Abstract
Primary immunodeficiencies are rare but serious diseases with diverse genetic causes. Accumulating evidence suggests that defects in DNA double-strand break (DSB) repair can underlie many of these syndromes. In this context, the nonhomologous end joining pathway of DSB repair is absolutely required for lymphoid development, but possible roles for the homologous recombination (HR) pathway have remained more controversial. While recent evidence suggests that HR may indeed be important to suppress lymphoid transformation, the specific relationship of HR to normal lymphocyte development remains unclear. We have investigated roles of the X-ray cross-complementing 2 (Xrcc2) HR gene in lymphocyte development. We show that HR is critical for normal B-cell development, with Xrcc2 nullizygosity leading to p53-dependent early S-phase arrest. In the absence of p53 (encoded by Trp53), Xrcc2-null B cells can fully develop but show high rates of chromosome and chromatid fragmentation. We present a molecular model wherein Xrcc2 is important to preserve or restore replication forks during rapid clonal expansion of developing lymphocytes. Our findings demonstrate a key role for HR in lymphoid development and suggest that Xrcc2 defects could underlie some human primary immunodeficiencies.
Human immunodeficiencies are complex, multivariate diseases, the underlying genetic causes of which remain mostly unknown (9, 11, 31, 32, 35). In the past decade, defects in DNA double-strand break (DSB) repair have been implicated in some lymphodeficiencies, such as radiosensitive severe combined immunodeficiency. To resist the deleterious effects of DSBs, cells must efficiently respond to, and repair, the damage and maintain genomic integrity. Most eukaryotes employ two predominant DSB repair pathways: nonhomologous end joining (NHEJ), which catalyzes the religation of cognate broken DNA ends irrespective of sequence homology; and homologous recombination (HR), which utilizes a homologous DNA template to effect error-free DSB repair. While all cells likely incur random DSBs as a result of endogenous metabolism or exposure to environmental agents, certain cell types also deliberately generate DSBs to effect specific developmental programs (for review, see references 18 and 20). In this context, NHEJ is required to resolve DNA breaks associated with V(D)J recombination in developing lymphocytes, failure of which can lead to lymphodeficiency (2, 20). In contrast, possible roles for the HR pathway, in either normal lymphocyte development or in lymphodeficiency, remain controversial. Recently the controversy has been rekindled by evidence that HR may be important to suppress T-cell lymphomagenesis (21).
Here we investigate the role of the X-ray cross-complementing 2 (Xrcc2) gene in B-cell development. Xrcc2 is a member of the Rad51 gene family and was initially identified by its ability to complement DNA damage sensitivity in mutant Chinese hamster ovary (CHO) cells (34). Subsequent studies have shown XRCC2 to participate in one or more unique, multiprotein complexes that regulate HR efficiency by modulating RAD51-dependent DNA heteroduplex formation (17, 33). In the chicken DT 40 B-cell line, complete deficiency in XRCC2 results in profound sensitivity to ionizing radiation, and other clastogenic insults, as well as a dramatic decrease in the immunoglobulin (Ig) pseudogene conversion rate (27, 33). Homozygous gene-targeted disruption of Xrcc2 in mice results in widespread apoptosis of fetal cells, culminating in embryonic lethality at mid to late gestation (6). In cell culture, Xrcc2-null cells show marked proliferation defects, hypersensitivity to DNA-damaging agents, and a high rate of spontaneous chromosomal abnormalities (7, 13).
Although inactivation of Xrcc2 leads to embryonic lethality around day 15 of embryonic development (E15), we were able to circumvent this phenotype to analyze normal lymphoid development in single- and double-mutant contexts, both in vitro and in vivo. We show that Xrcc2 is necessary for normal B-lymphocyte development and find that lack of Xrcc2 leads to p53-dependent early S-phase arrest, revealing a key role for HR. In the absence of p53 (encoded by Trp53), Xrcc2-null B cells developed to maturity but exhibited a high rate of chromosome or chromatid fragmentation. We present a working model in which XRCC2 functions at stalled or collapsed replication forks to effect their restart and suggest that restart failure in Xrcc2-null cells leads to abortive B-cell development. Finally, our data imply that mutations or hypomorphic alleles in human XRCC2 could be involved in some human lymphodeficiencies of currently unknown etiology.
MATERIALS AND METHODS
Mice.
Xrcc2-null and Trp53-null mice were previously described (8). Both Xrcc2 and Trp53 colonies were separately maintained by crossing heterozygotes of each strain (Xrcc2+/− or Trp53+/−); these were intercrossed to obtain single- and double-mutant genotypes, as well as controls. To generate fetal liver cells for both in vitro and in vivo lymphocyte development assays, embryos were isolated at E13 to E14, single-cell suspensions of fetal liver were prepared by dispersion through sterile nylon mesh, and residual tissue was collected for genotype analysis. All mice were housed in pressurized, individually ventilated cages and fed standard lab diet. All mouse work was carried out in accordance with IACUC-approved protocols.
Genotyping.
Small explants from weanling mouse tails or residual embryo-derived tissues were used for genotyping. Following extraction of total genomic DNA, PCRs were performed in a 25-μl reaction volume using 5-Prime Taq polymerase master mix (Fisher). For Xrcc2 genotyping, the following primers and reaction conditions were used: common forward, GGTTCTATCTTGTGCTTTTGTGTGTTTA; wild-type (WT) reverse, TCTGTTTTCCCCGTTCCTTCTG; and mutant reverse, GCATGCTCCAGACTGCCTTGG.
The cycling conditions were as follows: 94°C for 90 s followed by 25 cycles of 94°C for 35 s, 58°C for 1 min, and 72°C for 45 s and a final incubation at 72°C for 10 min. These conditions generated a 545-bp wild-type band and a 280-bp mutant band. For Trp53 genotyping, the following primers and reaction conditions were used: WT forward, GTGTTTCATTAGTTCCCCACCTTGAC; WT reverse, ATGGGAGGCTGCCAGTCCTAACCC; mutant forward, GTGGGAGGGACAAAAGTTCGAGGCC; and mutant reverse, TTTACGGAGCCCTGGCGCTCGATGT.
