Abstract
Histone methylation is crucial for transcriptional regulation and chromatin remodeling. It has been suggested that the SET domain containing protein RE-IIBP (interleukin-5 [IL-5] response element II binding protein) may perform a function in the carcinogenesis of certain tumor types, including myeloma. However, the pathogenic role of RE-IIBP in these diseases remains to be clearly elucidated. In this study, we have conducted an investigation into the relationship between the histone-methylating activity of RE-IIBP and transcriptional regulation. Here, we report that RE-IIBP is up-regulated in the blood cells of leukemia patients, and we characterized the histone H3 lysine 27 (H3-K27) methyltransferase activity of RE-IIBP. Point mutant analysis revealed that SET domain cysteine 483 and arginine 477 are critical residues for the histone methyltransferase (HMTase) activity of RE-IIBP. RE-IIBP also represses basal transcription via histone deacetylase (HDAC) recruitment, which may be mediated by H3-K27 methylation. In the chromatin immunoprecipitation assays, we showed that RE-IIBP overexpression induces histone H3-K27 methylation, HDAC recruitment, and histone H3 hypoacetylation on the IL-5 promoter and represses expression. Conversely, short hairpin RNA-mediated knockdown of RE-IIBP reduces histone H3-K27 methylation and HDAC occupancy around the IL-5 promoter. These data illustrate the important regulatory role of RE-IIBP in transcriptional regulation, thereby pointing to the important role of HMTase activity in carcinogenesis.
In eukaryotes, chromatin structure is modulated via the various posttranslational modifications of histone NH2-terminal tails. An accumulating body of evidence suggests that these histone modifications may play a role in a host of chromatin remodeling-associated processes, including replication, transcription, and DNA repair (7). One of the covalent modifications, histone lysine methylation, is facilitated by SET domain-harboring proteins and has been found to have important roles in chromatin remodeling and regulating gene expression (20). The SET domain-harboring proteins are a large family which was identified initially in three Drosophila proteins: Suppressor of variegation [Sur(var)3-9], Enhancer of zeste [E(z)], and Trithorax. Over 200 SET domain-harboring genes have been predicted to exist in various species, suggesting that this domain is highly conserved and that it may play critical roles in certain cellular functions. Within the last few decades, an increasing amount of evidence has been generated to support the notion that aberrant transcriptional regulation contributes to the development of human cancers. In fact, transcription regulatory proteins are often identified in oncogenic chromosomal rearrangements and have been shown to be overexpressed in a variety of malignancies. It has also been suggested that malfunctions in histone methylation may be linked to cancer as well as other diseases. When growth-regulatory genes are methylated in an aberrant manner, the relevant genes may be silenced, possibly resulting in tumor formation. However, in many of the SET domain-harboring proteins, the roles of proteins, including the enzymatic activity of histone lysine methyltransferase, have yet to be fully elucidated.
Via the application of a homology search to discover the SET domain-harboring proteins, we were able to collect different nucleotide sequences from humans and mice. Among these identified SET domain-harboring genes, we further investigated different transcripts of the WHSC1/MMSET/NSD2 gene, which has been determined to be involved in certain human malignancies, including leukemia and multiple myeloma (MM). Specifically, we have focused on the response element II binding protein (RE-IIBP) and further processed the protein by cloning the transcript. RE-IIBP has been reported to initiate translation at exon 15 of the transcript and possesses a SET domain within its C-terminal region (5). Among the WHSC1/MMSET/NSD2 splice variants, the expression of RE-IIBP appears to be universal and was shown to bind to RE-II of the human interleukin-5 (IL-5) promoter and to suppress its transcription (5, 8). However, despite the presence of a SET domain, the histone methyltransferase (HMTase) activity of RE-IIBP, in terms of its relation to transcriptional regulation, has yet to be clearly characterized.
In this study, we have attempted to characterize the HMTase activity of RE-IIBP via functional analyses. We determined that the SET domain-harboring RE-IIBP, which is upregulated in the blood cells of various leukemic patients, exhibits histone H3 lysine 27 (H3-K27) methyltransferase activity and exerts a transcriptional repression effect via the recruitment of histone deacetylase (HDAC). These results allowed us to elucidate the functional role of HMTase RE-IIBP in the development of human cancer or MM. We have demonstrated that RE-IIBP represses IL-5 expression through the histone H3-K27 hypermethylation around the promoter region and H3 hypoacetylation. These results imply that RE-IIBP-mediated histone methylation may constitute an important factor in the regulation of cancer-related target genes.
MATERIALS AND METHODS
Cloning of RE-IIBP.
SET domain-harboring proteins were listed using EnsMart, a data mining tool available at the Sanger website (www.ensembl.org) with a Pfam identification filter (PF00856). The proteins with a high probability of harboring SET domains were then selected via BLAST searches of a conserved domain database (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi). Among the WHSC1/MMSET/NSD2-related gene products, the SET domain-harboring RE-IIBP was amplified using specific primer sets. The full-length open reading frame of the RE-IIBP cDNA was inserted into the pGEX-4T1 bacterial expression vector (Amersham Biosciences) in order to construct the glutathione S-transferase (GST)-RE-IIBP, GST-RE-IIBP-1, and GST-RE-IIBP-2 fusion proteins. RE-IIBP (carrying the mutation R477A) and RE-IIBP (C483A) point mutants were introduced using a QuikChange site-directed mutagenesis kit (Stratagene). For the construction of mammalian expression vectors, we employed pcDNA3.1-HisTOPO (Invitrogen) and CMX-Gal4 in order to create the His-tagged RE-IIBP and Gal4-RE-IIBP proteins. The short hairpin RNA (shRNA) against human WHSC1/MMSET was purchased from Openbiosystem.
Northern blotting.
The adult human tissue samples blotted on membranes (Seegene) were incubated in 4 ml of prehybridization buffer (BD Biosciences) for 1 h at 58°C and then in hybridization buffer containing random-primed 32P-labeled 5′ untranslated region (UTR) RE-IIBP DNA. Hybridization was performed for 3 h at 58°C. The membranes were washed, and signals were visualized by a phosphorimager analyzer.
Immunofluorescence.
HeLa cells were seeded in four-well chamber slides and transiently transfected with enhanced green fluorescent protein-RE-IIBP, pcDNA3.1-HisTOPO-RE-IIBP, WHSC1/MMSET-shRNA and pcDNA3.0, as indicated (see Fig. 3D). After 48 h, the cells were washed in phosphate-buffered saline and fixed with 50% methanol-acetone. After being blocked with 1% bovine serum albumin, the cells were incubated with anti-histone antibodies (Chemicon), anti-dimethyl H3-K9, and anti-dimethyl H3-K27 antibodies (Upstate Biotechnology Inc.), followed by incubation with Cy3-conjugated anti-rabbit and Cy3-conjugated anti-mouse antibodies (Jackson ImmunoResearch Laboratories) and mounting with Aqua Poly Mount (Polysciences). Images were visualized with a confocal laser scanning microscope (Zeiss LSM 510).
FIG. 3.
