Abstract
TAZ is a WW domain containing a transcription coactivator that modulates mesenchymal differentiation and development of multiple organs. In this study, we show that TAZ is phosphorylated by the Lats tumor suppressor kinase, a key component of the Hippo pathway, whose alterations result in organ and tissue hypertrophy in Drosophila and contribute to tumorigenesis in humans. Lats phosphorylates TAZ on several serine residues in the conserved HXRXXS motif and creates 14-3-3 binding sites, leading to cytoplasmic retention and functional inactivation of TAZ. Ectopic expression of TAZ stimulates cell proliferation, reduces cell contact inhibition, and promotes epithelial-mesenchymal transition (EMT). Elimination of the Lats phosphorylation sites results in a constitutively active TAZ, enhancing the activity of TAZ in promoting cell proliferation and EMT. Our results elucidate a molecular mechanism for TAZ regulation and indicate a potential function of TAZ as an important target of the Hippo pathway in regulating cell proliferation tumorigenesis.
Genetic screens of flies over the past several years have delineated the new Hippo-signaling pathway, which controls tissue and organ size by regulating cell proliferation and apoptosis (23, 26). Two previously defined components of this pathway, Hippo (Hpo) and Warts (Wts), code for the serine/threonine kinases that belong to the Ste20 and the nuclear Dbf2-related protein kinase families, respectively (4, 30). Hpo complexes have the regulatory subunit Salvador (Sav) and activate Wts by phosphorylation, whereas Wts associates with its activating subunit Mats (mob as tumor suppressor) and phosphorylates Yorkie (Yki), which is a transcription coactivator (12, 36). High Yki activity induces the expression of cyclinE and Diap1 in Drosophila melanogaster, leading to increased cell proliferation and decreased apoptosis (8, 32). Merlin (Mer) and Expanded (Ex) are two cytoskeleton-associated proteins acting upstream of Hpo (8, 19). Recently, the tumor suppressor Fat, a cell surface protocadherin, has been added to the Hpo pathway, indicating a possible role for the Hippo pathway in cell contact inhibition (2, 27, 37). How the Hippo pathway regulates cytoskeleton organization and cell contact inhibition is not known.
Components of the Hippo pathway are found in all eukaryotes and are highly conserved in multiple cellular organisms. For example, Mst and Lats are human homologues of the Drosophila Hpo and Wts, respectively (7). The mammalian yes-associated protein (YAP) shows high sequence similarity with Yki and can functionally rescue the Drosophila yki mutation (7). These results suggest that the Hippo pathway is likely to function in mammals as well. A recent study has shown that YAP is indeed inhibited by the Hippo pathway in mammalian cells (6). Accumulating evidence supports an important role for this pathway in human cancer. YAP has recently been demonstrated to be the oncogene in human amplicon 11q22 (22). Moreover, Mer is encoded by the neurofibromatosis type 2 (NF2) tumor suppressor (19). Other components of the Hippo pathway, such as Sav, Mats, Lats1/Lats2, and Wts, have also been implicated in human cancer (14, 28, 29, 31).
The transcriptional coactivator with PDZ binding motif (TAZ), first reported as a 14-3-3 binding protein (13) and independently as a binding target of the oncogenic mouse polyomavirus T (tumor) antigens (34), is closely related to YAP. Both proteins share approximately 50% sequence identity, with a similar topology, containing two central WW domains and a C-terminal transactivation domain. TAZ has been reported to bind with a variety of transcription factors such as the RUNX family, TEAD, thyroid TF1 (TTF-1), TBX5, paired box homeotic gene 3 (Pax3), and PPARγ (3, 9, 10, 16, 20, 21, 24). Consistent with these diverse interactions, the TAZ function has been linked with the development of limb, heart, bone, muscle, fat, and lung tissues (9, 10, 16, 20, 24). TAZ has also been implicated in mesenchymal stem cell differentiation (9). However, whether TAZ plays a role in cancer is unclear. Additionally, it has also been reported that TAZ is a phosphoprotein, and phosphorylation likely regulates TAZ interaction with 14-3-3 and subcellular localization (13). The responsive TAZ kinase(s) has yet to be clearly defined.
In this study, we demonstrate that TAZ is regulated by the Hippo pathway. TAZ is inhibited by the Lats kinase via phosphorylation of the conserved HXRXXS motif. This phosphorylation creates the 14-3-3 binding site in TAZ, leading to the sequestration of TAZ from the cell nucleus. We also show that TAZ promotes cell proliferation and induces epithelial-mesenchymal transition (EMT), indicating a potential role for TAZ in tumorigenesis. Phosphorylation and inhibition of TAZ may represent a critical consequence of the Hippo pathway in tumor suppression.
MATERIALS AND METHODS
Cell culture and transfection.
293T, HeLa, and BOSC cells were cultured in Dulbecco's modified Eagle's medium (DMEM) (Invitrogen) supplemented with 10% fetal calf serum (HyClone), 100 U/ml penicillin, and streptomycin (Gibco). MCF10A cells were maintained in DMEM/F-12 medium (Invitrogen) supplemented with 5% horse serum (Invitrogen), 20 ng/ml epidermal growth factor, 0.5 μg/ml hydrocortisone, 10 μg/ml insulin, 100 ng/ml cholera toxin, 100 U/ml penicillin, and streptomycin (Gibco). Cell transfection was performed using Lipofectamine 2000 (Invitrogen) or calcium phosphate methods. Cells were harvested at 24 h posttransfection for protein analysis or luciferase activity assay.