These Xrcc2 primers were generated under the following cycling conditions: 94°C for 4 min, followed by 35 cycles of 94°C for 20 s, 55°C for 20 s, and 72°C for 40 s and a final incubation at 72°C for 10 min. These parameters produced a 320-bp wild-type band and a 150-bp mutant band. All PCR products were separated by agarose gel electrophoresis and visualized after staining with ethidium bromide.
Cell culture.
Following isolation of E13 to E14 fetal liver, single-cell suspensions were cocultured with T220 cells, an NIH 3T3-derived cell line secreting interleukin-7 (IL-7) (3), in growth medium comprising 30 to 50% fresh RPMI and 50 to 70% conditioned medium. Fresh RPMI 1640 growth medium contained 10% fetal bovine serum, 20 mM HEPES, 2 mM l-glutamate, 100 U/ml penicillin, 100 μg/ml streptomycin, and 25 ng/ml IL-7 (R&D Systems, Minneapolis, MN). Conditioned medium was generated by culturing T220 cells for 3 to 4 days in fresh RPMI 1640 medium and subsequently collecting the culture supernatant.
Flow cytometry and immunofluorescence.
For the in vitro studies, cells were brought to a concentration of 1 × 106 to 2 × 106 cells/ml and then stained with an antibody-fluorophore cocktail containing B220-phycoerythrin (PE)-Cy7, IgM-allophycocyanin (APC), CD43-PE, and CD43-fluorescein isothiocyanate (FITC) (eBioscience), on culture day 0, 6, or 9, to determine B-cell development. Flow cytometric detection of intercellularly activated caspase 3 was performed after fixation and staining as described below for immunofluorescence, except samples were kept in single-cell suspension until analysis. For the competitive reconstitution assays, cells were stained with an antibody-fluorophore cocktail containing polyclonal Ig-PE (eBioscience) and monoclonal antibodies IgM-APC and B220-AC7 (eBioscience), and green fluorescent protein (GFP) expression was detected in the FITC (FL1) channel.
To analyze cell cycle distribution, cells were fixed with 70% ethanol and resuspended in 0.1% Triton X-100 with 20 μg/ml propidium iodide (PI) for at least an hour before flow cytometry analysis. For the BrdU incorporation assay, cells were cultured for 7 days, supplemented with 1 mM BrdU, and harvested after 24 h. The cells were stained with directly conjugated anti-BrdU antibody and analyzed by flow cytometry, according to the manufacturer's suggested protocol (BD Bioscience catalog no. 559619). All flow cytometry procedures were performed using a FACSCalibur flow cytometer running Cell Quest Pro (BD Biosciences) acquisition software and analyzed using FlowJo software (V.8.4.6; Treestar, Inc.).
For immunofluorescence microscopy, cells were fixed with 3% formaldehyde-2% sucrose for 10 min at room temperature, permeabilized with 0.1% Triton X-100 in 1× phosphate-buffered saline, blocked with 5% fetal bovine serum, and stained for 16 to 18 h at 4°C with primary antibodies specific to one of the following targets, at the indicated dilution ratio: active caspase 3 (Abcam; 1:100), γ-H2AX (Bethyl; 1:400), PCNA (Abcam; 1:100), and γ-histone H3 (Upstate;1:200). Slides were then incubated for 30 min at room temperature in FITC-labeled goat anti-rabbit IgG (Vector Laboratories), and mounted with Vectashield mounting medium containing DAPI (4′,6′-diamidino-2-phenylindole) counterstain (Vector Laboratories). Images were captured by epifluorescence wide-field imaging on either a Nikon 90i, or a Leica DM-RE upright microscope. Images were analyzed using IPLab software (BD Biosciences, Rockville, MD) with minimal image processing.
Cytogenetic analyses.
To prepare metaphase chromosome spreads for cytogenetic analyses, cultured cells were incubated for 4 h in presence of 40 ng/ml Colcemid (Karyomax; Invitrogen), to enforce metaphase arrest, immediately prior to fixation. Cells were then transferred to prewarmed (37°C) hypotonic KCl solution (75 mM) for 7 min and fixed by two changes of cold 3:1 methanol-acetic acid. Metaphase chromosome preparations were adhered to slides and either stained with Vectashield containing DAPI (Vector Laboratories) to score for chromosomal structure abnormalities or further processed for spectral karyotyping (SKY) according to the manufacturer's protocols (Applied Spectral Imaging, Corona, CA). Imaging of DAPI-stained chromosomes was performed using Nikon 90i upright fluorescence microscope and analyzed using IPLab software. SKY imaging was performed using an ASI complete cytogenetics station (Applied Spectral Imaging, Corona, CA) and analyzed with dedicated analysis software (Applied Spectral Imaging, Corona, CA).
In vivo competitive repopulation assay.
Competitive hematopoietic reconstitution assays were performed by mixing 1 × 106 E13 to E14 fetal liver cells, of the appropriate genotype, with 1 × 106 competitor bone marrow cells derived from C57BL/6-Tg(UBC-GFP)30Scha/J female mice (JR# 004353; The Jackson Laboratory) ubiquitously expressing GFP, in sterile Hank's buffer. The mixed cell preparations, containing 2 × 106 total cells, were introduced via tail vein injection into lethally irradiated C57BL/6J recipient female mice, pretreated with split-dose gamma irradiation totaling 10 Gy. Each donor sample was introduced into three independent recipient mice. Peripheral blood was collected from recipient mice by retro-orbital bleed, beginning at 8 weeks postinjection. Prior to immunostaining for flow cytometry, red blood cells were lysed with hemolytic Geys’ buffer (10). At 8 weeks, a subset of the recipient mice were euthanized, and the spleen, bone marrow, thymus, and peripheral blood were collected for lymphocyte analysis by flow cytometry as described above.
