Abstract
Promoter clearance and transcriptional processivity in eukaryotic cells are fundamentally regulated by the phosphorylation of the carboxy-terminal domain of RNA polymerase II (RNAPII). One of the kinases that essentially performs this function is P-TEFb (positive transcription elongation factor b), which is composed of cyclin-dependent kinase 9 (CDK9) associated with members of the cyclin T family. Here we show that cellular GCN5 and P/CAF, members of the GCN5-related N-acetyltransferase family of histone acetyltransferases, regulate CDK9 function by specifically acetylating the catalytic core of the enzyme and, in particular, a lysine that is essential for ATP coordination and the phosphotransfer reaction. Acetylation markedly reduces both the kinase function and transcriptional activity of P-TEFb. In contrast to unmodified CDK9, the acetylated fraction of the enzyme is specifically found in the insoluble nuclear matrix compartment. Acetylated CDK9 associates with the transcriptionally silent human immunodeficiency virus type 1 provirus; upon transcriptional activation, it is replaced by the unmodified form, which is involved in the elongating phase of transcription marked by Ser2-phosphorylated RNAPII. Given the conservation of the CDK9 acetylated residues in the catalytic task of virtually all CDK proteins, we anticipate that this mechanism of regulation might play a broader role in controlling the function of other members of this kinase family.
One of the essential molecular events that regulate gene expression in eukaryotic cells is the phosphorylation of the carboxy-terminal domain (CTD) of RNA polymerase II (RNAPII), which is required for efficient promoter clearance and transcriptional processivity (29). Different members of the cyclin-dependent kinase (CDK) family, including CDK7, CDK8, and CDK9, are known to specifically induce this modification. In particular, CDK9 has been identified as the catalytic component of the P-TEFb (positive transcription elongation factor b) complex, which is composed of the kinase associated with cyclin T1 or one of the other related members of the cyclin T family (37, 38). A subset of cellular P-TEFb is found in an inactive form as a complex with the HEXIM1 protein (31, 52) and the 7SK snRNA (35, 51), while the enzymatically active complex associates with the bromodomain protein Brd4 (50).
In mammalian cells, the role of P-TEFb has been initially highlighted by the association of cyclin T1 with the human immunodeficiency virus type 1 (HIV-1) Tat transactivator (25, 47, 55). Following infection of susceptible cells, the HIV-1 provirus becomes integrated into the host genome and its long terminal repeat (LTR) is embedded into a chromatin conformation that has a repressive effect on gene expression (16, 27). Transcriptional activation is triggered by a variety of extracellular stimuli that lead to cellular activation and involves both the acetylation of the nucleosomes at the LTR and the recruitment of elongation-competent RNAPII complexes, the latter event being essentially catalyzed by P-TEFb (1, 12, 54). Whether these two concomitant events might be interconnected at the molecular level is yet unclear.
Studies performed using either RNA interference (RNAi) or highly specific inhibitors have indicated that P-TEFb, besides activating HIV-1 gene expression, acts as a global cofactor important for most RNAPII-mediated transcription (5, 41). Not surprisingly, therefore, its activity appears to be tightly regulated in different cell types during differentiation and as part of the cell response to various stimuli. Regulation is attained by controlling the levels of both cyclin T1 and CDK9 (14, 39, 48, 53), by modulating CDK9 posttranslational modification by phosphorylation (10) and ubiquitination (20), and by regulating CDK9 nucleocytoplasmic shuttling (34).
Here we describe a novel mechanism for regulation of the CDK9 function, which involves the acetylation of two specific lysine residues in the catalytic core of the enzyme. Members of the GCN5-related N-acetyltransferase (GNAT) family (GCN5 and P/CAF) of acetyltransferases are most effective in inducing this modification. In particular, GCN5-mediated in vivo acetylation preferentially targets one lysine (K48) that is essential for ATP coordination; this modification causes a profound inhibition of CDK9 catalytic and transcriptional activities and relocates the enzyme to the insoluble nuclear matrix compartment. Chromatin immunoprecipitation (ChIP) analyses showed that acetylated CDK9 associates with the latent HIV-1 proviral genome together with P/CAF and GCN5. Upon transcriptional activation, unmodified CDK9 predominates at the promoter region, concomitant with the onset of the elongating phase of transcription marked by RNAPII Ser2 phosphorylation.
MATERIALS AND METHODS
Plasmids.
pcDNA3-CDK9-his was kindly provided by D. H. Price (Iowa City, IA). From this template, CDK9 was subcloned into different expression vectors: pcDNA3 (Invitrogen) for in vitro translation, pGEX-2T (Amersham) for recombinant protein purification, and pFlag-CMV-2 (Sigma) and pEGFP-C1 (Clontech) for expression in eukaryotic cells.
pGEX-2T-CDK9 deletion mutants were prepared by PCR amplification of CDK9 with primers specific for the deleted versions. pGEX-2T and pFlag single and double CDK9 mutants were constructed using recombinant PCR starting from each original vector.
Gal4-CDK9 was generously provided by L. Lania (Napoli, Italy).
pG6(5′Pro) was kindly donated by B. M. Peterlin (San Francisco, CA).
Glutathione S-transferase (GST)-CTD-expressing bacteria were generously provided by R. Young (Cambridge, MA).
pGEX-GCN5 was a kind gift from M. Benkirane (Montpellier, France).
pGEX-2T-GCN5 deletion mutants were obtained by PCR amplification of GCN5 with primers specific for the deleted versions.
pcDNA3-HA-GCN5 was prepared by subcloning of GCN5 into the pcDNA3-HA vector.
Mutant pcDNA3-HA-GCN5 (Y260A/F261A), encoding catalytically inactive GCN5 (36), was constructed using recombinant PCR starting from the original vector.
pGEX-p300 HAT (aa 1195 to 1810) was a kind gift from E. Verdin (San Francisco, CA).
pCMVβ-p300 has been previously described (28).
The pCMV p300DY-myc plasmid, encoding mutant p300, was kindly provided by T. P. Yao (Durham, NC).
In vitro binding and acetylation assays.
[35S]-labeled CDK9 proteins used for in vitro binding assays were produced by using the TNT reticulocyte lysate system (Promega) and the corresponding pcDNA3 vectors as templates.
In vitro binding and histone acetyltransferase (HAT) assays were performed as previously described (28).
Transcription assays.
Plasmids encoding the reporter pG6(5′Pro), Gal4 fusion proteins, cofactors, or siRNAs against GCN5 or lucifarase were transfected in U2OS cells seeded in six-well plates with Polyfect (Qiagen) in the amounts indicated in Fig. 5. In each transfection 200 ng of pEGFP-C1 (Clontech) was cotransfected as an internal control. After 48 h, cells were harvested and some of them were analyzed by flow cytometry for transfection efficiency. The remaining cells were resuspended in 200 μl of 0.25 M Tris-HCl, pH 8.0, and lysed by repeated freeze-thaw cycles. The protein concentration of the extracts was determined by the Bradford assay (Bio-Rad), and then 2.5 μg of lysate was used to measure chloramphenicol acetyltransferase (CAT) expression with a CAT enzyme-linked immunosorbent assay kit (Roche) according to the manufacturer's instructions.
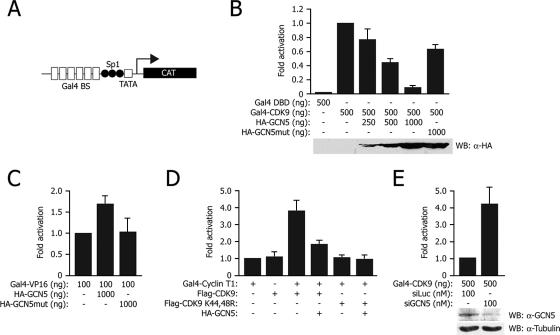
FIG. 5.
Acetylation impairs CDK9 transcriptional activity. (A) Schematic representation of the pG6(5′Pro) reporter vector used for the transcription assays. (B) Expression of wild-type GCN5, but not of its inactive mutant (mut), inhibits CDK9-dependent reporter gene transcription. The graph reports CAT analysis of extracts of U2OS cells transfected with pG6(5′Pro) and the indicated plasmids (means ± standard deviations); the bottom section shows a Western blot (WB) analysis of protein levels. All experiments were performed at least in triplicate. DBD, DNA binding domain; α, anti. (C) GCN5-mediated acetylation increases VP16-driven transcription. (D) Expression of GCN5 represses the positive effect that wild-type CDK9 exerts on cyclin T1-driven transcription. (E) Silencing of endogenous GCN5 by RNAi increases CDK9-driven transcription. si, small interfering; Luc, luciferase.
