Abstract
A common feature of nuclear receptor ligand binding domains (LBD) is a helical sandwich fold that nests a ligand binding pocket within the bottom half of the domain. Here we report that the ligand pocket of glucocorticoid receptor (GR) can be continuously extended into the top half of the LBD by binding to deacylcortivazol (DAC), an extremely potent glucocorticoid. It has been puzzling for decades why DAC, which contains a phenylpyrazole replacement at the conserved 3-ketone of steroid hormones that are normally required for activation of their cognate receptors, is a potent GR activator. The crystal structure of the GR LBD bound to DAC and the fourth LXXLL motif of steroid receptor coactivator 1 reveals that the GR ligand binding pocket is expanded to a size of 1,070 Å3, effectively doubling the size of the GR dexamethasone-binding pocket of 540 Å3 and yet leaving the structure of the coactivator binding site intact. DAC occupies only ∼50% of the space of the pocket but makes intricate interactions with the receptor around the phenylpyrazole group that accounts for the high-affinity binding of DAC. The dramatic expansion of the DAC-binding pocket thus highlights the conformational adaptability of GR to ligand binding. The new structure also allows docking of various nonsteroidal ligands that cannot be fitted into the previous structures, thus providing a new rational template for drug discovery of steroidal and nonsteroidal glucocorticoids that can be specifically designed to reach the unoccupied space of the expanded pocket.
Glucocorticoid receptor (GR) is a steroid hormone-regulated transcription factor that belongs to the nuclear receptor superfamily (1, 39). Upon ligand binding, GR regulates expression of an array of genes involved in glucose and lipid metabolism, bone turnover, lung maturation, and homeostasis of the immune, cardiovascular, and central nervous systems. GR ligands, including dexamethasone (DEX), fluticasone propionate, and other steroid analogs, are among the most effective agents for treating asthma, arthritis, leukemia, and various autoimmune diseases because of their potent anti-inflammatory and immunosuppressive effects. However, therapeutic use of glucocorticoids also induces a number of side effects including diabetes, bone loss, hypertension, and obesity (24). Although the molecular basis for these undesired side effects remains to be fully characterized (26), development of a GR ligand that can dissociate the therapeutic effects from the undesired adverse effects has been the subject of intense pharmaceutical research (23, 25).
The transcriptional function of GR is primarily controlled by ligand binding to its C-terminal ligand binding domain (LBD). In the absence of ligand, GR is retained in the cytoplasm by an association between the receptor LBD and the HSP90 chaperone complex (22). Ligand binding induces conformational changes in the LBD that lead to translocation of the receptor into the nucleus, where GR binds to DNA and regulates transcription of nearby genes. In addition to gene activation, ligand-bound GR also represses the transcription of genes that are activated by nuclear factor-κB and activator protein 1 (17). This GR-mediated repression has been considered to be a major basis for the anti-inflammatory and immunosuppressive effects of glucocorticoids. Both ligand-mediated activation and repression of GR require the integrated functions of the LBD. As such, there is great interest in the GR LBD structures and how they correlate with functional activities of diverse steroidal and nonsteroidal glucocorticoids.
Recent structures of the GR LBD bound to DEX and the antagonist RU486 provide the first template to understand structure-activity relationships for various synthetic steroids (3, 13). In both structures, the GR LBD adopts a helical bundle that embeds a cavity within the bottom half of the domain for ligand binding. In the DEX-bound structure, the ligand binding pocket is completely closed, with a volume of approximately 540 Å3. The C-3 ketone from the A-ring of the steroid makes a pair of hydrogen bonds with two conserved residues, R611 and Q570, which seal one side of the pocket along helices H3 and H7. The other side of the pocket is sealed by helix H12 (or the AF-2 helix), which makes direct contact with the C-11 hydroxyl and the C-18 methyl of the bound steroid. These interactions with DEX lock the AF-2 helix into the active conformation that is competent for the binding of the LXXLL motif of the TIF2 coactivator. In contrast, the large C-11 substitute in the RU486 structure pushes the AF-2 out of the active position, thus inactivating the receptor transcriptional function.
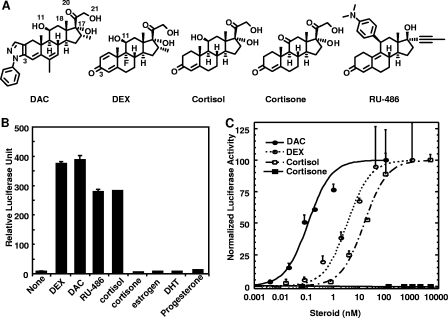
Although the DEX and RU486 structures provide a detailed mechanism of ligand-dependent regulation of the receptor, they fail to account for the binding of a large number of steroidal and nonsteroidal ligands reported in the literature (7, 25, 28, 35). One example is deacylcortivazol (DAC), which is a high-affinity glucocorticoid (27, 29). DAC has shown to be highly active against childhood acute leukemia that is resistant to treatment with other glucocorticoids (10, 30). The chemical structure of DAC contains a bulky phenylpyrazole group, replacing the C-3 ketone of the steroid A-ring (Fig. 1A). It is puzzling why DAC, with such a replacement, is 200-fold more potent than cortisol and 40-fold more potent than DEX (Fig. 1C) (27, 29, 32), since the 3-ketone is a critical group that mediates the conserved interactions observed for all steroid/receptor structures determined to date. This problem is only accentuated by the DEX-bound GR structure, because there is not enough space in the ligand binding cavity to accommodate the large phenylpyrazole group of DAC. Therefore, some rearrangement of the GR LBD has to occur upon the binding of DAC. The binding of other steroids with bulky substitutes usually causes structural changes of receptor LBDs which result in a reduction of gene activation. Therefore, a major unanswered question in glucocorticoid physiology is what structural reorganization of the DAC-bound GR LBD allows the receptor to function as a more potent activator.
FIG. 1.
SARs of GR ligands. (A) Chemical structures of DAC, DEX, RU486, cortisol, and cortisone. (B) Activation of an MMTV luciferase reporter by GR ligands and endogenous steroids at 10 nM in CHO cells. (C) Dose-response curves of DAC, DEX, cortisol, and cortisone for induction of GRE-tk Luc reporter in CV1 cells.
In this study, we have determined the DAC-bound GR LBD structure, which reveals an extra-large size of the GR ligand binding pocket that is expanded into the top half of the LBD, while the structure of the coactivator binding site remains unchanged. The expanded GR pocket allows docking of various steroidal and nonsteroidal GR ligands that contain a phenylpyrazole group. These results provide critical insight into the structure-activity relationship (SAR) of GR ligands as well as a new structural template for designing novel glucocorticoids with better therapeutic profiles.
