Abstract
Telomerase is a ribonucleoprotein reverse transcriptase that copies a short template within its integral telomerase RNA moiety (TER) onto eukaryotic chromosome ends, thus compensating for incomplete replication and degradation. The highly divergent yeast TER is structured in three long arms, with a catalytic core at its center. A binding site for the protein Ku80 is conserved within the 5′ arm of TER in Saccharomyces but not in Kluyveromyces budding yeast species. Consistently, KU80 deletion in Kluyveromyces lactis does not affect telomere length, while it causes telomere shortening in Saccharomyces cerevisiae. We found elements in the 5′ arm of K. lactis TER that are crucial for telomerase activity and stability. However, we found no indication of the association of Ku80 with this arm. Although the overexpression of Ku80 rescues a particular mutation in K. lactis TER1 that phenocopies a telomerase null mutation, this effect is indirect, caused by the repression of the recombination pathway competing for telomere maintenance. Interestingly, the overexpression of Est3, an essential telomerase protein whose function is still unknown, suppresses the phenotypes of mutations in this arm. These results indicate that the 5′ arm of K. lactis TER has critical roles in telomerase function, which may be linked to the function of Est3.
Telomerase is a ribonucleoprotein (RNP) that synthesizes short tandem telomeric repeats onto the chromosome ends of most eukaryotes to counterbalance the shortening caused by incomplete DNA replication and exonuclease action (reviewed in references 3 and 22). The essential telomerase catalytic core consists of telomerase RNA (TER, or TER1 in Kluyveromyces lactis and TLC1 in Saccharomyces cerevisiae) and telomerase reverse transcriptase protein (TERT, or Est2 in budding yeasts). TERT/Est2 is specialized in copying a relatively short template sequence within the much longer TER onto the telomere 3′ ends, thereby synthesizing species-specific telomeric repeats. In addition to TLC1 and Est2, the S. cerevisiae telomerase holoenzyme includes the proteins Est1, Est3, and Ku80 and the Sm complex. Both Est1 and Est3 are essential for telomerase activity in vivo but not in vitro (11). Est1 functions in the recruitment and/or activation of telomerase during the S phase of the cell cycle (5, 24), and the specific role of Est3 is still unknown. The Sm proteins are essential for the correct processing and stable accumulation of the telomerase complex (19). Finally, Ku80 functions in the recruitment of telomerase to the telomeres, especially during the G1 phase of the cell cycle (6).
TER was suggested to serve as a flexible scaffold for the binding of the telomerase proteins and the assembly of the telomerase holoenzyme (28). Secondary structure models for Kluyveromyces and Saccharomyces sensu stricto TERs revealed that, in both groups, TER is organized in three long arms, with a catalytic core at its center (1, 4, 10, 27). This catalytic core includes the template, the template boundary (18, 26), a critical pseudoknot element with conserved base triples (21), and a core-enclosing helix (10). The arm close to the 3′ end (terminal arm) contains the Sm binding site (19, 27) and a functional three-way junction (1), and the middle arm contains a conserved binding site for Est1 (Est1 arm) (17). In S. cerevisiae, the arm close to the 5′ end contains a binding site for Ku80, termed the 48-nucleotide (nt) stem-loop (Ku arm) (15). Deletion of the 48-nt stem-loop in TLC1 and mutation of the Ku80 residues that interact with this binding site resulted in comparable telomere shortening (23). Unlike in the case of S. cerevisiae, deletion of the KU80 gene in K. lactis did not affect telomere length (2), suggesting that Ku80 does not play a significant role in K. lactis telomerase function. Since the only known role of the Ku arm in S. cerevisiae is to bind Ku80, and Ku80 does not appear to be important for K. lactis telomerase function, we anticipated that the corresponding 5′ RNA arm would be dispensable for telomerase function in K. lactis. Strikingly, however, we found that it is absolutely critical. Here, we describe a detailed mutational analysis of the 5′ arm of TER1 and suggest that the telomerase protein Est3 is linked to the function of this arm.
MATERIALS AND METHODS
Yeast strains.
The K. lactis strain yJR27 (ter1Δ ura3 his2 trp1), based on the parental strain 7B520, was used for the CEN-ARS plasmid shuffling of the wild-type (WT) and mutant TER1 genes as described previously (16). The est3Δ strain yBZ1 (ter1Δ est3::hph ura3 his2 trp1) and the ku80Δ strain yBZ2 (ter1Δ ku80::hph ura3 his2 trp1) were based on the parental strain yJR27. The rad52Δ ter1Δ strain yND08 (ter1Δ rad52::hph ura3 his1 ade1) was made by mating strain GG1634 (rad52::hph) with GG1935 (ade2 ura3) and then with yJR27. Strains GG1634 and GG1935 were kindly given by Yde Steensma. All strains were grown in yeast nitrogen base drop-out medium at 30°C, unless otherwise indicated.
Genomic DNA preparation and Southern analysis.
Genomic DNA preparation and Southern analysis of telomeric restriction fragments were done as described previously (21). At least two independent clones were analyzed for each strain.
Total RNA extraction and real-time reverse transcription (RT)-PCR analysis of TER1 levels.
