Abstract
The c-Myb transcription factor regulates the proliferation and differentiation of hematopoietic cells, and activated alleles of c-myb induce leukemias and lymphomas in animals. Relatively minor changes in the structure of c-Myb protein change the genes that it regulates and can unleash its latent transforming activities. Here, quantitative assays were used to analyze the alternative splicing of human c-myb transcripts. We identified an array of variant transcripts, expressed in highly regulated, lineage-specific patterns, that were formed through the use of alternate exons 8A, 9A, 9B, 10A, 13A, and 14A. Expression levels of the different splice variant transcripts were regulated independently of one another during human hematopoietic cell differentiation, and the alternative splicing of c-myb mRNAs was increased in primary leukemia samples. The alternatively spliced c-myb transcripts were associated with polysomes and encoded a series of c-Myb proteins with identical DNA binding domains but unique C-terminal domains. In several types of assays, the variant c-Myb proteins exhibited quantitative and qualitative differences in transcriptional activities and specificities. The results suggest that the human c-myb gene encodes a family of related proteins with different transcriptional activities. Enhanced alternative splicing may be a mechanism for unmasking the transforming activity of c-myb in human leukemias.
The c-myb gene is the normal, cellular counterpart of v-myb, the oncogenic component of avian myeloblastosis virus (AMV) that transforms immature myeloid cells and induces myeloid and lymphoid leukemias in animals (1, 25, 26). Several types of evidence suggest that c-myb plays a critical role in regulating hematopoietic cell differentiation, proliferation, and function (25, 26). The c-myb gene is duplicated and is a target for chromosomal translocation in a subset of human leukemias, suggesting that c-myb is an oncogene in human cancers (4). The c-Myb protein is the founding member of a family of transcription factors that also includes the related proteins A-Myb (MYBL1) and B-Myb (MYBL2). All the Myb proteins are structurally similar and have nearly identical DNA binding domains near their N termini, but they are more divergent in their central transcriptional activation domains and C-terminal negative regulatory domains. Although c-Myb, A-Myb, and B-Myb can bind the same DNA sequences and activate the same promoters in reporter gene assays (31), the three proteins are differentially expressed, and mouse knockout experiments have shown that they have different biological roles (23, 36, 37). In addition, microarray assays showed that each protein activates a unique set of genes in human cells (30), and the differences in the activities of these proteins have been mapped primarily to differences in their unique transcriptional activation and C-terminal domains (18). Thus, posttranslational modifications or other changes that affect the transcriptional activation domains of Myb proteins may have profound effects on their abilities to regulate specific genes (20, 27).
Alternatively spliced c-myb transcripts have been detected in human, mouse, and chicken hematopoietic cells (33, 34, 40). The best-characterized of these transcripts includes an alternate exon that encodes a variant of c-Myb with a 121-amino-acid in-frame insertion in the transcriptional activation domain (7, 32, 34, 42). Although biological roles for the variant c-Myb proteins remain unclear, alternatively spliced forms of chicken c-myb have increased transcriptional and transforming potential (42). For example, a variant of mouse c-myb formed through alternative RNA splicing enhanced cell survival in hematopoietic cell assays in which wild-type c-myb accelerated cell death (15). Thus, alternative RNA splicing permits the c-myb gene to encode multiple versions of the c-Myb transcription factor with unique biological activities.
Several recent studies have suggested that relatively minor changes in the transcriptional activation domain of c-Myb can dramatically affect its transcriptional activity. Since the alternative splicing of c-myb gene products is a mechanism for producing variants of c-Myb with different transcriptional activation domains, we established qualitative and quantitative assays to begin investigating the extent and the significance of alternative RNA splicing for the human c-myb gene. We found that normal primary human hematopoietic and leukemia cells produce a large and diverse set of c-myb transcripts that show cell type-specific expression patterns and that the alternately spliced c-myb transcripts encode proteins with distinct activities. The results have significant implications for understanding the role of Myb proteins in the regulation of hematopoiesis and for determining how wild-type and variant c-Myb proteins contribute to leukemogenesis.
MATERIALS AND METHODS
Cell lines and primary hematopoietic cells.
The human hematopoietic cell lines K562 (ATCC CCL-240), THP-1 (ATCC TIB-202), Jurkat (ATCC TIB-152), and KG-1 (ATCC CCL-246) and the fibroblast cell line 293 (ATCC CRL-1573) were cultured as suggested by the American Type Culture Collection. The avian monocyte cell line HD11 was cultured in Dulbecco's modified Eagle's medium supplemented with 2% chicken serum, 8% fetal bovine serum, and 1× penicillin-streptomycin solution. Cell culture reagents were purchased from Invitrogen (Carlsbad, CA).
Buffy coats from healthy donors were obtained from United Blood Services (Albuquerque, NM). Peripheral blood mononuclear cells were isolated by Ficoll (Amersham, Piscataway, NJ) density centrifugation, and cell populations were depleted of monocytes by differential adherence. Protein or RNA was isolated from half of the nonadherent peripheral blood leukocytes (PBLs) immediately, while the remaining cells were cultured in Iscove's modified Dulbecco's medium (IMDM) with 10 U each of interleukin-2 (IL-2; PeproTech, Rocky Hill, NJ) and phytohemagglutinin (PHA; Gibco, Carlsbad, CA)/ml for 4 days prior to protein and RNA isolation.
Cryopreserved CD34+ cells derived from cytokine-mobilized peripheral blood were obtained from the Fred Hutchinson Cancer Research Center Large-Scale Cell Processing Core. For the CD34+ cell assays, 106 CD34+ cells per ml were cultured in IMDM supplemented with IL-3 (20 ng/ml), IL-6 (20 ng/ml), and stem cell factor and FLT-3 ligand (100 ng/ml each) (CC 100 cytokine cocktail; Stem Cell Technologies, Vancouver, Canada) for 4 days. Half of the cells were then cultured for 6 days in long-term myeloid medium (Stem Cell Technologies, Vancouver, Canada) supplemented with IL-3 and granulocyte-monocyte colony-stimulating factor (PeproTech, Rocky Hill, NJ) for myeloid differentiation as described previously (38). The remaining cells were cultured for 5 days in IMDM supplemented with IL-6, stem cell factor, and erythropoietin as described previously (14). Total RNA from CD34+ cell experiments was isolated using RNAzol Bee (Tel-Test Incorporated, Friends, TX) at 0, 4, and 10 days postculture. Pediatric T-cell acute lymphoblastic leukemia (T-ALL) samples and adult acute myeloid leukemia samples were obtained from the Children's Oncology Group and the Southwest Oncology Group, respectively.
RNA isolation and real-time quantitative PCR (qPCR) assays.
The isolation of total and cytoplasmic RNAs was performed using the RNeasy mini kit (Qiagen, Valencia, CA), and poly(A)+ RNA was purified from the cytoplasmic RNA using the poly(A)+ mRNA isolation kit (Miltenyi Biotech, Auburn, CA). cDNA synthesis using a first-strand cDNA synthesis kit (Invitrogen, Carlsbad, CA) and SYBR green-based real-time PCR using a Dynamo PCR kit (New England Biotech, Waltham, MA) were done as suggested by the manufacturers. Analyses of PCR efficiency and melting-point curves of PCR products for each SYBR green primer pair were performed to verify that the primers were specific (see Fig. S2 in the supplemental material), and PCR was performed in triplicate using primers described in Table S1 in the supplemental material. The results of relative gene expression assays were normalized to the level of β2-microglobulin gene expression, and the data were analyzed using the comparative threshold cycle method (21). Plasmid standard curves (106 to 102 copies) were generated for experiments measuring absolute (copy number) expression values.