MATERIALS AND METHODS
Protein preparation.
The human GR LBD (residues 525 to 777), containing an F602S mutation, was expressed as a 6× His-glutathione transferase (GST) fusion protein from the expression vector pET24a (Novagen). The modified protein contains a His6 tag (MKKGHHHHHHG) at the N terminus and a thrombin protease site between GST and the GR LBD. BL21(DE3) cells transformed with this expression plasmid were grown in LB broth at 16°C to an optical density at 600 nm of ∼1 and induced with 0.1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) and 50 μM DAC. Cells were harvested, resuspended in 400 ml extract buffer (50 mM Tris, pH 8.0, 150 mM NaCl, 2 M urea, 10% glycerol, and 1 μM DAC) per 24 liters of cells, and passed three times through a French press with pressure set at 1,000 Pa. The lysate was centrifuged at 20,000 rpm for 30 min, and the supernatant was loaded on a 25-ml nickel column. The column was washed with 900 ml extract buffer and eluted with 30% buffer B (25 mM Tris, pH 8.0, 500 mM imidazole, 10% glycerol, and 1 μM DAC). The GR LBD was cleaved overnight with thrombin at a protease/protein ratio of 1:1,000 in the cold room while being dialyzed against 20 mM Tris, pH 8.0, 500 mM NaCl, 10% glycerol, and 1 μM DAC. The His6-GST tag was removed by a pass through a nickel column. To prepare the protein-cofactor complex, we added a twofold excess of the peptide (SRC1-4, AQQKSLLQQLLDKDE) and further purified by a gel filtration column with a buffer containing 20 mM Tris, pH 8.0, 500 mM NaCl, 5 mM dithiothreitol, 10% glycerol, and 1 μM DAC. The final complexes were filter concentrated to 5 mg/ml, and the yield of the GR protein was 10 to 20 mg per 24 liters of cells.
Crystallization, data collection, structure determination, and refinement.
The GR/DAC/SRC1-4 crystals were grown at room temperature in hanging drops containing 1.0 μl of the above-described complex solution and 1.0 μl of well solution containing 1.3 M ammonium sulfate, 100 mM Tris, pH 8.5, 8% glycerol, and 3 mM n-hexadecyl-β-maltoside. Before data collection, crystals were cross-linked with glutaraldehyde for 5 min, transiently mixed with well buffer that contained additional 30% glycerol, and flash frozen in liquid nitrogen for data collection.
The GR/DAC/SRC1-4 crystals formed in the P62 space group, with values as follows: a, 93.8 Å; b, 93.8 Å; c, 130.0 Å; α and β, 90°; and γ, 120°. Each asymmetric unit cell contains one GR/ligand complex with 76% solvent content, which is relatively high compared to what is seen for most crystal packing. A full 360° data set was collected from a single crystal by use of 1° oscillation by a MAR 225 CCD detector at the ID line of sector-5 (DND) of the Advanced Photon Source. The observed reflections were reduced, merged, and scaled to 2.5 Å with DENZO and SCALEPACK in the HKL2000 package (21).
The structure was determined by molecular replacement using the GR/DEX/TIF2 structure (3) as a model with the AmoRe program (18). The phases from the molecular replacement solution were refined with solvent flattening and histogram matching as implemented in the CCP4 dm program. Manual model building was carried out with QUANTA (Accelrys Inc.), and structure refinement proceeded with CNS (5), using the maximum likelihood target. The pocket volume was calculated with Voidoo using the program default parameter and a 1.20-Å probe (14).
Binding assays.
GR proteins were prepared as 6× His-GST fusion proteins for the assays using an AlphaScreen kit for the detection of hexahistidine proteins (Perkin Elmer). The experiments were conducted with approximately 20 nM receptor LBD and 20 nM of biotinylated SRC2-3 peptide (QEPVSPKKKENALLRYLLDKDDTKD) in the presence of 5 μg/ml donor and acceptor beads in a buffer containing 50 nM MOPS (morpholinepropanesulfonic acid), pH 7.4, 50 mM NaF, 50 mM CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate}, and 0.1 mg/ml bovine serum albumin. The relative binding affinity of peptide binding motifs was determined by competition with unlabeled peptides at 500 nM to compete off the binding of biotinylated SRC2-3 to the GR LBD. Fifty-percent inhibitory concentration values for various coactivator LXXLL motifs were determined from a nonlinear least-squares fit of the data based on an average of three repeated experiments, with standard errors of typically less than 10% of the measurements. The sequences of unlabeled peptides are as follows: for SMRT-2, ASTNMGLEAIIRKALMGKYDQ; for SHP-1, ASHPTILYTLLSPGP; for SHP-2, APVPSILKKILLEEPNS; for SHP-3, ASQGRLARILLMAST; for DAX1, QWQGSILYNMLMSAK; for DAX2, PRQGSILYSMLTSAK; for DAX3, PRQGSILYSLLTSSK; for SRC1-1, SQTSHKLVQLLTTTA; for SRC1-2, TERHKILHRLLQESS; for SRC1-3, SKDHQLLRYLLDKDE; for SRC1-4, AQQKSLLQQLLTE; for SRC2-1, SKGQTKLLQLLTCSS; for SRC2-2, KEKHKILHRLLQDSS; for SRC2-3, KENALLRYLLDKDD; for SRC3-1, SKGHKKLLQLLTCSS; for SRC3-2, QEKHRILHKLLQNGN; for SRC3-3, KENNALLRYLLDRDD; for TRAP220-1, VSQNPILTSLLQITG; for TRAP220-2, KNIHPMLMNLLKDNP; for CBP-1, ASKHKQLSELLRGGS; for PGC1α-1, AEEPSLLKKLLLAPA; for PGC1α-2, RRPCSELLKYLTTND; for PGC1β-1, VDELSLLQKLLLATS; for PGC1β-2, WAEFSILRELLAQDV; for PRC, PREGSSLHKLLTLSR; for ARA70-1, QQQAQQLYSLLGQFN; for ARA70-2, RETSEKFKLLFQSYN; for ASC2-1, TLTSPLLVNLLQSDI; for ASC2-2, REAPTSLSQLLDNSG; for RIP140-2, KQDSTLLASLLQSFS; for RIP140-9, SKSFNVLKQLLLSEN; for PRIC285-1, NADDAILRELLDESQ; for PRIC285-2, NLPPAALRKLLRAEP; for PRIC285-3, FAGDEVLVQLLSGDK; for D30, HSSRLWELLMEAT; for ARN1, YRGAFQNLFQSVR; for ARN2, ASSSWHTLFTAEE; and for AR4-1, QPKHFTELYFKS.
