Abstract
We show that macroH2A1 histone variants are important for repressing the expression of endogenous murine leukemia viruses (MLVs) in mouse liver. Intact MLV proviruses and proviruses with deletions in env were nearly silent in normal mouse liver and showed substantial derepression in macroH2A1 knockout liver. In contrast, MLV proviruses with a deletion in the 5′ end of pro-pol were expressed in normal liver and showed relatively low levels of derepression in knockout liver. macroH2A1 nucleosomes were enriched on endogenous MLVs, with the highest enrichment occurring on the 5′ end of pro-pol. The absence of macroH2A1 also led to a localized loss of DNA methylation on the 5′ ends of MLV proviruses. These results demonstrate that macroH2A1 histones have a significant role in silencing endogenous MLVs in vivo and suggest that specific internal MLV sequences are targeted by a macroH2A1-dependent silencing mechanism.
macroH2A core histone variants have an N-terminal H2A domain that is ∼65% identical to conventional H2As and a C-terminal nonhistone domain that has been referred to as a macrodomain (30). These domains are connected by a linker segment that includes a basic region. Macrodomains occur in many proteins and as separate proteins in some bacteria (3, 31). The macrodomain of macroH2A1.1 binds the SirT1 metabolite O-acetyl-ADP-ribose (24). The significance of this binding activity for macroH2A function is not known. macroH2A variants appear to be highly conserved in vertebrate species and absent in most invertebrates (1, 31), although they are present in sea urchins. Three macroH2A variants have been identified in humans and mice. macroH2A1.1 and -1.2 are formed by alternate splicing of macroH2A1, and macroH2A2 is encoded by a separate gene (8, 15, 29, 32).
macroH2As are preferentially associated with transcriptionally repressed or silent chromatin domains, including the inactive X chromosome (8, 10, 14, 15), centromeric heterochromatin (16, 18), the XY body (21), and senescence-associated heterochromatic foci (40). This indicates that macroH2A nucleosomes have a role in repressing gene expression. A role for macroH2A1 in maintaining X inactivation is indicated by studies that used inhibitors of DNA methylation and histone deacetylation (20). A recent study with a cultured human T-cell line showed dramatic derepression of interleukin-8 in response to a macroH2A1 knockdown (2). In contrast, we did not find dramatic derepression of silenced genes in our study of gene expression in macroH2A1 knockout mouse liver (9). Instead, we found that macroH2A1 reduces the expression of specific genes that cluster in the area of lipid metabolism.
Our previous study of macroH2A distribution suggested that macroH2A1 might have a role in repressing the expression of endogenous retroviruses (10). Endogenous retroviruses can disrupt the structure and/or expression of normal cellular genes (27) and can contribute to the formation of pathological retroviruses (4, 11, 25). One important mechanism of controlling endogenous retroviral elements is silencing their expression (27). Here, we show that macroH2A1 histones have a role in silencing endogenous murine leukemia viruses (MLVs).
MATERIALS AND METHODS
Mice.
All animal protocols were approved by the University of Pennsylvania Institutional Animal Care and Use Committee. Mice used for organ harvest were 2 months of age and were killed between 8 and 11 a.m. The macroH2A1 knockout mutation was produced by removing the second exon of macroH2A1 and was bred into the C57BL/6 background for 10 generations (9). macroH2A1 knockout mice and their age- and sex-matched controls were raised in the same cage. All expression studies presented were done with female mice. Less complete studies done with male mice gave similar results (not shown).
Real-time PCR.
Real-time PCR was done as described previously (9). Denaturation was for 1 s at 95°C, annealing temperatures and times were primer specific, and elongation was at 72°C; time of elongation was primer specific (see Table S3 in the supplemental material for details). We averaged the results from two separate PCR runs for each sample. For gene expression studies, cDNAs were synthesized from total RNA by using an oligo(dT) primer. cDNA concentrations were equalized using glyceraldehyde phosphate dehydrogenase, and results from 13 macroH2A1 knockout livers were compared to those from 13 normal mouse livers (9). For macroH2A1 distribution studies, DNA isolated from macroH2A1-containing nucleosomes was compared to DNA isolated from bulk nucleosomes (10). macroH2A1-containing mono- and oligonucleosomes were purified from bulk chromatin by thiol-affinity chromatography (10). Results from four separate thiol-affinity preparations were averaged. Control thiol-affinity experiments done with macroH2A1 knockout chromatin showed that endogenous MLV sequences were essentially absent from the thiopropyl Sepharose-eluted fraction that normally contains macroH2A1 nucleosomes; real-time PCR showed that the abundance of these sequences in the knockout fraction was less than 1/1,000 of that obtained with normal liver.
Northern and Southern blots.
Northern blots and the hybridizations were done as described previously (9). DNA samples for Southern blots were run in 1% agarose gels and transferred onto Zeta-Probe GT membranes (Bio-Rad) with 0.4 M NaOH, following the standard protocol for Zeta-Probe membranes. The pol probe was a 362-bp fragment from the reverse transcriptase region of an endogenous MLV (10). The gag probe was a 408-bp fragment produced by PCR amplification from an endogenous MLV cDNA clone, gi 14810184 (see Table S3 in the supplemental material for primer information). The env probe was an ∼600-bp fragment produced by digesting an endogenous MLV cDNA clone, gi 14789949, with SalI and PstI.