Polyribosome analysis.
Transcripts associated with polyribosomes were analyzed as described previously (9). Briefly, K562 cell cytoplasmic fractions were layered onto 40 to 60% Nyzode (Sigma, St. Louis, MO) gradients containing either cycloheximide (for ribosome clamping) or EDTA (for ribosome release). After overnight centrifugation, 24 gradient fractions were recovered and RNA was isolated using RNAzol Bee (Tel-Test, Friends, TX). RNA from each fraction was analyzed using a total RNA Nano Chip kit (Agilent Technologies, San Jose, CA). Virtual gels showing tRNA and 18S and 28S rRNA were prepared using the Agilent software, and transcript levels were measured by qPCR as described above. The percentage of each transcript in each gradient fraction was calculated by dividing the number of transcript copies in the gradient fraction by the sum of the numbers of transcript copies in all the gradient fractions analyzed.
Expression plasmids, analysis of protein production, and transcription assays.
PCR products containing unique alternative exons were cloned in frame into a human c-Myb expression vector (pCDNA3; Invitrogen, Carlsbad, CA). For transfection assays, different amounts of expression plasmids (250 ng of the 14A variant c-Myb plasmid, the 13A variant c-Myb plasmid, and the 9B variant c-Myb plasmid, 180 ng of the wild-type c-Myb plasmid and the 8A variant c-Myb plasmid, and 90 ng of the 10A variant c-Myb plasmid and the 9A variant c-Myb plasmid), balanced by an empty vector, were used to equalize the levels of the proteins detected by Western blotting. The avian monocyte cell line HD11 was transfected using Superfect as recommended by the manufacturer (Qiagen, Valencia, CA). For endogenous mim-1 gene assays, HD11 cells were transfected as described above, total RNA was isolated 48 h later from half the cells, and the remainder of the cells were lysed in boiling 2× sodium dodecyl sulfate buffer for protein analysis. The expression of mim-1 RNA was analyzed using SYBR green-based real-time PCR, and relative mim-1 gene expression levels were calculated using glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene expression levels to normalize for equivalent cDNA levels. Primer sequences for mim-1 and the GAPDH gene are given in Table S1 in the supplemental material. Proteins were analyzed by Western blotting as described previously by using a polyclonal antibody raised against the conserved DNA binding domain of c-Myb (8). For reporter gene activation assays, triplicate samples of cells in 24-well dishes were transfected with 350 ng of the c-Myb-responsive EW5 reporter gene, along with 50 ng of the wild-type c-Myb expression plasmid (or the splice variant expression plasmids indicated in Fig. 6 using the same ratios listed above) and 10 ng of a renilla luciferase plasmid as a control for transfection efficiency. Luciferase levels were analyzed 2 days later with a dual luciferase kit (Promega, Madison, WI).
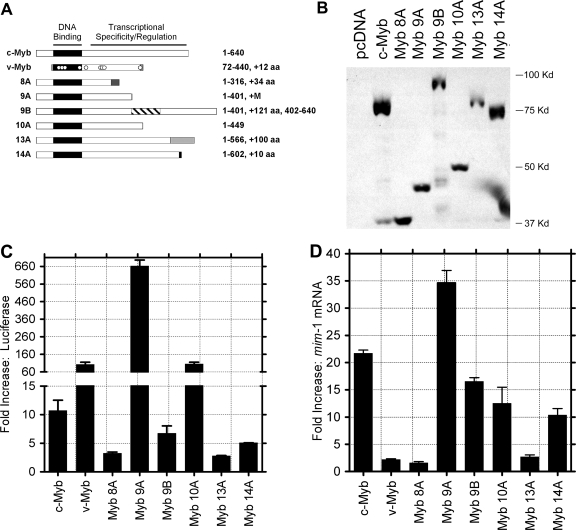
FIG. 6.
Variants of c-Myb display unique transcriptional activities. (A) Diagrams of c-Myb proteins encoded by normal c-myb, the v-myb oncogene from AMV, and the alternatively spliced forms of c-myb containing exon 8A, 9A, 9B, 10A, 13A, or 14A. The highly conserved Myb DNA binding domain is labeled and shaded black. The transcriptional specificity and regulation domain of c-Myb (labeled) is affected by the alternative exons, as indicated by shaded boxes. The numbers on the right indicate amino acid (aa) residues from c-Myb plus novel amino acids encoded by the alternative exons. (B) Western blot analysis. HD11 monocyte cells were transfected with plasmids expressing the indicated c-myb splice variants. Whole-cell extracts were isolated 2 days later for Western blot analysis using anti-Myb antibodies to detect the expressed proteins. The numbers on the right show the migration of molecular size markers. (C) Activation of a c-myb-responsive reporter gene. Cells were cotransfected with plasmids expressing the indicated Myb proteins and a Myb-responsive reporter plasmid, and extracts were prepared and assayed for Myb-dependent luciferase activity after 2 days. Results are plotted as the average normalized luciferase activities from triplicate measurements in a representative experiment performed twice. Error bars represent standard deviations. Note that the scale has been split to show the large variation in the activities of the different c-Myb proteins. (D) Activation of an endogenous target gene. Chicken monocyte HD11 cells were transfected with the indicated c-Myb expression plasmids. After 2 days, RNA was isolated and qPCR was used to measure the levels of the Myb-regulated mim-1 gene and the GAPDH control gene. The plots show the levels of induction (n-fold) of mim-1, normalized by comparison to the GAPDH gene, relative to the activation by an empty vector control. Error bars represent standard deviations of triplicate qPCR measurements. Similar results were obtained in two independent experiments.
Hep27, CXC chemokine receptor 4 (CXCR-4), cyclin A1, CFTR/MRP, and keratin-16 gene transcript levels in MCF-7 cells after adenoviral transduction were determined as previously described (30). Briefly, adenoviral vectors expressing the c-Myb splice variants indicated in Fig. 7 and coexpressing green fluorescent protein as a marker were constructed using the Adeasy system (22). More than 80% of MCF-7 cells were transduced, as assessed by green fluorescent protein expression. RNA was isolated from 90% of the cells by using an RNeasy mini kit (Qiagen, Valencia, CA), while the remaining 10% of the cells were boiled in sodium dodecyl sulfate buffer for Western blot analysis. Relative expression levels of the indicated genes were evaluated by quantitative reverse transcription-PCR using an assay-on-demand Taqman kit (Applied Biosystems, CA), and the results were analyzed as described above.
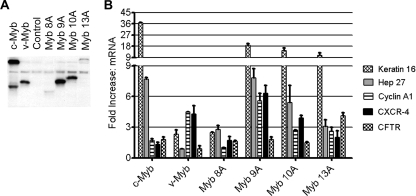
FIG. 7.
Variants of c-Myb display qualitatively different transcriptional properties. (A) Western blot analysis. MCF-7 cells were transduced with adenoviral vectors expressing the indicated c-Myb variants. Whole-cell extracts were isolated from 20% of the cells 18 h posttransduction for Western blot analysis using anti-Myb antibodies to detect the expressed proteins. (B) Activation of endogenous target genes. RNA was isolated from the remaining MCF-7 cells, and qPCR was used to measure the levels of expression of a panel of genes regulated by c-Myb (keratin-16 and Hep27 genes) or v-Myb (cyclin A1 and CXCR-4 genes) and the GAPDH control gene. The CFTR gene was activated by c-Myb in primary monocyte cells but not MCF-7 cells. The plot shows the levels of induction (n-fold) of the indicated genes, normalized by comparison to the GAPDH gene, relative to the activation by a control adenoviral vector. The data presented are representative of an experiment performed two times, and error bars represent standard deviations of triplicate qPCR measurements. Note that the scale has been split to show the large variation in activities of the different c-Myb proteins on the various target genes.