Transcriptional activation by VP16-GR and GR.
The VP16-GR chimera activator was constructed by fusing three copies of the recombinant VP16 minimal activation domain (34) to the N terminus of GR, which is under control of a cytomegalovirus (CMV) promoter (CMV-rVPGR). The mouse mammary tumor virus (MMTV)-Luc reporter was constructed by inserting three tandem repeats of a MMTV promoter upstream of the luciferase gene. Activation of the MMTV-Luc reporter by VP16-GR was performed with CHO-K1 cells, which were maintained in alpha minimal essential medium (MEM) containing 10% fetal bovine serum (FBS). Cells (50,000/well in a 24-well plate) were treated with 5% charcoal-stripped FBS 24 h prior to transfections. Cells were transfected in Opti-MEM with 200 ng of reporter plasmid, 5 ng of control plasmid phRLTK (constitutive expression of Renilla luciferase; Promega), and 200 ng of CMV-RVPGR by use of Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. Two hours after transfection, cells were induced with 10 nM of each ligand. Twenty-four hours after induction, cells were harvested and firefly and Renilla luciferase activities measured by the dual luciferase assay system (Promega). Luciferase data were normalized to Renilla luciferase as an internal control. All assays were performed in triplicate.
Dose curve experiments were performed with triplicate samples of CV-1 cells (20,000/well of 24-well plates in Dulbecco's MEM and 10% FBS), which were transfected 24 h later with 0.5 ng of pSG5-GR, 100 ng of GRE-tk Luc, and 10 ng of Renilla TS (gift from N. M. Ibrahim, O. Fröhlich, and S. R. Price; Emory University School of Medicine) by using FuGENE 6 reagent (Roche Molecular Biochemicals) according to the manufacturer's instructions. The total transfected DNA was adjusted to 300 ng/well with pBluescriptII SK+ (Stratagene). Twenty hours after transfection, cells were induced with steroid for 20 h and then lysed and assayed for reporter gene activity using dual luciferase assay reagents according to the manufacturer's instructions. The data were normalized for Renilla luciferase activity and expressed as percentages of the maximal response.
Modeling of nonsteroidal compounds.
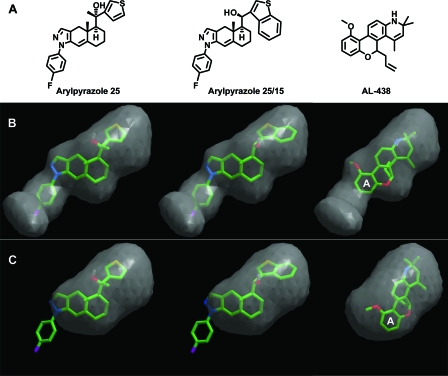
The structures of the GR/DAC and GR/DEX complexes (PDB code, 1M2Z.pdb) were superimposed based on Cα atoms. The ICM program (1) was used for protein and ligand preparation. The molecular structures of AL-438 (7) and the two arylpyrazole compounds (28, 31) (see Fig. 5) were constructed in the ICM interface and were optimized with an MMFF94 force field (11). These small molecules were docked into the ligand binding pocket of the GR/DAC or GR/DEX complex with default parameters implemented in the ICM program (9). The ligand binding pocket surfaces were produced by ICM PocketFinder.
FIG. 5.
Docking of various nonsteroidal ligands onto GR. (A) Chemical structures of three representative nonsteroidal compounds. (B) The docking model of the nonsteroidal compounds in the DAC-binding pocket. (C) The binding model of the nonsteroidal compounds in the DEX-binding pocket. The letter A in panels B and C indicates the A-ring of AL-438.
Protein structure accession number.
The PDB code for the GR/DAC/SRC1-4 ternary complex is 3BQD.
RESULTS AND DISCUSSION
SARs of GR ligands.
Synthetic GR ligands DAC, DEX, and RU486 and endogenous steroid hormones share a core four-ring structure linked with various functional groups (Fig. 1A). To determine the SARs of these ligands with GR, we have developed an MMTV luciferase reporter assay that is driven by a VP16-GR fusion activator. Because of the potent and constitutive activation domain of VP16, which is linked to the N terminus of GR, the activation of this reporter is based on the ligand binding event regardless of the antagonist property of RU486 or the agonist properties of DAC, DEX, and cortisol (Fig. 1B). Both GR agonists (DAC, DEX, and cortisol) and antagonist (RU486) induced 50- to 90-fold activation of the reporter (Fig. 1B). In this assay, GR displays stringent SAR toward endogenous steroid hormones: only cortisol (the physiological ligand of GR) achieved significant activation of the reporter at a 10 nM concentration, whereas cortisone, androgen, progesterone, and estrogen failed to activate GR to a detectable level (Fig. 1B). The activation properties of DAC, DEX, cortisol, and cortisone are further characterized with full dose-response curves (Fig. 1C), which give the activation potency of these ligands (50% effective concentrations of 0.1 nM, 4.0 nM, and 20 nM for DAC, DEX, and cortisol, respectively). These results recapitulate the fact that DAC is 40 times more potent than DEX and 200-fold more potent than cortisol (29), whereas cortisone did not bind or activate GR. Interestingly, the only difference between cortisol and cortisone is the C-11 hydroxyl in cortisol versus a ketone in cortisone; the difference between DAC and DEX is the phenylpyrazole in DAC versus a 3-ketone in DEX. The contrast between the stringent SAR between cortisol and cortisone at the C-11 position and the relaxed SAR between DAC and DEX at the C-3 position is intriguing, as the 3-ketone in steroids makes conserved interactions among 3-keto steroid receptors, whereas RU486, which has a large group attached to the C-11 position, still interacts well with GR.
Peptide profiling and crystallization of the GR LBD/DAC complex.