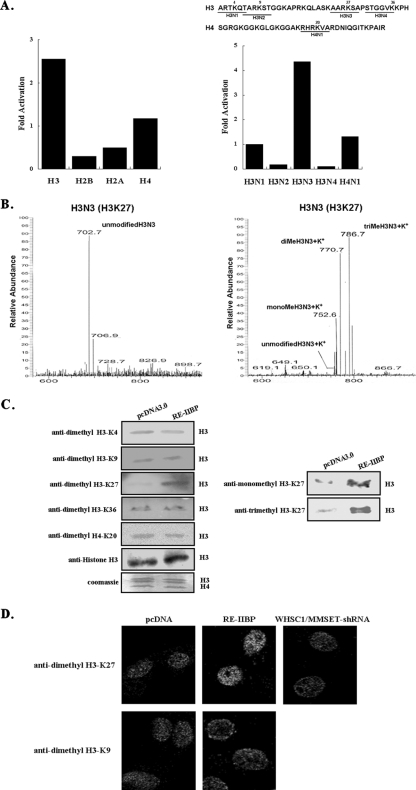
Histone H3-K27 methylation specificity of RE-IIBP. (A) HMTase assays were performed with individual histone subunits and five synthesized histone peptides, shown above as substrates. Reaction products were analyzed by the filter binding assay and scintillation counting. (B) Synthesized peptides (H3N3) were used as substrates in the HMTase assay with purified RE-IIBP. After 3 h, the proteins were precipitated with TCA buffer and removed by centrifugation. The methylated peptide samples were analyzed by LC-MS. (C) Transiently transfected cell extracts were immunoblotted against anti-dimethyl H3-K4, anti-dimethyl H3-K9, anti-monomethyl H3-K27, anti-dimethyl H3-K27, anti-trimethyl H3-K27, anti-dimethyl H3-K36, and anti-dimethyl H4-K20 antibodies. The equal amount of sample loading was confirmed by Western blotting with antibodies against histone H3. (D) Immunofluorescence staining of HeLa cells transfected with pcDNA3.1-HisTOPO-RE-IIBP and pSM2c-WHSC1/MMSET-shRNA using antihistone antibodies and anti-dimethyl H3-K9 and anti-dimethyl H3-K27 antibodies. Antibody staining was visualized using Cy3-conjugated antibodies. Me, methylated.
HMTase assays.
HMTase assays were carried out at 30°C for 2 h in 50-μl volumes containing 50 mM Tris-HCl, pH 8.5, 20 mM KCl, 10 mM MgCl2, 10 mM beta-mercaptoethanol, 1.25 M sucrose, 100 nCi of S-adenosyl-l-[methyl-14C]methionine ([14C]SAM) (Amersham Biosciences), 1 μg/μl core histones from calf thymus (Roche) or histone peptides, and 0.5 to 2.5 μg of GST-RE-IIBP, GST-RE-IIBP-1, GST-RE-IIBP-2 GST-RE-IIBP (C483A), GST-RE-IIBP (R477A), and GST. Peptides (H3N1, H3N2, H3N3, H3N4, and H4N1) were synthesized based on their N-terminal amino acid sequences of H3 and H4 histones (Peptron). Proteins/peptides were filtered using p81 filter paper (Upstate Biotechnology Inc.) and washed three times with cold 10% trichloroacetic acid (TCA) and 95% ethanol for 5 min at room temperature. The filters were allowed to air dry; 2 ml of Ultima Gold (Perkin Elmer) was added, and [14C]SAM was quantified using a scintillation counter. The HMTase assay using cell extracts incubated with RE-IIBP protein was conducted as previously reported, with minor modifications (12). The reaction products were separated via 14% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and visualized with a phosphorimage analyzer (Perkin Elmer).
LC-MS.
Small quantities (100 μM) of synthetic peptides (H3N1 and H3N3) were used as substrates in the HMTase assay with RE-IIBP; the reaction was stopped by 10% TCA precipitation. After the precipitates were removed by centrifugation, the supernatants were retrieved, and methylated peptides in the supernatants were analyzed by liquid chromatography-mass spectrometry (LC-MS) at the Korea Basic Science Institute.
Transfection assay.
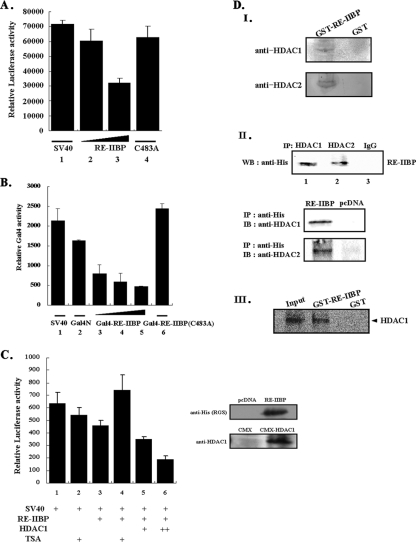
The transfection assay was conducted using CMX-Gal4-SV40 (where CMX is a cytomegalovirus-based construct and SV40 is simian virus 40) reporter and pcDNA3.1-HisTOPO-RE-IIBP or CMX-Gal4-RE-IIBP as internal controls where indicated. The quantity of DNA in each transfection was kept constant via the addition of pcDNA3.0. HeLa cells were transfected with CMX-Gal4-SV40 reporter (100 ng) and pcDNA3.1-HisTOPO-RE-IIBP (200 and 300 ng), CMX-HDAC1 (100 and 200 ng), CMX-Gal4N (100 ng), Gal4-RE-IIBP (100, 200, and 300 ng), or Gal4-RE-IIBP (C483) (200 ng). Cells were harvested and assayed for luciferase activity using a luciferase assay system (Promega). Each value is expressed as the mean of six replicates from a single assay, and the results were confirmed by at least three repetitions.
Immunoprecipitation.
For the HDAC interaction assays, the transfected cells were lysed in radioimmunoprecipitation assay lysis buffer and treated with DNase at 37°C for 30 min. After cells were immunoprecipitated with anti-His (Qiagen), anti-HDAC1, and anti-HDAC2 antibodies (Santa Cruz Biotechnology) and protein A agarose beads (Amersham Biosciences), the bound proteins were analyzed by immunoblotting with anti-His, anti-HDAC1, and anti-HDAC2 antibodies.
Chromatin immunoprecipitation ChIP and qPCR.
The HeLa cells were transfected with 4 μg of DNA and harvested after 48 h. Cells were cross-linked with 1% formaldehyde in the medium for 10 min at 37°C, followed by the addition of 10× glycine for 5 min at room temperature, after which they were scraped into SDS lysis buffer. The samples were further sonicated and diluted for immunoprecipitation with antibodies as indicated (see Fig. 5C). The immunoprecipitates were eluted and reverse cross-linked. The DNA fragments were purified and PCR amplified for quantification. Anti-dimethyl H3-K27, anti-acetyl histone H3, anti-acetyl histone H4 (Upstate Biotechnology Inc.), anti-HDAC1 (Santa Cruz Biotechnology), and anti-pan methyl lysine antibodies (Abcam) were employed for immunoprecipitations. The primers utilized for IL-5 promoter analysis were 5′-TATTAACCCAAAGATTCCTTC-3′ and 5′-GCAAAGAAAGTGCATAGTACAA-3′; for the IL-5 promoter distal region, 5′-ACCTTCCCTCTTTATCTTCA-3′ and 5′-TAAGGTAGACCACTAAACAG-3′ were used. Quantification of the chromatin immunoprecipitation (ChIP) DNA amount was confirmed with a Sybr Green PCR kit (Applied Biosystems, Inc.). Normalization was performed using input DNA levels as controls in the same reaction. Primer concentration for quantitative PCR (qPCR) was 0.2 μM/25 μl. The thermal cycler conditions were as follows: a hold for 10 min at 95°C, followed by three steps of PCR for 50 cycles of 94°C for 15 s, 60°C for 30 s, and 72°C for 30 s (RG-3000; Corbett Research). The relative expression ratio of input was calculated by the 2−ΔΔ(CT) (where CT is threshold cycle) method.