K. lactis strains were grown to an optical density at 600 nm of 1. Total RNA was extracted using an RNeasy mini kit (Qiagen) according to the manufacturer's instructions and treated with RNase-free RQ DNase (Promega). Four micrograms of total RNA was used for cDNA synthesis, using random hexamer primer and Superscript III (Invitrogen) at 50°C according to the manufacturer's instructions. Sybr green real-time PCRs were performed using ABI BioPrism 7000. TER1 levels were normalized to those of Msl1 mRNA, encoding a U2 snRNA binding protein. The 20-μl reaction mixture (in triplicates) contained 0.3 μM of each primer (see below), 10 μl of ABsolute Blue QPCR Sybr Green ROX mix (ABgene), and cDNA corresponding to 16 ng of total RNA. The PCR cycle was as follows: 2 min at 50°C, 15 min at 95°C, and then 40 cycles of 15 s at 95°C and 60 s at 60°C. The following primer sequences were used: AACCGCCAACAAAGAAACAA and TCCATAAGTGGCAAACAGCA for amplification of nt 29 to 180 of the Msl1 open reading frame and TCGACGTGGGTATGCTAGTG and AATGGCTTGACGGAAAAATG for amplification of TER1 nt 535 to 697.
Purification of telomerase extracts and in vitro telomerase assay.
Partially purified telomerase fractions were prepared and assayed for telomerase activity in vitro as described previously (1, 8).
RESULTS
The 5′ arm of TER1 is critical for telomerase function.
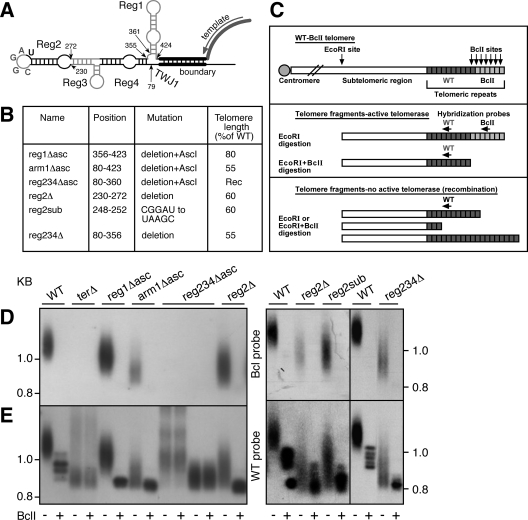
The binding of Ku80 to TLC1 was found to be important for the recruitment of the S. cerevisiae telomerase to the telomeres (6). The Ku80 binding site, a 48-nt stem-loop structure, is conserved at the distal end of the Ku arm in seven Saccharomyces sensu stricto species (4). We searched the sequences and predicted the structures of six Kluyveromyces TERs but failed to identify a conserved element that resembles the Saccharomyces Ku80 binding site, whether in the corresponding 5′ arm or elsewhere in these TERs. Consistently, it was recently found that Ku80 deletion in K. lactis does not affect telomere length (2). These observations suggest that in K. lactis, either Ku80 does not bind TER1 or its binding is normally redundant and not a limiting factor for telomere length regulation. If there were any functional role for the 5′ arm of TER1, it would therefore be independent of Ku80. However, the lack of apparent sequence conservation in this arm among the Kluyveromyces TERs suggests that it may be dispensable for telomerase function. To test this possibility, we deleted the entire 5′ arm, keeping the template boundary element (26) at the base of the arm intact (Fig. 1A and B, arm1Δ asc). The sequence of an AscI restriction site was introduced to facilitate easy cloning of other sequences in substitution for the 5′ arm. We replaced the WT TER1 gene in K. lactis cells with the mutant ter1 gene, using a plasmid-shuffling system described previously (16). A single-nucleotide mutation in the telomerase template is used to mark the nascent repeats incorporated by the investigated telomerase (16). This mutation results in the incorporation of BclI restriction sites into the telomeric repeats but is otherwise apparently silent. Genomic DNA was prepared from this strain and digested with either EcoRI or EcoRI-BclI restriction endonuclease. Telomere length and BclI repeat incorporation were analyzed by Southern blot hybridization first with a BclI-specific probe and then with a WT telomeric probe (see scheme in Fig. 1C). Although the telomerase mutant harboring a deletion of the 5′ arm was active, incorporating BclI telomeric repeats and maintaining stable telomeres, these telomeres were severely short (Fig. 1D and E, arm1Δ asc). In S. cerevisiae, most of the Ku arm can be replaced with the Ku80 binding 48-nt stem-loop, preserving (at least partially) the function of this arm (29). We introduced the S. cerevisiae 48-nt stem-loop into the arm1Δ mutant to test whether it can substitute for the 5′ arm. However, it failed to suppress the short telomere phenotype (not shown), indicating that either K. lactis Ku80 cannot bind the S. cerevisiae 48-nt stem-loop or such an interaction cannot rescue the impaired function of the arm1Δ asc mutant. Altogether, these experiments indicate that the 5′ arm of TER1 has a crucial role which, unlike in the case of S. cerevisiae, appears to be independent of Ku80.
FIG. 1.
Mutational analysis of the 5′ arm. (A) Scheme of the 5′ arm of TER1. Shown are the template, the template boundary, the various elements tested in this work, the nucleotide positions indicating the deletions (described in panel B), and the nucleotides of the Reg2 loop (conserved nucleotides in gray). (B) Table describing the mutations and the resulting average telomere length, presented as a percentage of the WT length. “AscI” indicates the insertion of the nucleotide sequence of the AscI restriction site. “Rec” indicates deregulated telomeres, maintained by the alternative recombination pathway. (C) Schematic representation showing telomeric fragments digested with the restriction endonuclease EcoRI or EcoRI-BclI and hybridization probes (WT and BclI specific). Plasmids carrying the ter1 genes with the template BclI mutation alone (WT), additional mutations, or an empty vector (terΔ), as indicated in panel B and above the lanes in panel D, were used to replace the WT TER1 gene encoded on a URA plasmid. Genomic DNA was prepared from the resulting strains in their sixth passage and analyzed by Southern blotting and hybridization first with a BclI-specific (D) and then with a WT (E) probe. Only the bottom portion of the gel is shown, which includes a group of 7 out of the 12 K. lactis telomeres (an example of the full gel is shown in Fig. 4).