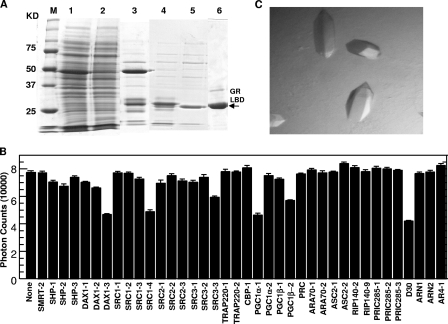
To determine the molecular basis for the high-affinity binding of DAC to GR, we expressed and purified the GR LBD for crystallographic studies using a protocol modified from previously reported procedures (3) (see Materials and Methods for details). Although the GR LBD (with the F602S mutation that was made to improve protein solubility) was purified to homogeneity (Fig. 2A), it remained unstable and could not be crystallized without the presence of a coactivator peptide. To identify peptide motifs for cocrystallization, we performed peptide-profiling experiments using a panel of 38 unlabeled peptides to compete off the binding of the third LXXLL motif of SRC2 (SRC2-3) to the GR LBD in the presence of DAC (Fig. 2B). The sequences of these 38 peptides, as reported previously (15), were selected from endogenous nuclear receptor coregulators, including the p160 family of coactivators: the PGC1, SHP, DAX1, and AR coactivators. In the peptide-profiling experiment, the amount of each unlabeled peptide used is identical at 500 nM; thus, the relative binding affinity of each peptide to GR can be measured by the degree of its inhibition of the binding of the biotinylated SRC2-3 motif to the GR LBD. Consistent with the agonistic properties of DAC, the corepressor motif from SMRT did not inhibit the binding of the SRC2-3 motif to the GR/DAC complex, while coactivator motifs exhibited various degrees of inhibition (Fig. 2B). Among these LXXLL motifs, DAX1-3, SRC1-4, SRC3-3, and PGC1α-1 appear to be the most potent competitors. The strong binding of the SRC1-4 and PGC1α-1 motif to GR is consistent with previous studies of the binding of the LXXLL motif to GR and mineralocorticoid receptor (MR) (12, 19, 37). Furthermore, the above peptide-profiling results for GR resemble those for MR, suggesting that these two receptors share an overlapped specificity for coactivator recruitment. Because the LXXLL motifs of DAX1-3, SRC1-4, and PGC1α-1 bind to GR with the highest affinities, these peptides were used for cocrystallization with the GR LBD/DAC complex. The crystals containing the SRC1-4 LXXLL motif were readily obtained (Fig. 2C), while complexes with other peptides failed to produce crystals.
FIG. 2.
Purification, peptide profiling, and crystallization of the GR LBD/DAC complex. (A) Purification of the GR LBD bound to DAC. The protein samples shown are crude extract, GST column flowthrough, GST bound, thrombin cut, nickel column flowthrough, and size column (lanes 1 to 6, respectively). Molecular mass markers are shown in lane M. (B) Relative binding affinities of various peptide motifs to the GR LBD/DAC complex as determined by peptide competitions in which various unlabeled peptides (500 nM) are used to compete off the binding of the SRC2-3 LXXLL motif to GR. All peptides have identical lengths of 15 residues, except for the SRC1-4 motif, which terminates at position +7 relative to the first leucines (L+1) in the LXXLL motif, and for the AR peptides and the corepressor motifs, which are longer than the coactivator motifs. The results shown are averages of triplicate experiments, with error bars showing standard deviations. Sequences of peptides are listed in Materials and Methods. (C) Crystals of the GR/DAC/SRC1-4 complex.
Structure of the GR LBD/DAC/SRC1-4 complex.
The GR/DAC/SRC1-4 complex was crystallized in the P62 space group with one LBD/ligand complex in each asymmetric unit. Diffraction data were collected to 2.5 Å, and the structure was determined by molecular replacement using the DEX-bound GR structure as the initial model (3). The statistics of the diffraction data and the refined structure are listed in Table 1. The current model includes the GR LBD (residues 525 to 777), the core helix of the SRC1-4 motif, and the bound DAC ligand.
TABLE 1.
Statistics of data and structure
| Parameter/statistic | Datum |
|---|---|
| Crystal identity | GR/DAC/SRC1-4 |
| PDB code | 3BQD |
| X-ray source | APS-5ID |
| Space group | P62 |
| Resolution (Å) | 50.0-2.50 |
| No. of unique reflections | 22,367 |
| Completeness (%) | 99.0 |
| I/σ | 32.1 |
| Rsyma (%) | 9.9 |
| Refinement statistics | |
| Rfactorb (%) | 21.53 |
| Rfree (%) | 24.76 |
| RMSD for bond lengths (Å) | 0.008 |
| RMSD for bond angles (degrees) | 1.568 |
| Total no. of nonhydrogen atoms | 2,248 |
Rsym = Σ|Iavg − Ii|/ΣIi.
Rfactor = Σ|FP − FPcalc|/ΣFp, where FP and FPcalc are observed and calculated structure factors, Rfree was calculated from a randomly chosen 8% of reflections excluded from refinement, and Rfactor was calculated for the remaining 92% of reflections.
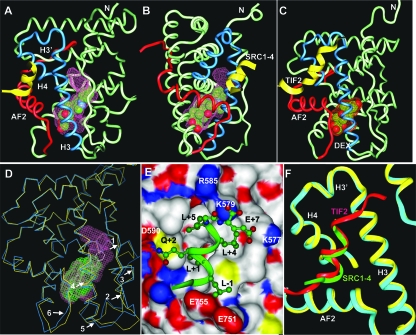
As expected, the overall structure of the GR/DAC/SRC1-4 complex resembles that of the DEX-bound structure, with 11 α-helices and 4 β-strands that fold into a three-layer helical sandwich bundle (Fig. 3A and B). The C-terminal AF-2 helix is also packed tightly against the main domain of the LBD, which assumes an active conformation. In this conformation, the AF-2 helix together with helices H3, H3′, and H4 forms a charge clamp pocket for the binding of the SRC1-4 LXXLL motif (Fig. 3E and F). The core SRC1-4 sequence (LLQKLL) adopts a two-turn α-helix that orients the hydrophobic leucine side chains into the center hydrophobic surface of the coactivator binding site (Fig. 3E). The charge clamp residues (E755 from AF2 and K579 from H3) form the capping interactions with the N and C termini of the coactivator helix. In addition, as reported previously (3), GR contains a second charge clamp (R585 from H3′ and D590 from H4) that mediates hydrogen bonds with Q+2 and the C-terminal acidic group of the SRC1-4 motif (Fig. 3E). In addition, the side chain E+7 of the SRC1-4 motif also forms a charge interaction with K577. The same network of interactions was also observed in the previous MR/SRC1-4 complex, indicating that GR and MR share a common structural mechanism for the preferential binding of the SRC1-4 motif. Importantly, despite different coactivator motifs and the bound ligands, the coactivator binding site in the GR/DAC/SRC1-4 complex is nearly identical to that of the GR/DEX/TIF2 complex (Fig. 3F), with the SRC1-4 helix adopting the same binding mode of the TIF2 motif.
FIG. 3.