FIG. 5.
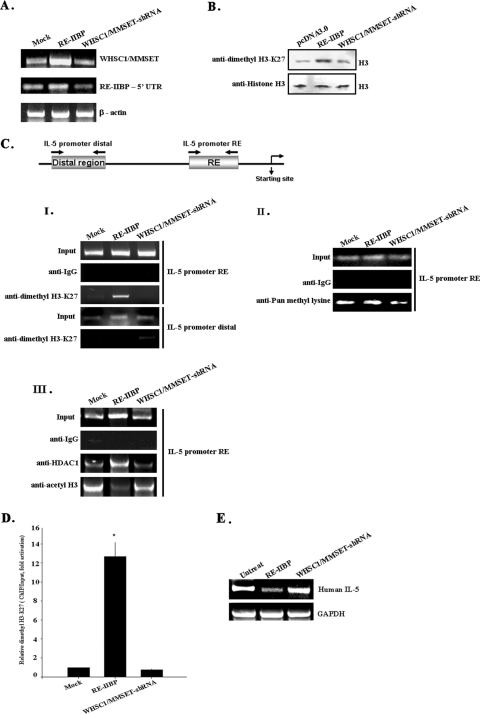
Effects of RE-IIBP transcript by RNA interference. (A) HeLa cells were mock transfected or transfected with pcDNA3.1-HisTOPO-RE-IIBP and pSM2c-WHSC1/MMSET-shRNA, and WHSC1/MMESET and RE-IIBP expression levels were confirmed by RT-PCR with specific primers. (B) The status of lysine methylation was determined using transiently transfected cells with RE-IIBP and WHSC1/MMSET-shRNA. Transfected cells were lysed and immunoblotted with anti-dimethyl H3-K27 antibodies. (C) HeLa cells were transfected with the pcDNA3.0, pcDNA3.1-HisTOPO-RE-IIBP, and pSM2c-WHSC1/MMSET-shRNA. Following transfection, ChIP assays were performed employing control IgG, anti-dimethyl H3-K27 (I), anti-pan methyl lysine antibodies (II), and anti-acetyl H3 and anti-HDAC1 antibodies (III). The immunoprecipitated DNA fragments were amplified by PCR from the proximal and distal promoter regions of the integrated IL-5. (D) ChIP analysis was performed with the indicated antibodies and examined by qPCR in the presence of proximal IL-5 promoter fragments. The dimethyl H3-K27 level was normalized by input. (E) Total RNA was isolated from HeLa cells. RT-PCR analysis was performed with primers for IL-5. Human glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an RNA loading control. The relative levels of gene expression of RE-IIBP mRNA in untreated cells and RE-IIBP- and WHSC1/MMSET-shRNA-transfected cells were compared by RT-PCR.
Patients' samples.
The mononuclear cells were obtained from bone marrow of eight leukemic patients at the time of diagnosis. Patients 1, 2, and 8 had acute lymphoblastic leukemia (ALL) of the precursor B cell type, and patients 3, 4, and 6 had acute lymphoblastic leukemia of the T cell type. Patients 5 and 7 had an acute myelogenous leukemia (AML) and M2-type AML, respectively. Informed consent was obtained from guardians of each patient, and study approval was obtained from the Institutional Review Board of the Chonnam National University Hospital.
Immunophenotype assay.
Mononuclear cells that were obtained from bone marrow were isolated by Ficoll-Hypaque density centrifugation. Immunophenotype analyses of lymphocyte markers were performed on a flow cytometer. Cells were stained using anti-CD2 and anti-CD7 for the detection of T-lineage cells; anti-CD10, anti-CD19, and anti-CD22 for the detection of B-lineage cells; and anti-CD13 and anti-CD33 for the detection of myeloid lineage cells.
RESULTS
Structural features of RE-IIBP protein.
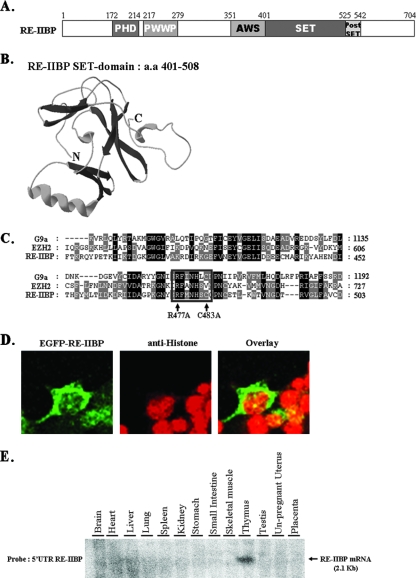
The structural characteristics of RE-IIBP protein were determined via protein sequence analysis. RE-IIBP was found to harbor highly conserved regions, including a PHD (plant homeodomain) domain, SET domain with a nearby AWS (associated with SET) domain, and a postSET domain (Fig. 1A). Structure prediction of the RE-IIBP SET domain was performed by using the stand-alone Swiss-PdbViewer program (11). Results of three-dimensional modeling indicated that the RE-IIBP SET domain (amino acids 401 to 508) is composed of several antiparallel beta sheets and an alpha helix which is similar to the oligonucleotide-oligosaccharide binding fold configuration shared by many SET domain proteins such as ALL-1, TRX, ASH1, and SET7 (11) (Fig. 1B). Proteins harboring a SET domain, including the G9a and Suv39h family proteins, have been shown to exhibit HMTase activity (17). Many SET domain-harboring proteins with HMTase activity also harbor two cysteine-rich regions located adjacent to the SET domain. RE-IIBP also possesses two cysteine-rich regions in its SET domain C terminus, although it does not share other structural motifs with G9a and Enhancer of Zest homolog 2 (EZH2) (Fig. 1C). Within the SET domain, an (H/R)ΦΦNHSC motif (where Φ indicates a hydrophobic residue) has been shown in previous works to be an important catalytic site for HMTase activity (15). Notably, RE-IIBP protein harbors an RΦΦNHSC motif (Fig. 1C, boxed), implying that with two cysteine-rich regions, the RE-IIBP SET domain may exhibit HMTase enzymatic activity. Next, we investigated the subcellular localization of ectopically introduced RE-IIBP. Similar to the previous studies, RE-IIBP localized both within cytoplasm and nucleoli compartments (8, 18) (Fig. 1D). In order to determine more precisely the expression pattern of the RE-IIBP gene in adult humans, multitissue samples were hybridized to a 32P-labeled 5′ UTR RE-IIBP DNA probe. The 2.1-kb RE-IIBP mRNA transcript was most abundant in the thymus, although it was detected ubiquitously (Fig. 1E).
FIG. 1.