The results were reported as percentages of CAT production with respect to the values obtained by transfecting only the pG6(5′Pro) reporter and the Gal4-CDK9 activator.
Each experiment was repeated at least three times.
Antibodies.
The following antibodies were used in this study: anti-acetylated lysine (Cell Signaling); anti-Flag (M2; Sigma), either free or bound to agarose beads; antihemagglutinin (anti-HA) (Y-11), anti-cyclin T1 (T-18), anti-CDK9 (C-20, D-7, and H-169, the last for ChIP analysis), anti-GCN5 (H-75), anti-p300 (N-15), anti-PCAF (E-8), anti-poly(ADP-ribose) polymerase 1 (anti-PARP-1) (H-250), anti-promyelocytic leukemia protein (PML) (PG-M3), and anti-RNAPlII (H-224), all purchased from Santa Cruz Biotech. Inc.; anti-phospho-Ser2 (H5) from Covance; anti-HEXIM1, kindly provided by O. Bensaude (Paris, France); anti-acetylated CDK9 (obtained as described above); and anti-5′-p-fluorosulfonylbenzoyladenosine (anti-FSBA), kindly provided by M. Wymann (Basel, Switzerland).
For the production of a polyclonal anti-acetylated CDK9 antibody, three rabbits were immunized with a 15-mer CDK9 peptide (residues 39 to 53 [see Fig. S2A in the supplemental material]) acetylated at positions 44 and 48 after conjugation with keyhole limpet hemocyanin (Sigma). The immunoglobulin G fraction was obtained from serum with the ImmunoPure (A) immunoglobulin G purification kit (Pierce). The antibody was characterized as shown in Fig. S2 in the supplemental material.
Immunoprecipitation and in vivo acetylation assay.
For immunoprecipitations, cell pellets were lysed 36 h after transfection in NHEN buffer (20 mM HEPES, pH 7.5, 300 mM NaCl, 0.5% NP-40, 20% glycerol, 1 mM EDTA) or radioimmunoprecipitation assay buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1% Triton X-100, 0.1% sodium dodecyl sulfate [SDS], 0.5% deoxycholic acid) containing 10 mM of sodium butyrate (Sigma) and protease inhibitors (Roche). The protein concentration of the extracts was determined by the Bradford assay (Bio-Rad). Specific antibodies were incubated overnight at 4°C with cell extracts (2 mg). After incubation, the immunocomplexes were extensively washed and then analyzed by Western blotting.
Immunoprecipitation kinase assay and FSBA labeling.
For the kinase assays, phosphatase inhibitors were added to the NHEN lysis buffer (20 mM NaF, 1 mM Na3VO4). Anti-Flag M2-agarose beads were incubated overnight at 4°C with 500 μg of cell extract and then extensively washed with NHEN plus 20 mM NaF. Immunoprecipitation beads were then washed twice with kinase buffer (50 mM Tris-HCl, pH 7.4, 5 mM MgCl2, 5 mM MnCl2, 20 mM NaF) and finally incubated in 20 μl reaction buffer (kinase buffer plus 10 μM ATP plus 2.5 mM dithiothreitol) with 5 μCi of [32P]ATP and GST-CTD beads (containing 1 μg of proteins) for 30 min at 30°C. Reactions were stopped by adding SDS sample buffer followed by boiling. Phosphorylated proteins were visualized by phosphorimaging (Cyclone) after separation by SDS-polyacrylamide gel electrophoresis (PAGE).
For FSBA labeling, Flag immunoprecipitation was conducted in the same manner. After extensive washing with NHEN buffer, Flag beads were incubated with 50 μM FSBA (alone or in the presence of 0.5 or 5 mM ATP) in phosphate-buffered saline (PBS) plus 0.1% Triton X-100 for 15 min at 30°C and then washed four times with PBS plus 0.5% Triton X-100. Proteins were resolved by SDS-PAGE and then analyzed by Western blotting using an anti-FSBA antibody. Incorporation of FSBA was then quantified by densitometric analysis. All quantification experiments were performed at least three times.
Immunofluorescence.
Following paraformaldehyde fixation, cells were washed with 100 mM glycine and permeabilized with 0.1% Triton X-100 for 5 min. Primary and secondary antibodies were incubated at 37°C for 1 h in a humidified chamber in PBS with the addition of 1% bovine serum albumin and 0.1% Tween 20. Images were acquired by TCS-SL Leica confocal microscopy.
Biochemical fractionation.
Biochemical fractionation assays with 293T cells were conducted as described by Fogal et al. (8). Briefly, at 36 h posttransfection cells were collected in PBS and nuclei were separated by lysis in buffer 1 (50 mM Tris-HCl, pH 7.9, 10 mM KCl, 1 mM EDTA, 0.2% NP-40, 10% glycerol) and centrifuged at 6,000 rpm for 3 min at 4°C. Pellets were washed in buffer 1 without detergent and lysed in buffer 2 (400 mM NaCl, 1% NP-40, 20% glycerol, 20 mM HEPES, pH 7.9, 10 mM KCl, 1 mM EDTA) for 20 min at 4°C. The insoluble and soluble nuclear fractions were separated by centrifugation at 14,000 rpm for 10 min.
Chromatin isolation and micrococcal nuclease treatment in U1 cells were performed essentially as described by Mendez and Stillman (30).
ChIP.
ChIP was performed essentially as described previously (23). Diluted concentrations of the input (1/10 and 1/100) chromatin were used as standards for each primer and TaqMan probe set. Real-time PCR amplifications were performed on an ABI Prism 7000 machine using the TaqMan technology (Applied Biosystems) with ChIP products standardized for primer efficiency. Enrichments (occupancy) for each of the antibodies were calculated as percentages of the input material and further normalized for the B13 genomic control region, as already described (23). Cells were treated (as indicated in Fig. 6) with 12-O-tetradecanoylphorbol 13-acetate (TPA) for 5 h at a concentration of 10−7 M.
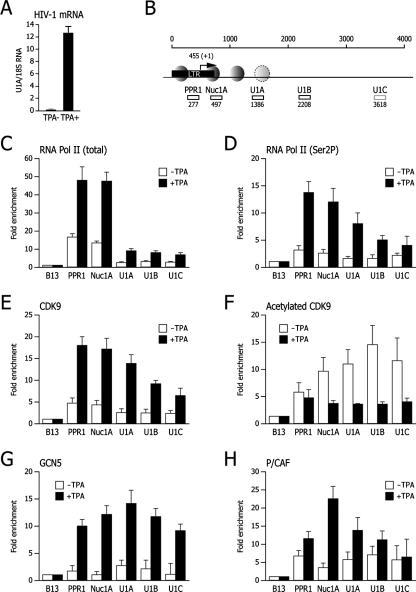
FIG. 6.
Acetylated CDK9 associates with the transcriptionally silent HIV-1 genome. (A) Induction of HIV-1 transcription in U1 cells after TPA stimulation. The levels of HIV-1 mRNA were measured by real-time PCR using TaqMan primers and a probe corresponding to the Nuc1A region; the results are expressed as a ratio between HIV-1 mRNA and the cellular 18S rRNA. (B) Positions of primers selected for the amplification of HIV-1 chromatin. The position of the LTR, including the transcription start site, is indicated, along with the known nucleosomal arrangement at the 5′ genome region. Numbers below investigated segments indicate the location of the 5′ primer used for amplification. (C to H) For each analyzed region, the amount of immunoprecipitated chromatin using the indicated antibodies was first normalized according to the input amount of chromatin and then expressed as enrichment over the amount in the B13 control genomic region. Each graph shows control (white bars) and TPA-induced (black bars) chromatin immunoprecipitations.