Crystal structure of the GR/DAC/SRC1-4 complex. (A and B) Two 90° views of the GR/DAC/SRC1-4 complex. The DAC-binding pocket is shown as a pink surface. The SRC1-4 peptide is in yellow and the bound DAC is shown in a space-filling representation with carbon and oxygen atoms depicted in green and red, respectively. Key structural elements are labeled. (C) Overall structure of the GR/DEX/TIF2 complex for comparison with the GR/DAC/SRC1-4 complex. The DEX-binding pocket is shown as a red surface. The TIF2/SRC2-3 peptide is in yellow and the bound DEX is in a space-filling representation with carbon and oxygen atoms depicted in green and red, respectively. (D) Overlay of the DAC and DEX-bound GR structures. The Cα atoms of the DAC-bound GR LBD are shown in cyan and the DEX-bound GR LBD is shown in yellow. The DAC- and DEX-binding pockets are in pink and green, respectively. Arrows indicate major differences between the DAC- and DEX-bound structures (residues E542 to D549, arrows 1 and 2; residues T556 to L566, arrows 4 and 5; residues L621 to A624, arrow 3; residues S746 and I747, arrow 6). (E) The binding mode of the SRC1-4 LXXLL (green) in the GR coactivator binding site. GR is shown as a protein surface that is colored by atom type as follows: carbon, white; oxygen, red; nitrogen, blue; sulfur, gold. SRC1-4 residues are labeled with black and the GR residues are labeled with white. (F) Structural comparison of the GR coactivator binding site between the DAC (cyan)- and DEX (yellow)-bound structures. The SRC1-4 and TIF2-4 helices are shown in green and red, respectively.
Unique features of the GR DAC-binding pocket.
The most pronounced feature of the DAC-bound GR structure is a dramatic expansion of the GR ligand binding pocket (Fig. 3A and B), which is completely enclosed by residues from helices 3, 4, 5, 6, 7, and 10 and the AF-2 helix as well as residues from the first two β strands. Compared with the DEX-binding pocket, the DAC-binding pocket also contains a core steroid shape pocket with a typical GR side pocket facing the C-17 hydroxyl. This GR side pocket has been demonstrated for the binding of GR ligands with large substitutes at the C-17α position (R. K. Bledsoe, M. H. Lambert, V. G. Montana, E. L. Stewart, and H. E. Xu, January 2004, European Patent Office, application 1375517). Strikingly, to accommodate the phenylpyrazole group, the DAC-binding pocket is extended beyond the traditional boundary by helix 5 on the top into helices 1 and 8 in the upper half of the LBD. This is a dramatic contrast to all nuclear receptor LBD structures reported to date, where the ligand binding pocket is strictly confined underneath helix 5 within the bottom half of the domain. Although the molecular size of DAC is only ∼25% larger than that of DEX, the DAC-binding pocket is expanded to a volume of 1,070 Å3, which is almost twice the size of the DEX-binding pocket (540 Å3).
Why does the GR pocket open so much upon binding to the slightly larger steroid DAC? Compared with the DEX-bound GR structure (Fig. 3C), the core helical domain of the DAC-bound structure is exceedingly similar, with the root mean square deviation (RMSD) of the Cα atoms being only 0.454 Å. The major differences are within the bottom half of the protein, particularly the following four amino acid stretches, which are immediately adjacent to the bound DAC ligand. The first region is from residues E542 to D549 and forms an extended loop between helices 1 and 3 (Fig. 3D). The Cα atoms of this region are moved out away from the ligand by as much as 2.5 Å at residue Y548. The second region is the N-terminal half of helix 3 (residues T556 to L566) (Fig. 3D), which is pushed downward by 0.7 to 0.8 Å, compared to a less-than-0.4-Å change in the C-terminal half of helix 3. The third region is β-strand 1 (residues L621 to A624), which contacts the phenylpyrazole moiety of DAC and is thus pushed out by the ligand by 0.8 to 1.4 Å. The fourth region is the loop preceding the AF-2 helix (residues S746 and I747), which is also pushed out by the ligand by 1.2 to 1.7 Å. Beside the above-described backbone changes, the conformational differences in the protein side chains also contribute significantly to the expanded GR pocket, as discussed in detail below.
Basis for the high-affinity binding of DAC by GR.
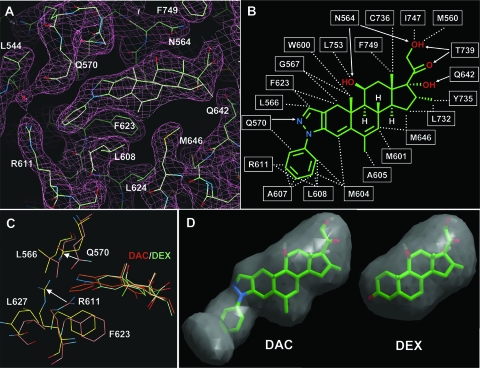
Within the expanded pocket, the bound conformation of DAC can be clearly defined by the excellent electron density (Fig. 4A). The steroid plane of DAC is placed at the center of the pocket with a 45° angle across the axis of helix 3. In this position, the hydrophobic ring of the steroid core is sandwiched by nonpolar residues W500, M601, M604, C736, F749, L753, and I754 on the top right side and L563, L608, F623, M646, and C643 from the bottom left side (Fig. 4B). The polar groups at the C-11, C-17, C-20, and C-21 positions also form an extensive network of hydrogen bonds with the receptor (Fig. 4B). It is worth noting that residues F749, L753, and I754 are from the AF-2 helix and the loop preceding the AF-2 helix. Their interactions with the steroid core may provide a mechanism of ligand-dependent activation of GR by stabilizing the AF-2 helix in the active conformation. Compared with the DEX-bound structure, the steroid core of DAC essentially occupies the same space as that of DEX, with an RMSD of the values for carbon atoms of less than 0.6 Å between the two structures, making the same network of interactions with the receptor. Thus, the overall binding mode and the mechanism of the receptor activation are conserved between the DAC- and DEX-bound structures.
FIG. 4.
Recognition of DAC by GR. (A) Electron density map showing the bound DAC ligand and the surrounding GR residues. (B) Schematic representation of GR/DAC interactions. Hydrophobic interactions are indicated by dashed lines and hydrogen bonds are indicated by arrows. (C) Conformational changes for the expanded DAC-binding pocket (pink). The carbons of the GR DAC structure are shown in cyan and the GR DEX structure is in gold, with arrows indicating movement of R611 and Q570. (D) A close-up comparison of the DAC- and DEX-binding pockets.