RESULTS
Alternative splicing of human c-myb RNA is frequent and cell type specific.
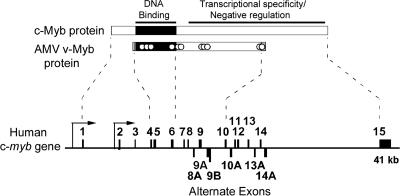
The human c-myb gene encompasses 15 exons on chromosome 6q22-q23 (Fig. 1). Exons 4 to 6 encode the highly conserved DNA binding domain (25), while exons 7 to 15 encode the C-terminal domains involved in transcriptional activation, specificity, and regulation (17, 18). The region encoded by exons 4 to 10 corresponds to the protein encoded by the v-myb oncogene from AMV and includes all the activities required for transformation and leukemogenesis. Several previous studies have investigated an alternatively spliced RNA containing exon 9B, which in its mouse and chicken forms is referred to as 9A (7, 34). However, a search of entries for the human c-myb gene in the GenBank database (accession number U22376) suggested the presence of additional alternative exons (8A, 9A, 10A, 13A, and 14A), some of which have also been detected by PCR methods or analyses of expressed sequence tag clones (5).
FIG. 1.
Structure of the human c-myb gene. The 640-amino-acid c-Myb protein (top) contains a highly conserved DNA binding domain (shaded black) near the N terminus and a large C-terminal domain that controls transcriptional specificity and is the target of numerous regulatory pathways. The chicken v-Myb protein is a truncated and mutated version of c-Myb that lacks the N and C termini and that harbors 11 point mutations (indicated by white dots). The human c-myb gene (bottom) spans 41,000 nucleotides and contains 15 exons (shaded black, above the line). The arrows represent the 5′ and intronic promoters. The alternative exons are shown below the line and encode proteins that differ in the transcriptional specificity and negative regulation domain of c-Myb (dotted lines).
Initial experiments using conventional reverse transcription-PCR assays with primers specific for the predicted alternative exons showed that each exon could be detected in RNA isolated from CD34+ cells and bone marrow aspirates from leukemia patients, suggesting that hematopoietic cells expressed a wider-than-expected range of c-myb gene products (see Fig. S1 in the supplemental material). Therefore, we utilized quantitative reverse transcription and real-time PCR assays with purified cytoplasmic, poly(A)+ mRNA as the template to obtain accurate expression data for various c-myb splice products. First, the absolute total levels of c-myb mRNAs were measured using primer sets designed to detect all variants of c-myb. As shown in Fig. 2A, Jurkat T-cell leukemia cells expressed the highest total levels of c-myb transcripts per nanogram of reverse-transcribed poly(A)+ RNA, followed by the myeloid leukemia cell line KG-1 and normal peripheral blood lymphocytes (PBLs) stimulated with IL-2 and PHA, each of which expressed c-myb at about one-third the level of expression in the Jurkat cells. The erythroid leukemia cell line K562 expressed sixfold less c-myb mRNA than the Jurkat cells.
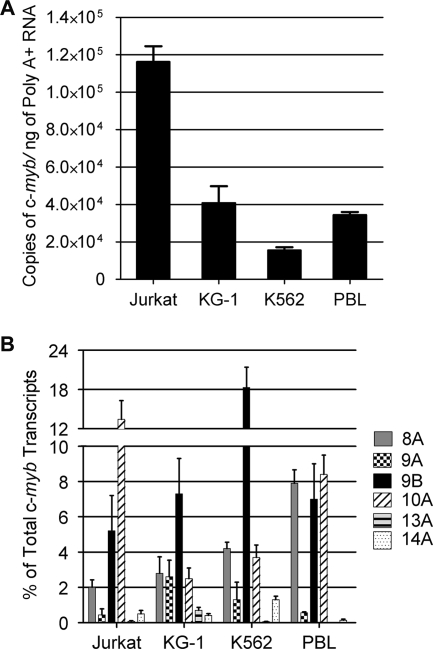
FIG. 2.
The c-myb splice variants exhibit a cell type-specific expression pattern. (A) Total numbers of c-myb mRNA copies from various hematopoietic cells. Absolute levels of c-myb RNA were detected by performing reverse transcription and SYBR green-based real-time PCR with primers from exons 14 and 15, starting with 20 ng of cytoplasmic, polyadenylated RNA from the Jurkat T-cell line, the KG-1 myeloid leukemia cell line, or the K562 erythroid leukemia cell line or from PBLs after 4 days of activation. Total numbers of c-myb copies were determined using a plasmid standard curve. The bar graph shows the average numbers of c-myb mRNA copies detected per nanogram of starting RNA in two independent experiments, each with triplicate assays. Error bars represent standard deviations. (B) Percentages of c-myb transcripts containing alternative exons. Real-time PCR was performed as described above using primers to detect individual alternative exons, and the results are plotted as percentages of total c-myb transcripts containing the indicated exons. Error bars represent standard deviations.
For transcripts containing alternative exons, specific primers were used in qPCR assays, and plasmid standard curves were used to measure the absolute amount of each type of transcript as a percentage of total c-myb transcripts. In these assays, the percentage of alternately spliced c-myb isoforms varied from ∼15% of total c-myb transcripts in KG-1 cells to ∼30% of total c-myb transcripts in K562 cells. Approximately 22% of the c-myb transcripts in PBLs and Jurkat cells contained alternative exons (Fig. 2B). While the total fraction of alternatively spliced c-myb isoforms did not vary dramatically among cell types, each type of cell expressed a unique, qualitatively different pattern of alternatively spliced c-myb products (Fig. 2B). For example, exon 9B-containing transcripts were about 5% of the total in Jurkat cells but more than 18% of the c-myb transcripts in K562 cells. In contrast, exon 10A was detected in about 12% of the Jurkat cell c-myb transcripts but in only 2, 4, and 8% of the transcripts in the KG-1 and K562 cells and the PBLs, respectively. The 8A exon was detected in about 8% of transcripts in PBLs but only 2 to 4% of those in the leukemia cell lines. In all cell types, transcripts containing the 9A, 13A, or 14A exon were present at very low levels. The qPCR results presented here were confirmed using a shotgun cloning and sequencing approach (see Table S2 in the supplemental material), which gave similar results. From these quantitative assays, it appears that approximately 15 to 20% of the c-myb transcripts in primary human hematopoietic cells and several common cell lines contain at least one alternative exon, and the relative levels of different transcripts were variable and cell type specific. Thus, the repertoire of c-myb transcripts is complex and varies in different hematopoietic cell types.
Alternative splicing of c-myb transcripts during hematopoietic cell differentiation.