RESULTS
Endogenous MLVs in C57BL/6 mice.
Our previous study of the distribution of macroH2A1 in mouse liver chromatin found that macroH2A1-containing nucleosomes were enriched on endogenous retroviral elements called RLTR4, which are derived from MLVs (10). There are 72 MLV proviruses in the genomic database for C57BL/6 mice (build 37.1). While the majority of these are free of large deletions compared to the RLTR4 consensus sequence, many have substantial deletions in one or more regions (Table 1). The most common deleted regions are env (23 proviruses) and the 5′ end of pro-pol (15 proviruses), and 10 proviruses have large deletions in both regions (see Fig. 1 for a diagram of an MLV provirus).
TABLE 1.
Endogenous MLVs in C57BL/6 micea
| Provirus classb | No. of proviruses | Genomic transcript size range (no. of bases) |
|---|---|---|
| Intact | 39 | ∼8,400 |
| gag deleted | 3 | 6,277-8,250 |
| pro-pol deleted; 5′ end | 5 | 6,378-7,042 |
| pro-pol deleted; middle region | 2 | 6,159-7,413 |
| env deleted | 11 | 6,434-7,113 |
| pro-pol 5′ and env deleted | 10 | 4,834-5,892 |
| pro-pol middle and env deleted | 2 | 5,883-5,932 |
These analyses were based on the public genomic database for C57BL/6 mice, build 37.1.
Sequences were compared to the consensus sequence for RLTR4. Only gaps greater than 100 bp were counted as deletions.
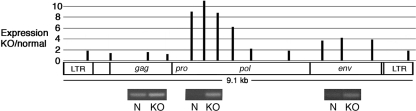
FIG. 1.
macroH2A1 represses the expression of endogenous MLVs. The expression of specific endogenous MLV sequences in normal liver and macroH2A1 knockout liver was determined by real-time PCR. Representative samples of PCR products from normal (N) and knockout (KO) liver are shown directly below three of the sites that were probed. The P values for the two-tailed t test were 0.02 or less for all expression comparisons except for those at the beginning and end of gag (see Table S1 in the supplemental material for all P values). The MLV diagram is based on the annotation of RLTR4 in the Repbase Update (23) and alignment with Moloney MLV sequences.
There are two major types of transcripts expected from MLV proviruses. Genomic transcripts initiate in the 5′ long terminal repeat (LTR), terminate in the 3′ LTR, and include all of the sequences in between. Genomic transcripts from intact proviruses should be ∼8.4 kb and proportionately smaller for proviruses with deletions of internal sequences (Table 1). Subgenomic transcripts splice out the gag and pro-pol sequences to form env-containing transcripts of ∼3 kb (26).
Analysis of endogenous MLV expression by real-time PCR.
We used real-time PCR to examine the expression of the four major regions of endogenous MLV proviruses: LTR, gag, pro-pol, and env. The primer sequences were based on the consensus sequence for RLTR4 obtained from Repbase Update (23). Most MLV proviruses are more than 95% identical to this consensus sequence, which allowed us to probe many proviruses at a time. In agreement with our hypothesis that macroH2A1 suppresses transcription, all four MLV regions showed increased expression in the livers of macroH2A1 knockout mice in comparison to levels in normal mouse liver (Fig. 1). Surprisingly, however, the magnitudes of the increases were very different for different regions. A high level of macroH2A1-dependent repression, ∼10-fold, was seen with probes in the 5′ end of pro-pol. In contrast, macroH2A1-dependent repression was less than twofold when we probed gag or the 3′ end of pro-pol. An intermediate level of macroH2A1-dependent repression, ∼4-fold, was observed with probes in env.
A possible explanation for the large differences in apparent repression among different proviral regions lies in the variations in structure among the 72 endogenous MLV proviruses in C57BL/6 mice. Fifteen proviruses have a substantial deletion in the 5′ end of pro-pol. Although the exact positions of these deletions differ between proviruses, they are all within the region that showed a large increase in expression in knockout liver. If these proviruses escape macroH2A1-dependent repression and are expressed in normal liver, then the level of macroH2A1-dependent repression that we observe for the MLV sequences outside the 5′ end of pro-pol will be reduced.
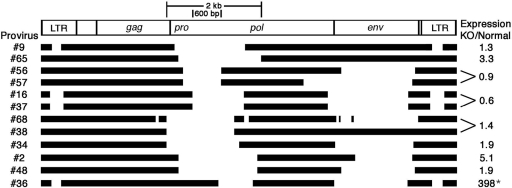
To test this idea, we designed PCR primers that bridge the pro-pol deletions present in specific proviruses. Most of these deletions are more than 1 kb, which made it possible to selectively amplify cDNAs from pro-pol-deleted proviruses by using a very short elongation time. Each primer pair was designed to specifically probe the expression of one or in some cases two pro-pol-deleted proviruses. This approach succeeded with eight primer pairs, as judged by the production of a single DNA fragment of the correct size, and allowed us to probe the expression of 11 of the 15 proviruses that have a deletion in the 5′ end of pro-pol. Most of these proviruses showed a <2-fold increase in expression in macroH2A1 knockout liver (Fig. 2), in contrast to the ∼10-fold increase that we observed when we probed the 5′ end of pro-pol (Fig. 1). A region of ∼600 bp is common to all of the 5′ pro-pol deletions that appear to reduce macroH2A-dependent repression (Fig. 2). The deletion in provirus no. 36 (shown at the bottom of Fig. 2) starts toward the middle of pro-pol and does not include the 600-bp region that is deleted in the other proviruses shown in Fig. 2. This provirus had a very high level of derepression in knockout liver, indicating that the middle of pro-pol is not critical for macroH2A1-dependent repression.