Primers for HIV-1 (U1 cells) chromatin immunoprecipitation were the following: for PPR1, 5′-GCCTCCTAGCATTTCGTCAC (forward), 5′-AGAGCTGCATCCGGA (probe), and 5′-CTCGATGTCAGCAGTTCTTTGTAGT (reverse); for Nuc1A, 5′-GCTAGCTAGGGAACCCACTGCTTA (forward), 5′-CCTCAATAAAGCTTGCC (probe), and 5′-CTACAAGTAGTGTGTGCCCGTCT (reverse); for U1A, 5′-ACATCAAGCAGCCATGCAAAA (forward), 5′-AAGAGACCATCAATGAGGAA (probe), and 5′-CAGAATGGGATAGATTGCATCCA (reverse); for U1B, 5′-TCAGAAGCAGGACCCGATAGA (forward), 5′-ACTGTATCCTTTAGCTTCCCT (probe), and 5′-TCACTCTTTGGCAGCGACC (reverse); for U1C, 5′-ATGAAGGGTGCCCACACTAATG (forward), 5′ TGAAACAATTAACAGAGGCAGT (probe), and 5′ CAAAAAATAGCCACAGAAAGCATAGTAA (reverse).
RESULTS
CDK9 is acetylated by cellular acetyltransferases.
By using a polyclonal antibody to acetyllysine, we initially observed that exogenously expressed CDK9 was acetylated inside the cells (Fig. 1A) and that the levels of acetylated protein were further increased by trichostatin A (TSA), an inhibitor of histone deacetylases (Fig. 1B). To identify which HAT was responsible for CDK9 acetylation, we tested the levels of CDK9 acetylation upon expression of representative members of the two best-characterized families of HATs involved in transcriptional coactivation (49), namely, GCN5 and P/CAF for the GNAT family and p300 for the p300-CBP family. Expression of both GCN5 and P/CAF remarkably increased CDK9 acetylation, while p300 had a more modest effect (Fig. 1C). The increase in CDK9 acetylation was specifically due to the HAT activity of the cotransfected proteins, since enzymatically inactive mutants of GCN5 (GCN5 Y260A/F261A) and p300 (p300DY) were completely ineffective, albeit being expressed at the same levels as their wild-type counterparts (Fig. 1D).
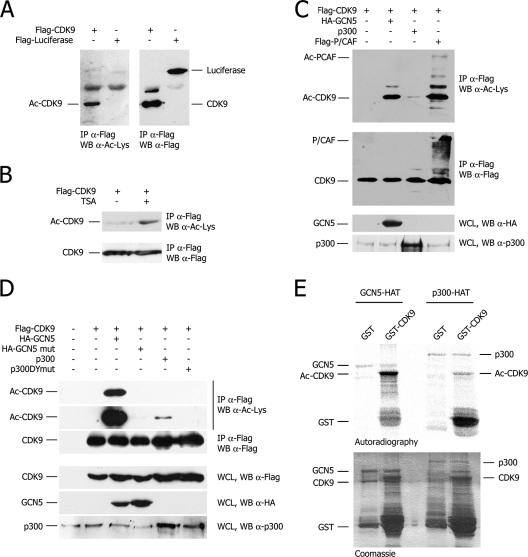
FIG. 1.
CDK9 acetylation in vitro and in vivo. (A) In vivo acetylation. (Left) Western blot analysis of Flag immunoprecipitates (IP) from 293T cells transfected with the indicated plasmids using an anti-acetylated lysine antibody. (Right) The same immunoprecipitates analyzed with an anti-Flag antibody as the loading control. WB, Western blot; Ac, acetylated; α, anti. (B) Same as in panel A except for cell treatment with TSA. The exposure time is shorter than that for panel A. (C) Acetylation of CDK9 upon expression of various HATs. (Top two sections) Same conditions as those for panel A. The basal level of CDK9 acetylation is not appreciable in these blots since the exposure time is significantly shorter than that for panel A. (Bottom two sections) Western blot analysis to show GCN5 and p300 transfected protein levels. WCL, whole-cell lysate. (D) Acetylation of CDK9 depends on the integrity of the GCN5 and p300 catalytic domains. The experiment was performed as in panel C upon transfection of the indicated plasmids. The top two sections show different exposures of the same blot. mut, mutant. (E) In vitro CDK9 acetylation. Autoradiography (top) and Coomassie blue staining (bottom) of products of in vitro HAT assays with GST-CDK9 and GST-GCN5 or GST-p300 HATs.
The HAT activities of GCN5 and p300 on CDK9 were further comparatively assessed in vitro using the recombinant enzymes. Recombinant full-length GST-CDK9 was incubated with the same amount of recombinant GST-p300 or GST-GCN5 HATs in the presence of [14C]acetyl-coenzyme A and then resolved by SDS-PAGE followed by autoradiography (Fig. 1E). In contrast to the GST control, GST-CDK9 scored positive with both HATs, the acetylation signal being remarkably higher with GCN5. In addition to CDK9, p300 and GCN5 were also positive for acetylation due to the autocatalytic activity of the enzymes (6, 44), thus demonstrating that the same amounts of HATs had been used in each assay.
We also observed that CDK9 specifically bound GCN5 both by coimmunoprecipitation in vivo and GST pulldown assays in vitro. This interaction was found to require the integrity of amino acids (aa) 41 to 50 at the N terminus of CDK9 and to involve the bromo- and HAT domains of GCN5. Of interest, the catalytically inactive GCN5 mutant (Y260A/F261A) was still able to bind CDK9 as strongly as the wild-type enzyme, suggesting that the interaction between the two proteins did not depend on the HAT activity of GCN5 (see Fig. S1 in the supplemental material).
Collectively, these results show that CDK9 can be modified by acetylation both in vitro and in vivo by different HATs and that the GNAT family member GCN5 specifically interacts with CDK9 and is remarkably effective at inducing this modification.
GCN5 acetylates lysines 44 and 48 of CDK9.
To map the residues of CDK9 that were acetylated by GCN5, we obtained a series of truncated or point-mutated versions of the kinase (Fig. 2A) and tested their acetylation in vitro using recombinant GCN5. Starting from the N terminus, deletion of the first 40 aa did not affect the levels of CDK9 acetylation; however, all mutants with deletions downstream of aa 50 were not further acetylated. Thus, the target region for GCN5 acetylation lies between aa 41 and 50, namely, the same region that interacted with GCN5. All three lysines in this region (K44, K48, and K49) were mutated, either one at a time or in combination, into arginines, since this amino acid is characterized by the same positive charge as lysine but cannot be modified by acetylation. All three single mutants as well as the double mutant K44,49R were still positive for acetylation; in contrast, the double mutant K44,48R was negative (Fig. 2B). We concluded that lysines at positions 44 and 48 are both substrates for GCN5-mediated acetylation in vitro.
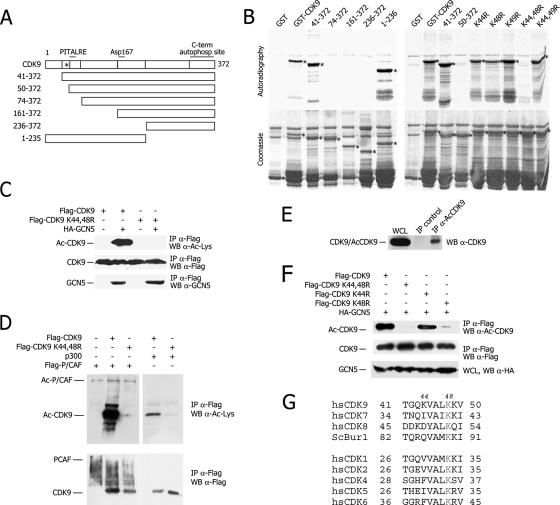
FIG. 2.
GCN5-mediated acetylation of CDK9 at lysines 44 and 48 in vitro and vivo. (A) GST-CDK9 fragments used for the acetylation assay shown in panel B. The position of lysines found positive for acetylation is indicated by an asterisk. (B) In vitro mapping of CDK9 acetylated lysines. Autoradiography (top) and Coomassie blue staining (bottom) of products of in vitro HAT assays with recombinant GCN5 and CDK9 full-length or mutant proteins are shown. (C) Integrity of K44 and K48 is essential for GCN5-mediated CDK9 acetylation in vivo. A Western blot (WB) analysis of Flag immunoprecipitates from 293T transfected with the indicated plasmids is shown. IP, immunoprecipitation; Ac-CDK9, acetylated CDK9; α, anti. (D) Integrity of K44 and K48 is essential for p300- and P/CAF-mediated CDK9 acetylation in vivo. (E) Anti-Ac-CDK9 antibody detects endogenous Ac-CDK9. HeLa whole-cell lysates (WCL) were immunoprecipitated with the anti-Ac-CDK9 antibody and immunoblotted with an anti-CDK9 antibody. Control IP was performed with the preimmune serum. (F) Levels of acetylation of CDK9 variants with individual substitutions at K44 and K48. The experiment was performed as in panel C. (G) Conservation of K48 in other CDKs.