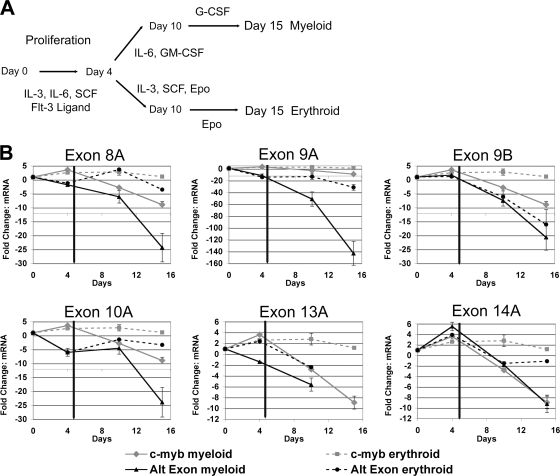
The comparisons of different hematopoietic cell lines described above suggested that the alternative splicing of c-myb transcripts was regulated in a lineage-specific manner. We next analyzed the expression of various splice variants to determine whether c-myb alternative splicing was regulated during normal hematopoiesis. Primary human hematopoietic CD34+ cells were cultured in vitro using a protocol that stimulated proliferation for several days, and then the cells were divided and transferred to conditions designed to stimulate differentiation toward granulocytes or erythrocytes for an additional 11 days (Fig. 3A). This standard CD34+ differentiation protocol was previously used to investigate changes in gene expression and alternative splicing (6, 38). At various time points, the relative total c-myb transcript levels or levels of variant transcripts containing alternate exons were analyzed by qPCR. Differentiation-specific cell surface markers were assayed using flow cytometry in order to monitor the effectiveness of the differentiation protocols (data not shown). The results are shown in Fig. 3B, where the relative levels of each type of c-myb variant are compared to the changes in total c-myb transcript levels and plotted as the amounts of change (n-fold) compared to the starting levels in the unstimulated CD34+ cells. (Note that, for illustrative purposes, each panel in Fig. 3B is plotted with a different scale, and the total c-myb transcript levels are plotted in all the panels.) As shown in Fig. 3B, the total levels of c-myb transcripts increased severalfold during the initial proliferation phase of the experiment (days 1 to 4) and remained fairly constant when the cells were switched to erythroid differentiation medium. However, total c-myb levels in the cells that were switched to granulocyte differentiation conditions (days 4 to 15) declined about ninefold, which is consistent with data in previously published reports (2).
FIG. 3.
Independent and lineage-specific expression of c-myb transcripts containing alternate exons during in vitro differentiation of CD34+ hematopoietic cells. (A) Differentiation protocol for CD34+ cells. Populations of primary human CD34+ cells were expanded (days 1 to 4) and then differentiated toward the myeloid or erythroid lineage (days 5 to 15) by using the indicated cytokines. SCF, stem cell factor; G-CSF, granulocyte colony-stimulating factor; Epo, erythropoietin. (B) Results from exon-specific qPCR analysis of differentiating CD34+ cells. RNA was isolated at the indicated time points, and qPCR was used to measure the levels of c-myb transcripts containing the indicated alternate exons. The graphs show the change (n-fold) in mRNA levels for the alternatively spliced c-myb variants, normalized by comparison to β2-microglobulin gene expression levels, relative to the mRNA levels in unstimulated CD34+ (day 0) cells. The solid vertical line on day 4 represents the transition of the cells from proliferation toward differentiation. The solid black lines indicate splice variant levels in cells undergoing myeloid differentiation, while the dashed black lines represent those in cells undergoing erythroid differentiation. For comparison purposes, the relative change (n-fold) in total c-myb mRNA levels was plotted on each graph and is represented by the gray lines. Exon 13A was not detectable after 10 days in culture. Error bars represent standard deviations of triplicate qPCR measurements. Similar results were obtained in two independent experiments. Note that the scale on the y axis is different in each graph to show the large variation in relative mRNA levels for the different c-myb splice variants. Alt exon, alternative exon.
The analysis of the alternatively spliced variants showed that each was expressed in a unique pattern during this proliferation and differentiation protocol. For example, the initial proliferation phase led to increases in the levels of transcripts containing exon 9B, 13A, or 14A (twofold, threefold, or sixfold, respectively), similar to the increase in total c-myb transcript levels. In contrast, during the same period, levels of transcripts containing exon 8A, 9A, or 10A decreased 2-, 13-, and 6-fold, respectively, suggesting that the levels of the individual alternative transcripts were regulated independently. Even bigger differences were noted when the cells were stimulated to differentiate. Granulocyte differentiation conditions caused the levels of all the alternative exon-containing transcripts to decrease at least 2-fold (for those containing exon 13A or 14A) and as much as 50-fold (for those containing exon 9A) by day 10, although total c-myb transcript levels declined only about 2-fold during that time period. The erythroid differentiation conditions, which led to very little change in total levels of c-myb transcripts, caused the most unique changes in the levels of some of the variants. For example, by day 15 of erythroid differentiation, levels of transcripts containing exon 9A or 9B had decreased about 15- to 30-fold while those of transcripts containing exon 8A, 10A, or 14A had decreased only 1.5- to 3-fold. These results suggest that changes in the patterns of c-myb alternative RNA splicing occur during normal hematopoietic cell differentiation, at least under these in vitro conditions, and occur in a lineage- and differentiation-specific manner. The results also imply that different types of hematopoietic cells express different combinations of normal and variant forms of c-Myb proteins, which may have unique transcriptional activities.
Altered levels of c-myb transcripts in primary leukemia samples.
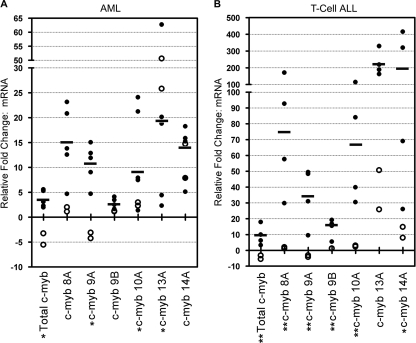
Truncated versions of c-myb have oncogenic activity in animals. Since the alternatively spliced forms of c-myb transcripts had the potential to encode truncated versions of the protein (see below), we tested whether expression levels of the alternative c-myb transcripts were altered in leukemia samples. RNA was prepared from normal bone marrow samples or from bone marrow samples collected from patients with pediatric T-ALL or adult acute myelogenous leukemia (AML), and then total levels of c-myb transcripts and levels of the splice variants were measured using qPCR assays and compared to the levels observed in PHA-activated PBLs. Consistent with data in previous reports (4, 16), total c-myb mRNA was overexpressed 13- and 40-fold in AML and T-ALL, respectively, compared to the level of expression in normal bone marrow samples (Fig. 4). Transcripts containing exon 8A or 10A were overexpressed at levels similar to the total c-myb overexpression levels in both AML and T-ALL, whereas relative levels of transcripts containing exon 9A were significantly increased in T-ALL samples (as much as 180-fold compared to those in normal bone marrow samples). In contrast, levels of transcripts containing exon 9B, 13A, or 14A relative to total c-myb transcript levels in leukemia samples were decreased compared to those in normal bone marrow samples. A leukemia-specific pattern of c-myb splice variant expression was also observed in a pediatric B-cell ALL sample compared to the expression patterns in CD34+ and PHA-activated PBLs by using a shotgun cloning and sequencing approach (see Table S2 in the supplemental material). The data suggest that the c-myb gene may be a target for aberrant splicing during leukemogenesis, leading to the expression of a leukemia-specific signature of c-myb transcripts.
FIG. 4.