FIG. 2.
Deletions in the 5′ end of pro-pol are associated with decreased macroH2A1-dependent repression. The deletions in specific endogenous MLV proviruses are shown. The 2-kb segment indicates the range of the deletions associated with reduced macroH2A1-dependent repression. The 600-bp segment indicates the region that is common to all of these deletions; note that the deletion in provirus no. 36 does not include this 600-bp segment and that this provirus shows a high level of macroH2A1-dependent repression (*). Relative expression was determined by real-time PCR. The statistical analyses of these comparisons are shown in Table S1 in the supplemental material. Proviruses were numbered sequentially according to their occurrence in the genome of C57BL/6 mice, genome build 37.1 (see Table S2 in the supplemental material).
Most of the proviruses with a deletion in pro-pol also have a deletion in env (Table 1). This suggested that env might also contribute to macroH2A1-dependent repression. However, two of the pro-pol-deleted proviruses that we probed by real-time PCR did not have an env deletion, and they still showed relatively low levels of macroH2A1-dependent repression, 1.3-fold for provirus no. 9 and 3.3-fold for provirus no. 65 (Fig. 2). In addition, provirus no. 36, which has a very high level of macroH2A1-dependent repression, has a large deletion in env. These results indicate that the env region is not critical for macroH2A1-dependent repression.
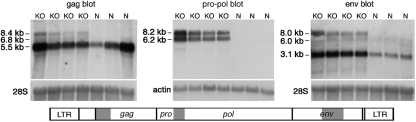
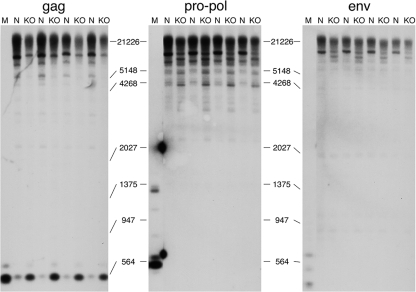
Northern blot analysis of endogenous MLV expression.
Transcripts that hybridized to a probe from the 5′ end of pro-pol were nearly undetectable on Northern blots of normal mouse liver but were readily detected in macroH2A1 knockout liver (Fig. 3, pro-pol blot). Two major pol-containing transcript sizes were evident in the knockouts, one at ∼8.2 kb and the other at ∼6.2 kb. The 8.2-kb transcripts correspond in size to genomic transcripts from intact proviruses (Table 1). We believe that the 6.2-kb band primarily contains genomic transcripts from proviruses with env deletions (see sequencing section below), although transcripts of this size could also come from one provirus with a deletion in gag and one provirus with a deletion in the middle of pro-pol (Table 1).
FIG. 3.
Northern blot analyses of endogenous MLV expression. Total mouse liver RNA was used. The locations of the gag, pro-pol, and env hybridization probes are shown in gray on the diagram below the blots. Hybridization with a 28S rRNA or β-actin probe was used to show equal loading of the lanes. Band sizes were estimated from a standard curve constructed from an RNA standard. Lanes: KO, RNA from female macroH2A1 knockout liver; N, RNA from normal female liver.
Transcripts hybridizing with a gag probe were readily detected in normal mouse liver (Fig. 3, gag blot). The major gag-containing band was centered at ∼5.5 kb, indicating that these transcripts came from proviruses with deletions in both pro-pol and env (Table 1). Consistent with this identification, transcripts of this size and abundance were not seen on the blots hybridized with the pro-pol or env probes. The gag probe should also detect the genomic transcripts from the intact and env-deleted proviruses that were detected on the pol blot. Indeed, two fainter bands of the appropriate sizes are present in the knockout lanes of the gag blot.
env-containing transcripts were substantially more abundant in macroH2A1 knockout liver than in normal liver (Fig. 3, env blot), consistent with our real-time PCR results shown in Fig. 1. The major band on the env blot is ∼3 kb in size, as expected for subgenomic transcripts that have spliced-out gag and pro-pol sequences (26). The upper band in the knockout lanes of the env blot corresponds in size to genomic transcripts from intact proviruses. The heterogeneous bands below this band appear to be genomic transcripts from proviruses with a deletion in the 5′ end of pro-pol, based on their sizes (Table 1) and their absence in the normal lanes of the pol blot. Unlike the transcripts from intact proviruses, some of these transcripts were detected in normal liver. This is consistent with the idea that deletions in the 5′ end of pro-pol can relieve macroH2A1-dependent repression.
Sequence analysis of proviral transcripts.