This finding was also confirmed in vivo. Wild-type CDK9 or its double mutant K44,48R was expressed in the cells together with GCN5, and, after immunoprecipitation, CDK9 acetylation was visualized with an antiacetyllysine antibody. In contrast to wild-type CDK9, its K44,48R mutant scored completely negative for acetylation, even if the two proteins were expressed and immunoprecipitated to the same extent (Fig. 2C). Of interest, the K44,48R mutant still coimmunoprecipitated GCN5 as wild-type CDK9 did, indicating that the loss of acetylation of the K44,48R mutant does not result from its inability to associate with GCN5.
Next, acetylation of the K44,48R mutant was also tested upon expression of p300 and P/CAF. In both cases, mutation of K44 and K48 almost abolished acetylation (Fig. 2D), indicating that the same lysines are targets for all analyzed HATs.
An antibody against acetylated CDK9 detects endogenous cellular CDK9 acetylated at K44 and K48.
On the basis of the above-described results, we raised an antibody that specifically recognizes CDK9 acetylated at K44 and K48. For the characterization of this antibody see Fig. S2 in the supplemental material. This antibody bypassed the sensitivity limits of standard antiacetyllysine antibodies and also detected endogenous acetylated CDK9 inside the cells (Fig. 2E).
This antibody was used to confirm that K44 and K48 were the major targets of GCN5-induced CDK9 acetylation in vivo. For this purpose, wild-type CDK9, its K44,48R double mutant, and the single mutants K44R and K48R were expressed in the cells together with GCN5 and, after immunoprecipitation, CDK9 acetylation was visualized. The results of this experiment confirmed that the double mutant was completely negative for acetylation and indicated that, while the K44R mutant could be still partially acetylated, acetylation of K48R was grossly impaired in vivo (Fig. 2F).
Taken together, these results indicate that CDK9 K44 and K48 are targets for GCN5-, P/CAF-, and p300-mediated acetylation inside the cells and that K48, in particular, is a specific in vivo substrate for GCN5.
Acetylation of CDK9 inhibits its enzymatic activity.
The acetylated lysines of CDK9 lie in close proximity to the PITALRE amino acid sequence (positions 60 to 66). This region is involved in the formation of the catalytic pocket of CDK9, and its sequence is highly conserved in all members of the CDK family (Fig. 2G). In particular, K48 corresponds to K33 of CDK2; structural data indicate that this residue is essentially involved in orienting the ATP phosphate residues as well as in magnesium binding within the catalytic pocket of the enzyme (7).
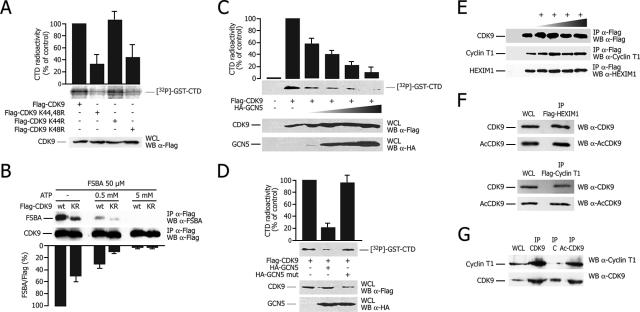
This observation prompted us to analyze the effect of the K44R and K48R mutations on CDK9 activity. Wild-type CDK9 or its mutant derivatives were immunoprecipitated from transfected cells and then incubated with recombinant GST-CTD and [γ-32P]ATP. Both K48R and the double mutant had markedly reduced kinase activity in comparison to the wild-type enzyme (Fig. 3A).
FIG. 3.
Acetylation of CDK9 inhibits kinase activity. (A) Mutation K48R inhibits CDK9 kinase activity. An immunoprecipitation kinase assay from 293T cells transfected with Flag-tagged wild-type CDK9, K44R, K48R, and K44,48R is shown. The graph reports the quantification of labeled GST-CTD (means ± standard deviations [SD] of three independent experiments), and the picture at the bottom shows a representative experiment. The Western blot (WB) in the lower panel shows the levels of protein expression. WCL, whole-cell lysates; α, anti. (B) CDK9 K44,48R (KR) reduced ATP binding activity. Transfected wild-type (wt) or K44,48R Flag-CDK9 was immunoprecipitated (IP) and labeled with the ATP analog FSBA in the presence or absence of excess ATP. The graph shows the ratio between FSBA-labeled protein and total immunoprecipitated Flag protein, determined after densitometric quantification (means ± SD from three independent experiments). The gels show the results of a representative experiment. (C) GCN5-mediated acetylation inhibits CDK9 kinase activity. Immunoprecipitation kinase assay of 293T cells transfected with the indicated plasmids. The experiment was performed as in panel A. (D) Same procedure as in panel C, also using the catalytically inactive GCN5 mutant (mut). (E) Inhibition of CDK9 enzymatic activity by GCN5 does not impair binding to cyclin T1 and HEXIM1. Shown is a Western blot analysis of Flag immunoprecipitated from the same extracts as in panel C. (F) Cyclin T1 and HEXIM1 show indistinguishable binding to either endogenous total CDK9 or acetylated CDK9 (AcCDK9). Lysates from cells transfected with Flag-HEXIM1 (top) or Flag-cyclin T1 (bottom) were immunoprecipitated for the respective proteins and probed by Western blotting using either anti-total CDK9 or anti-acetylated CDK9 antibodies. (G) Cyclin T1 shows indistinguishable binding to either total or acetylated CDK9. Cells transfected with CDK9 and HA-GCN5 were immunoprecipitated with anti-total CDK9 or anti-acetylated CDK9 antibodies or preimmune serum as a negative control (see panel C) and then analyzed by Western blotting with anti-cyclin T1 or anti-CDK9 antibodies.
To examine whether the loss of kinase activity was due to a failure to bind ATP, we utilized the ATP analog FSBA to measure the ATP binding capacities of the different CDK9 mutants. In keeping with the predicted essential role of K48 in ATP binding, the K44,48R mutant showed markedly reduced affinity for FSBA (Fig. 3B). FSBA labeling was specific for the nucleotide binding site of CDK9, since the addition of a molar excess of ATP in the reaction efficiently competed with FSBA labeling of the kinase.
These experiments clearly indicated that the integrity of the acetylated lysines was essential for CDK9-mediated catalysis and thus suggested that acetylation might directly control enzyme activity. To tackle this issue, we performed the immunoprecipitation kinase assay with cells transfected with CDK9 and GCN5 in order to induce CDK9 acetylation. We found that GCN5 transfection impaired CDK9 kinase activity on the GST-CTD substrate in a dose-dependent manner (Fig. 3C). This effect was clearly due to the acetyltransferase activity of GCN5 since the mutant HA-GCN5 (Y260A/F261A), which is HAT defective but still binds CDK9, had no effect (Fig. 3D).
We also wondered whether CDK9 acetylation might modify its interaction with cyclin T1 and HEXIM1. For this purpose, we immunoprecipitated CDK9 from the same extracts used in the kinase assay (in which the CDK9 enzymatic activity is almost completely impaired as shown in Fig. 3C) and tested the levels of endogenous interacting cyclin T1 and HEXIM1 by immunoblotting. No difference was detected for either protein (Fig. 3E). The same conclusion was also reached by observing that the amount of acetylated CDK9 coimmunoprecipitating with either cyclin T1 or HEXIM1 was proportionally similar to total CDK9 (Fig. 3F). Finally, we also observed that the amounts of cyclin T1 that coimmunoprecipitated with either total or acetylated CDK9 in cells overexpressing GCN5 were proportional to the levels of immunoprecipitated CDK9 (Fig. 3G).