Functional stem-loop element in TER1.
An analysis of the 5′ arm of TER1 and the identification of functional elements require a secondary structure model. In the absence of sequence conservation, we could not use phylogenetic covariation to assist with secondary structure prediction. We therefore used the computer program mfold (31) to predict the secondary structure of the 5′ arm in each of the six Kluyveromyces species based on free energy calculations (data not shown). Since the template boundary stem was confirmed experimentally (26), we could use the sequence between (and including) the two strands of the template boundary element to predict the structures of the independent structural domain of the 5′ arm in each species. Since mfold predicts suboptimal structures as well, we compared all structures within 10% of the lowest energy (percentage of suboptimality). The lowest-energy predictions for five of the species revealed similar structures of a long stem and one to two smaller stem-loops branching off of it, as shown schematically in Fig. 1A. In Kluyveromyces marxianus, a suboptimal structure with a 4% higher free energy was similar to that predicted for the rest of the species. We defined the stem-loop structure at the distal end of the long stem (the location corresponding to the Ku80 binding site in TLC1) as Reg2 and the short stem-loop branching in the middle as Reg3. Another stem-loop closer to the template boundary in four of the species was termed Reg1, and a three-way junction was termed TWJ1 (Fig. 1A). While the predictions were based on free energy calculations, the similarity between the six species supported them. Therefore, we used the predicted structure for TER1 as a working model, which enabled us to investigate specific functional elements in the 5′ arm, as described below.
Notably, a CGGA sequence is conserved in the loop of Reg2 in all six species (Fig. 1A), suggesting that it has functional importance. To analyze it, we first deleted the 42 nt forming the stem and loop of Reg2 and analyzed the resulting telomere phenotype. Interestingly, this deletion resulted in telomere shortening almost as severe as that caused by the much larger deletion of the entire arm (60% of the WT length for the reg2Δ mutant, compared to 55% for the arm1Δ asc mutant; Fig. 1). Consistently, the reg2Δ mutant showed a rough colony phenotype, a hallmark of impaired telomere maintenance in K. lactis (see Fig. 3C). We also made a five-nucleotide transition mutation in the loop of Reg2, which included the conserved CGGA motif (CGGAU to UAAGC). Strikingly, this substitution had the same effect as the deletion of Reg2 (Fig. 1, compare reg2sub and reg2Δ), suggesting that the conserved sequence of the loop is essential for the function of Reg2. Altogether, these results reveal a novel stem-loop element, which is located within TER1 at the position corresponding to the Ku80 binding site in TLC1 and is important for telomerase function in vivo. Unlike in the case of S. cerevisiae, the Reg2 stem-loop element was not sufficient to substitute for the entire arm (not shown), suggesting that its proper function requires either the correct positioning within the telomerase complex or additional elements within this arm.
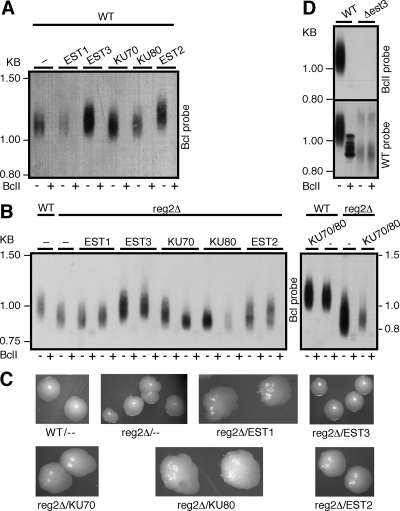
FIG. 3.
Overexpression of telomerase proteins in the WT and the reg2Δ mutant. Genes encoding the K. lactis telomerase proteins Est1, Est2, Est3, Ku70, and Ku80, including their endogenous promoters, were cloned into a K. lactis 2μm, high-copy-number plasmid (pKDU7) and introduced into K. lactis cells containing WT (A) or reg2Δ ter1 mutants (B) on CEN-ARS plasmids. Cells were grown for six passages and analyzed by Southern blot hybridization as described in the legend of Fig. 1. Only BclI-specific hybridization is shown. (C) Typical colony phenotypes of K. lactis strains taken at their fourth passage. Impaired telomere maintenance is associated with rough colonies, as opposed to the smooth appearance of WT colonies. (D) Genomic DNA was prepared from an est3Δ strain in its sixth passage and analyzed by Southern blot hybridization as described in the legend of Fig. 1.
Incremental deletions reveal a second functional element close to the base of the 5′ arm.
To search for additional functional elements in the 5′ arm, we made incremental deletions in the long stem ending with Reg2. These deletions only slightly exacerbated the short telomere phenotype of the reg2Δ mutant (see Table S1 in the supplemental material), even when the long arm (nt 79 to 356) was removed down to TWJ1 (Fig. 1, reg234Δ). Surprisingly, however, the removal of only five additional nucleotides (nt 356 to 360) and the introduction of the sequence of an AscI restriction site (GGCGCGCC) instead reduced the in vivo telomerase activity below the limit of detection, as revealed by the lack of BclI repeat incorporation and the long smears observed by Southern analysis (Fig. 1, reg234Δ asc). These smears indicated the deregulation of telomere length, which is typical of the alternative, Rad52-dependent recombination pathway for telomere maintenance (12). This mutation was more detrimental than the larger deletion of the entire arm (Fig. 1, arm1Δ asc), suggesting that it caused an alteration of the TER1 structure (possibly in the position of Reg1 with respect to the rest of the RNA) that is disruptive to telomerase stability, assembly, or function. Finally, deleting Reg1 while keeping Reg234 intact caused moderate telomere shortening (80% of the WT length [Fig. 1, reg1Δ asc]), suggesting that Reg1 is important, at least to some extent, for telomerase function.