The expression of the c-myb splice variants is altered in primary leukemia bone marrow samples. qPCR assays using primers for the indicated alternative exons were performed on bone marrow aspirates from patients with AML (n = 5; filled circles) or pediatric T-ALL (n = 4; filled circles) or from normal donors (n = 2; empty circles). Each dot represents the average of triplicate measurements from a single sample, and the horizontal bars represent the median values for the leukemia samples. The plots show the differences (n-fold) in the levels of each transcript compared to the levels in PHA-activated PBLs. Statistical analysis was performed using the two-sample F-test for variances. The c-myb splice variants that gave significantly different results for leukemia samples and samples from normal donors are labeled with * (P ≤ 0.05), and highly significant differences are indicated with ** (P ≤ 0.01).
Alternately spliced c-myb transcripts are associated with polyribosomes.
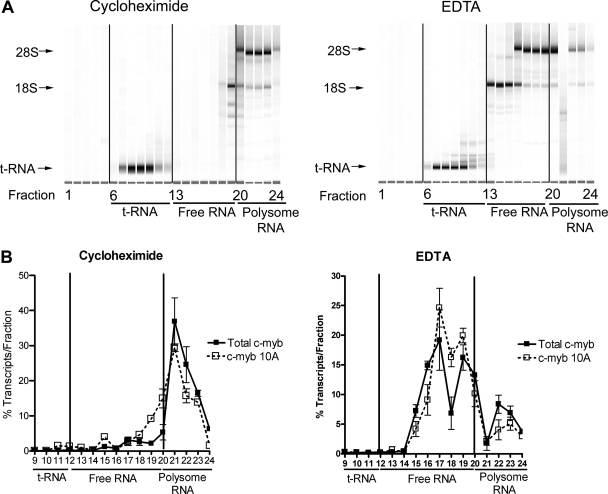
The expression profiles of the alternatively spliced c-myb transcripts suggested that the transcripts were individually regulated in specific patterns in different lineages and during hematopoietic cell differentiation. We next investigated whether the alternatively spliced c-myb variants were associated with polyribosomes, which would indicate that they are likely to be translated to produce different forms of c-Myb protein. Nyzode density gradients were used to fractionate the polysome-containing cytoplasmic extracts derived from K562 cells. In order to accurately assess polysome-associated mRNA and non-polysome-associated (free) mRNA density fractions, we compared samples analyzed on gradients containing cycloheximide, which prevents ribosome dissociation from mRNA, to samples run on gradients containing EDTA, which disrupts the ribosome-RNA complexes. Actively translated mRNAs are expected to shift to lower-density fractions in EDTA gradients than in cycloheximide gradients, since EDTA causes the more rapidly sedimenting polysomes to dissociate into mRNAs and free ribosomes. RNA samples from the gradient fractions were analyzed by electrophoresis and qPCR. As shown in Fig. 5A, RNA fractionated on the cycloheximide gradient yielded three distinct sets: tRNA (fractions 7 to 12), free RNA (fractions 13 to 19), and polysome-associated RNA (fractions 20 to 24). Treatment with EDTA did not change the migration of the tRNA, but it did cause a dramatic shift of the 18S and 28S rRNAs to lower-density fractions (fractions 13 to 16), confirming that the EDTA treatment disrupted the polysomes.
FIG. 5.
The c-myb splice variants associate with polysomes. Cytoplasmic extracts isolated from K562 cells were fractionated on Nyzode density gradients containing cycloheximide (to inhibit polysome-RNA dissociation) or EDTA (to release polysomes from RNA). The gradients were separated into 24 fractions (fraction 1, top; fraction 24, bottom). (A) RNA profiles. RNA samples isolated from the gradients containing cycloheximide (left panel) or EDTA (right panel) were analyzed as described in Materials and Methods. Fractions 20 to 24 contained polysomes, as indicated by the presence of both the 18S and 28S rRNAs. Non-polysome-associated RNA was present in fractions 13 to 19, between the tRNA in fractions 7 to 12 and the ribosome-containing fractions. Note the shift toward lower-density fractions for the 18S ribosomal subunit in the EDTA gradient (compare fractions 13 to 19 in EDTA and cycloheximide gradients), confirming that the EDTA treatment disrupted the polysomes. (B) Results from qPCR analysis. Relative total c-myb transcript levels or levels of transcripts containing exon 10A were detected in the gradient fractions by qPCR. Copy numbers of each transcript were determined using plasmid standard curves. The plots show the percentages of transcripts recovered from each fraction. Note the shift in both total c-myb and exon 10A transcripts in the EDTA gradient. Nearly identical results were obtained for transcripts containing exons 8A, 9A, 9B, 13A, and 14A (not shown).
When the cycloheximide gradient fractions were analyzed by qPCR, the majority of total c-myb transcripts and the alternative exon-containing transcripts were found in the polysome-associated fractions (Fig. 5B, top panel). However, both types of transcripts shifted to the lower-density fractions containing the free mRNAs in the EDTA gradients. Although Fig. 5B shows the data only for exon 10A-containing transcripts, nearly identical results were obtained with the other alternative exon-containing transcripts (data not shown). Overall, a vast majority of the alternatively spliced transcripts (68 to 94%) were associated with polysomes in human K562 cells. Even the rarest c-myb splice variants were predominantly associated with polyribosomes. The results suggest that the c-myb splice variants are likely to be translated into proteins and imply that human hematopoietic cells produce a mixture of different c-Myb proteins, which may have unique activities.
Alternative splicing produces c-Myb proteins with different activities.
All of the alternative c-myb exons are in the region of the gene that encodes the transcriptional activation, specificity, and regulatory functions of c-Myb (18, 30). That part of c-Myb has been shown to be very sensitive to mutations: numerous studies have shown that c-Myb proteins with parts of that domain mutated or truncated have dramatically altered transcriptional activities (18, 20, 30). The c-myb splice variants are predicted to encode proteins with altered transcriptional activation and specificity domains, raising the possibility that they may display unique transcriptional activities.
Figure 6A summarizes the differences in the proteins encoded by the normal c-myb transcript, by the v-myb oncogene from AMV, and by the variants of c-myb containing the alternative exons described in this paper. Compared to c-Myb, the AMV v-Myb protein is truncated at both the N and C termini and has numerous point mutations, many of which are important for its transforming activity and which affect its ability to activate specific target genes (18). Each of the alternatively spliced c-myb variants described above is predicted to encode a different version of c-Myb, with the same N terminus and DNA binding domain as those of the normal c-Myb but with a unique C terminal domain. The transcript containing exon 8A would encode a protein containing the first 316 amino acids of c-Myb, followed by 34 novel amino acids. The 9A variant-encoded protein includes the first 401 amino acids of c-Myb, plus one additional methionine residue. Exon 9B leads to the insertion of 121 additional amino acids in the middle of the transcriptional activation and specificity domain. The exon 10A version includes 449 amino acids of c-Myb, prematurely truncated without any new amino acids. The addition of exon 13A produces a protein with 566 amino acids from c-Myb fused to a novel 100-amino-acid domain at the C terminus. Finally, adding exon 14A leads to a protein with the first 602 amino acids of c-Myb fused to 10 novel amino acids at the C terminus.
The activities of the normal and variant proteins were tested in several types of transcription assays to determine whether the altered C-terminal domains affected these activities. First, plasmids expressing normal c-Myb and the six variant proteins produced through alternative splicing were constructed. As shown in Fig. 6B, when HD11 monocyte cells were transfected with the plasmid vectors expressing the different splice variants, Myb proteins of the expected sizes, ranging from the 40,000-molecular-weight protein encoded by the 8A variant (lane 3) to the 83,600-molecular-weight protein encoded by the 9B variant (lane 5), were produced.