We sought to identify some of the pro-pol-containing MLV proviruses that are derepressed in macroH2A1 knockout liver by sequencing cloned cDNAs that were amplified by PCR. Although proviruses are very similar to one another, many can be uniquely identified by nucleotide sequence comparisons. We amplified a segment of more than 700 bp that starts in pol just 3′ of the 600-bp region marked in Fig. 2. Real-time PCR indicated that this segment was ∼11 times more abundant in knockout liver than in normal liver. To minimize the formation of hybrid duplexes from different proviruses, we stopped the PCR at an early stage when most duplexes will be formed by replication, not annealing. In assigning a sequence to a provirus, we allowed one mismatch, provided that the mismatch appeared to be a sequencing error or a PCR mutation, i.e., the mismatched nucleotide did not match any provirus in the genomic database. Using this approach, we assigned ∼70% of the sequences to a specific provirus.
Sequence analysis of 22 clones from normal mouse liver identified five different expressed proviruses. Three intact proviruses were detected, and two of these are in active genes (Table 2). The other two proviruses that we identified have deletions in the 5′ end of pro-pol. Although the primers that we used cannot amplify cDNAs from most pro-pol-deleted proviruses, three proviruses with deletions in the 5′ end of pro-pol can be amplified. The most frequently detected provirus has a deletion in the 5′ end of pro-pol and a deletion in env. Neither of the pro-pol-deleted proviruses identified in this analysis would be detected on our pol Northern blot shown in Fig. 3.
TABLE 2.
Identification of some expressed MLV provirusesa
| cDNA group and provirus no.b | Provirus type | Frequency (no. of hits) |
|---|---|---|
| cDNAs from normal liver | ||
| 1 | Intact, in Ctse | 2 |
| 22 | Intact, in Galnt11 | 4 |
| 32 | Intact | 2 |
| 38 | pro-pol deletedc | 1 |
| 68 | pro-pol and env deletedc | 7 |
| Not assignedd | 6 | |
| cDNAs from macroH2A1 knockout liver | ||
| 32 | Intact | 2 |
| 62 | Intact | 1 |
| 38 | pro-pol deletedc | 1 |
| 29 | env deleted | 2 |
| 39 | env deleted | 5 |
| 70 | env deleted | 4 |
| 68 | pro-pol and env deletedc | 1 |
| Not assignedd | 7 |
Cloned MLV cDNAs from the pro-pol region were sequenced and compared to the public genomic database for C57BL/6 mice (build 37.1). See Table S3 in the supplemental material for primer sequences.
Proviruses were numbered according to their occurrence in the public genomic database for C57BL/6 mice (see Table S2 in the supplemental material).
These pro-pol deletions ended before the start of the pro-pol segment that we amplified for this analysis.
Sequences were not assigned to a provirus when they matched multiple proviruses or had more than one mismatch to their best hit.
Sequence analysis of 23 clones from macroH2A1 knockout liver identified seven different expressed proviruses (Table 2). The most frequently detected proviruses were three env-deleted proviruses that were not identified in our analysis of normal liver. This supports the idea that env-deleted proviruses are strongly derepressed in knockout liver. pol-env-deleted provirus no. 68, which had seven hits in our analysis of normal liver, had only one hit with knockout liver. This indicates that its expression does not increase dramatically in knockout liver, consistent with our PCR analysis of this provirus shown in Fig. 2.
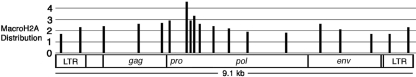
Distribution of macroH2A1 on MLV proviruses.
We examined the distribution of macroH2A1 nucleosomes on MLV sequences in normal mouse liver chromatin to assess the possibility that macroH2A1 histones have a direct role in repressing the expression of the endogenous MLVs. We used real-time PCR to determine the abundance of specific proviral sequences in DNA isolated from macroH2A1-containing nucleosomes relative to that in DNA from bulk nucleosomes (10). macroH2A1 was enriched on all of the MLV sequences that we probed (Fig. 4). Interestingly, the highest concentration of macroH2A1 occurred in the 5′ end of pro-pol, coincident with the region of pro-pol that is deleted in proviruses with reduced macroH2A-dependent repression. Analyzing proviral LTRs is complicated by the presence of many solo LTRs. However, we found two PCR primer pairs that should distinguish MLV proviral LTRs from most solo LTRs.
FIG. 4.
macroH2A1 nucleosomes are enriched on endogenous MLVs. The relative concentration of macroH2A1 was determined by real-time PCR using DNA from macroH2A1 nucleosomes that were purified by thiol-affinity chromatography (10). A value of 1 indicates a macroH2A1 concentration equal to that of the bulk chromatin applied to the thiol-affinity procedure. The values shown are the averages determined from four independent preparations. The P values for a t test of the enrichment were <0.02 at all sites (see Table S1 in the supplemental material for specific P values and the standard deviation analysis).
DNA methylation.
DNA methylation is an important mechanism for repressing retroelements such as endogenous MLVs (35, 39). We used the methylation-sensitive restriction nuclease HpaII to examine whether the absence of macroH2A1 affects the methylation of endogenous MLVs. Total genomic DNA from normal and macroH2A1 knockout liver was digested to completion with HpaII and run on Southern blots. The blots were hybridized with the same gag, pol, and env probes used on the Northern blots (Fig. 3). For comparison, these DNA samples were digested with MspI, which has the same recognition sequence as HpaII but is not sensitive to methylation. While evidence for reduced methylation was evident with all three probes, only the gag blot showed evidence for a substantial loss of methylation in the immediate vicinity of the probe (Fig. 5). This probe is located at the beginning of gag, less than 500 bases downstream of the 5′ LTR. Because of the numerous solo MLV-related LTRs, we did not use this approach to analyze the methylation of MLV proviral LTRs. The HpaII digestion patterns of bulk DNA, LINE L1 DNA, and gamma satellite DNA were not substantially altered by a macroH2A1 knockout (see Fig. S1 in the supplemental material), indicating that there is no generalized reduction of DNA methylation in knockout liver.