Reduced TER1 levels in the three-way junction mutants.
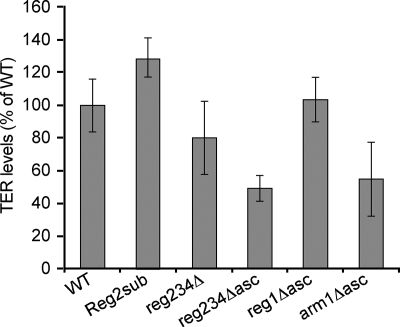
To test whether the telomere shortening observed in the 5′ arm mutants was caused by the reduced stability of telomerase RNA, we used real-time RT-PCR to measure the relative amounts of TER1 in the various 5′ arm mutants. We found normal or slightly elevated TER1 levels in the reg1Δ asc and reg2sub mutants (Fig. 2; see also Table S1 in the supplemental material). However, in the reg234Δ asc and arm1Δ asc mutants, TER1 levels were reduced to 50 and 55% of the WT levels, respectively. Comparable results were obtained by Northern analysis (data not shown). These results suggest that Reg2 is important for telomerase activity rather than stability, while TWJ1 is important for the stability of TER1. The reduction of TER1 levels could contribute to the short telomere phenotypes of the reg234Δ and arm1Δ asc mutants. However, it could not fully account for the null phenotype of the reg234Δ asc mutant, since the arm1Δ asc mutant had comparable TER1 levels but was active.
FIG. 2.
TER1 levels in mutants of the 5′ arm. The levels of TER1 in the mutant strains (indicated on the x axis) were determined using real-time RT-PCR, as described in Materials and Methods, normalized to the mRNA levels of MSL1, a protein component of U2 snRNP, and presented as a percentage of the WT TER1 levels. Error bars indicate the standard errors calculated for three measurements of each cDNA.
Overexpression of Est3 suppresses the phenotypes of mutations in Reg2.
To gain insight into the function of Reg2, we investigated whether any of the telomerase proteins are genetically or physically associated with this element. If a mutation affected the binding site of a protein, increased expression of this protein may compensate for the reduced affinity and suppress the telomere phenotype, as was shown for S. cerevisiae Est1 and its binding site in TLC1 (17). The genes encoding the K. lactis putative orthologs of Est1, Est2, Est3, Ku70, and Ku80 were identified in the K. lactis genome by their sequence homology to the S. cerevisiae proteins (1). The K. lactis Est1, Est2, and Est3 protein sequences showed 27, 36, and 36% identities and 50, 55, and 51% similarities, respectively, to their S. cerevisiae orthologs (see Fig. S1 in the supplemental material for multiple sequence alignment of Est3 proteins). Notably, the K. lactis EST3 gene contains an intact open reading frame, and thus, apparently does not require a translational frameshifting for the expression of a full-length Est3 protein, as does the S. cerevisiae EST3 gene (13). We deleted the EST3 gene from the K. lactis genome and analyzed the resulting telomere phenotype. As shown in Fig. 3D, the est3Δ mutation abolished telomerase function, indicating that Est3 is essential for K. lactis telomerase function in vivo, as is the case for the S. cerevisiae and Candida albicans Est3 orthologs (9, 11).
To overexpress these proteins, their genes, including the endogenous promoters, were cloned into pKDU7, a K. lactis 2μm-based high-copy-number plasmid, as described previously (1). Since Ku70 and Ku80 are known to function as a heterodimer, their genes were also cloned together onto one plasmid. These plasmids, along with the parent pKDU7 plasmid as a control, were introduced into K. lactis cells containing a WT or a mutant ter1 gene on a CEN-ARS plasmid. The overexpression of these genes was verified by Northern analysis (data not shown). First, we examined the effect of the overexpression of these genes in a WT TER1 background. As shown in Fig. 3A, the overexpression of Est3 and Est2 caused a slight and a moderate increase in telomere length (105 and 120% of the WT telomere length, respectively). The overexpression of Est1, Ku70, Ku80, and Ku70/Ku80 had no apparent effect. When combined with the reg2Δ and reg2sub mutations, the overexpression of Est1, Ku80, and Ku70/Ku80 did not affect telomere length (Fig. 3B and data not shown). One clone overexpressing Ku70 seemed to partially suppress the short telomere phenotype of the reg2Δ mutant, but this effect was not reproduced in another clone of this strain, as well as in two clones overexpressing Ku70 in the reg2sub mutant (Fig. 3B and data not shown). Est2 overexpression partially suppressed the short telomere and rough colony phenotypes of the reg2Δ and reg2sub mutants (Fig. 3B and C and data not shown). However, since Est2 overexpression caused telomere elongation in the WT TER1 strain (Fig. 3A), this effect may result from a general increase in the amount of assembled telomerase and is not specifically related to the Reg2 mutations. Remarkably, Est3 overexpression fully suppressed both the short telomere and rough colony phenotypes of the reg2Δ and reg2sub mutants without significantly affecting telomere length in the WT TER1 strain (Fig. 3 and data not shown).
Overexpression of Est2, Est3, and Ku80 rescues the activity of the reg234Δ asc mutant.