In summary, these results indicate that the CDK9 residues that are targeted by acetylation are essential for CDK9 enzymatic activity and that their modification suppresses catalytic function independent of the interaction of the kinase with its known regulatory partners.
Acetylated CDK9 shows subnuclear localization different from that of total CDK9.
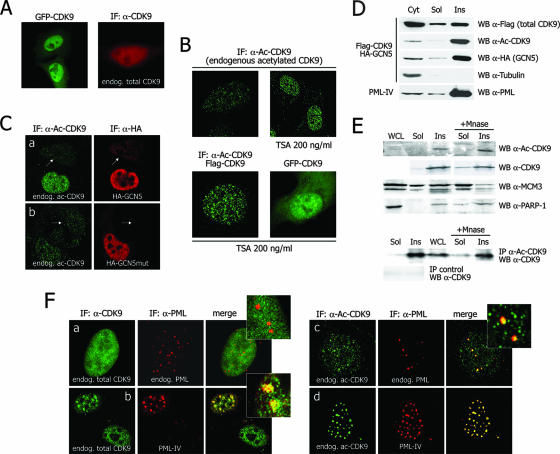
Next we took advantage of our novel anti-acetylated CDK9 antibody to assess the localization of the modified protein inside the cells. Total CDK9 was distributed diffusely throughout the nucleoplasm, with lower levels of fluorescence also detectable in the cytoplasm (Fig. 4A). In sharp contrast, acetylated CDK9 was exclusively found in the nucleus (with the exclusion of the nucleoli), where it generated a speckled pattern, indicative of distribution within specific subnuclear compartments (Fig. 4B). The levels of acetylated protein within these CDK9 foci increased upon cell treatment with the histone deacetylase inhibitor TSA. Of notice is the fact that the same treatment also determined the redistribution of transfected green fluorescent protein (GFP)-CDK9 from its diffuse nucleoplasmic localization to form a punctuated pattern, further suggesting that CDK9 acetylation might control its localization (Fig. 4B). Expression of GCN5 (Fig. 4C, row a), but not of its catalytically inactive mutant (row b), determined a remarkable increase in the levels of endogenously acetylated CDK9 having a speckled localization. Figure 4C shows cells that were not transfected with GCN5 and in which the levels of acetylated CDK9 were markedly reduced.
FIG. 4.
Subnuclear localization of acetylated CDK9. (A) Total endogenous CDK9 (detected with an anti-CDK9 antibody) and transfected GFP-CDK9 show a diffuse nuclear localization in HeLa cells. IF, immunofluorescence; α, anti. (B) Both endogenous and transfected acetylated CDK9 (Ac-CDK9; detected with the novel anti-Ac-CDK9 antibody) have a speckled nuclear localization; the amount of acetylated protein detectable in these speckles increases upon cell treatment with TSA, which also determines a relocalization of GFP-CDK9 into speckles. (C) Transfection of enzymatically active GCN5 increases the amount of Ac-CDK9. Cells were transfected with either wild-type HA-GCN5 (row a) or its catalytically inactive mutant (row b), followed by immunofluorescence with the anti-Ac-CDK9 antibody and with an anti-HA antibody. Cells not transfected with GCN5 (not visible with the anti-HA antibody) are indicated by arrows. In these cells, as well as in cells transfected with mutant GCN5, the levels of acetylated CDK9 are remarkably reduced compared to those in cells transfected with enzymatically active GCN5. (D) Ac-CDK9 fractionates in the insoluble nuclear fraction together with PML. Shown is a Western blot (WB) analysis after biochemical fractionation of 293T cells transfected with Flag-CDK9 and HA-GCN5. PML localization in the insoluble fraction was verified. (E) Endogenous Ac-CDK9 resides in the nuclear matrix fraction. Monocytic U1 whole-cell lysates (WCL) were fractionated to obtain the soluble (Sol) and insoluble (Ins) nuclear fractions, and the levels of endogenous Ac-CDK9 were analyzed directly by Western blotting (top) or after immunoprecipitation (IP) with the anti-Ac-CDK9 antibody followed by Western blotting with an anti-total CDK9 antibody (bottom). Partitioning of Ac-CDK9 in the insoluble compartment did not change after micrococcal nuclease (Mnase) addition, a treatment known to solubilize loosely attached factors, such as MCM-3, used as a control. PARP-1, poly(ADP-ribose) polymerase 1. (F) Most endogenous CDK9 does not colocalize with endogenous PML (row a); however, overexpression of PML-IV forces its redistribution into nuclear bodies (row b). In contrast, Ac-CDK9 spontaneously colocalizes with both endogenous (row c) and transfected (row d) PML in nuclear bodies.
To further explore the distribution of acetylated CDK9, we biochemically fractionated extracts from 293T cells that were transfected with CDK9 and GCN5 in order to enhance acetylation of the former protein. Total CDK9 partitioned in the soluble and insoluble nuclear fractions, as well as in the cytoplasmic fraction. In contrast, acetylated CDK9 was almost exclusively present in the insoluble nuclear matrix fraction, together with transfected GCN5 (Fig. 4D). PML, previously shown to associate with the insoluble nuclear matrix (8), was also present in the same fraction.
These findings were further reinforced by biochemical fractionation of endogenous proteins in HIV-1-infected U1 monocytic cells, followed by micrococcal nuclease treatment to release factors loosely attached to chromatin (30). Acetylated CDK9 was found to reside exclusively in the chromatin/nuclear matrix fraction, as concluded from the analysis of acetylated CDK9 levels by both direct Western blotting of cell lysates and immunoprecipitation (Fig. 4E, top and bottom, respectively). Partitioning of the protein in the insoluble compartment was not modified after micrococcal nuclease addition, indicating tight association of the modified protein with the nuclear matrix compartment. In these experiments, total CDK9 was also predominantly found in the insoluble nuclear fraction, but a vast part of the protein was released by micrococcal nuclease. To prove the efficiency of the enzymatic treatment in releasing proteins loosely attached to chromatin, the localization of two other cellular proteins was also assessed. Most of MCM3 was effectively released by the enzyme, as described previously (30), while poly(ADP-ribose) polymerase 1 behaved similarly to total CDK9.
The above-described biochemical fractionation experiment indicates that acetylated CDK9 partitions in the insoluble nuclear compartment together with PML. We therefore sought to verify whether this association might be also visualized by immunofluorescence. We indeed observed that the localization of total endogenous CDK9 (which is mainly not acetylated) was remarkably different from that of endogenous PML, the former being diffused and the latter localized (Fig. 4F, row a). In sharp contrast, a remarkable proportion of endogenous acetylated CDK9 localized within endogenous PML bodies (row c). Overexpression of PML determined the relocalization of total CDK9 into these bodies (row b) and markedly enhanced the colocalization of endogenous acetylated CDK9 with PML (row d).
Collectively, these results indicate that acetylated CDK9 resides in specific subnuclear compartments within the nucleus that correspond to PML bodies.
Acetylated CDK9 has impaired transcriptional activity.
Next we wanted to investigate the function of CDK9 acetylation in transcriptional regulation. We initially tested the effect of acetylation when the kinase was directly tethered onto a promoter. For this purpose we exploited the pG6(5′Pro) reporter system (43), in which the expression of the CAT gene is controlled by an HIV-1-derived minimal promoter with an upstream cassette containing six Gal4-binding sites (Fig. 5A). When tethered onto this promoter by fusion to the Gal4 DNA binding domain, CDK9 induces transcription of the reporter gene. This assay system appears particular convenient for the analysis of the effects of CDK9 acetylation, since it allows the avoidance of other pleiotropic effects that acetylation exerts on HIV-1 chromatin (6, 23). We observed that promoter activation by CDK9 was inhibited by the expression of increasing amounts of enzymatically active GCN5 but not of its inactive mutant (Fig. 5B). In sharp contrast, the expression of GCN5 had an opposite, activating effect when transcription was directed by the Gal4-VP16 transactivator, as expected (4) (Fig. 5C). Expression of GCN5 also repressed the positive effect that wild-type CDK9 had on transcription when cyclin T1 was tethered to the promoter but was ineffective in repressing the effect of the nonacetylatable K44,48R CDK9 mutant (Fig. 5D). Moreover, silencing GCN5 by RNAi significantly increased CDK9-driven transcription (Fig. 5E).