RESULTS
B-cell developmental failure is observed in Xrcc2-null cells.
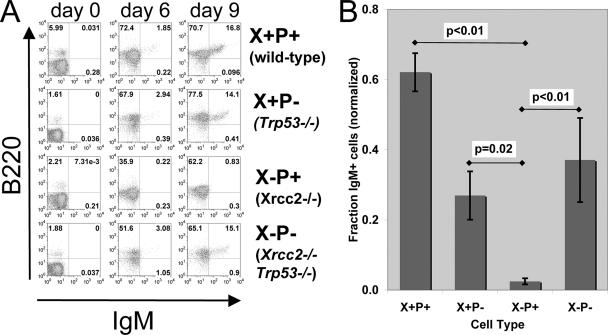
Because nullizygosity for Xrcc2 in mice leads to embryonic lethality after the onset of fetal hematopoiesis (data available upon request), we established a cell culture system, based on fetal liver cell cocultures, that permitted analysis of B-cell development and genome stability (1, 6). This system allowed us to analyze the details of B-cell development in both single- and double-mutant contexts (see below), which has not previously been possible (Fig. 1). To evaluate the role of Xrcc2 in developing B-lymphocytes, we obtained fetal liver cells from E13 to E14 Xrcc2−/− and littermate control embryos. These cells were cocultured with an NIH 3T3 cell line expressing IL-7 (T220) to support B-cell development (3). Fetal liver cells were cocultured for up to 9 days and assessed for B-cell development at various time points by fluorescence-based flow cytometry. To measure the efficiency of B-lineage commitment, cells were stained with antibodies specific for the B-cell lineage marker CD45R/B220 and for the B-cell development markers CD19, CD43, and IgM. This analysis revealed a significant B-cell developmental failure in Xrcc2-null cultures, relative to their normal counterparts (Fig. 1A and B). Following 6 days of coculture, all samples showed accumulation of B220-positive (B220+) cells (Fig. 1A), demonstrating that Xrcc2−/− cells are not overtly compromised in their ability to initiate B-cell development. However, a profound defect was observed between 6 and 9 days of coculture. After 9 days, WT cultures readily produced mature B220+ IgM+ B cells (Fig. 1B). In striking contrast, Xrcc2−/− cultures showed little, if any, progression past the B220+ IgM− stage, exhibiting, on average, a 25-fold less-efficient production of mature IgM+ B cells (Fig. 1B).
FIG. 1.
Xrcc2-null B cells are developmentally deficient in vitro. (A) Flow cytometric analysis of fetal liver cell cultures. Assays were performed after 0, 6, or 9 days in culture, and cells were immunostained for B220 and IgM and quantitatively analyzed by flow cytometry. Representative plots for each genotype are shown. (B) Graph showing expression of B220 and IgM on fetal liver culture day 9, normalized to WT littermate control values. The numbers of samples are as follows: WT (also denoted as X+P+), 38; Xrcc2−/− (X−P+), 4; Trp53−/− (X+P−), 9; and Xrcc2−/− Trp53−/− (X−P−), 3. Error bars indicate standard error; P values were determined using Student's t test.
We reasoned that the developmental failure of Xrcc2-null B cells might be dependent on p53 function, perhaps owing to genotoxic stress. To test this notion, we generated Xrcc2−/− Trp53−/− (double null) and control littermate E13 to E14 embryos, and isolated fetal liver cells for coculture assays, as described above. Xrcc2−/− Trp53−/− fetal liver cells, unlike the Xrcc2 single-mutant cells, were capable of development to the B220+ IgM+ B-cell stage, albeit with reduced efficiency. On average, Xrcc2−/− Trp53−/− cultures showed B-cell development that was approximately 1.7-fold less efficient than that of the WT but 15-fold more efficient than that of counterpart Xrcc2 single-mutant cultures, as measured by the percentage of B220+ IgM+ cells after 9 days of culture (Fig. 1B). These data establish that the B-cell developmental failure, seen in the absence of Xrcc2, is largely dependent on p53.
Xrcc2 Trp53 double-null cells show an in vivo B-cell development defect.
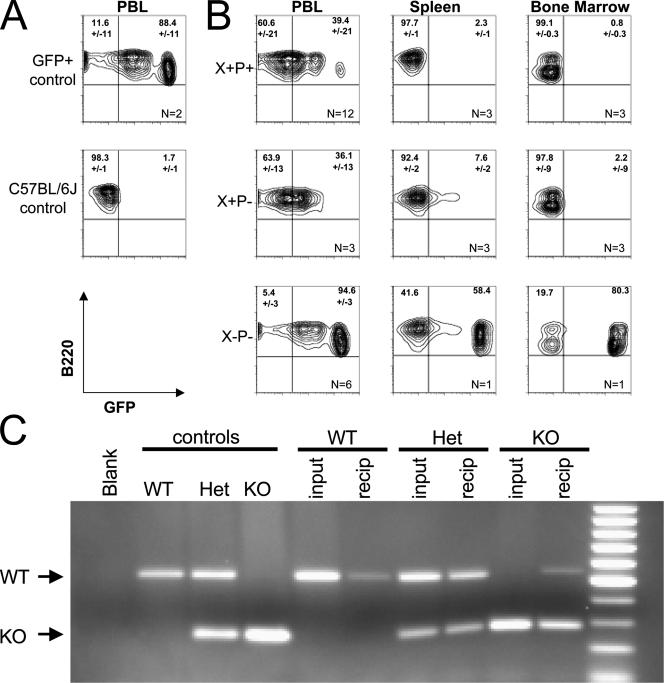
Because Xrcc2−/− Trp53−/− B-cell development was mildly impaired in vitro, we next determined whether Xrcc2 Trp53 double deficiency would lead to more pronounced in vivo effects, via competitive repopulation assays. Fetal liver cells were mixed at a 1:1 ratio with GFP-marked WT competitor bone marrow. Mixed donor cells were introduced into lethally irradiated C57BL/6J recipients and monitored for hematopoietic reconstitution beginning 6 weeks later, measuring for GFP-positive versus GFP-negative B cells in blood or lymphoid organs (Fig. 2). Both WT and Trp53−/− fetal liver efficiently reconstituted B cells, with recipients showing greater than 60% circulating GFP− B220+ lymphocytes and greater than 90% GFP− B220+ cells in spleen and bone marrow (Fig. 2B). In contrast, Xrcc2−/− Trp53−/− donors were substantially outcompeted by the normal cells, with 95% of the circulating B cells and 60 to 80% of the spleen and bone marrow B cells being GFP+. To assess whether the small fraction of GFP− cells represented Xrcc2−/− Trp53−/− donor cells versus background from the irradiated recipient, GFP+ and GFP− B-cell fractions were separated by flow sorting and were tested for the presence of the Xrcc2-null allele in the purified fractions (Fig. 2C). PCR genotype showed that the GFP-negative fraction contained cells harboring the Xrcc2-null allele, confirming that Xrcc2−/− hematopoietic cells can successfully engraft but are significantly outcompeted by WT cells during subsequent lymphocyte development (Fig. 2C).