We first tested the activities of the various Myb proteins by using a well-established cotransfection assay incorporating a Myb-responsive reporter gene. Myb-negative HD11 cells were transfected with plasmids expressing normal and variant c-Myb proteins, along with a Myb-responsive reporter plasmid containing five high-affinity Myb binding sites from the Myb-regulated mim-1 gene promoter upstream of a minimal promoter and a luciferase reporter gene. As shown in Fig. 6C, the expression of wild-type c-Myb resulted in an approximately 10-fold increase in reporter gene activity compared to the vector (cDNA) control. In contrast, the expression of the v-Myb protein from AMV led to >100-fold activation, illustrating that the oncogenic mutations in v-Myb, including the changes in the C-terminal domain, lead to greater activity in this assay. Although they were expressed at similar levels, the six variant forms of c-Myb, produced from transcripts incorporating alternative exons, had widely different transcriptional activities. The proteins encoded by the 9B and 14A variants activated the reporter gene sevenfold and fivefold, respectively, and so had activities similar to that of normal c-Myb. The 8A variant protein was less active than normal c-Myb and activated the reporter gene only about 2.5-fold. Weakest of all was the 13A variant protein, which had little detectable activity in this assay. However, two proteins, the 10A and 9A variants, activated much better than normal c-Myb, stimulating the reporter gene about 10-fold and nearly 60-fold better than normal c-Myb and, thus, exhibiting activity similar to that of the oncogenic v-Myb protein. Since protein expression levels in the transfected HD11 cells were nearly equivalent (Fig. 6B), the nearly 600-fold range in transcriptional activities observed is likely due to differences in the C-terminal domains of the c-Myb splice variants.
The mim-1 gene is a well-characterized Myb-regulated target gene that can be activated by the ectopic expression of c-Myb, but not the oncogenic v-Myb allele, in avian myeloid cells (11, 19, 20, 28, 29). Interestingly, c-Myb, but not v-Myb, is able to activate the endogenous mim-1 gene, while both c-Myb and v-Myb are able to activate plasmid-based reporter genes containing the mim-1 promoter, suggesting that the regulation of the endogenous gene is more restrictive, possibly requiring protein-protein interactions that are disrupted by the mutations in v-Myb (11, 18, 19, 20). We next tested the abilities of the variant c-Myb proteins to activate the mim-1 gene to see if they behaved qualitatively more like c-Myb or v-Myb. Briefly, the HD11 myeloid cell line was transfected with the various Myb expression vectors, and the resulting mim-1 gene expression was measured by qPCR 2 days later. As shown in Fig. 6D, the endogenous mim-1 gene was activated about 22-fold in cells transfected with the vector expressing wild-type c-Myb compared to mim-1 expression in cells transfected with a v-Myb expression vector or a control vector. The c-Myb splice variants had very different activities in this assay, ranging from little or no ability to activate the mim-1 target gene (8A and 13A variants) to activity much higher than that of c-Myb (the 9A variant gave about 35-fold activation). Several variants, those corresponding to exons 9B, 10A, and 14A, activated the mim-1 gene between 10- and 18-fold and so had activities that were comparable to that of normal c-Myb.
Recently, microarray assays have identified a number of human genes that are activated or repressed by different Myb proteins in mamma-derived cells (18, 20, 30). To investigate if the unique transcriptional properties observed for the c-myb splice variants in hematopoietic cells also occurred in other c-myb-expressing cell types, we tested the abilities of the variant proteins to activate several target genes in the MCF-7 mammary cell line. Briefly, adenoviral vectors expressing v-Myb, c-Myb, and the splice variants corresponding to exons 8A, 9A, 10A, and 13A were constructed and used to transduce the MCF-7 mammary cell line. After 18 h, Western blot analysis was used to check protein levels and qPCR was performed to test the expression of genes regulated by c-Myb (Hep27 and keratin-16 genes) and those regulated by v-Myb (cyclin A1 and CXCR-4 genes) (20, 30). Figure 7A shows that the 9A and 10A variants were expressed at levels comparable to those of wild-type c-Myb and v-Myb and that the 8A and 13A variants were expressed at somewhat lower levels in MCF-7 cells. When the gene expression assays were performed, each of the Myb proteins displayed a unique pattern of target gene activation. For example, the 9A variant activated the keratin-16 and Hep27 genes like c-Myb but also strongly activated the cyclin A1 and CXCR-4 genes, which c-Myb did not activate but which were activated by v-Myb. Thus, the 9A variant appeared to have some activities that resembled those of normal c-Myb and others like those of v-Myb. The 10A variant had activities similar to those of the 9A variant but had less activity toward the cyclin A1 and CFTR target genes. Although the 13A variant had no detectable activity in the reporter gene or mim-1 gene activation assays summarized in Fig. 6, it was a strong activator of the keratin-16 and CFTR target genes in MCF-7 cells. Thus, the additional protein sequences at the C-terminal end of the 13A variant (Fig. 6A) had a dramatic effect on its activities. Taken together, these results suggest that the variant c-Myb proteins encoded as a result of alternative RNA splicing have unique transcriptional activities and regulate distinct sets of target genes.
DISCUSSION
The expression of the c-myb gene is required for normal hematopoiesis (23) and for T-cell development (3), and this gene is expressed in all proliferating hematopoietic cells, where it presumably regulates genes involved in proliferation and differentiation. However, the mechanisms of c-myb regulation of multiple cell fate pathways and the cell type-specific genes required for such complex regulation are not yet understood. In this report, we demonstrate that the human c-myb gene generates several different transcripts through alternative RNA splicing whose protein products have diverse transcriptional properties. Quantitative assays showed dramatically different expression patterns and relative levels of c-myb variants in PBLs, CD34+ cells, and primary leukemia samples, implying that the splicing of c-myb transcripts is highly regulated and lineage specific and that the ratios or the relative levels of alternatively spliced c-Myb variants play a role in the lineage-specific functions of hematopoietic cells. Furthermore, the ratios of alternately spliced c-myb transcripts changed dramatically in a lineage-specific manner following the in vitro differentiation of CD34 cells (Fig. 3). Thus, the regulation of c-myb alternative splicing may play an important role in hematopoietic cell differentiation.
Recent reports suggest that human gene transcripts undergo extensive alternative splicing (12), providing an important mechanism for creating diversity from a limited number of genes. Here, we categorized a variety of human c-myb splice variants containing alternative exons. During these studies, we also discovered additional alternatively spliced c-myb transcripts from primary hematopoietic cells that were formed by a wide range of complex splicing events, including the combinatorial use of alternative exons, the use of alternative 3′ splice donor sites in some exons, and exon skipping (see Table S2 in the supplemental material). Others have speculated that the variant c-myb gene products, many of which contain premature stop codons, are likely to be subject to nonsense-mediated RNA decay (5). However, our results for hematopoietic cell lines indicate that the stabilities of the alternatively spliced c-myb transcripts were not significantly affected by cycloheximide treatment and that the variant transcripts had rates of decay similar to that of the wild-type transcript (data not shown). Transcripts subject to nonsense-mediated decay are generally rapidly degraded following cycloheximide treatment, suggesting that most of the c-myb transcripts are not significantly affected by that regulatory mechanism. Furthermore, the alternatively spliced c-myb transcripts were found to be associated with polyribosomes (Fig. 5), suggesting that most of the variants are stable and are likely to encode functional proteins with altered C termini. Interestingly, Western blot analyses of hematopoietic cell lines with the use of an antibody to the c-Myb DNA binding domain detected multiple immunoreactive bands whose protein sizes were consistent with the predicted sizes of some of the variants (see Fig. S3 in the supplemental material). Antibodies specific for novel peptides in the variant proteins will be required to confirm that these proteins are stably expressed in primary cells.