Taken together, these findings clearly indicate that CDK9 acetylation inhibits the transcriptional activity of the enzyme.
Acetylated CDK9 associates with the transcriptionally silent HIV-1 genome.
The relevance of CDK9 acetylation in the context of whole-virus transcription was assessed in the U1 monocytic cell line, a well-defined model of HIV-1 postintegration latency (9). This cell line contains two silent copies of the provirus, which are markedly induced by different mitogens, including TPA (Fig. 6A). ChIP was performed in these cells using antibodies directed against RNAPII, the Ser2-phosphorylated (Ser2P) CTD of RNAPII, GCN5, P/CAF, total CDK9, and acetylated CDK9. Six different genomic sites were investigated; five of these map to contiguous regions in the HIV-1 proviral DNA (two in the promoter and three in the coding region at the 5′ end of the genome); the sixth one (B13) maps to an unrelated region on chromosome 19 (45) and was used as a reference control to express all data (Fig. 6B). In uninduced conditions, RNAPII was present on the promoter region but not inside the HIV-1 genome (Fig. 6C); most of this polymerase was not phosphorylated on Ser2 (Fig. 6D). Following TPA induction, the levels of both total RNAPII and Ser2P-RNAPII on the promoter region increased; Ser2P-RNAPII was also found distributed inside the proviral genome.
Phosphorylation of RNAPII on Ser2 is known to be essentially catalyzed by CDK9 (37). Consistently, the levels of CDK9 associated with both the promoter and the proviral genome were significantly increased upon TPA induction, thus paralleling those of Ser2P-RNAPII (Fig. 6E). Most strikingly, in uninduced cells, catalytically inactive, acetylated CDK9 was associated with both the HIV-1 promoter and the HIV-1 genome (Fig. 6F). The levels of acetylated CDK9 significantly decreased after TPA stimulation. This decrease was apparently more pronounced inside the coding region than inside the promoter; however, if the levels of acetylated CDK9 are normalized to those of total CDK9, which peaked over the promoter region, it can be concluded that the decrease in acetylated CDK9 involved all the analyzed regions of the HIV-1 genome.
In accordance with our previous observations (23), both GCN5 and P/CAF bound the promoter region in cells stimulated with TPA, in agreement with their role in acetylating the promoter chromatin (Fig. 6G and H, respectively). Of interest, however, low levels of GCN5 and, in particular, of P/CAF were also found to be associated with the proviral DNA in the absence of stimulation when high levels of acetylated CDK9 were also detected at the same location. This coincidence is remarkable and strongly consistent with the possibility that the two HATs inactivate CDK9 by acetylation.
DISCUSSION
This work describes a novel mechanism of P-TEFb regulation that involves the inhibition of CDK9 kinase activity by posttranslational modification of its catalytic core. Among the different HATs that were proven to acetylate CDK9, members of the GNAT family were found to be the most effective. In particular, GCN5 was found to acetylate one lysine (K48) that is conserved in the so-called subdomain II of almost all the eukaryotic protein kinases (conserved in over 95% of 370 sequences) and is hence essential for enzyme function (18). Structural data on CDK2 indicate that the conserved subdomain II lysine is essentially involved in orienting the ATP phosphate residues as well as in magnesium binding within the catalytic pocket of the enzyme (7). Not surprisingly, we found that the CDK9 K44,48R mutant had reduced affinity for the ATP analogue FSBA, which was previously used to map the ATP-binding sites of various proteins, including protein kinase A (56). The FSBA reactive residues are indeed known to play crucial roles in the phosphotransfer reaction (24). The residual kinase and FSBA-binding activities of the K44,48R mutant suggest that other lysines might inefficiently compensate for the K48 mutation, as occurs with analogous mutations in other kinases (15).
Our experiments indicate that lysines 44 and 48 of CDK9 are the substrates for acetylation by the GNAT acetyltransferase family members GCN5 and P/CAF and, to a much lower extent, by p300. Very recent work has indeed shown that p300 might preferentially target K44 and that histone deacetylase proteins negatively regulate CDK9 function, thus implying a positive effect of acetylation (11). The contradiction between those findings and the data presented here might be only apparent. Our work shows that GCN5-mediated acetylation, essentially occurring on K48, is repressive of transcription, whereas Fu et al. analyzed the effects of histone deacetylase 3, which presumably acts on K44. Thus, it might well be envisaged that CDK9, while kept in an inactive state on the transcriptionally silent provirus by GCN5- and P/CAF-mediated K48 acetylation, might become selectively acetylated on K44 by p300 only when the promoter becomes activated. Our previous ChIP experiments on the HIV-1 promoter have indeed revealed that p300, together with other HATs, is recruited onto the LTR upon transcriptional activation (23, 28). Other proteins, such as the viral transactivator Tat (21), are known to be acetylated on different lysines by different HATs, with different functional consequences.
Protein modification by acetylation, as well as the function of cellular HATs, is commonly associated with increased transcriptional activity, mainly due to the positive effect that this modification has on chromatin. However, in several instances factor acetylation is inhibitory of transcription. The acetylation of NF-κB p65 (19), HMGI(Y) (33), IRF7 (3), AFX (Foxo4) (13), and Brm (2) has been reported to repress gene expression; for some of these factors, acetylation might operate as a feedback mechanism to control the duration of transcription. In addition, GCN5-mediated acetylation has been recently shown to inhibit and relocalize the transcriptional coactivator PGC-1α (22). Thus, CDK9 is not the only transcriptional regulator that is negatively regulated by this posttranslational modification.
Our ChIP experiments indicate that the latent state of HIV-1 is characterized by the binding, within the promoter region, of low levels of RNAPII, which is not phosphorylated on Ser2 since CDK9 is kept in an enzymatically inactive state by acetylation. Transcriptional activation of the latent provirus is paralleled by an increase in the total levels of recruited CDK9 and, most notably, in a marked reduction of its acetylated form. These results are consistent with the conclusion that acetylation of CDK9 participates in maintaining the transcriptional latency of the HIV-1 genome. Consistent with this notion, the latent state is also characterized by the presence, on the provirus, of detectable levels of the P/CAF and GCN5 HATs, which are likely responsible for CDK9 acetylation and thus cooperate to maintain promoter latency.
The observation that, in contrast to total CDK9, its acetylated form is detectable, both biochemically and by immunofluorescence, in a specific subnuclear compartment that corresponds to the insoluble nuclear fraction deserves further comments. This is the same compartment in which, together with the PML protein, both GCN5 (A. Sabò and M. Giacca, unpublished observations) and unphosphorylated RNAPII (46) reside, along with a large fraction of cyclin T1, the main CDK9 partner (26). Forcing protein accumulation into this compartment exerts a repressive role on HIV-1 transcription (26). Based on these findings, we propose a model according to which the latent provirus is held in a transcriptionally inactive state when complexed with several of these interactors, which would modify its subnuclear localization and bring it into a transcriptionally repressive environment. According to this model, transcriptional activation would be concomitant with a modification of the subnuclear localization of proviral DNA with respect to the repressive matrix domain. This modification would be paralleled by the replacement of acetylated CDK9 with its unacetylated form, the consequent phosphorylation of RNPII, and the onset of processive transcription. In this scenario, the same HATs that exert a repressive role by acetylating CDK9 once in the repressive compartment would become transcriptional coactivators when outside this compartment by acetylating the proviral chromatin. Besides being compatible with all available experimental observations, this model would also eventually provide a molecular connection between the mechanisms that govern the rate of transcriptional initiation and those involved in regulating transcriptional elongation at the HIV-1 promoter. Further experiments will verify this testable hypothesis.
Finally, to our knowledge, this work reports the first example in which a eukaryotic HAT is found to specifically regulate the activity of a serine/threonine kinase by acetylating the conserved lysine positioned in its catalytic core. Indeed, only a couple of examples of HATs that regulate enzymatic function have very recently been discovered. In both Salmonella enterica (42) and mammals (17, 40), acetyl coenzyme A synthetase is inhibited by the acetylation of the catalytic site of the enzyme. The Yersinia effector protein YopJ acetylates conserved serine and threonine residues in the activation loop of eukaryotic mitogen-activated protein kinase kinase 6 (32); in this case, acetylation inhibits kinase activation since these residues must be phosphorylated for the kinase to become active. The strict conservation of CDK9 K48 in all the CDKs suggests that this mechanism of regulation might play a broader role in controlling the function of other members of this kinase family. Preliminary observations in our laboratory indeed indicate that some CDKs involved in cell cycle control are also acetylated by GCN5.