FIG. 5.
DNA methylation of endogenous MLVs is reduced in macroH2A1 knockout liver. DNA isolated from mouse liver was digested to completion with HpaII and analyzed on Southern blots by using the gag, pro-pol, and env probes shown in Fig. 3. Lanes: N, DNA from normal mouse liver; KO, DNA from macroH2A1 knockout liver; M, DNA digested with MspI. Positions of marker bands are indicated between the blots. The hybridization seen between the M lane and the first N lane of the pol blot is spillover from coincidental hybridization of the probe to fragments in the marker lane.
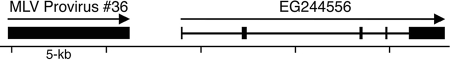
Effect of proviral derepression on the expression of a neighboring gene.
Provirus no. 36, which is strongly derepressed in knockout liver (Fig. 2), is less than 3 kb upstream of a gene called EG244556 (Fig. 6). We examined the expression of this gene to see whether it is affected by the derepression of this provirus. Using real-time PCR to amplify a sequence in the 3′ end of this gene, we found a 1.7-fold increase in expression in female macroH2A1 knockout liver (P < 10−5; two-tailed t test). This gene encodes a KRAB domain zinc finger protein of an unknown function. This gene would not have been detected in our previous analysis of gene expression (9), because it is not represented on the microarray that we used.
FIG. 6.
Close proximity of derepressed MLV provirus to gene EG244556. The diagram shows the position of MLV provirus no. 36 relative to that of EG244556 on mouse chromosome 8. Provirus no. 36 is strongly derepressed in macroH2A1 knockout liver (Fig. 2), and EG244556 expression is increased 70% in knockout liver. Arrows indicate direction of transcription.
Expression of other repetitive elements in macroH2A1 knockout liver.
We used real-time PCR to examine whether the expression of other repetitive elements is derepressed in macroH2A1 knockout liver (Table 3). We observed significant macroH2A1-dependent repression of MusD retroviral elements and ERVβ7 endogenous beta retroviral elements. We found no increase in the expression of gamma satellite (a centromeric satellite), LINE L1 elements, the mammalian apparent LTR retrotransposon ORR1a, and the retroviral elements IAP and RLTR6. These results indicate that macroH2A1 histones have a role in repressing some, but not all, endogenous retroviral elements.
TABLE 3.
Expression of repetitive elements in macroH2A1 knockout liver
| Element | Expression level (P)a |
|---|---|
| ERVβ7; pol1 | 2.0 (4 × 10−8) |
| ERVβ7; pol2 | 2.4 (3 × 10−9) |
| MusD; pol | 1.9 (3 × 10−7) |
| Gamma satellite | 1 |
| IAP; pol | 0.9 |
| LINE L1; 5′ end | 0.9 |
| LINE L1; 3′ end | 0.9 |
| ORR1a; internal | 1 |
| RLTR6; pol | 1 |
Expression was determined by real-time PCR and is given as expression levels in macroH2A1 knockout liver divided by those in normal liver. The P values of a two-tailed t test are shown in parentheses. Primer sequences were based on the consensus sequences for these elements from Repbase (23). See Table S3 in the supplemental material for primer sequences.
DISCUSSION
Our results demonstrate that macroH2A1 histones have a significant role in repressing the expression of endogenous MLVs. While it is possible that a macroH2A1 knockout has indirect effects on the stability of MLV RNAs, the distribution of macroH2A1 nucleosomes on endogenous MLVs, the effects of the knockout on proviral DNA methylation (Fig. 5), and the expression of a neighboring gene (Fig. 6) all suggest a direct effect on proviral transcription. macroH2A1 nucleosomes are enriched on endogenous MLV proviruses in mouse liver, with the highest enrichment occurring on the 5′ end of pro-pro (Fig. 4), a region that appears to be important for macroH2A1-dependent repression (Fig. 2). macroH2A1 nucleosomes are not limited to the 5′ end of pro-pol, which suggests that this region might promote the formation of macroH2A1 nucleosomes throughout the provirus rather than at a specific site. This could potentially be achieved by recruiting a nucleosome-remodeling complex that exchanges macroH2A1 into nucleosomes. macroH2A1 nucleosomes could then inhibit initiation of transcription (17). The presence of macroH2A1 nucleosomes on the transcribed regions of MLV proviruses leaves open the possibility that these nucleosomes might inhibit elongation or even RNA processing events, but there is no direct evidence for either of these mechanisms.
There is long-standing evidence that DNA methylation has a role in repressing the expression of endogenous retroviral elements (19, 22, 28, 35). Our results provide the first evidence that macroH2As have a role in promoting the methylation of a specific genomic region. Interestingly, the absence of macroH2A1 does not reduce DNA methylation of the entire provirus but rather affects a specific area toward the 5′ end. This could be a direct effect of macroH2A1 on DNA methylation of this region or an indirect effect that occurs after transcriptional repression.