FIG. 2.
Xrcc2 Trp53 double-null B cells are developmentally deficient in vivo. (A) Flow cytograms showing B220 staining versus GFP fluorescence in peripheral blood from C57BL/6-Tg(UBC-GFP)30Scha/J (GFP+ control) and wild-type (C57GL/6J control) mice. (B) Flow cytometric analysis of B220 staining versus GFP fluorescence in adoptive transfer recipients receiving mixed GFP+ competitor bone marrow plus fetal liver cells from donors of the indicated genotypes. Cells for analysis were isolated from peripheral blood (PBL), spleen, or bone marrow, as indicated. The number of recipient mice for each genotype and sample type is indicated (n) in the lower right quadrant, and the average percentage and value range are indicated for each relevant quadrant. (C) PCR detection of WT and Xrcc2-null (KO) alleles, indicated by arrows, in WT, heterozygous (Het), or Xrcc2-null (KO) control mice; in fetal liver cells (input) of the indicated genotypes; and in sorted GFP-negative peripheral blood cells from adoptive transfer recipients (recip) of the corresponding input fetal liver cells.
Accumulation of cells with unrepaired DSBs in Xrcc2-null B-cell cultures.
We next tested whether the defect observed in Xrcc2-null B-cell cultures was accompanied by an increase in unrepaired DNA DSBs, as might be predicted for HR-deficient cells. Phosphorylated histone H2AX (γ-H2AX) has been extensively used as a marker for the presence of unrepaired DSBs (22-26). To evaluate the extent of unrepaired DSBs, B-cell cultures of each genotype were analyzed for the presence of spontaneously occurring γ-H2AX foci. As controls for this assay, freshly isolated primary splenocytes were subjected to either 0 or 5 Gy of ionizing radiation and processed identically to the B-cell cultures. Untreated WT cultures comprised approximately 50% cells with more than one γ-H2AX focus, roughly equivalent to untreated WT splenocytes (Fig. 3A and B). Xrcc2−/− B-cell cultures, which fail to develop to the IgM+ stage, comprised roughly 80% focus-positive cells, a significant increase in the percentage of cells with spontaneously occurring foci relative to WT (Fig. 3B). Conversely, Xrcc2−/− Trp53−/− cells showed a moderate decrease in the proportion (to roughly 30%) of focus-positive cells relative to WT (Fig. 3B). These data indicated that the fraction of cells in the Xrcc2-deficient cultures containing unrepaired DSBs was significantly higher than in the WT and that this effect was dependent on p53. Importantly, the average number of foci per damaged Xrcc2−/− cell was similar to WT. The latter finding implies that in Xrcc2−/− cultures, naturally occurring DSBs do not properly resolve, and thus damaged cells accumulate (Fig. 3C).
FIG. 3.
Increased incidence of cells with DSBs in Xrcc2-null cultures. (A) Immunostaining of γ-H2AX foci in untreated developing B cells with indicated genotypes. (B) Fraction of developing B cells of indicated genotypes with γ-H2AX foci. For controls, WT splenocytes were irradiated at 5 Gy (IR) or left untreated (Non-). Error bars indicate standard error. (C) Fraction of cells exhibiting 0 or 1, 2 to 5, 6 to 10, 11 to 15, or >15 γ-H2AX foci for each of the indicated genotypes. The mean and the median number of foci, in cells with two or more foci, are indicated. The numbers of cells scored for each genotype are as follows: WT, 122; Xrcc2−/−, 202; Xrcc2−/− Trp53−/−, 38; untreated splenocytes, 192; and irradiated splenocytes, 206. Error bars indicate standard error.
Xrcc2-null B cells exhibit a p53-dependent cell cycle arrest.