Supplementary Material
Acknowledgments
We are grateful to V. Liverani for excellent technical assistance.
This work was supported by grants from the National Research Program on AIDS of the Istituto Superiore di Sanità, Italy, and from the Fondazione CRTrieste of Trieste, Italy.
M.G. is on a leave of absence from the Department of Biomedicine, Faculty of Medicine, University of Trieste, Italy.
Footnotes
Published ahead of print on 4 February 2008.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Bieniasz, P. D., T. A. Grdina, H. P. Bogerd, and B. R. Cullen. 1998. Recruitment of a protein complex containing Tat and cyclin T1 to TAR governs the species specificity of HIV-1 Tat. EMBO J. 177056-7065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bourachot, B., M. Yaniv, and C. Muchardt. 2003. Growth inhibition by the mammalian SWI-SNF subunit Brm is regulated by acetylation. EMBO J. 226505-6515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Caillaud, A., A. Prakash, E. Smith, A. Masumi, A. G. Hovanessian, D. E. Levy, and I. Marie. 2002. Acetylation of interferon regulatory factor-7 by p300/CREB-binding protein (CBP)-associated factor (PCAF) impairs its DNA binding. J. Biol. Chem. 27749417-49421. [DOI] [PubMed] [Google Scholar]
- 4.Candau, R., P. A. Moore, L. Wang, N. Barlev, C. Y. Ying, C. A. Rosen, and S. L. Berger. 1996. Identification of human proteins functionally conserved with the yeast putative adaptors ADA2 and GCN5. Mol. Cell. Biol. 16593-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chao, S. H., and D. H. Price. 2001. Flavopiridol inactivates P-TEFb and blocks most RNA polymerase II transcription in vivo. J. Biol. Chem. 27631793-31799. [DOI] [PubMed] [Google Scholar]
- 6.Col, E., C. Caron, D. Seigneurin-Berny, J. Gracia, A. Favier, and S. Khochbin. 2001. The histone acetyltransferase, hGCN5, interacts with and acetylates the HIV transactivator, Tat. J. Biol. Chem. 27628179-28184. [DOI] [PubMed] [Google Scholar]
- 7.De Bondt, H. L., J. Rosenblatt, J. Jancarik, H. D. Jones, D. O. Morgan, and S. H. Kim. 1993. Crystal structure of cyclin-dependent kinase 2. Nature 363595-602. [DOI] [PubMed] [Google Scholar]
- 8.Fogal, V., M. Gostissa, P. Sandy, P. Zacchi, T. Sternsdorf, K. Jensen, P. P. Pandolfi, H. Will, C. Schneider, and G. Del Sal. 2000. Regulation of p53 activity in nuclear bodies by a specific PML isoform. EMBO J. 196185-6195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Folks, T. M., J. Justement, A. Kinter, C. A. Dinarello, and A. S. Fauci. 1987. Cytokine-induced expression of HIV-1 in a chronically infected promonocyte cell line. Science 238800-802. [DOI] [PubMed] [Google Scholar]
- 10.Fong, Y. W., and Q. Zhou. 2000. Relief of two built-in autoinhibitory mechanisms in P-TEFb is required for assembly of a multicomponent transcription elongation complex at the human immunodeficiency virus type 1 promoter. Mol. Cell. Biol. 205897-5907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fu, J., H. G. Yoon, J. Qin, and J. Wong. 2007. Regulation of P-TEFb elongation complex activity by CDK9 acetylation. Mol. Cell. Biol. 274641-4651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fujinaga, K., T. P. Cujec, J. Peng, J. Garriga, D. H. Price, X. Grana, and B. M. Peterlin. 1998. The ability of positive transcription elongation factor B to transactivate human immunodeficiency virus transcription depends on a functional kinase domain, cyclin T1, and Tat. J. Virol. 727154-7159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fukuoka, M., H. Daitoku, M. Hatta, H. Matsuzaki, S. Umemura, and A. Fukamizu. 2003. Negative regulation of forkhead transcription factor AFX (Foxo4) by CBP-induced acetylation. Int. J. Mol. Med. 12503-508. [PubMed] [Google Scholar]
- 14.Garriga, J., J. Peng, M. Parreno, D. H. Price, E. E. Henderson, and X. Grana. 1998. Upregulation of cyclin T1/CDK9 complexes during T cell activation. Oncogene 173093-3102. [DOI] [PubMed] [Google Scholar]
- 15.Gibbs, C. S., and M. J. Zoller. 1991. Rational scanning mutagenesis of a protein kinase identifies functional regions involved in catalysis and substrate interactions. J. Biol. Chem. 2668923-8931. [PubMed] [Google Scholar]
- 16.Greene, W. C., and B. M. Peterlin. 2002. Charting HIV's remarkable voyage through the cell: basic science as a passport to future therapy. Nat. Med. 8673-680. [DOI] [PubMed] [Google Scholar]
- 17.Hallows, W. C., S. Lee, and J. M. Denu. 2006. Sirtuins deacetylate and activate mammalian acetyl-CoA synthetases. Proc. Natl. Acad. Sci. USA 10310230-10235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hanks, S. K., and T. Hunter. 1995. Protein kinases 6. The eukaryotic protein kinase superfamily: kinase (catalytic) domain structure and classification. FASEB J. 9576-596. [PubMed] [Google Scholar]
- 19.Kiernan, R., V. Bres, R. W. Ng, M. P. Coudart, S. El Messaoudi, C. Sardet, D. Y. Jin, S. Emiliani, and M. Benkirane. 2003. Post-activation turn-off of NF-kappa B-dependent transcription is regulated by acetylation of p65. J. Biol. Chem. 2782758-2766. [DOI] [PubMed] [Google Scholar]
- 20.Kiernan, R. E., S. Emiliani, K. Nakayama, A. Castro, J. C. Labbe, T. Lorca, K. Nakayama, and M. Benkirane. 2001. Interaction between cyclin T1 and SCF(SKP2) targets CDK9 for ubiquitination and degradation by the proteasome. Mol. Cell. Biol. 217956-7970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kiernan, R. E., C. Vanhulle, L. Schiltz, E. Adam, H. Xiao, F. Maudoux, C. Calomme, A. Burny, Y. Nakatani, K. T. Jeang, M. Benkirane, and C. Van Lint. 1999. HIV-1 Tat transcriptional activity is regulated by acetylation. EMBO J. 186106-6118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lerin, C., J. T. Rodgers, D. E. Kalume, S. H. Kim, A. Pandey, and P. Puigserver. 2006. GCN5 acetyltransferase complex controls glucose metabolism through transcriptional repression of PGC-1alpha. Cell Metab. 3429-438. [DOI] [PubMed] [Google Scholar]
- 23.Lusic, M., A. Marcello, A. Cereseto, and M. Giacca. 2003. Regulation of HIV-1 gene expression by histone acetylation and factor recruitment at the LTR promoter. EMBO J. 226550-6561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Madhusudan, E. A. Trafny, N. H. Xuong, J. A. Adams, L. F. Ten Eyck, S. S. Taylor, and J. M. Sowadski. 1994. cAMP-dependent protein kinase: crystallographic insights into substrate recognition and phosphotransfer. Protein Sci. 3176-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mancebo, H. S., G. Lee, J. Flygare, J. Tomassini, P. Luu, Y. Zhu, J. Peng, C. Blau, D. Hazuda, D. Price, and O. Flores. 1997. P-TEFb kinase is required for HIV Tat transcriptional activation in vivo and in vitro. Genes Dev. 112633-2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marcello, A., A. Ferrari, V. Pellegrini, G. Pegoraro, M. Lusic, F. Beltram, and M. Giacca. 2003. Recruitment of human cyclin T1 to nuclear bodies through direct interaction with the PML protein. EMBO J. 222156-2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marcello, A., M. Lusic, G. Pegoraro, V. Pellegrini, F. Beltram, and M. Giacca. 2004. Nuclear organization and the control of HIV-1 transcription. Gene 3261-11. [DOI] [PubMed] [Google Scholar]