Interestingly, the effect of a macroH2A1 knockout on specific MLV proviruses ranges from essentially no effect to dramatic derepression. Deletions in the 5′ end of pro-pol appear to be important for allowing some proviruses to escape macroH2A1-dependent repression. We examined the expression of 11 proviruses with deletions in the 5′ end of pro-pol by real-time PCR, and most of them showed <2-fold derepression in macroH2A1 knockout liver (Fig. 2). Northern blots showed that transcripts from proviruses that have deletions in this region can be detected in normal mouse liver and that the expression of these transcripts is little changed in macroH2A1 knockouts (Fig. 3, gag and env blots). In contrast, the expression of transcripts that contain the 5′ end of pro-pol was very low in normal liver and dramatically increased in macroH2A1 knockout liver (Fig. 1 and 3). While the expression of individual proviruses is likely affected by their local chromatin environment and other factors, our results indicate that the region on the 5′ end of pro-pol is specifically important for macroH2A1-dependent repression of these elements. Consistent with this idea, the highest concentration of macroH2A1 nucleosomes occurs on this region (Fig. 4).
In contrast, proviruses that have a deletion only in env appear to have enhanced derepression in macroH2A1 knockout liver. The pro-pol Northern blot indicates that transcripts from env-deleted proviruses in macroH2A1 knockout liver are similar in abundance to transcripts from intact proviruses (Fig. 3), even though there are many more intact proviruses in the genome (Table 1). This preference for the derepression of env-deleted proviruses in macroH2A1 knockouts was also seen in our sequence analysis (Table 2). None of the env-deleted proviruses in C57BL/6 mice are in a known gene, so it does not appear that the preferential derepression of these proviruses is related to their presence in active genes. These results suggest that MLV env is targeted by a repressive mechanism that does not require macroH2A1.
The abundant expression of proviruses that have deletions in the 5′ ends of both pro-pol and env may be due to their escape from both a macroH2A1-dependent repressive mechanism that targets pro-pol and a macroH2A1-independent repressive mechanism that targets env. While factors such as local chromatin environment probably also contribute to expression, deletion of these two regions appears to allow significant expression of some proviruses in normal liver. There are other studies that indicate that endogenous MLVs with pro-pol and env deletions may escape silencing. Burn injury induces the expression of endogenous pol-env-deleted MLV proviruses in a variety of mouse tissues, including liver (12, 13). Increased expression of proviruses with deletions in pro-pol and env could increase the chance of generating pathological viruses. Spontaneous retrovirally induced leukemia in BXH-2 mice frequently involves a defective MLV with deletions in pro-pol and env (11). A murine immunodeficiency syndrome also involves an MLV that has deletions in pro-pol and env (4). In all of these cases, the pro-pol deletion involves the 5′ end of pro-pol and the env deletion involves a large segment of env similar to those shown in Fig. 2.
Studies with retroviral vectors indicate that retroviral LTRs and the primer-binding site can trigger genomic silencing in the absence of internal retroviral sequences (5, 7, 38). Our studies indicate that repressive mechanisms targeted to internal retroviral sequences can also be important. There are other examples that suggest the preferential expression and spread of retroviral elements that have lost specific internal sequences (27). Early transposon (ETn) elements are LTR retrotransposons that lack nearly all of the typical internal retroviral sequences. Retrotransposition of ETn elements is dependent on MusD elements, which are endogenous beta retroviral elements (33, 34). Transcripts from ETn elements are much more abundant than MusD transcripts, despite the much greater abundance of MusD elements in the genome and the near identity of the LTR sequences in these two elements (6). In humans, human endogenous retrovirus H elements that have deletions in pol and env are more abundant in the genome than intact human endogenous retrovirus H proviruses, and the deleted forms appear to be expressed at much higher levels than intact elements (36, 37). We suggest that in at least some cases, deletion of internal retroviral sequences allows expression of the element by relieving specific repressive mechanisms. Increased expression could promote the proliferation of the defective elements in the genome by retrotransposition or infection using retroviral proteins produced by other proviruses. Based on their relative abundance, pol-env-deleted MLVs could be at an early stage of expansion in the mouse genome (Table 1). There appears to have been at least one recent retrotransposition of a pol-env-deleted provirus involving MLV proviruses no. 16 and no. 37, which are identical except for a single nucleotide.
Endogenous retroviruses can affect the expression of neighboring genes, and we identified one example where derepression of an endogenous MLV in macroH2A1 knockout liver appears to increase the expression of a neighboring gene. Such effects could potentially account for the changes in gene expression that we previously documented for macroH2A1 knockout liver (9). However, endogenous MLVs would not appear to be involved in the genes that we documented in that study, because there are no endogenous MLVs in the proximity of those genes.
A wide variety of mechanisms are used to protect the genome from retroviral elements (27). Our results indicate that macroH2A1 histones are a part of a system that protects the genome by suppressing the expression of endogenous MLVs and some other retroelements. Because macroH2As show very little evolutionary variation in structure (31) and endogenous retroviral sequences are highly variable, we believe that it is likely that other factors participate in the targeting of macroH2A1-dependent repression to these genomic elements.
Supplementary Material
Acknowledgments
We thank Dannee Chen, Adrian Leu, and Carl Costanzi for assistance with the experiments; Sandra Martin for providing the LINE L1 probe; and Richard Katz for critical reading of the manuscript.