Structural features of the RE-IIBP. (A) Schematic view of RE-IIBP domains: amino acids 172 to 214 were predicted to be the PHD domain; amino acids 217 to 279 were predicted to be the PWWP (proline-tryptophan-tryptophan-proline) domain; amino acids 401 to 525 were predicted to be the SET domain; and amino acids 526 to 542 were predicted to be the postSET domain. (B) Three-dimensional modeling of RE-IIBP SET domain was performed by using the stand-alone Swiss-PdbViewer with an installed protein loop database. (C) Multiple alignments of the SET domains of G9a, EZH2, and RE-IIBP proteins. RE-IIBP protein harbors an RΦΦNHSC motif (where Φ indicates a hydrophobic residue; boxed). The arrows indicate the R477 and C483 residues of the RΦΦNHSC motif that are mutated for testing the HMTase activity of RE-IIBP protein. Conserved amino acid residues between three sequences are shown in black, and similar residues between these two sequences are shaded in gray. (D) HeLa cells were transfected with enhanced green fluorescent protein (EGFP)-RE-IIBP, stained with anti-histone antibodies, and detected with Cy3-conjugated secondary antibodies. (E) The adult human tissue blot was hybridized to a 32P-labeled 5′ UTR RE-IIBP DNA probe. The 2.1-kb RE-IIBP mRNA transcript is indicated by an arrow.
RE-IIBP is a novel SET-dependent HMTase.
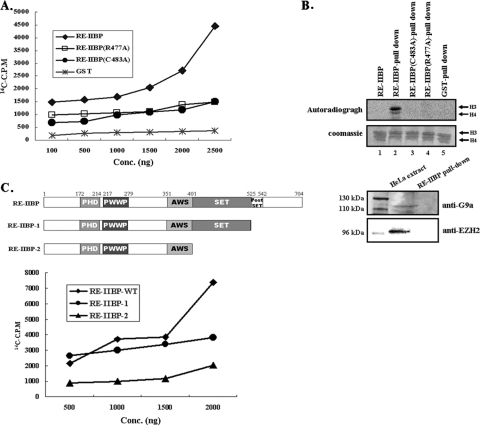
In order to determine whether RE-IIBP might have HMTase activity, we first conducted an HMTase assay with a purified GST-RE-IIBP fusion protein. Core histones and [14C]SAM were utilized as the substrate and methyl donor, respectively. As shown in Fig. 2B, lane 1, we failed to detect the histone-methylating activity of GST-RE-IIBP under our assay conditions using an SDS-PAGE gel developed by phosphorimager. Therefore, we performed more a sensitive in vitro HMTase assay using scintillation counting. Using an HMTase assay kit, we were able to detect the intrinsic HMTase activity of RE-IIBP on core histones in a dose-dependent manner (Fig. 2A). When we tested the HMTase activity of purified RE-IIBP on nucleosomal substrates, we could detect enzymatic strength similar to that detected for core histones in a scintillation counting assay (data not shown). Amino acid sequence alignment of the mammalian SET domain-harboring proteins from EZH2 and G9a revealed that the positions of the cysteine and arginine residues are highly conserved (Fig. 1C, boxed). Previous reports have shown that the replacement of the conserved cysteine and arginine residues abolished HMTase activity of ESET and G9a (17, 19). Therefore, we created point mutants in which the cysteine (amino acid 483) or arginine (amino acid 477) was replaced with alanine and then examined the changes in the enzyme activities. No HMTase activities were observed with either the RE-IIBP C483A or R477A point mutant, indicating the importance of the two conserved residues in the SET domain for the HMTase activity (Fig. 2A).
FIG. 2.
HMTase activity of RE-IIBP. (A) Core histones were used as substrates in the HMTase assay with GST-RE-IIBP and GST-RE-IIBP point mutants (C483A and R477A). Methylation levels were quantified via filter binding assay, and data are represented as raw counts per minute incorporated. (B) Purified GST-RE-IIBP, GST-RE-IIBP point mutants, and GST proteins were incubated with HeLa cell extracts and subjected to HMTase assays. GST-bound beads were used as the negative control. The lower panel represents GST-RE-IIBP pulled down and immunoblotted with anti-G9a and anti-EZH2 antibodies. HeLa cell extracts were used as the positive control. (C) A similar HMTase assay as that described in panel A was performed with RE-IIBP deletion mutants (GST-RE-IIBP-1 and GST-RE-IIBP-2). A schematic representation of the domain structure of the recombinant RE-IIBP deletion mutants is shown. Conc, concentration; WT, wild type.
Next, we assumed that the apparent absence of strong signal in the in vitro assay using the phosphorimager might have been caused by the lack of certain associating cellular factor(s) for HMTase activity. Therefore, we designed and conducted an HMTase assay in the presence of cellular components, mimicking the in vivo status of the protein via RE-IIBP GST pull-down assays. When GST-RE-IIBP was incubated with HeLa cell extracts and washed precipitates were added in the HMTase assay, we observed strong HMTase activity (Fig. 2B, upper panel, lane 2). The incubation of GST alone with cell extracts evidenced no enzyme activity (Fig. 2B, upper panel, lane 5). To rule out the possibility that a different histone H3-methylating HMTase might copurify with RE-IIBP during incubation, immunoblotting using anti-EZH2 and anti-G9a antibodies was performed and proved that histone H3-methylating enzymes EZH2 and G9a were not pulled down during the incubation (Fig. 2B, lower panel). These results suggest that RE-IIBP exhibited stronger HMTase activity when incubated with cell extracts, possibly via the aid of associating factors, under in vivo conditions. Again, the HMTase activities were completely abolished by mutations of C to A and R to A (Fig. 2B, upper panel, lanes 3 and 4). The point mutant analysis further indicates that the HMTase activity of RE-IIBP might be from the RE-IIBP itself rather than from other HMTases associated during the incubation. The importance of the postSET domain in HMTase activity has been reported previously (16). Deletion of the postSET domain drastically reduced the HMTase activity of RE-IIBP, and deletion of the SET domain completely abolished enzyme activity, as expected (Fig. 2C, RE-IIBP-1 and -2). Taken together, these results prove the presence of SET domain-dependent intrinsic HMTase activity of RE-IIBP and show further that enhanced enzyme activity when incubated with cell extracts.
RE-IIBP selectively transferred methyl groups to K27 of histone H3.
It has been generally accepted that the targeting of lysine residues is a unique property of certain HMTases and that the identification of methylated lysine might allow us to predict its role in transcriptional regulation. Therefore, we attempted to determine which lysine was methylated by RE-IIBP HMTase activity. First, we performed an HMTase assay with individual histone subunits. The HMTase activity of RE-IIBP appears to be very specific for histone H3 and to a lesser extent for histone H4 (Fig. 3A, left panel). This is consistent with the in vitro HMTase assay result in phosphorimager analysis, which showed that RE-IIBP had strong methylation activity toward histone H3 and exerted poor activity toward H4 (Fig. 2B, lane 2). Then, we conducted a more specific HMTase assay with histone H3 and H4 peptides, in which each six-amino-acid peptide contains one lysine residue. In a finding consistent with the assay using individual histone subunits, incubation with peptide H3N3 enhanced strong methylation activity, which clearly indicates that histone H3-K27 is the major methylation target residue for RE-IIBP (Fig. 3A, right panel). To further confirm the lysine specificity of RE-IIBP, histone peptides were analyzed by LC-MS after the HMTase assay. The calculated molecular mass of the H3N3 peptide is 702 Da, and each addition of a methyl group increases the mass by 14 to 15 Da. The nonmethylated H3N3 peptide had its main peak at 702.7 Da, while the mono-, di-, and trimethylated peptides appeared at 752.6, 770.7, and 786.7 Da, respectively, with the 37-Da mass of K+ incorporated during the peptide sample preparation after the HMTase assay (Fig. 3B). The spectroscopy result clearly indicates that histone peptides H3N3 were mono-, di-, and trimethylated at H3-K27. No peaks corresponding to mono-, di-, or trimethylated forms of the H4N1 (H4-K20) peptides were observed (data not shown).