The only difference between DAC and DEX is the phenylpyrazole of DAC versus the 3-ketone of DEX, and the higher affinity of DAC binding by GR can be readily accounted for by additional interactions of GR with the phenylpyrazole group of DAC. In the DEX structure, the 3-ketone forms a network of hydrogen bonds with Q570 and R611, which are conserved in all structures of 3-ketone steroid receptors AR, PR, and MR (15, 16, 36). In the DAC structure, Q570 and R611 are moved 3 to 4 Å away from the DEX structure to accommodate the phenylpyrazole group (Fig. 4C). In this position, the amide side chain of Q570 forms a direct hydrogen bond with the nitrogen atom of the pyrazole ring of DAC, and the side chain R611 packs against the benzyl ring of DAC. The hydrogen bond network between DAC and Q570 and R611 appears to be less ideal than that formed by the 3-ketone of DEX. Dynamic simulations also indicate that Q570 and R611 in the DEX structure are in a lower energy conformation than they are in the DAC structure (data not shown). However, the phenylpyrazole group forms extensive tight packing interactions with L566, Q570, M604, A607, L608, R611, and F623, including a π-π interaction between the benzyl ring of F623 and the pyrazole ring of DAC. These additional hydrophobic interactions in the DAC structure increase the receptor/ligand interface by 40%, therefore overcoming the less favorable hydrogen bonds between DAC and GR to increase the overall binding affinity of DAC to the receptor.
Unexpectedly, the conformational changes in Q570 and R611 open up a new cavity that is not occupied by the ligand (Fig. 4D and 3A and D). The top of this empty cavity is capped by K667 from helix 8, which forms a cation-π interaction with W577 from helix 3 and a charge interaction with E540, the residue immediately following helix 1. Interestingly, these three residues are 100% conserved in all steroid receptors and they form a conserved structure component in the top half of the LBD besides capping the empty pocket. The rest of this pocket is surrounded by hydrophobic residues L544, A574, L603, M604, A607, and L608 and the two hydrophilic residues Q570 and R611. The continuity of this empty pocket with the main steroid pocket suggests the possibility of designing a larger ligand that would occupy this new pocket.
A new template for modeling nonsteroidal glucocorticoids.
As the result of intense pharmaceutical efforts, a number of nonsteroidal ligands that display preferential activation of GR-mediated transrepression over transactivation have been reported (7, 25, 28, 31, 35). These ligands are termed dissociated glucocorticoids, and there is hope that they can retain the beneficial anti-inflammatory effects that are postulated to derive from transrepression pathways and reduce the side effects derived from transactivation pathways. Understanding the binding mode of these nonsteroidal ligands has been challenging, as their distinct chemical structures cannot be easily docked onto the DEX-binding pocket. The availability of the DAC-bound GR structure provides an opportunity for docking of these compounds onto the expanded GR pocket. Figure 5A shows the chemical structures of three nonsteroidal glucocorticoids that selectively modulate GR transrepression and transactivation activity (7, 28, 31, 35). All three compounds are readily docked onto the DAC-binding pocket but not the DEX-binding pocket (Fig. 5B and C). In the case of arylpyrazole compounds, the fluorophenylpyrazole group mediates the same interactions with GR as phenylpyrazole in DAC, and the only hydroxyl mimics the C-11 hydroxyl of steroidal GR ligands, where the other C-11 substituents dock onto the space occupied by the D-ring and C-19 to C-21 of DAC. In the case of AL-438, its A aromatic ring occupies the same space as phenylpyrazole of DAC, where it is capable of forming an amide-π interaction with Q570 while maintaining the key hydrogen bond between the D-ring amine with the side chain of N564. This is the same conserved hydrogen bond formed by the C-11 hydroxyl of all steroid glucocorticoids. However, both arylpyrazole and AL-438 compounds lack the same interactions with the AF-2 helix and the loop preceding the AF2 as mediated by the C-18, C-20, and C-21 groups of DAC (Fig. 1A and 4B), and as such these dissociated compounds are not optimal for stabilizing GR in the active conformation, therefore explaining their preferential loss of transcriptional activity. The SARs of the arylpyrazole compounds and AL-438 have been studied extensively, and our current ability to dock these compounds onto GR should provide a structural framework for understanding the SARs of these compounds and designing new GR ligands with better therapeutic profiles.
Concluding remarks.
In summary, the crystal structure of the GR LBD bound to DAC and the SRC1-4 LXXLL motif provides important insights into the conformational adaptability of GR to accommodate ligand binding and coactivator recognition. Most remarkably, the binding of DAC effectively doubles the size of the GR ligand binding pocket and yet does not affect the structure of the coactivator binding site. Conformational flexibility has been characterized for a number of adopted orphan nuclear receptors, including proxisome proliferator-activated receptors (38), the pregnane X receptor (6), and the liver X receptor (8), as well as the Drosophila ecdysone receptor (2). The structural flexibility of these receptors has allowed them to interact promiscuously with a wide range of low-affinity metabolic ligands. Recent structural studies of estrogen receptors and thyroid hormone receptors also reveal a great range of conformational flexibility in their ligand binding pocket upon the binding of distinct ligands (4, 20, 33), in contrast to a well-accepted perception that these classic endocrine receptors possess a relatively well-defined pocket to account for the high-affinity and specific binding of a unique endogenous ligand. However, conformational changes in these receptors have resulted only in an incremental increase in their ligand binding pocket that is strictly confined within the bottom half of the LBD. The size increase of the GR pocket and its expansion to the upper half of the LBD described here provide an extreme example of the fact that nuclear receptors may have an even greater degree of conformational capacity to adopt a wide range of synthetic or natural ligands.
The dramatic expansion of the GR pocket and the detailed GR/DAC interactions also have important implications in drug discovery, as DAC has been shown to be highly active against childhood acute lymphoblastic leukemia that is resistant to DEX (10, 30). However, the therapeutic application of DAC has been limited due to its toxicity. The binding mode of DAC in the expanded GR pocket will provide a template to design DAC derivatives with better toxicity profiles. Finally, as glucocorticoids remain the most effective immunosuppressive agents but have adverse side effects, the discovery of dissociated glucocorticoids that can separate the two effects described above will continue to be a major challenge for pharmaceutical research (25). The ability of the expanded GR pocket to dock steroidal and nonsteroidal compounds helps to pave an avenue to uncover the molecular mechanism of dissociated glucocorticoids, which ultimately will aid the discovery of better and safer GR drugs.
Acknowledgments
We thank Z. Wawrzak and J. S. Brunzelle for assistance in data collection at the DND-CAT (sector 5) of the Advance Photo Source, D. Petillo for DNA sequencing, and W. Minor for the HKL2000 package.