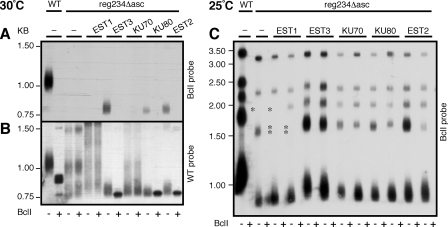
As described previously, the reg234Δ asc mutation reduced telomerase activity below the limit of detection. We examined the ability of the putative telomerase proteins to rescue the activity of this telomerase mutant when overexpressed. In this case, in addition to that of Est3 and Est2, the overexpression of Ku80 (but not Ku70 or Ku70/Ku80) restored the ability of telomerase to incorporate BclI repeats and maintain stable, even if severely short, telomeres (Fig. 4A and B and data not shown). This is consistent with the possibility that Ku80 binds a yet-unidentified site impaired by the deletion. While this interaction is not normally a limiting factor for telomerase action, it may become critical in the background of the reg234Δ asc mutant. Alternatively, the suppression effect of Ku80 may be indirect, resulting from its ability to repress telomeric recombination (2, 30), which in turn may uncover the action of a barely active telomerase. It is not clear why the overexpression of Ku80 rescued telomerase activity while the combination of Ku70 and Ku80 did not. It is possible that Ku80 has a Ku70-independent function that is lost in the context of the heterodimer. Alternatively, the overexpression of Ku80 alone may have unique consequences for the stability or nuclear localization of the Ku heterodimer or for the depletion of negative regulators of telomerase.
FIG. 4.
Overexpression of telomerase proteins in the reg234Δ asc mutant. High-copy-number plasmids harboring telomerase proteins were introduced into the reg234Δ asc mutant, as described in the legend of Fig. 3, grown at 30°C (A and B) or 25°C (C) for six passages, and analyzed by Southern blot hybridization, as described in the legend of Fig. 1. In panel C, the entire gel, including the 12 K. lactis telomeres, is shown. Asterisks indicate missing telomeric fragments, presumably due to subtelomeric recombination. Only BclI-specific hybridization is shown for the 25°C experiment.
Growing the reg234Δ asc strain at 25°C instead of 30°C showed that it is a temperature-sensitive mutation. At the lower temperature, the reg234Δ asc mutant was active and able to incorporate BclI repeats and maintain severely short telomeres (25 to 30% of the WT length; Fig. 4C). However, these telomeres were not stable, as revealed by the loss of several telomere restriction fragments (Fig. 4C), presumably caused by recombination events at the subtelomeric sequences. The overexpression of Est3, Ku70, Ku80, and Est2, but not Est1, prevented the appearance of the subtelomeric recombination events. However, only the overexpression of Est3 was clearly able to elongate the severely short telomeres of this mutant when grown at 25°C (up to 40% of the WT length).
Blocking recombination rescues the telomerase activity of the reg234Δ asc mutant.
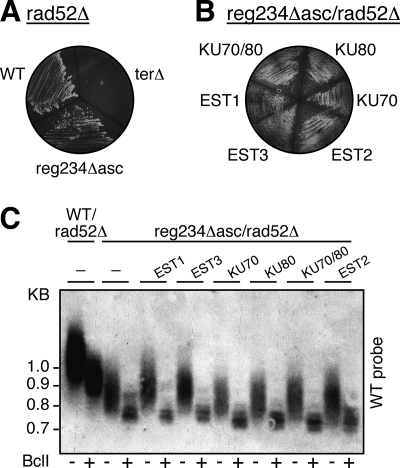
To test whether the alternative recombination pathway is inhibiting the action of the reg234Δ asc telomerase mutant, we introduced it into a rad52Δ strain, using the same plasmid-shuffling system used before. The deletion of RAD52 blocks the alternative recombination pathway and therefore is synthetically lethal with telomerase null mutations (12). Indeed, the ter1Δ rad52Δ double mutant grew only for two passages after selection against the TER-encoding plasmid (by 5-fluoroorotic acid treatment) and then completely lost viability (Fig. 5A). In contrast, the reg234Δ asc rad52Δ strain was viable for at least six passages, at 25°C, 30°C, and 34°C (Fig. 5 and data not shown). Consistently, the reg234Δ asc telomerase mutant was active in the absence of Rad52, even at 30°C, as revealed by the stable maintenance of short telomeres and the incorporation of BclI repeats (Fig. 5C). These results indicate that the reg234Δ asc telomerase mutant is potentially active but is inhibited in a WT Rad52 strain by the alternative recombination pathway. In addition, the overexpression of Ku70, Ku80, and Ku70/Ku80 did not have any effect on the telomere length of the reg234Δ asc rad52Δ mutant grown at 30°C, while Est1 and Est3 overexpression slightly elongated the telomeres (Fig. 5C). Therefore, the overexpression of Ku80, similarly to the deletion of the RAD52 gene, may uncover the activity of the reg234Δ asc telomerase mutant by repressing telomeric recombination.
FIG. 5.
Overexpression of telomerase proteins in a reg234Δ asc rad52Δ double mutant. (A) Deletion of the RAD52 gene was combined with the reg234Δ asc, WT, and ter1Δ strains and grown at 30°C (shown is the third passage). (B) High-copy-number plasmids harboring telomerase proteins were introduced into the reg234Δ asc rad52Δ double mutant and grown at 30°C (shown is the sixth passage). (C) Genomic DNA was prepared from these strains at their sixth passage and analyzed by Southern blot hybridization, as described in the legend of Fig. 1. Only hybridization to the WT telomeric probe is shown.
In summary, the overexpression of Est3 fully suppressed the phenotypes of the reg2Δ and reg2sub mutants (Fig. 3 and data not shown) and partially suppressed the phenotypes of the reg234Δ asc, reg23Δ, and arm1Δ asc mutants (Fig. 4 and data not shown). Common to all of them is the substitution or deletion of Reg2. In contrast, Est3 overexpression did not at all suppress the mild phenotype of the reg1Δ asc mutant, in which Reg2 is intact (data not shown). These results suggest that the function of Reg2 is linked to the telomerase protein Est3 rather than to Ku80.
Est3 and Reg2 are dispensable for telomerase activity in vitro.