The methylation of H3-K27 by RE-IIBP was further confirmed in vivo via the immunoblotting of the RE-IIBP-overexpressed cells with lysine methylation-specific antibodies. After the HeLa cells had been transiently transfected with RE-IIBP, total cell lysates were immunoblotted against antibodies specific to dimethylated K4, K9, K27, K36 of H3, and K20 of H4. Consistent with the results of the in vitro peptide study and MS analysis, H3-K27 methylation increased significantly in the RE-IIBP-overexpressing cell lysates compared to the other tested lysine residues (Fig. 3C, left panel). When we asked whether RE-IIBP might mono- or trimethylate H3-K27, we determined that RE-IIBP can induce both mono- and trimethylation of H3-K27 as well, which is consistent with the MS results (Fig. 3C, right panel). Collectively, these results show that RE-IIBP methylates histones H3 and H4 and specifically H3-K27 of histone H3 under both in vitro and in vivo conditions. The methylation of histone H4 was much weaker than that of histone H3 but was consistently observed in both assays with GST-RE-IIBP alone and GST-RE-IIBP pull-down precipitates. These results suggest that RE-IIBP might target other lysine residue(s) in histone H4 besides H4-K20.
To further investigate the H3-K27 specificity of RE-IIBP, we performed immunofluorescence experiments with anti-dimethylated H3-K9 and H3-K27 antibodies in RE-IIBP-transfected HeLa cells. As shown in Fig. 3D, overexpression of RE-IIBP results in significant increase in H3-K27 dimethylation compared to that of pcDNA3.0-treated cells. There were no apparent differences for the H3-K9 dimethylation pattern between RE-IIBP- and control vector-transfected cells (Fig. 3D). WHSC1/MMSET-shRNA knock-down resulted in the selective loss of H3-K27 dimethylation (Fig. 3D).
RE-IIBP represses transcription through HDAC recruitment.
The methylation of histone H3-K27 has been reported to be involved in the transcriptional repression of the Hox gene (13). In order to determine more precisely whether the methylation of H3-K27 of RE-IIBP can be attributed to the repression of general transcription, we conducted a transient transfection assay using the CMX-Gal4-SV40 reporter system. The transient transfection of the SV40 promoter-driven reporter itself elicited certain levels of basal transcriptional activity (Fig. 4A). Transfection with an increasing amount of RE-IIBP resulted in the repression of luciferase activity in a dose-dependent manner, ultimately reducing the activity level to 47% of basal transcriptional activity (Fig. 4A, lanes 2 and 3). When HMTase activity was determined in cells transfected with the RE-IIBP (C483A), a significant decrease in transcriptional repression was observed (Fig. 4A, lane 4). These experiments allowed us to conclude that HMTase activity is important for transcriptional repression by RE-IIBP. We next asked whether direct recruitment of RE-IIBP to the promoter could mediate transcriptional repression. The Gal4-RE-IIBP fusion protein repressed the basal transcription of the reporter gene in a dose-dependent manner (Fig. 4B, lanes 3 to 5). Again, the RE-IIBP (C483A) point mutant failed to repress basal transcription, which strongly indicated that HMTase activity of RE-IIBP is critical for the transcriptional repression activity (Fig. 4B, lane 6). In order to characterize in more detail the manner in which RE-IIBP-associated transcriptional repression could be mediated via HDAC, we conducted a series of SV40-driven promoter assays, using HDAC and the HDAC inhibitor, trichostatin A (TSA). The overexpression of RE-IIBP repressed basal transcription and the addition of TSA significantly ameliorated the transcriptional repression effected by RE-IIBP (Fig. 4C, lanes 3 and 4). This TSA-associated effect suggests strongly that HDAC is involved in RE-IIBP-induced transcriptional repression. We also conducted cotransfection experiments with increasing amounts of HDAC1, and, as expected, a further repression of transcription was detected, occurring in a dose-dependent manner (Fig. 4C, lanes 5 and 6).
FIG. 4.
RE-IIBP represses transcription and associates with HDAC. (A) HeLa cells were transfected with Gal4-SV40 and increasing concentrations of RE-IIBP constructs, as indicated. Following transfection of the pcDNA3.1-HisTOPO-RE-IIBP and the point mutant (C483A), cell extracts were assayed for luciferase activity. (B) Cotransfections of a Gal4-SV40 luciferase reporter with the GAL4 DNA binding domain alone or Gal4-RE-IIBP and Gal4-RE-IIBP point mutant (C483A) are shown. (C) HeLa cells were transfected with Gal4-SV40 reporter vector, pcDNA3.1-HisTOPO-RE-IIBP, and CMX-HDAC1. TSA was added, as indicated, 24 h after transfection. Expression of RE-IIBP and HDAC1 used in transfection assays was confirmed by immunoblotting using anti-His and anti-HDAC1 antibodies. (D) GST-RE-IIBP and GST were incubated in HeLa cell extract and immunoblotted with anti-HDAC1 and anti-HDAC2 antibodies (I). HeLa cells were transfected with pcDNA3.1-HisTOPO-RE-IIBP (RE-IIBP) and pcDNA3.0 (pcDNA) constructs as indicated (II). After cell lysates were treated with DNase, immunoprecipitation was performed with anti-HDAC1, anti-HDAC2, anti-His, and anti-IgG antibodies; immunoblot analysis was then performed with anti-His (RE-IIBP) or anti-HDAC1 or anti-HDAC2 antibodies. (III) CMX-HDAC1 was in vitro transcribed and translated and incubated with purified GST-RE-IIBP and GST bound to beads. Pulled-down proteins were analyzed by SDS-PAGE and phosphorimager. IP, immunoprecipitation; IB, immunoblotting; WB, Western blotting.
On the basis of the above observation that RE-IIBP-associated transcriptional repression is dependent upon HDAC activity, we attempted to characterize the interaction occurring between RE-IIBP and HDACs. We pulled down endogenous HDACs from a HeLa cell extract using GST-RE-IIBP, followed by immunoblotting with anti-HDAC1 and -2 antibodies. Compared to the pull-down incubation with the GST alone, HDAC1 and -2 were clearly detected (Fig. 4D, panel I). We then overexpressed RE-IIBP in HeLa cells and conducted immunoprecipitations using anti-HDAC1 and -2 antibodies. Western blotting with the anti-His antibodies of the immunoprecipitates clearly indicated the occurrence of interactions between RE-IIBP and HDAC1 and -2 (Fig. 4D, panel II). Then, we performed the immunoprecipitation assay with anti-His antibodies and immunoblotted with anti-HDAC1 and -2 antibodies and confirmed the interactions between RE-IIBP and HDAC1 and -2 (Fig. 4D, panel II). Again, HDAC1 was transcribed and translated in vitro and incubated with purified GST-RE-IIBP or GST alone. As had been expected, HDAC1 interacted with the GST-RE-IIBP but not with the GST alone, further evidencing a direct interaction between RE-IIBP and HDAC1 (Fig. 4D, panel III). Collectively, these results suggest that RE-IIBP and HDACs 1 and 2 are physically associated in vitro and in vivo and mediate the observed transcriptional repression effected by RE-IIBP.
Targeted interferences of WHSC1/MMSET transcripts by shRNA.
