hormone pathway through an antiestrogenic action. Mol. Cell. Biol. 262012-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.El-Deiry, W. S., S. E. Kern, J. A. Pietenpol, K. W. Kinzler, and B. Vogelstein. 1992. Definition of a consensus binding site for p53. Nat. Genet. 145-49. [DOI] [PubMed] [Google Scholar]
- 12.Espinosa, J. M., and B. M. Emerson. 2001. Transcriptional regulation by p53 through intrinsic DNA/chromatin binding and site-directed cofactor recruitment. Mol. Cell 857-69. [DOI] [PubMed] [Google Scholar]
- 13.Frederick, J. P., N. T. Liberati, D. S. Waddell, Y. Shi, and X.-F. Wang. 2004. Transforming growth factor β-mediated transcriptional repression of c-myc is dependent on direct binding of Smad3 to a novel repressive Smad binding element. Mol. Cell. Biol. 242546-2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Germain, S., M. Howell, G. M. Esslemont, and C. S. Hill. 2000. Homeodomain and winged-helix transcription factors recruit activated Smads to distinct promoter elements via a common Smad interaction motif. Genes Dev. 14435-451. [PMC free article] [PubMed] [Google Scholar]
- 15.Ho, J., and S. Benchimol. 2003. Transcriptional repression mediated by the p53 tumour suppressor. Cell Death Differ. 10404-408. [DOI] [PubMed] [Google Scholar]
- 16.Jones, T. A., J. Y. Zou, S. W. Cowan, and M. Kjeldgaard. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47(Pt 2)110-119. [DOI] [PubMed] [Google Scholar]
- 17.Kang, Y., C. R. Chen, and J. Massague. 2003. A self-enabling TGFbeta response coupled to stress signaling: Smad engages stress response factor ATF3 for Id1 repression in epithelial cells. Mol. Cell 11915-926. [DOI] [PubMed] [Google Scholar]
- 18.Kretzschmar, M., J. Doody, and J. Massague. 1997. Opposing BMP and EGF signalling pathways converge on the TGF-beta family mediator Smad1. Nature 389618-622. [DOI] [PubMed] [Google Scholar]
- 19.Lee, K. C., A. J. Crowe, and M. C. Barton. 1999. p53-mediated repression of alpha-fetoprotein gene expression by specific DNA binding. Mol. Cell. Biol. 191279-1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Levy, L., M. Howell, D. Das, S. Harkin, V. Episkopou, and C. S. Hill. 2007. Arkadia activates Smad3/Smad4-dependent transcription by triggering signal-induced SnoN degradation. Mol. Cell. Biol. 276068-6083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lozano, G., and R. Montes de Oca Luna. 1998. MDM2 function. Biochim. Biophys. Acta 1377M55-M59. [DOI] [PubMed] [Google Scholar]
- 22.Luo, K. 2004. Ski and SnoN: negative regulators of TGF-β signaling. Curr. Opin. Genet. Dev. 1465-70. [DOI] [PubMed] [Google Scholar]
- 23.Lyons, J. P., U. W. Mueller, H. Ji, C. Everett, X. Fang, J. C. Hsieh, A. M. Barth, and P. D. McCrea. 2004. Wnt-4 activates the canonical beta-catenin-mediated Wnt pathway and binds Frizzled-6 CRD: functional implications of Wnt/beta-catenin activity in kidney epithelial cells. Exp. Cell Res. 298369-387. [DOI] [PubMed] [Google Scholar]
- 24.Macias-Silva, M., W. Li, J. I. Leu, M. A. Crissey, and R. Taub. 2002. Up-regulated transcriptional repressors SnoN and Ski bind Smad proteins to antagonize transforming growth factor-beta signals during liver regeneration. J. Biol. Chem. 27728483-28490. [DOI] [PubMed] [Google Scholar]
- 25.Massague, J. 1998. TGF-beta signal transduction. Annu. Rev. Biochem. 67753-791. [DOI] [PubMed] [Google Scholar]
- 26.Massague, J., J. Seoane, and D. Wotton. 2005. Smad transcription factors. Genes Dev. 192783-2810. [DOI] [PubMed] [Google Scholar]
- 27.Massague, J., and D. Wotton. 2000. Transcriptional control by the TGF-beta/Smad signaling system. EMBO J. 191745-1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matsuura, I., N. G. Denissova, G. Wang, D. He, J. Long, and F. Liu. 2004. Cyclin-dependent kinases regulate the antiproliferative function of Smads. Nature 430226-231. [DOI] [PubMed] [Google Scholar]
- 29.Matsuura, I., G. Wang, D. He, and F. Liu. 2005. Identification and characterization of ERK MAP kinase phosphorylation sites in Smad3. Biochemistry 4412546-12553. [DOI] [PubMed] [Google Scholar]
- 30.Minsky, N., and M. Oren. 2004. The RING domain of Mdm2 mediates histone ubiquitylation and transcriptional repression. Mol. Cell 16631-639. [DOI] [PubMed] [Google Scholar]
- 31.Mizejewski, G. J. 2002. Biological role of alpha-fetoprotein in cancer: prospects for anticancer therapy. Expert Rev. Anticancer Ther. 2709-735. [DOI] [PubMed] [Google Scholar]