This work was supported by Public Health Service grant GM49351 from the National Institute of General Medical Sciences, a grant from the University of Pennsylvania Research Foundation, and USDA Formula Funds.
Footnotes
Published ahead of print on 14 January 2008.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Abbott, D. W., M. Laszczak, J. D. Lewis, H. Su, S. C. Moore, M. Hills, S. Dimitrov, and J. Ausio. 2004. Structural characterization of macroH2A containing chromatin. Biochemistry 431352-1359. [DOI] [PubMed] [Google Scholar]
- 2.Agelopoulos, M., and D. Thanos. 2006. Epigenetic determination of a cell-specific gene expression program by ATF-2 and the histone variant macroH2A. EMBO J. 254843-4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allen, M. D., A. M. Buckle, S. C. Cordell, J. Lowe, and M. Bycroft. 2003. The crystal structure of AF1521 a protein from Archaeoglobus fulgidus with homology to the non-histone domain of macroH2A. J. Mol. Biol. 330503-511. [DOI] [PubMed] [Google Scholar]
- 4.Aziz, D. C., Z. Hanna, and P. Jolicoeur. 1989. Severe immunodeficiency disease induced by a defective murine leukaemia virus. Nature 338505-508. [DOI] [PubMed] [Google Scholar]
- 5.Barklis, E., R. C. Mulligan, and R. Jaenisch. 1986. Chromosomal position or virus mutation permits retrovirus expression in embryonal carcinoma cells. Cell 47391-399. [DOI] [PubMed] [Google Scholar]
- 6.Baust, C., L. Gagnier, G. J. Baillie, M. J. Harris, D. M. Juriloff, and D. L. Mager. 2003. Structure and expression of mobile ETnII retroelements and their coding-competent MusD relatives in the mouse. J. Virol. 7711448-11458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bestor, T. H. 2000. Gene silencing as a threat to the success of gene therapy. J. Clin. Investig. 105409-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chadwick, B. P., and H. F. Willard. 2001. Histone H2A variants and the inactive X chromosome: identification of a second macroH2A variant. Hum. Mol. Genet. 101101-1113. [DOI] [PubMed] [Google Scholar]
- 9.Changolkar, L. N., C. Costanzi, N. A. Leu, D. Chen, K. J. McLaughlin, and J. R. Pehrson. 2007. Developmental changes in histone macroH2A1-mediated gene regulation. Mol. Cell. Biol. 272758-2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Changolkar, L. N., and J. R. Pehrson. 2006. macroH2A1 histone variants are depleted on active genes but concentrated on the inactive X. Mol. Cell. Biol. 264410-4420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cho, B. C., J. D. Shaughnessy, Jr., D. A. Largaespada, H. G. Bedigian, A. M. Buchberg, N. A. Jenkins, and N. G. Copeland. 1995. Frequent disruption of the Nf1 gene by a novel murine AIDS virus-related provirus in BXH-2 murine myeloid lymphomas. J. Virol. 697138-7146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cho, K., L. K. Adamson, and D. G. Greenhalgh. 2002. Induction of murine AIDS virus-related sequences after burn injury. J. Surg. Res. 10453-62. [DOI] [PubMed] [Google Scholar]
- 13.Cho, K., and D. Greenhalgh. 2003. Injury-associated induction of two novel and replication-defective murine retroviral RNAs in the liver of mice. Virus Res. 93189-198. [DOI] [PubMed] [Google Scholar]
- 14.Costanzi, C., and J. R. Pehrson. 1998. Histone macroH2A1 is concentrated in the inactive X chromosome of female mammals. Nature 393599-601. [DOI] [PubMed] [Google Scholar]
- 15.Costanzi, C., and J. R. Pehrson. 2001. MACROH2A2, a new member of the MACROH2A core histone family. J. Biol. Chem. 27621776-21784. [DOI] [PubMed] [Google Scholar]
- 16.Costanzi, C., P. Stein, D. M. Worrad, R. M. Schultz, and J. R. Pehrson. 2000. Histone macroH2A1 is concentrated in the inactive X chromosome of female preimplantation embryos. Development 1272283-2289. [DOI] [PubMed] [Google Scholar]
- 17.Doyen, C. M., W. An, D. Angelov, V. Bondarenko, F. Mietton, V. M. Studitsky, A. Hamiche, R. G. Roeder, P. Bouvet, and S. Dimitrov. 2006. Mechanism of polymerase II transcription repression by the histone variant macroH2A. Mol. Cell. Biol. 261156-1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grigoryev, S. A., T. Nikitina, J. R. Pehrson, P. B. Singh, and C. L. Woodcock. 2004. Dynamic relocation of epigenetic chromatin markers reveals an active role of constitutive heterochromatin in the transition from proliferation to quiescence. J. Cell Sci. 1176153-6162. [DOI] [PubMed] [Google Scholar]
- 19.Groudine, M., R. Eisenman, and H. Weintraub. 1981. Chromatin structure of endogenous retroviral genes and activation by an inhibitor of DNA methylation. Nature 292311-317. [DOI] [PubMed] [Google Scholar]