In order to further define the general role of the WHSC1/MMSET transcripts, including RE-IIBP, WHSC1/MMSET was silenced using shRNA. In order to determine whether the WHSC1/MMSET-shRNA was specific for the WHSC1/MMSET transcripts, including RE-IIBP, we introduced pSM2c-WHSC1/MMSET-shRNA into HeLa cells. Interestingly, reverse transcription-PCR (RT-PCR) analysis using primers specific for WHSC1/MMSET and the RE-IIBP 5′ UTR revealed that WHSC1/MMSET-shRNA had a stronger effect on RE-IIBP than WHSC1/MMSET (Fig. 5A). This suggests that the WHSC1/MMSET-shRNA construct efficiently reduced the levels of RE-IIBP transcripts and was appropriate for further RE-IIBP knock-down experiments. Next, we conducted a comparison of the histone H3-K27 methylation status of RE-IIBP and WHSC1/MMSET-shRNA to that of the pcDNA3.0-transfected HeLa cell extracts using anti-dimethyl H3-K27 antibodies. As expected, we determined that RE-IIBP overexpression effected an increase in the dimethylation of histone H3-K27 compared to the pcDNA3.0 control, a result which was consistent with the previous results in this study (Fig. 5B). When the WHSC1/MMSET-shRNA was transfected, we noted a significant decrease in H3-K27 dimethylation, which indicated that WHSC1/MMSET-shRNA did, indeed, specifically block the HMTase activity of RE-IIBP (Fig. 5B).
RE-IIBP has been demonstrated to interact with its specific binding region, encompassing position −123 to −92 at the human IL-5 gene promoter, and also repressed the transcription of the IL-5 gene (5). Because RE-IIBP associates with HDAC1 and -2 and promotes the methylation of H3-K27, we reasoned that RE-IIBP should target histone H3-K27 hypermethylation at the promoter region of IL-5. To evaluate this hypothesis, we conducted ChIP assays using anti-dimethyl H3-K27 antibodies and determined the histone methylation status of the RE-IIBP binding region of the IL-5 promoter. We transfected the HeLa cells with pcDNA3.0 vector as well as with RE-IIBP and WHSC1/MMSET-shRNA constructs. Immunoprecipitations were conducted with anti-dimethyl H3-K27 antibodies, and nonspecific immunoglobulin G (IgG) was used as a negative control. The precipitated DNA fragments were then purified and amplified via PCR with primers specific to the IL-5 promoter regions. A constant level of input DNA was observed from the cells transfected with pcDNA3.0, RE-IIBP, and WHSC1/MMSET-shRNA (Fig. 5C, panel I). The control IgG antibodies revealed a background of the PCR-amplified IL-5 promoter region (Fig. 5C, panel I). As had been expected, we observed a marginal increase in H3-K27 dimethylation in the RE-IIBP-transfected cells compared to the mock-transfected cells (Fig. 5C, panel I). When RE-IIBP was silenced by WHSC1/MMSET-shRNA, we observed a significant reduction in the levels of H3-K27 dimethylation (Fig. 5C, panel I, IL-5 promoter RE). On the contrary, there were no changes in histone H3-K27 dimethylation levels in the distal IL-5 promoter region. Consistent results were obtained from the ChIP analysis using anti-pan methyl lysine antibodies (Fig. 5C, panel II). To further test whether RE-IIBP-mediated IL-5 repression is facilitated by direct HDAC recruitment to the promoter and causes histone hypoacetylation, we conducted ChIP analysis with anti-HDAC1 and anti-acetylated histone H3 and H4 antibodies. The results indicated that HDAC1 is recruited to the IL-5 promoter region when RE-IIBP is overexpressed (Fig. 5C, panel III). Again, WHSC/MMSET-shRNA significantly reduced the HDAC1 recruitment to the IL-5 promoter (Fig. 5C, panel III). Consistent with this study, we detected a significantly reduced histone H3 acetylation level around the IL-5 promoter region (Fig. 5C, panel III). We could not observe significant histone H4 acetylation level changes, which is consistent with our previous results showing primary RE-IIBP effects on histone H3 (data not shown). When RE-IIBP was knocked down by shRNA, the occupancy by HDAC1 was reduced, and histone H3 acetylation levels were increased around the IL-5 promoter (Fig. 5C, panel III, WHSC1/MMSET-shRNA). On the basis of these results, we provide evidence that RE-IIBP-mediated histone H3-K27 methylation recruited HDAC1, promoted targeted histone H3 hypoacetylation around the IL-5 promoter, and repressed its transcription. To further clarify the role of RE-IIBP in H3-K27 methylation around the IL-5 promoter, we performed a ChIP assay with real-time PCR. Using anti-dimethyl H3-K27 antibodies, RE-IIBP- and WHSC1/MMSET-shRNA-transfected cells were immunoprecipitated. An apparent increase in dimethylated H3-K27 level around the IL-5 promoter region was detected upon RE-IIBP transfection, and a substantial decrease in methylation level was seen when WHSC1/MMSET-shRNA was transfected (Fig. 5D). In order to further determine whether RE-IIBP actually repressed the expression of IL-5, we conducted RT-PCR analysis on the same RE-IIBP- and WHSC1/MMSET-shRNA-transfected HeLa cells. In the RE-IIBP-overexpressing HeLa cells, IL-5 expression was reduced, whereas the knock-down of RE-IIBP induced an increase in IL-5 expression (Fig. 5E, WHSC1/MMSET-shRNA). Altogether, these data support the suggestion that RE-IIBP actively represses IL-5 expression via the histone H3-K27 methylation and the H3 hypoacetylation of the IL-5 promoter region via the recruitment of HDAC1.
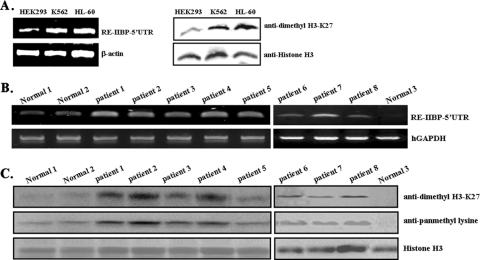
RE-IIBP expression and histone methylation are increased in leukemia patients.
Considering the known relationship of the RE-IIBP-related protein family with human carcinoma including leukemia, the levels of RE-IIBP expression and histone H3-K27 dimethylation were compared among different leukemic cell lines, K562 (erythroleukemia) and HL-60 (myelomonocytic leukemia). Compare to expression levels of HEK293 cell lines, both RE-IIBP expression and dimethylated histone H3-K27 levels were notably increased to similar levels among different leukemia cell lines (Fig. 6A). Next, we further investigated whether RE-IIBP levels were differently regulated in leukemia patients by measuring the levels of RE-IIBP expression in bone marrow mononuclear cells from leukemic patients. The histones were isolated from healthy subjects and from the white blood cells obtained from patients with different leukemia types. After the isolation of mononuclear cells, cell populations were immunophenotyped by flow cytometry, and their characteristics are shown in Table 1. Two patients had diagnosed precursor B-type ALL, three had T-cell ALL, one had AML, one had AML with M2, and one had precursor B-type ALL and TEL-AML(+). Using semiquantitative RT-PCR analysis with primers specific to the 5′ UTR RE-IIBP region, it has been demonstrated that RE-IIBP expression tends to be elevated in leukemia patients compared to what is seen in normal cell preparations (Fig. 6A, leukemia patients 1 to 8). The levels of histone methylation in each of the patients were detected using both anti-pan methyl lysine and anti-dimethyl H3-K27 antibodies. Because of the histone H3-K27 methylation specificity of RE-IIBP, we focused on the methylation status of histone H3. Using anti-dimethyl H3-K27 antibodies, significantly increased H3-K27 dimethylation levels were confirmed in leukemia patients although slight differences of H3-K27 dimethylation levels among the samples were observed (Fig. 6C, upper panel). Elevated levels of histone H3 pan-methylation were also detected in the cells acquired from the leukemia patients (Fig. 6C, middle panel). Consistent with previous results in this study, the histone H4 methylation status increases were also detected in leukemia patients' histone H4 though these were less dramatic than increases of histone H3 (data not shown). These results suggest that the increased RE-IIBP expression observed in leukemia patients may be responsible, in part, for the increased methylation status of histones H3 including H3-K27 dimethylation.