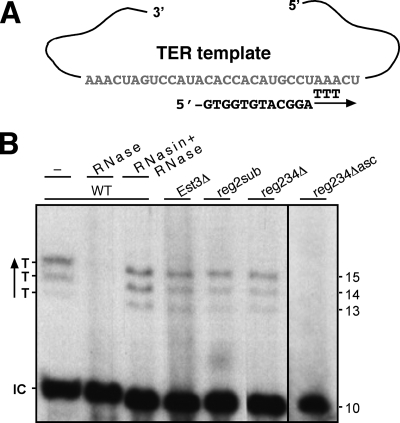
Est3 is dispensable for the in vitro activity of S. cerevisiae telomerase (11) but important for the in vitro activity of C. albicans telomerase (9). To test whether Est3 and the elements of the 5′ arm are important for the in vitro activity of K. lactis telomerase, we prepared partially purified fractions from the WT, est3Δ, reg2sub, reg234Δ, and reg234Δ asc strains, as described in reference 8. We tested these fractions for telomerase activity by using an oligonucleotide that pairs to the telomerase template and radioactively labeled dTTP in the absence of the other three nucleotides. Under these conditions, telomerase can add only three nucleotides to the oligonucleotide substrate (Fig. 6). These products of the WT telomerase were RNase sensitive, confirming that they were specific telomerase products. The est3Δ telomerase synthesized products comparable to those of the WT, indicating that Est3 is not important for telomerase in vitro activity, as observed for the S. cerevisiae telomerase (11). Interestingly, the reg2sub and reg234Δ strains produced telomerase products comparable to those of the WT, indicating that these elements are dispensable for telomerase in vitro activity and consistent with a functional linkage between Est3 and Reg2. In contrast, the reg234Δ asc strain reduced telomerase activity below the limit of detection, suggesting that this mutant is impaired in the assembly or activity of the telomerase catalytic core.
FIG. 6.
Est3 and Reg2 are dispensable for telomerase activity in vitro. (A) Scheme showing the telomerase template and a 5′-biotin-modified 12-mer oligonucleotide, which base pairs with its 3′ end at position 25 of the template and serves as a telomerase substrate. Above the arrow indicating the direction of polymerization, are three T nucleotides that are added in a T-only reaction. (B) Partially purified telomerase fractions were prepared from WT, est3Δ, reg2sub, reg234Δ, and reg234Δ asc strains and assayed (10 μl each) for telomerase activity in a reaction mixture containing [α-32P]dTTP and no other deoxyribonucleotide, using the conditions described in reference 1. After inactivation at 95°C for 5 min, a 5′ radioactively labeled, 3′-biotin-modified, 10-mer control oligonucleotide (IC) was added and purified together with the reaction products by the affinity of the biotin to streptavidin-coated magnetic beads (1). “RNase” indicates pretreatment of the telomerase fraction with 10 μg/ml RNase A for 5 min at 25°C, followed by the addition of 50 units of RiboLock RNase inhibitor and additional incubation for 5 min at 25°C. “RNasin+RNase” indicates that the RNase inhibitor treatment preceded that of the RNase. The levels of TER1 in the fractions were measured by slot blot hybridization with a TER1 probe and found to be comparable to each other.
DISCUSSION
Yeast telomerase RNAs are relatively large in size and highly divergent in sequence even among closely related species. Nevertheless, secondary structure models revealed that Saccharomyces and Kluyveromyces TERs are similarly organized in three long arms, with a catalytic core at their centers (1, 4, 10). Previous work identified a conserved Sm site and a functional three-way junction in the terminal arm (1, 19) and an Est1 binding site in the middle arm of budding yeast TERs (Est1 arm) (17). A conserved Ku80 binding site was found in the 5′ arm of Saccharomyces TERs (Ku arm) (15). The human Ku70/Ku80 heterodimer was also reported to bind the human TER (25), but the functional significance of this interaction is not clear. In contrast, no conserved Ku80 binding site is apparent in the 5′ arm or elsewhere in the Kluyveromyces TERs. Furthermore, the deletion of KU80 in K. lactis did not affect telomere length (2), suggesting that the binding of Ku80, if it occurs, is not normally a limiting factor for K. lactis telomere synthesis.
To test whether the 5′ arm of TER1 is important at all for telomerase function, we deleted the entire arm and tested the function of telomerase in vivo. This deletion caused severe telomere shortening, indicating that the 5′ arm has a crucial role (Fig. 1). We next used the free-energy-based computer program mfold to predict a secondary structure working model for this arm. We defined several structural elements in this arm (Reg1-4 and TWJ1) and analyzed them in detail (Fig. 1; see also Table S1 in the supplemental material).
Reg2 was found to be crucial for telomerase function in vivo but not in vitro (Fig. 1 and 6). Within the loop of Reg2, we identified a sequence motif (CGGA) conserved in all Kluyveromyces TERs. Strikingly, a transition substitution of this sequence resulted in severe telomere shortening, indicating that this apical loop is essential for the function of this element (Fig. 1). Another critical element is a three-way junction (TWJ1) formed by Reg1, Reg4, and the template boundary stem. This element was found to be important for the stable accumulation of TER1 in vivo and for telomerase activity in vitro. A combination of a deletion in Reg2-4 and a substitution in TWJ1 completely reduced telomerase activity below the limit of detection when the yeasts were grown at 30°C (Fig. 3, reg234Δ asc), suggesting that it caused an alteration of the TER1 structure that is even more disruptive to telomerase function than the deletion of the entire 5′ arm.