Use of the Advanced Photon Source was supported by the Office of Science of the U.S. Department of Energy. H.E.X. acknowledges the generosity of the Jay and Betty Van Andel Foundation, the Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor (grant 085P1000817), the Department of Defense (W81XWH0510043), and the National Institutes of Health (DK066202 and DK071662).
Footnotes
Published ahead of print on 26 December 2007.
REFERENCES
- 1.Beato, M., P. Herrlich, and G. Schutz. 1995. Steroid hormone receptors: many actors in search of a plot. Cell 83851-857. [DOI] [PubMed] [Google Scholar]
- 2.Billas, I. M., T. Iwema, J. M. Garnier, A. Mitschler, N. Rochel, and D. Moras. 2003. Structural adaptability in the ligand-binding pocket of the ecdysone hormone receptor. Nature 42691-96. [DOI] [PubMed] [Google Scholar]
- 3.Bledsoe, R. K., V. G. Montana, T. B. Stanley, C. J. Delves, C. J. Apolito, D. D. McKee, T. G. Consler, D. J. Parks, E. L. Stewart, T. M. Willson, M. H. Lambert, J. T. Moore, K. H. Pearce, and H. E. Xu. 2002. Crystal structure of the glucocorticoid receptor ligand binding domain reveals a novel mode of receptor dimerization and coactivator recognition. Cell 11093-105. [DOI] [PubMed] [Google Scholar]
- 4.Borngraeber, S., M. J. Budny, G. Chiellini, S. T. Cunha-Lima, M. Togashi, P. Webb, J. D. Baxter, T. S. Scanlan, and R. J. Fletterick. 2003. Ligand selectivity by seeking hydrophobicity in thyroid hormone receptor. Proc. Natl. Acad. Sci. USA 10015358-15363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brunger, A. T., P. D. Adams, G. M. Clore, W. L. DeLano, P. Gros, R. W. Grosse-Kunstleve, J. S. Jiang, J. Kuszewski, M. Nilges, N. S. Pannu, R. J. Read, L. M. Rice, T. Simonson, and G. L. Warren. 1998. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. Sect. D 54905-921. [DOI] [PubMed] [Google Scholar]
- 6.Chrencik, J. E., J. Orans, L. B. Moore, Y. Xue, L. Peng, J. L. Collins, G. B. Wisely, M. H. Lambert, S. A. Kliewer, and M. R. Redinbo. 2005. Structural disorder in the complex of human pregnane X receptor and the macrolide antibiotic rifampicin. Mol. Endocrinol. 191125-1134. [DOI] [PubMed] [Google Scholar]
- 7.Coghlan, M. J., P. B. Jacobson, B. Lane, M. Nakane, C. W. Lin, S. W. Elmore, P. R. Kym, J. R. Luly, G. W. Carter, R. Turner, C. M. Tyree, J. Hu, M. Elgort, J. Rosen, and J. N. Miner. 2003. A novel antiinflammatory maintains glucocorticoid efficacy with reduced side effects. Mol. Endocrinol. 17860-869. [DOI] [PubMed] [Google Scholar]
- 8.Farnegardh, M., T. Bonn, S. Sun, J. Ljunggren, H. Ahola, A. Wilhelmsson, J. A. Gustafsson, and M. Carlquist. 2003. The three-dimensional structure of the liver X receptor beta reveals a flexible ligand-binding pocket that can accommodate fundamentally different ligands. J. Biol. Chem. 27838821-38828. [DOI] [PubMed] [Google Scholar]
- 9.Fernandez-Recio, J., M. Totrov, and R. Abagyan. 2003. ICM-DISCO docking by global energy optimization with fully flexible side-chains. Proteins 52113-117. [DOI] [PubMed] [Google Scholar]
- 10.Gaynon, P. S., and A. L. Carrel. 1999. Glucocorticosteroid therapy in childhood acute lymphoblastic leukemia. Adv. Exp. Med. Biol. 457593-605. [DOI] [PubMed] [Google Scholar]
- 11.Halgren, T. A. 1999. MMFF. VI. MMFF94s option for energy minimization studies. J. Comp. Chem. 20720-729. [DOI] [PubMed] [Google Scholar]
- 12.Hultman, M. L., N. V. Krasnoperova, S. Li, S. Du, C. Xia, J. D. Dietz, D. S. Lala, D. J. Welsch, and X. Hu. 2005. The ligand-dependent interaction of mineralocorticoid receptor with coactivator and corepressor peptides suggests multiple activation mechanisms. Mol. Endocrinol. 191460-1473. [DOI] [PubMed] [Google Scholar]
- 13.Kauppi, B., C. Jakob, M. Farnegardh, J. Yang, H. Ahola, M. Alarcon, K. Calles, O. Engstrom, J. Harlan, S. Muchmore, A. K. Ramqvist, S. Thorell, L. Ohman, J. Greer, J. A. Gustafsson, J. Carlstedt-Duke, and M. Carlquist. 2003. The three-dimensional structures of antagonistic and agonistic forms of the glucocorticoid receptor ligand-binding domain: RU-486 induces a transconformation that leads to active antagonism. J. Biol. Chem. 27822748-22754. [DOI] [PubMed] [Google Scholar]
- 14.Kleywegt, G. J., and T. A. Jones. 1994. Detection, delineation, measurement and display of cavities in macromolecular structures. Acta Crystallogr. Sect. D 50178-185. [DOI] [PubMed] [Google Scholar]
- 15.Li, Y., K. Suino, J. Daugherty, and H. E. Xu. 2005. Structural and biochemical mechanisms for the specificity of hormone binding and coactivator assembly by mineralocorticoid receptor. Mol. Cell 19367-380. [DOI] [PubMed] [Google Scholar]
- 16.Matias, P. M., P. Donner, R. Coelho, M. Thomaz, C. Peixoto, S. Macedo, N. Otto, S. Joschko, P. Scholz, A. Wegg, S. Basler, M. Schafer, U. Egner, and M. A. Carrondo. 2000. Structural evidence for ligand specificity in the binding domain of the human androgen receptor. Implications for pathogenic gene mutations. J. Biol. Chem. 27526164-26171. [DOI] [PubMed] [Google Scholar]
- 17.McKay, L. I., and J. A. Cidlowski. 1999. Molecular control of immune/inflammatory responses: interactions between nuclear factor-kappa B and steroid receptor-signaling pathways. Endocr. Rev. 20435-459. [DOI] [PubMed] [Google Scholar]
- 18.Navaza, J., S. Gover, and W. Wolf. 1992. AMoRe: a new package for molecular replacement, p. 87-90. In E. J. Dodson (ed.), Molecular replacement: proceedings of the CCP4 study weekend. SERC, Daresbury, United Kingdom.