FIG. 6.
RE-IIBP expression and histone methylation levels are increased in leukemia patient samples. (A) Nuclear extracts from leukemic cell lines K562 and HL-60 amplified with primers for the 5′ UTR RE-IIBP region were immunoblotted with anti-dimethyl H3-K27 antibodies. (B) Semiquantitative RT-PCR analysis was performed with primers for the 5′ UTR RE-IIBP region. Human glyceraldehyde-3-phosphate dehydrogenase (hGAPDH) was used as an RNA loading control. (C) Histones extracted from blood cells of healthy individuals and from leukemia patients were separated by 18% SDS-PAGE and immunoblotted against anti-dimethyl H3-K27 and anti-pan methyl lysine antibodies. The loading of equal amounts of sample was confirmed by Western blotting with antibodies against histone H3.
TABLE 1.
Clinical data for patients with leukemia
| Patient no. | Sex | Diagnosis | Amount (%) of surface antigen in samples used for immunophenotypinga
|
||
|---|---|---|---|---|---|
| CD19 | CD7 | CD13 | |||
| 1 | F | ALL, precursor B-cell type | 88 | 10 | |
| 2 | F | ALL, precursor B-cell type | 99.6 | 0.4 | 0.5 |
| 3 | F | ALL, T-cell type | 10 | 93 | 23 |
| 4 | M | ALL, T-cell type | 2 | 98 | 1.4 |
| 5 | F | AML | 0.6 | 2 | 91 |
| 6 | F | ALL, T-cell type | 12 | 85 | 3.5 |
| 7 | F | AML, M2 type | 0.2 | 93 | 24.5 |
| 8 | F | ALL, precursor B-cell type; TEL-AML(+) | 92 | 3 | 1.4 |
CD19 is a general B-lineage surface antigen; CD7 is found on T-lineage and ALL, precursor T-cell type, cells; CD13 is found on myeloid lineage cells.
DISCUSSION
While searching for uncharacterized proteins with SET domains, we have identified several different transcripts of the WHSC1/MMSET/NSD2 gene, which is involved in certain types of leukemia. Among these, we focused on MMSET II and RE-IIBP and elected to clone the transcripts. Via either alternative splice or some different transcriptional initiation protocol, MMSET generates four different products, MMSET I, MMSET II, MMSET III, and RE-IIBP. Two of the transcripts, MMSET II and RE-IIBP, are expected to harbor an intact SET domain at the C-terminal region. As the principal purpose of this study was to evaluate the HMTase activity in the SET domain-harboring proteins and to further investigate its role in human disease, particularly carcinoma, we elected to amplify the shortest transcript variant with an intact SET domain, namely RE-IIBP. The close genetic and biochemical relationship of RE-IIBP with the MM-related WHSC1/MMSET protein clearly indicates its role in certain types of human diseases, including MM. However, the molecular mechanism by which lysine methylation of histones by RE-IIBP contributes to chromatin remodeling and transcription of target genes in these diseases remains unknown. Due to the potential HMTase activity centered on the SET domain of RE-IIBP, we attempted specifically to identify the HMTase activity of RE-IIBP and to determine its effects on the transcriptional regulation of the target gene, among other related activities. The relevance of histone lysine methylation to transcriptional regulation strongly indicates that the presence or absence of the SET domain in these transcripts may exert a crucial impact on the regulation of leukemia pathogenesis. For example, the N terminus of mixed-lineage leukemia becomes fused to a wide variety of proteins via chromosomal translocation (1, 3). The fusion proteins lose the C-terminal SET domain with H3-K4 HMTase activity, which affects specific transcriptional regulation in patients suffering from myeloid leukemia.
In this study, we have characterized the SET domain-harboring protein RE-IIBP, which is a member of the WHSC1/MMSET transcript family. The significance of this study is in the demonstration that RE-IIBP exhibits histone H3-K27-specific methyltransferase activity and also the finding that the RE-IIBP is overexpressed in leukemia patients. We have shown that RE-IIBP expression is significantly increased in leukemia patients, hence, the observed elevation in the levels of histone H3 methylation, especially the H3-K27 methylation level. It is intriguing that similar levels of RE-IIBP overexpression and increased histone H3-K27 dimethylation were observed among patients with different types of leukemia. Interestingly, the analyses among different leukemia cell lines further suggest that elevated expression of RE-IIBP and the histone H3-K27 methylation level might be common aspects among many types of leukemia. Based on our results, we postulate that the aberrant transcriptional regulation of RE-IIBP might be responsible for the elevated expression of RE-IIBP in leukemia patients. Furthermore, the increased histone methylation status in leukemic patients as a result of RE-IIBP further affects transcription of target genes such as IL-5 through histone methylation-mediated chromatin remodeling, and this might have clinical implications in leukemic diseases. Previous reports have suggested that the methylation of histones H3-K9, H3-K27, and H4-K20 is correlated with chromatin repression (4). There have been reports to the effect that the Drosophila ESC-E(Z) complex and its human counterpart EZH2, both components of the Polycomb group, specifically methylate H3-K27 and contribute to Polycomb group-mediated target gene silencing and the progression of prostate cancer (2, 9, 14). EZH2 and histone H3 trimethyl K27 have been previously associated with the silencing of IL-4 and IL-13 in T-helper cell differentiation (10). The abundant expression of RE-IIBP in thymus and its IL-5 regulatory role suggest that RE-IIBP may have involved in T-helper cell-mediated humoral immunity and hormonal regulation.
Consistent with the above reports, H3-K27-methylating RE-IIBP was found in our study to repress basal transcription levels when overexpressed. In addition, RE-IIBP interacts with HDAC1 and -2, which suggests that transcriptional repression is mediated via the recruitment of HDAC. Via analyses of the promoter region of IL-5, one of the specific target genes of RE-IIBP, we further determined that the IL-5 proximal promoter region is hyper-H3-K27 methylated and hypo-H3 acetylated as a consequence of RE-IIBP overexpression, resulting in the transcriptional repression of IL-5 possibly by the recruitment of HDACs. Using WHSC1/MMSET-shRNA, which knocked down RE-IIBP transcripts, we were able to confirm the H3-K27 methylation, HDAC1 recruitment, and H3 hypoacetylation activities of RE-IIBP. Furthermore, point mutant analysis revealed that HMTase activities and transcriptional repression of RE-IIBP are dependent on the cysteine 483 and arginine 477 residues of SET domain. It is interesting that HMTase activity of RE-IIBP is further enhanced when incubated with cell extracts. In fact, HMTase activity of SMYD3 was enhanced in the presence of cofactor HSP90A, which suggests the existence of possible effector proteins for RE-IIBP HMTase activity (6). The identification of the associating factors for the enhanced HMTase enzyme activity of RE-IIBP and the transcriptional repression complex encompassing HDACs and other related corepressors will provide us with the detailed mechanisms underlying the transcriptional regulation of RE-IIBP. Taken together, these results suggest that histone H3-K27 methylation and H3 hypoacetylation can occur simultaneously in RE-IIBP-overexpressing cells and that they contribute together to transcriptional repression of IL-5. The information we have provided in this study as well as data gleaned from further research into RE-IIBP will generate substantial detail regarding the connection between transcriptional regulation via chromatin modification and leukemogenesis.