- 20.Hernandez-Munoz, I., A. H. Lund, P. van der Stoop, E. Boutsma, I. Muijrers, E. Verhoeven, D. A. Nusinow, B. Panning, Y. Marahrens, and M. van Lohuizen. 2005. Stable X chromosome inactivation involves the PRC1 Polycomb complex and requires histone MACROH2A1 and the CULLIN3/SPOP ubiquitin E3 ligase. Proc. Natl. Acad. Sci. USA 1027635-7640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoyer-Fender, S., C. Costanzi, and J. R. Pehrson. 2000. Histone macroH2A1.2 is concentrated in the XY-body by the early pachytene stage of spermatogenesis. Exp. Cell Res. 258254-260. [DOI] [PubMed] [Google Scholar]
- 22.Jaenisch, R., A. Schneike, and K. Harbers. 1985. Treatment of mice with 5-azacytidine efficiently activates silent retroviral genomes in different tissues. Proc. Natl. Acad. Sci. USA 821451-1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jurka, J., V. V. Kapitonov, A. Pavlicek, P. Klonowski, O. Kohany, and J. Walichiewicz. 2005. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet. Genome Res. 110462-467. [DOI] [PubMed] [Google Scholar]
- 24.Kustatscher, G., M. Hothorn, C. Pugieux, K. Scheffzek, and A. G. Ladurner. 2005. Splicing regulates NAD metabolite binding to histone macroH2A. Nat. Struct. Mol. Biol. 12624-625. [DOI] [PubMed] [Google Scholar]
- 25.Laigret, F., R. Repaske, K. Boulukos, A. B. Rabson, and A. S. Khan. 1988. Potential progenitor sequences of mink cell focus-forming (MCF) murine leukemia viruses: ecotropic, xenotropic, and MCF-related viral RNAs are detected concurrently in thymus tissues of AKR mice. J. Virol. 62376-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lazo, P. A., V. Prasad, and P. N. Tsichlis. 1987. Splice acceptor site for the env message of Moloney murine leukemia virus. J. Virol. 612038-2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maksakova, I. A., M. T. Romanish, L. Gagnier, C. A. Dunn, L. N. van de Lagemaat, and D. L. Mager. 2006. Retroviral elements and their hosts: insertional mutagenesis in the mouse germ line. PLoS Genet. 2e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Niwa, O., and T. Sugahara. 1981. 5-Azacytidine induction of mouse endogenous type C virus and suppression of DNA methylation. Proc. Natl. Acad. Sci. USA 786290-6294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pehrson, J. R., C. Costanzi, and C. Dharia. 1997. Developmental and tissue expression patterns of histone macroH2A1 subtypes. J. Cell. Biochem. 65107-113. [DOI] [PubMed] [Google Scholar]
- 30.Pehrson, J. R., and V. A. Fried. 1992. MacroH2A, a core histone containing a large nonhistone region. Science 2571398-1400. [DOI] [PubMed] [Google Scholar]
- 31.Pehrson, J. R., and R. N. Fuji. 1998. Evolutionary conservation of macroH2A subtypes and domains. Nucleic Acids Res. 262837-2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rasmussen, T. P., T. Huang, M. A. Mastrangelo, J. Loring, B. Panning, and R. Jaenisch. 1999. Messenger RNAs encoding mouse histone macroH2A1 isoforms are expressed at similar levels in male and female cells and result from alternative splicing. Nucleic Acids Res. 273685-3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ribet, D., M. Dewannieux, and T. Heidmann. 2004. An active murine transposon family pair: retrotransposition of “master” MusD copies and ETn trans-mobilization. Genome Res. 142261-2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ribet, D., F. Harper, M. Dewannieux, G. Pierron, and T. Heidmann. 2007. Murine MusD retrotransposon: structure and molecular evolution of an “intracellularized” retrovirus. J. Virol. 811888-1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Walsh, C. P., J. R. Chaillet, and T. H. Bestor. 1998. Transcription of IAP endogenous retroviruses is constrained by cytosine methylation. Nat. Genet. 20116-117. [DOI] [PubMed] [Google Scholar]
- 36.Wilkinson, D. A., J. D. Freeman, N. L. Goodchild, C. A. Kelleher, and D. L. Mager. 1990. Autonomous expression of RTVL-H endogenous retroviruslike elements in human cells. J. Virol. 642157-2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wilkinson, D. A., N. L. Goodchild, T. M. Saxton, S. Wood, and D. L. Mager. 1993. Evidence for a functional subclass of the RTVL-H family of human endogenous retrovirus-like sequences. J. Virol. 672981-2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wolf, D., and S. P. Goff. 2007. TRIM28 mediates primer binding site-targeted silencing of murine leukemia virus in embryonic cells. Cell 13146-57. [DOI] [PubMed] [Google Scholar]
- 39.Yoder, J. A., C. P. Walsh, and T. H. Bestor. 1997. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 13335-340. [DOI] [PubMed] [Google Scholar]
- 40.Zhang, R., M. V. Poustovoitov, X. Ye, H. A. Santos, W. Chen, S. M. Daganzo, J. P. Erzberger, I. G. Serebriiskii, A. A. Canutescu, R. L. Dunbrack, J. R. Pehrson, J. M. Berger, P. D. Kaufman, and P. D. Adams. 2005. Formation of MacroH2A-containing senescence-associated heterochromatin foci and senescence driven by ASF1a and HIRA. Dev. Cell 819-30. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.