- 32.Murphy, M., J. Ahn, K. K. Walker, W. H. Hoffman, R. M. Evans, A. J. Levine, and D. L. George. 1999. Transcriptional repression by wild-type p53 utilizes histone deacetylases, mediated by interaction mSin3a. Genes Dev. 132490-2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nead, M. A., L. A. Baglia, M. J. Antinore, J. W. Ludlow, and D. J. McCance. 1998. Rb binds c-Jun and activates transcription. EMBO J. 172342-2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nguyen, T. T., K. Cho, S. A. Stratton, and M. C. Barton. 2005. Transcription factor interactions and chromatin modifications associated with p53-mediated, developmental repression of the alpha-fetoprotein gene. Mol. Cell. Biol. 252147-2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nishita, M., M. K. Hashimoto, S. Ogata, M. N. Laurent, N. Ueno, H. Shibuya, and K. W. Cho. 2000. Interaction between Wnt and TGF-beta signalling pathways during formation of Spemann's organizer. Nature 403781-785. [DOI] [PubMed] [Google Scholar]
- 36.Nomura, T., M. M. Khan, S. C. Kaul, H. D. Dong, R. Wadhwa, C. Colmenares, I. Kohno, and S. Ishii. 1999. Ski is a component of the histone deacetylase complex required for transcriptional repression by Mad and thyroid hormone receptor. Genes Dev. 13412-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oka, T., H. Tsuda, A. Scarpa, M. Sakamoto, and S. Hirohashi. 1992. p53 gene mutation spectrum in hepatocellular carcinoma. Cancer Res. 526358-6364. [PubMed] [Google Scholar]
- 38.Schmierer, B., and C. S. Hill. 2005. Kinetic analysis of Smad nucleocytoplasmic shuttling reveals a mechanism for transforming growth factor β-dependent nuclear accumulation of Smads. Mol. Cell. Biol. 259845-9858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sengupta, N., and E. Seto. 2004. Regulation of histone deacetylase activities. J. Cell. Biochem. 9357-67. [DOI] [PubMed] [Google Scholar]
- 40.Stroschein, S. L., W. Wang, S. Zhou, Q. Zhou, and K. Luo. 1999. Negative feedback regulation of TGF-beta signaling by the SnoN oncoprotein. Science 286771-774. [DOI] [PubMed] [Google Scholar]
- 41.Takebayashi-Suzuki, K., J. Funami, D. Tokumori, A. Saito, T. Watabe, K. Miyazono, A. Kanda, and A. Suzuki. 2003. Interplay between the tumor suppressor p53 and TGF beta signaling shapes embryonic body axes in Xenopus. Development 1303929-3939. [DOI] [PubMed] [Google Scholar]
- 42.Ueki, N., and M. J. Hayman. 2003. Direct interaction of Ski with either Smad3 or Smad4 is necessary and sufficient for Ski-mediated repression of transforming growth factor-beta signaling. J. Biol. Chem. 27832489-32492. [DOI] [PubMed] [Google Scholar]
- 43.Um, S. H., C. Mulhall, A. Alisa, A. R. Ives, J. Karani, R. Williams, A. Bertoletti, and S. Behboudi. 2004. Alpha-fetoprotein impairs APC function and induces their apoptosis. J. Immunol. 1731772-1778. [DOI] [PubMed] [Google Scholar]
- 44.van Grunsven, L. A., G. Verstappen, D. Huylebroeck, and K. Verschueren. 2005. Smads and chromatin modulation. Cytokine Growth Factor Rev. 16495-512. [DOI] [PubMed] [Google Scholar]
- 45.Wilkinson, D. S., S. K. Ogden, S. A. Stratton, J. L. Piechan, T. T. Nguyen, G. A. Smulian, and M. C. Barton. 2005. A direct intersection between p53 and transforming growth factor β pathways targets chromatin modification and transcription repression of the α-fetoprotein gene. Mol. Cell. Biol. 251200-1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wrana, J. L., and L. Attisano. 2000. The Smad pathway. Cytokine Growth Factor Rev. 115-13. [DOI] [PubMed] [Google Scholar]
- 47.Wu, J. W., A. R. Krawitz, J. Chai, W. Li, F. Zhang, K. Luo, and Y. Shi. 2002. Structural mechanism of Smad4 recognition by the nuclear oncoprotein Ski: insights on Ski-mediated repression of TGF-beta signaling. Cell 111357-367. [DOI] [PubMed] [Google Scholar]
- 48.Zawel, L., J. L. Dai, P. Buckhaults, S. Zhou, K. W. Kinzler, B. Vogelstein, and S. E. Kern. 1998. Human Smad3 and Smad4 are sequence-specific transcription activators. Mol. Cell 1611-617. [DOI] [PubMed] [Google Scholar]
- 49.Zhu, Q., A. R. Krakowski, E. E. Dunham, L. Wang, A. Bandyopadhyay, R. Berdeaux, G. S. Martin, L. Sun, and K. Luo. 2007. Dual role of SnoN in mammalian tumorigenesis. Mol. Cell. Biol. 27324-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.