Acknowledgments
We thank D. Chakravarti for important suggestions on the manuscript.
This work was supported by a grant (R01-2004-000-10121-0) from the Basic Research Program of the KOSEF and by a Molecular and Cellular Bio Discovery Research Program grant (2004M1AN01000125000) from the MOST in South Korea. Hyun Kook and Hae Jin Kee were supported in part by the Korea Science and Engineering Foundation through the MRC for Gene Regulation (R13-2002-013-03002-0) at Chonnam National University.
Footnotes
Published ahead of print on 2 January 2008.
REFERENCES
- 1.Ayton, P. M., and M. L. Cleary. 2001. Molecular mechanisms of leukemogenesis mediated by MLL fusion proteins. Oncogene 205695-5707. [DOI] [PubMed] [Google Scholar]
- 2.Cao, R., L. Wang, H. Wang, L. Xia, H. Erdjument-Bromage, P. Tempst, R. S. Jones, and Y. Zhang. 2002. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 2981039-1043. [DOI] [PubMed] [Google Scholar]
- 3.Daser, A., and T. H. Rabbitts. 2004. Extending the repertoire of the mixed-lineage leukemia gene MLL in leukemogenesis. Genes Dev. 18965-974. [DOI] [PubMed] [Google Scholar]
- 4.Fischle, W., Y. Wang, and C. D. Allis. 2003. Binary switches and modification cassettes in histone biology and beyond. Nature 425475-479. [DOI] [PubMed] [Google Scholar]
- 5.Garlisi, C. G., A. S. Uss, H. Xiao, F. Tian, K. E. Sheridan, L. Wang, M. Motasim Billah, R. W. Egan, K. S. Stranick, and S. P. Umland. 2001. A unique mRNA initiated within a middle intron of WHSC1/MMSET encodes a DNA binding protein that suppresses human IL-5 transcription. Am. J. Respir. Cell Mol. Biol. 2490-98. [DOI] [PubMed] [Google Scholar]
- 6.Hamamoto, R., Y. Furukawa, M. Morita, Y. Iimura, F. P. Silva, M. Li, R. Yagyu, and Y. Nakamura. 2004. SMYD3 encodes a histone methyltransferase involved in the proliferation of cancer cells. Nat. Cell Biol. 6731-740. [DOI] [PubMed] [Google Scholar]
- 7.Jenuwein, T., and C. D. Allis. 2001. Translating the histone code. Science 2931074-1080. [DOI] [PubMed] [Google Scholar]
- 8.Keats, J. J., C. A. Maxwell, B. J. Taylor, M. J. Hendzel, M. Chesi, P. L. Bergsagel, L. M. Larratt, M. J. Mant, T. Reiman, A. R. Belch, and L. M. Pilarski. 2005. Overexpression of transcripts originating from the MMSET locus characterizes all t(4;14)(p16;q32)-positive multiple myeloma patients. Blood 1054060-4069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kirmizis, A., S. M. Bartley, A. Kuzmichev, R. Margueron, D. Reinberg, R. Green, and P. J. Farnham. 2004. Silencing of human Polycomb target genes is associated with methylation of histone H3 Lys 27. Genes Dev. 181592-1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koyanagi, M., A. Baguet, J. Martens, R. Margueron, T. Jenuwein, and M. Bix. 2005. EZH2 and histone 3 trimethyl lysine 27 associated with Il4 and Il13 gene silencing in Th1 cells. J. Biol. Chem. 28031470-31477. [DOI] [PubMed] [Google Scholar]
- 11.Krajewski, W. A., T. Nakamura, A. Mazo, and E. Canaani. 2005. A motif within SET-domain proteins binds single-stranded nucleic acids and transcribed and supercoiled DNAs and can interfere with assembly of nucleosomes. Mol. Cell. Biol. 251891-1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li, B., L. Howe, S. Anderson, J. R. Yates, 3rd, and J. L. Workman. 2003. The Set2 histone methyltransferase functions through the phosphorylated carboxyl-terminal domain of RNA polymerase II. J. Biol. Chem. 2788897-8903. [DOI] [PubMed] [Google Scholar]
- 13.Martin, C., and Y. Zhang. 2005. The diverse functions of histone lysine methylation. Nat. Rev. Mol. Cell Biol. 6838-849. [DOI] [PubMed] [Google Scholar]
- 14.Muller, J., C. M. Hart, N. J. Francis, M. L. Vargas, A. Sengupta, B. Wild, E. L. Miller, M. B. O'Connor, R. E. Kingston, and J. A. Simon. 2002. Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell 111197-208. [DOI] [PubMed] [Google Scholar]
- 15.Rea, S., F. Eisenhaber, D. O'Carroll, B. D. Strahl, Z. W. Sun, M. Schmid, S. Opravil, K. Mechtler, C. P. Ponting, C. D. Allis, and T. Jenuwein. 2000. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 406593-599. [DOI] [PubMed] [Google Scholar]
- 16.Sun, X. J., J. Wei, X. Y. Wu, M. Hu, L. Wang, H. H. Wang, Q. H. Zhang, S. J. Chen, Q. H. Huang, and Z. Chen. 2005. Identification and characterization of a novel human histone H3 lysine 36-specific methyltransferase. J. Biol. Chem. 28035261-35271. [DOI] [PubMed] [Google Scholar]
- 17.Tachibana, M., K. Sugimoto, T. Fukushima, and Y. Shinkai. 2001. Set domain-containing protein, G9a, is a novel lysine-preferring mammalian histone methyltransferase with hyperactivity and specific selectivity to lysines 9 and 27 of histone H3. J. Biol. Chem. 27625309-25317. [DOI] [PubMed] [Google Scholar]
- 18.Todoerti, K., D. Ronchetti, L. Agnelli, S. Castellani, S. Marelli, G. L. Deliliers, A. Zanella, L. Lombardi, and A. Neri. 2005. Transcription repression activity is associated with the type I isoform of the MMSET gene involved in t(4;14) in multiple myeloma. Br. J. Haematol 131214-218. [DOI] [PubMed] [Google Scholar]
- 19.Yang, L., L. Xia, D. Y. Wu, H. Wang, H. A. Chansky, W. H. Schubach, D. D. Hickstein, and Y. Zhang. 2002. Molecular cloning of ESET, a novel histone H3-specific methyltransferase that interacts with ERG transcription factor. Oncogene 21148-152. [DOI] [PubMed] [Google Scholar]
- 20.Zhang, Y., and D. Reinberg. 2001. Transcription regulation by histone methylation: interplay between different covalent modifications of the core histone tails. Genes Dev. 152343-2360. [DOI] [PubMed] [Google Scholar]