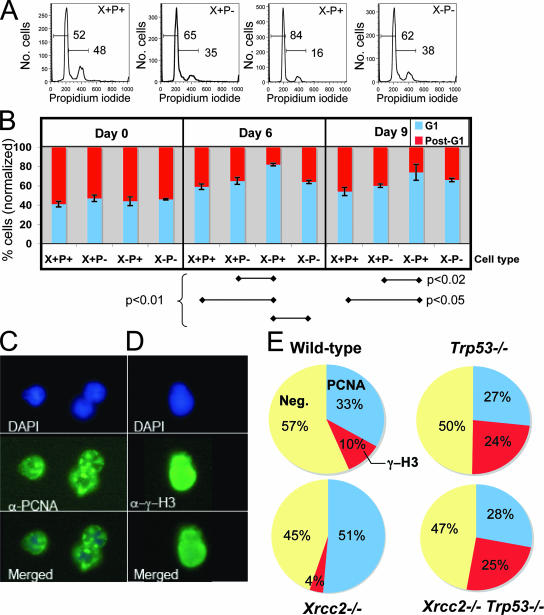
Spontaneous γ-H2AX foci can occur as a natural consequence of S-phase passage (16), so we considered the possibility that the increased incidence of focus-positive cells in Xrcc2-null cultures might be related to cell cycle control. As a first test, asynchronous cultures of each genotype were evaluated for cell cycle distribution via flow cytometric analysis following staining with the DNA binding dye PI. We quantified the percentage of cells showing pre/early replication (G1/early S) DNA content, versus a late or postreplication (late S/G2) DNA content after 0, 6, or 9 days in culture (Fig. 4A and B). Initially, on culture day 0, the cell cycle distributions of Xrcc2−/− and Xrcc2−/− Trp53−/− cultures were indistinguishable from either WT or Trp53-null controls, each showing roughly 40% cells with a G1/early-S-phase DNA content (Fig. 4B). However, by day 6 and continuing through day 9, Xrcc2-null cultures showed significantly higher percentages of cells with a G1/early-S-phase DNA content and concomitantly lower percentages of late-S/G2 cells than either WT or Trp53−/− controls (Fig. 4B). Importantly, the difference between Xrcc2−/− and WT cell cycle distributions was largely dependent on p53, because Xrcc2−/− Trp53−/− cell cycle profiles resembled those of the WT at all culture time points (Fig. 4A and B).
FIG. 4.
Altered cell cycle distribution in developing Xrcc2-null B cells. (A) Representative flow cytometry histograms of PI-stained cells for the indicated genotypes. Overlaid horizontal bars denote the sub-G1/G1 and post-G1 histogram peaks, with corresponding percentages indicated above. (B) Chart showing the normalized percentage of B cells in the G1 and post-G1 peaks, as shown in panel A, on culture days 0, 6, and 9. Error bars indicate standard error. (C) Representative positive immunostaining for PCNA in WT developing B cells. Shown are the images from the DAPI staining (upper panel), anti-PCNA (α-PCNA) staining (middle panel), and merged images (lower panel). α-γ-H3, anti-γ-H3. The numbers of cells scored for each genotype are as follows: WT, 111; Xrcc2−/−, 113; Trp53−/−, 107; and Xrcc2−/− Trp53−/−, 85. (D) Representative positive immunostaining of phosphorylated H3 (γ-H3) in developing B cells. The numbers of cells scored for each genotype are as follows: WT, 106; Xrcc2−/−, 75; Trp53−/−, 83; and Xrcc2−/− Trp53−/−, 110. (E) Percentages of developing B cells, of the indicated genotypes, showing PCNA-positive staining, γ-H3 staining, or neither (Neg).
Because cells in G1 can be difficult to distinguish from cells in early S phase, by PI staining, we performed two additional measures of cell cycle distribution: immunostaining for PCNA and phosphorylated histone H3 (γ-H3), for the S and M phases, respectively. (Fig. 4C and D). Xrcc2-null cultures showed a substantially higher fraction of PCNA+ cells (51%) than cultures of either WT (32%) or Xrcc2−/− Trp53−/− (29%) cells, indicating a higher-than-WT fraction of cells that had committed to, but not completed, S phase (Fig. 4E). Consistent with this interpretation, Xrcc2- null cultures exhibited the lowest percentage of cells in M phase, as detected by γ-H3 immunostaining (Fig. 4E). Together with the PI-staining data (above), these results suggest that developing Xrcc2−/− B cells can initiate, but fail to complete, S phase.
Developing, Xrcc2-deficient B cells arrest early in S phase.
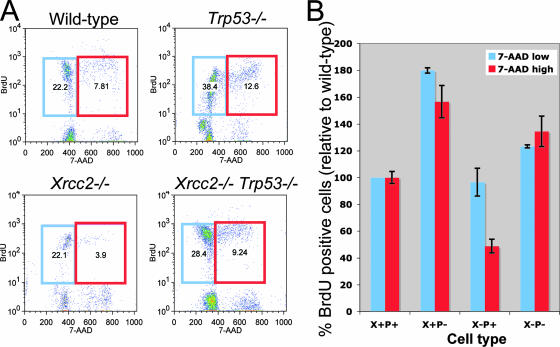
To critically test this idea, we conducted quantitative BrdU incorporation assays (Fig. 5). Fetal liver cells were cocultured for 7 days with T220 cells, switched to medium supplemented with BrdU, and cultured for an additional day prior to analysis. Confirming our PI and immunostaining results, the percentage of BrdU+ G1/early-S-phase cells in Xrcc2−/− cultures was similar to WT, but there was a significant decrease in the percentage of BrdU+ late S/G2 cells, to ∼50% WT levels (Fig. 5B). In contrast, Xrcc2−/− Trp53−/− cultures showed an increase in the percentage of BrdU+ G1/early S (∼120% of WT) and BrdU+ late S/G2 (∼130% of WT) cell fractions, indicative of overall increased cycling and demonstrating that S-phase arrest in Xrcc2−/− B-lineage cells required p53 activity (Fig. 5B).
FIG. 5.
p53-dependent early S-phase arrest in developing Xrcc2-null B cells. (A) Representative flow cytometry scatter plots of BrdU uptake of developing B cells. The blue gate represents early-S-phase cells that have incorporated BrdU, and the red gate represents late-S-phase cells that have incorporated BrdU. (B) Percentages of cells within the indicated gates are shown. (A) Chart showing percentages of BrdU plus 7-AAD-low (early S-phase; blue bars) and BrdU plus 7-AAD-high (late S-phase; red bars) normalized to WT. Error bars indicate standard error.
Developing Xrcc2 Trp53 double-null B cells exhibit chromosomal instability.