One of the most interesting conclusions from our studies is that the different c-Myb splice variants display qualitative differences in their transcriptional activities. While all of the splice variants contained identical DNA binding domains, they had different C termini. Recent studies from our laboratory demonstrate that small changes in the C-terminal domains influence target gene specificity (18, 20, 30). In this study, we showed that each splice variant product can activate a unique set of genes, suggesting that the alternate splicing of c-myb produces a family of transcription factors with unique biological activities. Therefore, it may be the ratios of alternatively spliced c-Myb proteins present in a specific cell type that allow c-Myb to regulate hematopoietic cell function in a lineage-specific manner.
In order for c-Myb to be transforming, it must be activated to unmask its transforming potential. For example, the c-myb gene is a common target of retroviral insertion leading to leukemia formation in mouse models (24, 41). Recently, two genetic abnormalities in the human c-myb gene in T-ALL patients were found, a duplication of the c-myb gene and a reciprocal translocation of c-myb and T-cell receptor beta loci, with both of these abnormalities leading to the overexpression of c-Myb (4, 16). Our results show that the c-myb gene may also be a target for deregulated or aberrant splicing in leukemia. An association between cancer and the deregulated alternative splicing of many different genes has been well documented previously (35, 39). However, recent reports suggest that the alternately spliced forms of genes such as those for the tyrosine kinase receptor RON (10) and the kinase SK61 (13) are themselves oncogenic when expressed inappropriately. Also, the overexpression of splicing factor 2/alternative splicing factor, which changes alternative splicing in a limited cohort of genes, can transform fibroblast cells, further linking aberrant splicing and oncogenesis (13). Our results show that the c-myb splicing patterns were different in bone marrow samples from control subjects and leukemia patients and, in particular, that exon 9A-containing transcripts were overexpressed nearly 120-fold in T-ALL samples. The inclusion of the alternate exon 9A produces a protein that is truncated similarly to the AMV v-Myb protein, which transforms immature myeloid cells and induces myeloid leukemias in animals. It is interesting that in our transcriptional assays, the exon 9A variant of c-Myb activated both c- and v-Myb-regulated genes (Fig. 7). These results suggest that aberrant or deregulated alternative splicing of the c-myb gene may be an additional mechanism to unmask its transforming potential. Additional experiments will be required to determine how different versions of c-Myb contribute to normal hematopoiesis and leukemogenesis and to identify the signaling pathways that regulate the alternative splicing of c-myb transcripts in human cells.
Supplementary Material
Acknowledgments
This work was supported by Public Health Service grants CA058443 and CA105257 from the National Cancer Institute (to S.A.N.) and American Cancer Society institutional research grant IRG-92-024 (to J.P.O.). Some experiments used the facilities or services provided by the Keck-University of New Mexico Genomics Resource, a facility supported by a grant from the W. M. Keck Foundation as well as the State of New Mexico, the University of New Mexico Cancer Research and Treatment Center, and the New Mexico Center for Environmental Health Sciences. Purified CD34+ cells were provided by the Fred Hutchinson Cancer Research Center Large-Scale Cell Processing Core, supported by NIDDK Core Center of Excellence in Hematology grant DK56465.
Footnotes
Published ahead of print on 14 January 2008.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Badiani, P. A., D. Kioussis, D. M. Swirsky, I. A. Lampert, and K. Weston. 1996. T-cell lymphomas in v-Myb transgenic mice. Oncogene 132205-2212. [PubMed] [Google Scholar]
- 2.Bellon, T., D. Perrotti, and B. Calabretta. 1997. Granulocytic differentiation of normal hematopoietic precursor cells induced by transcription factor PU.1 correlates with negative regulation of the c-myb promoter. Blood 901828-1839. [PubMed] [Google Scholar]
- 3.Bender, T. P., C. S. Kremer, M. Kraus, T. Buch, and K. Rajewsky. 2004. Critical functions for c-Myb at three checkpoints during thymocyte development. Nat. Immunol. 5721-729. [DOI] [PubMed] [Google Scholar]
- 4.Clappier, E., W. Cuccuini, A. Kalota, A. Crinquette, J. M. Cayuela, W. A. Dik, A. W. Langerak, B. Montpellier, B. Nadel, P. Walrafen, O. Delattre, A. Aurias, T. Leblanc, H. Dombret, A. M. Gewirtz, A. Baruchel, F. Sigaux, and J. Soulier. 2007. The C-MYB locus is involved in chromosomal translocation and genomic duplications in human T-cell acute leukemia (T-ALL), the translocation defining a new T-ALL subtype in very young children. Blood 1101251-1261. [DOI] [PubMed] [Google Scholar]
- 5.Close, J., L. Game, B. Clark, J. Bergounioux, A. Gerovassili, and S. L. Thein. 2004. Genome annotation of a 1.5 Mb region of human chromosome 6q23 encompassing a quantitative trait locus for fetal hemoglobin expression in adults. BMC Genomics 533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dao, M. A., and J. A. Nolta. 2007. Cytokine and integrin stimulation synergize to promote higher levels of GATA-2, c-myb, and CD34 protein in primary human hematopoietic progenitors from bone marrow. Blood 1092373-2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dasgupta, P., and E. P. Reddy. 1989. Identification of alternatively spliced transcripts for human c-myb: molecular cloning and sequence analysis of human c-myb exon 9A sequences. Oncogene 41419-1423. [PubMed] [Google Scholar]
- 8.Dash, A. B., F. C. Orrico, and S. A. Ness. 1996. The EVES motif mediates both intermolecular and intramolecular regulation of c-Myb. Genes Dev. 101858-1869. [DOI] [PubMed] [Google Scholar]
- 9.del Prete, M. J., R. Vernal, H. Dolznig, E. W. Mullner, and J. A. Garcia-Sanz. 2007. Isolation of polysome-bound mRNA from solid tissues amenable for RT-PCR and profiling experiments. RNA 13414-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ghigna, C., S. Giordano, H. Shen, F. Benvenuto, F. Castiglioni, P. M. Comoglio, M. R. Green, S. Riva, and G. Biamonti. 2005. Cell motility is controlled by SF2/ASF through alternative splicing of the Ron protooncogene. Mol. Cell 20881-890. [DOI] [PubMed] [Google Scholar]
- 11.Introna, M., J. Golay, J. Frampton, T. Nakano, S. A. Ness, and T. Graf. 1990. Mutations in v-myb alter the differentiation of myelomonocytic cells transformed by the oncogene. Cell 631289-1297. [DOI] [PubMed] [Google Scholar]