To gain insight into the role of the 5′ arm, we combined the various TER1 mutants with the overexpressed K. lactis proteins Est1, Est2, Est3, Ku70, Ku80, and Ku70/Ku80. Interestingly, the overexpression of Est3 fully suppressed the telomerase deficiency of the reg2Δ and reg2sub mutants (Fig. 3 and data not shown), restoring both colony morphology and telomere length to normal phenotypes. The overexpression of Est3 also rescued the in vivo telomerase activity of the reg234Δ asc telomerase mutant. However, in the case of this mutant, Est3 overexpression only partially suppressed the telomere and colony phenotypes. In addition, the overexpression of Est2 and Ku80, but not Ku70 or Ku70/Ku80, also rescued the telomerase activity of this mutant. This observation raised two possible explanations. The first is that Ku80 does have a role in K. lactis telomerase action. However, this role is redundant, and therefore, Ku80 deletion does not affect telomere length in a normal background. Alternatively, the effect of Ku80 overexpression may be indirect, resulting from the repression of the alternative recombination pathway, which in turn enables telomere synthesis by telomerase. Indeed, the Ku heterodimer was shown to suppress telomeric recombination in K. lactis and Arabidopsis thaliana (2, 30), and our observations are also consistent with the latter hypothesis. First, at the growth temperature of 25°C, in which the reg234Δ asc telomerase mutant is active, Est3 was the only protein able to partially elongate the severely short telomeres of this mutant. Second, the overexpression of Ku70 or Ku80, although it did not affect telomere length, suppressed subtelomeric recombination in this mutant when grown at 25°C (Fig. 4C). Third, the deletion of RAD52, encoding a central recombination protein that is essential for elongating telomeres by the alternative recombination pathway, was not lethal in combination with the reg234Δ asc mutant even at 30°C or higher growth temperatures (Fig. 5A), indicating that the reg234Δ asc mutant is potentially active but its activity is repressed when the recombination pathway is activated. Indeed, BclI incorporation at 30°C in a rad52Δ background shows that the reg234Δ asc telomerase mutant is sufficiently active to maintain short but stable telomeres when the recombination pathway is blocked (Fig. 5C). Interestingly, these observations indicate that the two pathways for telomere maintenance are competing when telomeres are critically short, and the recombination-based pathway has an inhibitory effect on telomerase action. Therefore, the rescue of the reg234Δ asc telomerase activity by Ku80 overexpression is likely to be a result of the repression of telomeric recombination by Ku80 rather than of an increase in Ku80 binding to TER1.
The overexpression of Est3 suppressed, at least partially, the phenotypes of all mutants containing a deletion or substitution in Reg2, while it did not significantly affect telomere length in the reg1Δ asc mutant or the WT TER1 strains. Consistent with the functional linkage between Est3 and Reg2, both are dispensable for the in vitro activity of telomerase (Fig. 6). Whether Est3 physically interacts with Reg2 and therefore the increased concentration of Est3 compensates for the reduced affinity has yet to be tested. In S. cerevisiae, Est3 was found to interact with Est1 and Est2 but not with TLC1 (7, 14). C. albicans Est1 and Est3 have been suggested to be mutually dependent for their inclusion in the telomerase complex (9). Furthermore, it is suggested in this study that a network of low-affinity interactions among multiple telomerase components is required for the proper assembly of the telomerase holoenzyme. Although S. cerevisiae Est3 has not been shown to bind directly to TLC1, it was shown to be capable of binding RNA (20). Our results are consistent with a scenario in which the telomerase proteins interact with each other and with TER1. Their interaction with TER1 may contribute to the stability of the complex and/or be important for telomerase function.
Supplementary Material
Acknowledgments
We thank Yde Steensma for K. lactis strains, Yogev Brown, Noa Gil, Martin Kupiec, and Nikolai Ulyanov for critical reading of the manuscript, and the members of the Tzfati laboratory for stimulating conversations.
This work was supported in part by Israel Science Foundation grant 676/02, United States-Israel Binational Science Foundation grants 2001065 and 2005088, and German-Israeli Foundation grant I-849-253.13/2004.
Footnotes
Published ahead of print on 14 January 2008.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Brown, Y., M. Abraham, S. Pearl, M. M. Kabaha, E. Elboher, and Y. Tzfati. 2007. A critical three-way junction is conserved in budding yeast and vertebrate telomerase RNAs. Nucleic Acids Res. 356280-6289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carter, S. D., S. Iyer, J. Xu, M. J. McEachern, and S. U. Astrom. 2007. The role of nonhomologous end-joining components in telomere metabolism in Kluyveromyces lactis. Genetics 1751035-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Collins, K. 2006. The biogenesis and regulation of telomerase holoenzymes. Nat. Rev. Mol. Cell Biol. 7484-494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dandjinou, A. T., N. Levesque, S. Larose, J. F. Lucier, S. Abou Elela, and R. J. Wellinger. 2004. A phylogenetically based secondary structure for the yeast telomerase RNA. Curr. Biol. 141148-1158. [DOI] [PubMed] [Google Scholar]
- 5.Evans, S. K., and V. Lundblad. 1999. Est1 and Cdc13 as comediators of telomerase access. Science 286117-120. [DOI] [PubMed] [Google Scholar]