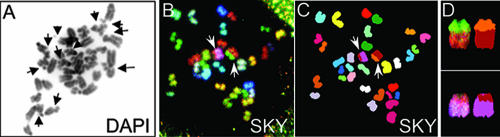
Finally, to investigate whether the lack of Xrcc2 would lead to increased chromosomal instability, karyotype analysis was performed (Fig. 6 and Table 1). Metaphase spreads were generated from 6-day cocultures and processed for either simple or spectral karyotyping. First, DAPI-stained metaphase spreads were scored for structural abnormalities, including fragmentation (Fig. 6A and Table 1). Unexpectedly, Xrcc2-null cells were indistinguishable from WT cells, but Xrcc2−/− Trp53−/− cells showed chromosome fragmentation rates significantly higher than WT or Trp53−/− cells (Table 1). These data reaffirm prior results showing that Xrcc2 and Trp53 likely cooperate to maintain genome stability (21). To determine whether the increased fragmentation in Xrcc2−/− Trp53−/− cells also related to increased translocations, SKY analysis was performed (Figure 6B to D and Table 1). Consistent with the fragmentation data, Xrcc2−/− cells showed a low overall translocation rate of 4%, whereas Trp53−/− cells exhibited a 10% translocation rate. Finally, Xrcc2−/− Trp53−/− cells exhibited a translocation rate (12%) comparable to that of Trp53−/− cells (Table 1). Together, the DAPI and SKY analyses suggest that Xrcc2 is important to prevent chromosome fragmentation but may be largely dispensable for suppression of subsequent translocations.
FIG. 6.
Xrcc2−/− Trp53−/− B-cell cultures exhibit chromosomal instability. (A) Analysis of structural abnormalities in DAPI-stained metaphase spread. Chromosome or chromatid fragments are indicated by arrows. (B to D) SKY analysis of metaphase chromosome spread showing two translocations. Shown are the spectral image (B) and the chromosome classified image (C). Translocations are indicated by arrows (B and C) and are shown enlarged in the far-right panels (D).
TABLE 1.
Chromosome instability in developing B-cell cultures
| Assay and parameter | Result for genotype:
|
|||
|---|---|---|---|---|
| WT | Trp53−/− | Xrcc2−/− | Xrcc2−/−Trp53−/− | |
| DAPI staining | ||||
| No. of metaphases | 30 | 84 | 80 | 21 |
| No. of cells with fragments (no. of fragments observed) | 0 | 87 (12) | 0 | 5 (6) |
| % of cells with chromosome fragments | 0 | 8 | 0 | 24 |
| No. of fragments/damaged nucleus | 0 | 1.7 | 0 | 1.2 |
| No. of other aberrations/nucleus | 0 | 0.01a | 0.01b | 0 |
| No. of chromosomes/ nucleus | 43 | 41.4 | 39.7 | 38.4 |
| SKY | ||||
| No. of metaphases | NDc | 21 | 26 | 33 |
| % of cells with translocations | ND | 10 | 4 | 12 |
| No. of translocations/ damaged nucleus | ND | 1.5 | 1 | 1.5 |
Ring chromosome.
Dicentric chromosome.
ND, not determined.
DISCUSSION
Here we demonstrate a critical role for HR in lymphocyte development. In the absence of Xrcc2, B-cell development fails due to p53-dependent early S-phase arrest. This developmental arrest likely occurs during proliferation accompanying Ig heavy chain rearrangement. Importantly, we demonstrate that lack of p53 in Xrcc2-null B-lymphoid cells restores cell cycle passage and B-cell development to the IgM+ stage of maturation, but that Xrcc2−/− Trp53−/− cultures incur substantial chromosome and chromatid fragmentation. These are the first direct data that HR is an essential pathway for normal—as opposed to malignant—mammalian lymphoid development.
Human immunodeficiencies represent a diverse and pleiotropic group of diseases (9, 11, 20, 31). While the genetic origins for some rare primary immunodeficiencies have been identified in the past decade, the genetic bases for many more remain unknown. For example, the genes involved in the most frequently diagnosed primary immunodeficiency, common variable immunodeficiency, have been identified in only 10 to 15% of all cases (31). Our results now suggest that deficient alleles of homologous recombination genes could underlie some human lymphodeficiencies of unknown genetic etiology, such as common variable immunodeficiency. In addition to developmental defects, Xrcc2 mutations or hypomorphic alleles likely predispose cells to malignant transformation. Given the central role of Rad51 and related proteins in maintaining genome integrity via HR-mediated DSB repair, Rad51 family genes should act as tumor suppressors. Indeed, one recent report indicated that Xrcc2 may modify the T-cell lymphoma phenotype associated with p53 deficiency (21). Our genome instability results now implicate the B-lymphoid lineage as another probable target for transformation in the context of Xrcc2 deficiency.
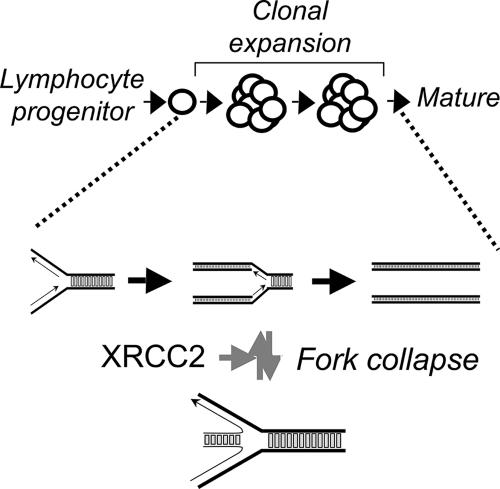
Our data demonstrate an early-S-phase arrest, indicating that the developmental blockade in Xrcc2-null B cells probably occurs during proliferative expansion. In rapidly proliferating cells, prompt completion of DNA replication is mandatory but replication forks can collapse or regress (4, 12). Annealing of two complementary nascent strands to each other can result in reversal of fork direction, producing a dead-end structure with a single DNA break (5, 36). The resultant cruciform DNA resembles Holliday junction intermediates for homologous recombination (28, 36). We thus propose a molecular model wherein XRCC2 restarts naturally stalled or collapsed DNA replication forks occurring in rapidly dividing lymphocytes and suggest that restart failure leads to interruption of the normal developmental program (Fig. 7). In response to fork collapse or regression, we speculate that XRCC2 binds to the cruciform “chicken foot,” perhaps also recruiting other HR factors, catalyzing recombination-mediated fork restart to ensure timely completion of DNA replication. This model explains the observed B-cell development defect in Xrcc2−/− cells, as well as the early S-phase cell cycle arrest, and is supported by several additional lines of evidence. First, there is accumulating data that HR factors are required for replication fork restart following stalling or regression (28-30). Second, Xrcc2 is required, at least in some cells, for resistance to hydroxyurea-induced replication stress (14). Third, in mammals, the XRCC2 binding partner RAD51C is required for processing structures resembling collapsed replication forks in mammalian cells (15). Finally, a tumor-specific Xrcc2 mutant protein has shown the ability to bind to Holliday junctions (19).