- 28.Marzio, G., M. Tyagi, M. I. Gutierrez, and M. Giacca. 1998. HIV-1 Tat transactivator recruits p300 and CREB-binding protein histone acetyltransferases to the viral promoter. Proc. Natl. Acad. Sci. USA 9513519-13524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meinhart, A., T. Kamenski, S. Hoeppner, S. Baumli, and P. Cramer. 2005. A structural perspective of CTD function. Genes Dev. 191401-1415. [DOI] [PubMed] [Google Scholar]
- 30.Mendez, J., and B. Stillman. 2000. Chromatin association of human origin recognition complex, Cdc6, and minichromosome maintenance proteins during the cell cycle: assembly of prereplication complexes in late mitosis. Mol. Cell. Biol. 208602-8612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Michels, A. A., V. T. Nguyen, A. Fraldi, V. Labas, M. Edwards, F. Bonnet, L. Lania, and O. Bensaude. 2003. MAQ1 and 7SK RNA interact with CDK9/cyclin T complexes in a transcription-dependent manner. Mol. Cell. Biol. 234859-4869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mukherjee, S., G. Keitany, Y. Li, Y. Wang, H. L. Ball, E. J. Goldsmith, and K. Orth. 2006. Yersinia YopJ acetylates and inhibits kinase activation by blocking phosphorylation. Science 3121211-1214. [DOI] [PubMed] [Google Scholar]
- 33.Munshi, N., T. Agalioti, S. Lomvardas, M. Merika, G. Chen, and D. Thanos. 2001. Coordination of a transcriptional switch by HMGI(Y) acetylation. Science 2931133-1136. [DOI] [PubMed] [Google Scholar]
- 34.Napolitano, G., P. Licciardo, R. Carbone, B. Majello, and L. Lania. 2002. CDK9 has the intrinsic property to shuttle between nucleus and cytoplasm, and enhanced expression of cyclin T1 promotes its nuclear localization. J. Cell. Physiol. 192209-215. [DOI] [PubMed] [Google Scholar]
- 35.Nguyen, V. T., T. Kiss, A. A. Michels, and O. Bensaude. 2001. 7SK small nuclear RNA binds to and inhibits the activity of CDK9/cyclin T complexes. Nature 414322-325. [DOI] [PubMed] [Google Scholar]
- 36.Paulson, M., C. Press, E. Smith, N. Tanese, and D. E. Levy. 2002. IFN-stimulated transcription through a TBP-free acetyltransferase complex escapes viral shutoff. Nat. Cell Biol. 4140-147. [DOI] [PubMed] [Google Scholar]
- 37.Peterlin, B. M., and D. H. Price. 2006. Controlling the elongation phase of transcription with P-TEFb. Mol. Cell 23297-305. [DOI] [PubMed] [Google Scholar]
- 38.Price, D. H. 2000. P-TEFb, a cyclin-dependent kinase controlling elongation by RNA polymerase II. Mol. Cell. Biol. 202629-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sanó, M., M. Abdellatif, H. Oh, M. Xie, L. Bagella, A. Giordano, L. H. Michael, F. J. DeMayo, and M. D. Schneider. 2002. Activation and function of cyclin T-Cdk9 (positive transcription elongation factor-b) in cardiac muscle-cell hypertrophy. Nat. Med. 81310-1317. [DOI] [PubMed] [Google Scholar]
- 40.Schwer, B., J. Bunkenborg, R. O. Verdin, J. S. Andersen, and E. Verdin. 2006. Reversible lysine acetylation controls the activity of the mitochondrial enzyme acetyl-CoA synthetase 2. Proc. Natl. Acad. Sci. USA 10310224-10229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shim, E. Y., A. K. Walker, Y. Shi, and T. K. Blackwell. 2002. CDK-9/cyclin T (P-TEFb) is required in two postinitiation pathways for transcription in the C. elegans embryo. Genes Dev. 162135-2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Starai, V. J., I. Celic, R. N. Cole, J. D. Boeke, and J. C. Escalante-Semerena. 2002. Sir2-dependent activation of acetyl-CoA synthetase by deacetylation of active lysine. Science 2982390-2392. [DOI] [PubMed] [Google Scholar]
- 43.Taube, R., X. Lin, D. Irwin, K. Fujinaga, and B. M. Peterlin. 2002. Interaction between P-TEFb and the C-terminal domain of RNA polymerase II activates transcriptional elongation from sites upstream or downstream of target genes. Mol. Cell. Biol. 22321-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thompson, P. R., D. Wang, L. Wang, M. Fulco, N. Pediconi, D. Zhang, W. An, Q. Ge, R. G. Roeder, J. Wong, M. Levrero, V. Sartorelli, R. J. Cotter, and P. A. Cole. 2004. Regulation of the p300 HAT domain via a novel activation loop. Nat. Struct. Mol. Biol. 11308-315. [DOI] [PubMed] [Google Scholar]
- 45.Todorovic, V., S. Giadrossi, C. Pelizon, R. Mendoza-Maldonado, H. Masai, and M. Giacca. 2005. Human origins of DNA replication selected from a library of nascent DNA. Mol. Cell 19567-575. [DOI] [PubMed] [Google Scholar]
- 46.von Mikecz, A., S. Zhang, M. Montminy, E. M. Tan, and P. Hemmerich. 2000. CREB-binding protein (CBP)/p300 and RNA polymerase II colocalize in transcriptionally active domains in the nucleus. J. Cell Biol. 150265-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wei, P., M. E. Garber, S. M. Fang, W. H. Fischer, and K. A. Jones. 1998. A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell 92451-462. [DOI] [PubMed] [Google Scholar]
- 48.Yang, X., M. O. Gold, D. N. Tang, D. E. Lewis, E. Aguilar-Cordova, A. P. Rice, and C. H. Herrmann. 1997. TAK, an HIV Tat-associated kinase, is a member of the cyclin-dependent family of protein kinases and is induced by activation of peripheral blood lymphocytes and differentiation of promonocytic cell lines. Proc. Natl. Acad. Sci. USA 9412331-12336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang, X. J. 2004. The diverse superfamily of lysine acetyltransferases and their roles in leukemia and other diseases. Nucleic Acids Res. 32959-976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang, Z., J. H. Yik, R. Chen, N. He, M. K. Jang, K. Ozato, and Q. Zhou. 2005. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol. Cell 19535-545. [DOI] [PubMed] [Google Scholar]
- 51.Yang, Z., Q. Zhu, K. Luo, and Q. Zhou. 2001. The 7SK small nuclear RNA inhibits the CDK9/cyclin T1 kinase to control transcription. Nature 414317-322. [DOI] [PubMed] [Google Scholar]
- 52.Yik, J. H., R. Chen, R. Nishimura, J. L. Jennings, A. J. Link, and Q. Zhou. 2003. Inhibition of P-TEFb (CDK9/cyclin T) kinase and RNA polymerase II transcription by the coordinated actions of HEXIM1 and 7SK snRNA. Mol. Cell 12971-982. [DOI] [PubMed] [Google Scholar]
- 53.Zhou, M., M. A. Halanski, M. F. Radonovich, F. Kashanchi, J. Peng, D. H. Price, and J. N. Brady. 2000. Tat modifies the activity of CDK9 to phosphorylate serine 5 of the RNA polymerase II carboxyl-terminal domain during human immunodeficiency virus type 1 transcription. Mol. Cell. Biol. 205077-5086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhou, Q., D. Chen, E. Pierstorff, and K. Luo. 1998. Transcription elongation factor P-TEFb mediates Tat activation of HIV-1 transcription at multiple stages. EMBO J. 173681-3691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhu, Y., T. Pe'ery, J. Peng, Y. Ramanathan, N. Marshall, T. Marshall, B. Amendt, M. B. Mathews, and D. H. Price. 1997. Transcription elongation factor P-TEFb is required for HIV-1 tat transactivation in vitro. Genes Dev. 112622-2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zoller, M. J., N. C. Nelson, and S. S. Taylor. 1981. Affinity labeling of cAMP-dependent protein kinase with p-fluorosulfonylbenzoyl adenosine. Covalent modification of lysine 71. J. Biol. Chem. 25610837-10842. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.