- 6.Fisher, T. S., A. K. Taggart, and V. A. Zakian. 2004. Cell cycle-dependent regulation of yeast telomerase by Ku. Nat. Struct. Mol. Biol. 111198-1205. [DOI] [PubMed] [Google Scholar]
- 7.Friedman, K. L., J. J. Heit, D. M. Long, and T. R. Cech. 2003. N-terminal domain of yeast telomerase reverse transcriptase: recruitment of Est3p to the telomerase complex. Mol. Biol. Cell 141-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fulton, T. B., and E. H. Blackburn. 1998. Identification of Kluyveromyces lactis telomerase: discontinuous synthesis along the 30-nucleotide-long templating domain. Mol. Cell. Biol. 184961-4970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hsu, M., E. Y. Yu, S. M. Singh, and N. F. Lue. 2007. Mutual dependence of Candida albicans Est1p and Est3p in telomerase assembly and activation. Eukaryot. Cell 61330-1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin, J., H. Ly, A. Hussain, M. Abraham, S. Pearl, Y. Tzfati, T. G. Parslow, and E. H. Blackburn. 2004. A universal telomerase RNA core structure includes structured motifs required for binding the telomerase reverse transcriptase protein. Proc. Natl. Acad. Sci. USA 10114713-14718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lingner, J., T. R. Cech, T. R. Hughes, and V. Lundblad. 1997. Three Ever Shorter Telomere (EST) genes are dispensable for in vitro yeast telomerase activity. Proc. Natl. Acad. Sci. USA 9411190-11195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McEachern, M. J., and E. H. Blackburn. 1996. Cap-prevented recombination between terminal telomeric repeat arrays (telomere CPR) maintains telomeres in Kluyveromyces lactis lacking telomerase. Genes Dev. 101822-1834. [DOI] [PubMed] [Google Scholar]
- 13.Morris, D. K., and V. Lundblad. 1997. Programmed translational frameshifting in a gene required for yeast telomere replication. Curr. Biol. 7969-976. [DOI] [PubMed] [Google Scholar]
- 14.Osterhage, J. L., J. M. Talley, and K. L. Friedman. 2006. Proteasome-dependent degradation of Est1p regulates the cell cycle-restricted assembly of telomerase in Saccharomyces cerevisiae. Nat. Struct. Mol. Biol. 13720-728. [DOI] [PubMed] [Google Scholar]
- 15.Peterson, S. E., A. E. Stellwagen, S. J. Diede, M. S. Singer, Z. W. Haimberger, C. O. Johnson, M. Tzoneva, and D. E. Gottschling. 2001. The function of a stem-loop in telomerase RNA is linked to the DNA repair protein Ku. Nat. Genet. 2764-67. [DOI] [PubMed] [Google Scholar]
- 16.Roy, J., T. B. Fulton, and E. H. Blackburn. 1998. Specific telomerase RNA residues distant from the template are essential for telomerase function. Genes Dev. 123286-3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seto, A. G., A. J. Livengood, Y. Tzfati, E. H. Blackburn, and T. R. Cech. 2002. A bulged stem tethers Est1p to telomerase RNA in budding yeast. Genes Dev. 162800-2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seto, A. G., K. Umansky, Y. Tzfati, A. J. Zaug, E. H. Blackburn, and T. R. Cech. 2003. A template-proximal RNA paired element contributes to Saccharomyces cerevisiae telomerase activity. RNA 91323-1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Seto, A. G., A. J. Zaug, S. G. Sobel, S. L. Wolin, and T. R. Cech. 1999. Saccharomyces cerevisiae telomerase is an Sm small nuclear ribonucleoprotein particle. Nature 401177-180. [DOI] [PubMed] [Google Scholar]
- 20.Sharanov, Y. S., M. I. Zvereva, and O. A. Dontsova. 2006. Saccharomyces cerevisiae telomerase subunit Est3p binds DNA and RNA and stimulates unwinding of RNA/DNA heteroduplexes. FEBS Lett. 5804683-4690. [DOI] [PubMed] [Google Scholar]
- 21.Shefer, K., Y. Brown, V. Gorkovoy, T. Nussbaum, N. B. Ulyanov, and Y. Tzfati. 2007. A triple helix within a pseudoknot is a conserved and essential element of telomerase RNA. Mol. Cell. Biol. 272130-2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smogorzewska, A., and T. de Lange. 2004. Regulation of telomerase by telomeric proteins. Annu. Rev. Biochem. 73177-208. [DOI] [PubMed] [Google Scholar]
- 23.Stellwagen, A. E., Z. W. Haimberger, J. R. Veatch, and D. E. Gottschling. 2003. Ku interacts with telomerase RNA to promote telomere addition at native and broken chromosome ends. Genes Dev. 172384-2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Taggart, A. K., S. C. Teng, and V. A. Zakian. 2002. Est1p as a cell cycle-regulated activator of telomere-bound telomerase. Science 2971023-1026. [DOI] [PubMed] [Google Scholar]
- 25.Ting, N. S., Y. Yu, B. Pohorelic, S. P. Lees-Miller, and T. L. Beattie. 2005. Human Ku70/80 interacts directly with hTR, the RNA component of human telomerase. Nucleic Acids Res. 332090-2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tzfati, Y., T. B. Fulton, J. Roy, and E. H. Blackburn. 2000. Template boundary in a yeast telomerase specified by RNA structure. Science 288863-867. [DOI] [PubMed] [Google Scholar]
- 27.Tzfati, Y., Z. Knight, J. Roy, and E. H. Blackburn. 2003. A novel pseudoknot element is essential for the action of a yeast telomerase. Genes Dev. 171779-1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zappulla, D. C., and T. R. Cech. 2004. Yeast telomerase RNA: a flexible scaffold for protein subunits. Proc. Natl. Acad. Sci. USA 10110024-10029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zappulla, D. C., K. Goodrich, and T. R. Cech. 2005. A miniature yeast telomerase RNA functions in vivo and reconstitutes activity in vitro. Nat. Struct. Mol. Biol. 121072-1077. [DOI] [PubMed] [Google Scholar]
- 30.Zellinger, B., S. Akimcheva, J. Puizina, M. Schirato, and K. Riha. 2007. Ku suppresses formation of telomeric circles and alternative telomere lengthening in Arabidopsis. Mol. Cell 27163-169. [DOI] [PubMed] [Google Scholar]
- 31.Zuker, M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 313406-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.