FIG. 7.
Model for XRCC2 function in B-cell development. In lymphocytes undergoing rapid clonal expansion, we propose that replication fork collapse prompts a p53-dependent cell cycle arrest. XRCC2 binding at or near the cruciform structure of the collapsed DNA replication forks initiates recombination-mediated restart. Following restoration of the fork structure, replication, and thus development, resumes normally.
In summary, we have demonstrated that HR plays a key role in B-cell development by facilitating S-phase progression and document a p53-dependent early S-phase arrest that is activated in Xrcc2-deficient B-lineage cells. This genetic interaction between Xrcc2 and Trp53 is likely critical for proliferative expansion accompanying B-cell development. These are the first results to implicate HR as an important pathway with a defined role in lymphocyte differentiation or maturation and suggest that XRCC2 and p53 coordinately orchestrate normal B-cell development. It will be important, in this context, to determine if and how these roles for XRCC2 are related to specific human lymphodeficiencies, or possibly to tumor suppression in rapidly proliferating cells, including those of the immune system.
Acknowledgments
We thank Susan Ackerman, MaryAnn Handel, David Harrison, and Sarah Maas for critical readings and helpful suggestions and Yiguo Hu, and Bonnie Lyons for technical assistance.
This work was supported by grant T32 DK07449 (M.G.H.) and a Scholar grant from the V Foundation for Cancer Research (K.D.M.).
Footnotes
Published ahead of print on 22 January 2008.
REFERENCES
- 1.Adam, J., B. Deans, and J. Thacker. 2007. A role for Xrcc2 in the early stages of mouse development. DNA Repair (Amsterdam) 6224-234. [DOI] [PubMed] [Google Scholar]
- 2.Bassing, C. H., W. Swat, and F. W. Alt. 2002. The Mechanism and regulation of chromosomal V(D)J recombination. Cell 109S45-S55. [DOI] [PubMed] [Google Scholar]
- 3.Borzillo, G. V., K. Endo, and Y. Tsujimoto. 1992. Bcl-2 confers growth and survival advantage to interleukin 7-dependent early pre-B cells which become factor independent by a multistep process in culture. Oncogene 7869-876. [PubMed] [Google Scholar]
- 4.Branzei, D., and M. Foiani. 2005. The DNA damage response during DNA replication. Curr. Opin. Cell Biol. 17568-575. [DOI] [PubMed] [Google Scholar]
- 5.Courcelle, J., J. R. Donaldson, K. H. Chow, and C. T. Courcelle. 2003. DNA damage-induced replication fork regression and processing in Escherichia coli. Science 2991064-1067. [DOI] [PubMed] [Google Scholar]
- 6.Deans, B., C. S. Griffin, M. Maconochie, and J. Thacker. 2000. Xrcc2 is required for genetic stability, embryonic neurogenesis and viability in mice. EMBO J. 196675-6685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deans, B., C. S. Griffin, P. O'Regan, M. Jasin, and J. Thacker. 2003. Homologous recombination deficiency leads to profound genetic instability in cells derived from Xrcc2-knockout mice. Cancer Res. 638181-8187. [PubMed] [Google Scholar]
- 8.Donehower, L., M. Harvey, B. Slagle, M. McArthur, C. Montgomery, J. Butel, and A. Bradley. 1992. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature 356215-221. [DOI] [PubMed] [Google Scholar]
- 9.Fischer, A., M. Cavazzana-Calvo, G. De Saint Basile, J. P. DeVillartay, J. P. Di Santo, C. Hivroz, F. Rieux-Laucat, and F. Le Deist. 1997. Naturally occurring primary deficiencies of the immune system. Annu. Rev. Immunol. 1593-124. [DOI] [PubMed] [Google Scholar]
- 10.Julius, M., and L. Herzenberg. 1974. Isolation of antigen-binding cells from unprimed mice. J. Exp. Med. 140904-920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kopecky, O., and S. Lukesova. 2007. Genetic defects in common variable immunodeficiency. Int. J. Immunogenet. 34225-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Labib, K., and B. Hodgson. 2007. Replication fork barriers: pausing for a break or stalling for time? EMBO Rep. 8346-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu, N., J. E. Lamerdin, R. S. Tebbs, D. Schild, J. D. Tucker, M. R. Shen, K. W. Brookman, M. J. Siciliano, C. A. Walter, W. Fan, L. S. Narayana, Z. Q. Zhou, A. W. Adamson, K. J. Sorensen, D. J. Chen, N. J. Jones, and L. H. Thompson. 1998. XRCC2 and XRCC3, new human Rad51-family members, promote chromosome stability and protect against DNA cross-links and other damages. Mol. Cell 1783-793. [DOI] [PubMed] [Google Scholar]
- 14.Liu, N., and C. S. Lim. 2005. Differential roles of XRCC2 in homologous recombinational repair of stalled replication forks. J. Cell Biochem. 95942-954. [DOI] [PubMed] [Google Scholar]
- 15.Liu, Y., J. Y. Masson, R. Shah, P. O'Regan, and S. C. West. 2004. RAD51C is required for Holliday junction processing in mammalian cells. Science 303243-246. [DOI] [PubMed] [Google Scholar]