- 12.Johnson, J. M., J. Castle, P. Garrett-Engele, Z. Kan, P. M. Loerch, C. D. Armour, R. Santos, E. E. Schadt, R. Stoughton, and D. D. Shoemaker. 2003. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science 3022141-2144. [DOI] [PubMed] [Google Scholar]
- 13.Karni, R., E. de Stanchina, S. W. Lowe, R. Sinha, D. Mu, and A. R. Krainer. 2007. The gene encoding the splicing factor SF2/ASF is a proto-oncogene. Nat. Struct. Mol. Biol. 14185-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Komor, M., S. Guller, C. D. Baldus, S. de Vos, D. Hoelzer, O. G. Ottmann, and W. K. Hofmann. 2005. Transcriptional profiling of human hematopoiesis during in vitro lineage-specific differentiation. Stem Cells 231154-1169. [DOI] [PubMed] [Google Scholar]
- 15.Kumar, A., S. J. Baker, C. M. Lee, and E. P. Reddy. 2003. Molecular mechanisms associated with the regulation of apoptosis by the two alternatively spliced products of c-Myb. Mol. Cell. Biol. 236631-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lahortiga, I., K. De Keersmaecker, P. Van Vlierberghe, C. Graux, B. Cauwelier, F. Lambert, N. Mentens, H. B. Beverloo, R. Pieters, F. Speleman, M. D. Odero, M. Bauters, G. Froyen, P. Marynen, P. Vandenberghe, I. Wlodarska, J. P. Meijerink, and J. Cools. 2007. Duplication of the MYB oncogene in T cell acute lymphoblastic leukemia. Nat. Genet. 39593-595. [DOI] [PubMed] [Google Scholar]
- 17.Lei, W., F. Liu, and S. A. Ness. 2005. Positive and negative regulation of c-Myb by cyclin D1, cyclin-dependent kinases, and p27 Kip1. Blood 1053855-3861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lei, W., J. J. Rushton, L. M. Davis, F. Liu, and S. A. Ness. 2004. Positive and negative determinants of target gene specificity in myb transcription factors. J. Biol. Chem. 27929519-29527. [DOI] [PubMed] [Google Scholar]
- 19.Leverson, J. D., and S. A. Ness. 1998. Point mutations in v-Myb disrupt a cyclophilin-catalyzed negative regulatory mechanism. Mol. Cell 1203-211. [DOI] [PubMed] [Google Scholar]
- 20.Liu, F., W. Lei, J. P. O'Rourke, and S. A. Ness. 2006. Oncogenic mutations cause dramatic, qualitative changes in the transcriptional activity of c-Myb. Oncogene 25795-805. [DOI] [PubMed] [Google Scholar]
- 21.Livak, K. J., and T. D. Schmittgen. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25402-408. [DOI] [PubMed] [Google Scholar]
- 22.Luo, J., Z. L. Deng, X. Luo, N. Tang, W. X. Song, J. Chen, K. A. Sharff, H. H. Luu, R. C. Haydon, K. W. Kinzler, B. Vogelstein, and T. C. He. 2007. A protocol for rapid generation of recombinant adenoviruses using the AdEasy system. Nat. Protoc. 21236-1247. [DOI] [PubMed] [Google Scholar]
- 23.Mucenski, M. L., K. McLain, A. B. Kier, S. H. Swerdlow, C. M. Schreiner, T. A. Miller, D. W. Pietryga, W. J. J. Scott, and S. S. Potter. 1991. A functional c-myb gene is required for normal murine fetal hepatic hematopoiesis. Cell 65677-689. [DOI] [PubMed] [Google Scholar]
- 24.Mukhopadhyaya, R., and L. Wolff. 1992. New sites of proviral integration associated with murine promonocytic leukemias and evidence for alternate modes of c-myb activation. J. Virol. 666035-6044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ness, S. A. 1996. The Myb oncoprotein: regulating a regulator. Biochim. Biophys. Acta 1288F123-F139. [DOI] [PubMed] [Google Scholar]
- 26.Ness, S. A. 1999. Myb binding proteins: regulators and cohorts in transformation. Oncogene 183039-3046. [DOI] [PubMed] [Google Scholar]
- 27.Ness, S. A. 2003. Myb protein specificity: evidence of a context-specific transcription factor code. Blood Cells Mol. Dis. 31192-200. [DOI] [PubMed] [Google Scholar]
- 28.Ness, S. A., E. Kowenz-Leutz, T. Casini, T. Graf, and A. Leutz. 1993. Myb and NF-M: combinatorial activators of myeloid genes in heterologous cell types. Genes Dev. 7749-759. [DOI] [PubMed] [Google Scholar]
- 29.Ness, S. A., A. Marknell, and T. Graf. 1989. The v-myb oncogene product binds to and activates the promyelocyte-specific mim-1 gene. Cell 591115-1125. [DOI] [PubMed] [Google Scholar]
- 30.Rushton, J. J., L. M. Davis, W. Lei, X. Mo, A. Leutz, and S. A. Ness. 2003. Distinct changes in gene expression induced by A-Myb, B-Myb and c-Myb proteins. Oncogene 22308-313. [DOI] [PubMed] [Google Scholar]
- 31.Rushton, J. J., and S. A. Ness. 2001. The conserved DNA binding domain mediates similar regulatory interactions for A-Myb, B-Myb, and c-Myb transcription factors. Blood Cells Mol. Dis. 27459-463. [DOI] [PubMed] [Google Scholar]
- 32.Schuur, E. R., P. Dasgupta, E. P. Reddy, J. M. Rabinovich, and M. A. Baluda. 1993. Alternative splicing of the chicken c-myb exon 9A. Oncogene 81839-1847. [PubMed] [Google Scholar]
- 33.Schuur, E. R., J. M. Rabinovich, and M. A. Baluda. 1994. Distribution of alternatively spliced chicken c-myb exon 9A among hematopoietic tissues. Oncogene 93363-3365. [PubMed] [Google Scholar]
- 34.Shen-Ong, G. L., R. M. Skurla, Jr., J. D. Owens, and J. F. Mushinski. 1990. Alternative splicing of RNAs transcribed from the human c-myb gene. Mol. Cell. Biol. 102715-2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Srebrow, A., and A. R. Kornblihtt. 2006. The connection between splicing and cancer. J. Cell Sci. 1192635-2641. [DOI] [PubMed] [Google Scholar]
- 36.Tanaka, Y., N. P. Patestos, T. Maekawa, and S. Ishii. 1999. B-myb is required for inner cell mass formation at an early stage of development. J. Biol. Chem. 27428067-28070. [DOI] [PubMed] [Google Scholar]
- 37.Toscani, A., R. V. Mettus, R. Coupland, H. Simpkins, J. Litvin, J. Orth, K. S. Hatton, and E. P. Reddy. 1997. Arrest of spermatogenesis and defective breast development in mice lacking A-myb. Nature 386713-717. [DOI] [PubMed] [Google Scholar]
- 38.Tschan, M. P., K. M. Fischer, V. S. Fung, F. Pirnia, M. M. Borner, M. F. Fey, A. Tobler, and B. E. Torbett. 2003. Alternative splicing of the human cyclin D-binding Myb-like protein (hDMP1) yields a truncated protein isoform that alters macrophage differentiation patterns. J. Biol. Chem. 27842750-42760. [DOI] [PubMed] [Google Scholar]
- 39.Venables, J. P. 2004. Aberrant and alternative splicing in cancer. Cancer Res. 647647-7654. [DOI] [PubMed] [Google Scholar]
- 40.Westin, E. H., K. M. Gorse, and M. F. Clarke. 1990. Alternative splicing of the human c-myb gene. Oncogene 51117-1124. [PubMed] [Google Scholar]
- 41.Wolff, L., R. Koller, J. Bies, V. Nazarov, B. Hoffman, A. Amanullah, M. Krall, and B. Mock. 1996. Retroviral insertional mutagenesis in murine promonocytic leukemias: c-myb and Mml1. Curr. Top. Microbiol. Immunol. 211191-199. [DOI] [PubMed] [Google Scholar]
- 42.Woo, C. H., L. Sopchak, and J. S. Lipsick. 1998. Overexpression of an alternatively spliced form of c-Myb results in increases in transactivation and transforms avian myelomonoblasts. J. Virol. 726813-6821. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.