- 19.Needham, M., S. Raines, J. McPheat, C. Stacey, J. Ellston, S. Hoare, and M. Parker. 2000. Differential interaction of steroid hormone receptors with LXXLL motifs in SRC-1a depends on residues flanking the motif. J. Steroid Biochem. Mol. Biol. 7235-46. [DOI] [PubMed] [Google Scholar]
- 20.Nettles, K. W., J. B. Bruning, G. Gil, E. O'Neill, E., J. Nowak, Y. Guo, Y. Kim, E. R. Desombre, R. Dilis, R. N. Hanson, A. Joachimiak, and G. L. Greene. 2007. Structural plasticity in the oestrogen receptor ligand-binding domain. EMBO Rep. 8610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Otwinowski, Z., and W. Minor. 1997. Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 276307-326. [DOI] [PubMed] [Google Scholar]
- 22.Pratt, W. B., and D. O. Toft. 1997. Steroid receptor interactions with heat shock protein and immunophilin chaperones. Endocr. Rev. 18306-360. [DOI] [PubMed] [Google Scholar]
- 23.Rosen, J., and J. N. Miner. 2005. The search for safer glucocorticoid receptor ligands. Endocr. Rev. 26452-464. [DOI] [PubMed] [Google Scholar]
- 24.Schacke, H., W. D. Docke, and K. Asadullah. 2002. Mechanisms involved in the side effects of glucocorticoids. Pharmacol. Ther. 9623-43. [DOI] [PubMed] [Google Scholar]
- 25.Schacke, H., and H. Rehwinkel. 2004. Dissociated glucocorticoid receptor ligands. Curr. Opin. Investig. Drugs 5524-528. [PubMed] [Google Scholar]
- 26.Schacke, H., H. Rehwinkel, K. Asadullah, and A. C. Cato. 2006. Insight into the molecular mechanisms of glucocorticoid receptor action promotes identification of novel ligands with an improved therapeutic index. Exp. Dermatol. 15565-573. [DOI] [PubMed] [Google Scholar]
- 27.Schlechte, J. A., S. S. Simons, Jr., D. A. Lewis, and E. B. Thompson. 1985. [3H]cortivazol: a unique high affinity ligand for the glucocorticoid receptor. Endocrinology 1171355-1362. [DOI] [PubMed] [Google Scholar]
- 28.Shah, N., and T. S. Scanlan. 2004. Design and evaluation of novel nonsteroidal dissociating glucocorticoid receptor ligands. Bioorg. Med. Chem. Lett. 145199-5203. [DOI] [PubMed] [Google Scholar]
- 29.Simons, S. S., Jr., E. B. Thompson, and D. F. Johnson. 1979. Anti-inflammatory pyrazolo-steroids: potent glucocorticoids containing bulky A-ring substituents and no C3-carbonyl. Biochem. Biophys. Res. Commun. 86792-800. [DOI] [PubMed] [Google Scholar]
- 30.Styczynski, J., A. Kurylak, and M. Wysocki. 2005. Cytotoxicity of cortivazol in childhood acute lymphoblastic leukemia. Anticancer Res. 252253-2258. [PubMed] [Google Scholar]
- 31.Thompson, C. F., N. Quraishi, A. Ali, R. T. Mosley, J. R. Tata, M. L. Hammond, J. M. Balkovec, M. Einstein, L. Ge, G. Harris, T. M. Kelly, P. Mazur, S. Pandit, J. Santoro, A. Sitlani, C. Wang, J. Williamson, D. K. Miller, T. T. Yamin, C. M. Thompson, E. A. O'Neill, D. Zaller, M. J. Forrest, E. Carballo-Jane, and S. Luell. 2007. Novel glucocorticoids containing a 6,5-bicyclic core fused to a pyrazole ring: synthesis, in vitro profile, molecular modeling studies, and in vivo experiments. Bioorg. Med. Chem. Lett. 173354-3361. [DOI] [PubMed] [Google Scholar]
- 32.Thompson, E. B., D. Srivastava, and B. H. Johnson. 1989. Interactions of the phenylpyrazolo steroid cortivazol with glucocorticoid receptors in steroid-sensitive and -resistant human leukemic cells. Cancer Res. 492253s-2258s. [PubMed] [Google Scholar]
- 33.Togashi, M., S. Borngraeber, B. Sandler, R. J. Fletterick, P. Webb, and J. D. Baxter. 2005. Conformational adaptation of nuclear receptor ligand binding domains to agonists: potential for novel approaches to ligand design. J. Steroid Biochem. Mol. Biol. 93127-137. [DOI] [PubMed] [Google Scholar]
- 34.Urlinger, S., U. Baron, M. Thellmann, M. T. Hasan, H. Bujard, and W. Hillen. 2000. Exploring the sequence space for tetracycline-dependent transcriptional activators: novel mutations yield expanded range and sensitivity. Proc. Natl. Acad. Sci. USA 977963-7968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang, J. C., N. Shah, C. Pantoja, S. H. Meijsing, J. D. Ho, T. S. Scanlan, and K. R. Yamamoto. 2006. Novel arylpyrazole compounds selectively modulate glucocorticoid receptor regulatory activity. Genes Dev. 20689-699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Williams, S. P., and P. B. Sigler. 1998. Atomic structure of progesterone complexed with its receptor. Nature 393392-396. [DOI] [PubMed] [Google Scholar]
- 37.Wu, J., Y. Li, J. Dietz, and D. S. Lala. 2004. Repression of p65 transcriptional activation by the glucocorticoid receptor in the absence of receptor-coactivator interactions. Mol. Endocrinol. 1853-62. [DOI] [PubMed] [Google Scholar]
- 38.Xu, H. E., T. B. Stanley, V. G. Montana, M. H. Lambert, B. G. Shearer, J. E. Cobb, D. D. McKee, C. M. Galardi, K. D. Plunket, R. T. Nolte, D. J. Parks, J. T. Moore, S. A. Kliewer, T. M. Willson, and J. B. Stimmel. 2002. Structural basis for antagonist-mediated recruitment of nuclear co-repressors by PPARalpha. Nature 415813-817. [DOI] [PubMed] [Google Scholar]
- 39.Yamamoto, K. R. 1995. Multilayered control of intracellular receptor function. Harvey Lect. 911-19. [PubMed] [Google Scholar]