- 16.MacPhail, S. H., J. P. Banath, Y. Yu, E. Chu, and P. L. Olive. 2003. Cell cycle-dependent expression of phosphorylated histone H2AX: reduced expression in unirradiated but not X-irradiated G1-phase cells. Radiat. Res. 159759-767. [DOI] [PubMed] [Google Scholar]
- 17.Masson, J. Y., M. C. Tarsounas, A. Z. Stasiak, A. Stasiak, R. Shah, M. J. McIlwraith, F. E. Benson, and S. C. West. 2001. Identification and purification of two distinct complexes containing the five RAD51 paralogs. Genes Dev. 153296-3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mills, K. D., D. O. Ferguson, and F. W. Alt. 2003. The role of DNA breaks in genomic instability and tumorigenesis. Immunol. Rev. 19477-95. [DOI] [PubMed] [Google Scholar]
- 19.Mohindra, A., E. Bolderson, J. Stone, M. Wells, T. Helleday, and M. Meuth. 2004. A tumour-derived mutant allele of XRCC2 preferentially suppresses homologous recombination at DNA replication forks. Hum. Mol. Genet. 13203-212. [DOI] [PubMed] [Google Scholar]
- 20.O'Driscoll, M., and P. Jeggo. 2006. The role of double-strand break repair-insights from human genetics. Nat. Rev. Genet. 745-54. [DOI] [PubMed] [Google Scholar]
- 21.Orii, K., Y. Lee, N. Kondo, and, P. J. McKinnon. 2006. Selective utilization of nonhomologous end-joining and homologous recombination DNA repair pathways during nervous system development. Proc. Natl. Acad. Sci. USA 10310017-10022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paull, T. T., E. P. Rogakou, V. Yamazaki, C. U. Kirchgessner, M. Gellert, and W. M. Bonner. 2000. A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr. Biol. 10886-895. [DOI] [PubMed] [Google Scholar]
- 23.Redon, C., D. Pilch, E. Rogakou, O. Sedelnikova, K. Newrock, and W. Bonner. 2002. Histone H2A variants H2AX and H2AZ. Curr. Opin. Genet. Dev. 12162-169. [DOI] [PubMed] [Google Scholar]
- 24.Rogakou, E. P., C. Boon, C. Redon, and W. M. Bonner. 1999. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J. Cell Biol. 146905-916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rogakou, E. P., W. Nieves-Neira, C. Boon, Y. Pommier, and W. M. Bonner. 2000. Initiation of DNA fragmentation during apoptosis induces phosphorylation of H2AX histone at serine 139. J. Biol. Chem. 2759390-9395. [DOI] [PubMed] [Google Scholar]
- 26.Rogakou, E. P., D. R. Pilch, A. H. Orr, V. S. Ivanova, and W. M. Bonner. 1998. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 2735858-5868. [DOI] [PubMed] [Google Scholar]
- 27.Sale, J. E., D. M. Calandrini, M. Takata, S. Takeda, and M. S. Neuberger. 2001. Ablation of XRCC2/3 transforms immunoglobulin V gene conversion into somatic hypermutation. Nature 412921-926. [DOI] [PubMed] [Google Scholar]
- 28.Saleh-Gohari, N., H. E. Bryant, N. Schultz, K. M. Parker, T. N. Cassel, and T. Helleday. 2005. Spontaneous homologous recombination is induced by collapsed replication forks that are caused by endogenous DNA single-strand breaks. Mol. Cell. Biol. 257158-7169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saleh-Gohari, N., and T. Helleday. 2004. Conservative homologous recombination preferentially repairs DNA double-strand breaks in the S phase of the cell cycle in human cells. Nucleic Acids Res. 323683-3688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saleh-Gohari, N., and T. Helleday. 2004. Strand invasion involving short tract gene conversion is specifically suppressed in BRCA2-deficient hamster cells. Oncogene 239136-9141. [DOI] [PubMed] [Google Scholar]
- 31.Schaffer, A. A., U. Salzer, L. Hammarstrom, and B. Grimbacher. 2007. Deconstructing common variable immunodeficiency by genetic analysis. Curr. Opin. Genet. Dev. 17201-212. [DOI] [PubMed] [Google Scholar]
- 32.Sobacchi, C., V. Marrella, F. Rucci, P. Vezzoni, and A. Villa. 2006. RAG-dependent primary immunodeficiencies. Hum. Mutat. 271174-1184. [DOI] [PubMed] [Google Scholar]
- 33.Takata, M., M. S. Sasaki, S. Tachiiri, T. Fukushima, E. Sonoda, D. Schild, L. H. Thompson, and S. Takeda. 2001. Chromosome instability and defective recombinational repair in knockout mutants of the five Rad51 paralogs. Mol. Cell. Biol. 212858-2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thacker, J., C. Tambini, P. Simpson, L. Tsui, and S. Scherer. 1995. Localization to chromosome 7q36.1 of the human XRCC2 gene, determining sensitivity to DNA-damaging agents. Hum. Mol. Genet. 4113-120. [DOI] [PubMed] [Google Scholar]
- 35.Villa, A., C. Sobacchi, and P. Vezzoni. 2002. Omenn syndrome in the context of other B cell-negative severe combined immunodeficiencies. Isr. Med. Assoc. J. 4218-221. [PubMed] [Google Scholar]
- 36.Wu, L., and I. D. Hickson. 2003. The Bloom's syndrome helicase suppresses crossing over during homologous recombination. Nature 426870-874. [DOI] [PubMed] [Google Scholar]