To establish stable TAZ-expressing cells, pBabe-TAZ retroviruses were generated and used to infect MCF10A cells. Stable pools were selected with puromycin for 5 days. TAZ stable pool cells were seeded in 6-well plates with 1 × 105 cells/well in triplicates. Cell growth was counted every day for 7 days.
Immunofluorescence staining.
Cells were fixed in 4% paraformaldehyde for 1 h, followed by washing in phosphate-buffered saline (PBS). The endogenous peroxidase activity was inactivated in a solution containing 3% hydrogen peroxide (H2O2) in methanol. The following detection and visualization procedures were performed according to the manufacturer's protocols. Negative control slides were performed without primary antibody. Control slides known to be positive for each antibody were incorporated. For double-fluorescence staining, pretreated sections were first blocked with goat serum at a 1:20 dilution and 1% bovine serum albumin blocking and then incubated with mouse antibody hemagglutinin (HA) (catalog no. sc-9353; Santa Cruz Biotechnology) and rabbit antibody Flag (product no. F3165; Sigma) at 4°C overnight, followed by fluorescein isothiocyanate (FITC)-conjugated anti-mouse 488 (catalog no. A11001; Invitrogen) and anti-rabbit 594 (catalog no. A11012; Invitrogen) secondary antibody.
Luciferase activity assay.
For the luciferase reporter assay, BOSC cells were seeded in 24-well plates. A mixture of 5× upstream activating sequence (UAS)-luciferase reporter, Renilla, and the indicated plasmids were cotransfected. Twenty-four hours after cells were transfected, they were lysed, and luciferase activity was measured using a dual-luciferase reporter assay system (catalog no. E1960; Promega) following the manufacturer's instructions. The luciferase activity was measured by a luminometer (model TD-20/20). Transfection efficiency was normalized to thymidine kinase-driven Renilla luciferase activity.
BrdU labeling and flow cytometric analysis.
For cell cycle analysis, cells were cultured to the desired confluence and then labeled with 5-bromo-2-deoxyuridine (BrdU) and analyzed by flow cytometry, using an FITC BrdU flow kit obtained from BD Biosciences (San Jose, CA), following the manufacturer's instructions. Briefly, cells were pulse-labeled with 15 μM BrdU in the culture medium for 30 min. After cells underwent trypsinization and a PBS wash, they were fixed and permeabilized. After DNase treatment, cells were stained with FITC-conjugated anti-BrdU antibody. Total DNA was stained by propidium iodide. Data were collected with a BD FACSCalibur and analyzed with Cell Quest Pro software.
Immunoprecipitation and kinase assay.
For the Lats2 and Mst2 kinase assays, 293T cells were transfected with HA-Lats2 or Flag-Mst2. Thirty-six hours posttransfection, cells were lysed with lysis buffer (50 mM HEPES [pH 7.5], 150 mM NaCl, 1 mM EDTA, 1% NP-40, 50 mM NaF, 1.5 mM Na3VO4, protease inhibitor cocktail [Roche], 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride [PMSF]) and immunoprecipitated with anti-HA or anti-Flag antibodies. The immunoprecipitates were washed three times with lysis buffer, followed by a single wash with wash buffer (40 mM HEPES, 200 mM NaCl) and a single wash with kinase assay buffer (30 mM HEPES, 50 mM potassium acetate, 5 mM MgCl2). The immunoprecipitated Lats2 or Mst2 was subjected to a kinase assay in the presence of 500 μM cold ATP and 1 μg His-TAZ expressed and purified from Escherichia coli as the substrate. The reaction mixtures were incubated at 25°C for 50 min, terminated with sodium dodecyl sulfate (SDS) sample buffer, and subjected to SDS-polyacrylamide gel electrophoresis (PAGE) and enhanced chemiluminescence development.
RNA isolation and real-time PCR.
Total RNA was isolated from cultured cells using Trizol reagent (Invitrogen). cDNA was synthesized by reverse transcription using oligo(dT) as the primer and proceeded to real-time PCR with gene-specific primers in the presence of SYBR Premix Ex Taq (catalog no. DRR041A; TaKaRa). The relative abundance of mRNA was calculated by normalization to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA.
Western blotting analysis.
Protein lysates were prepared from transfected 293T, BOSC, MCF10A, and MCF10A TAZ stable pool cells in a buffer containing 50 mM Tris-HCl (pH 8.0), 150 mM NaCl, 0.1% SDS, 0.5% deoxycholate, 1% NP-40, 1 mM EDTA, 1 mM PMSF, 25 mM NaF, and cocktail protease inhibitors (Roche). Tissue lysate (40 μg) was resolved by SDS-PAGE, followed by Western blotting analysis using HA (catalog no. A00168; GenScript), FLAG (catalog no. A00170; GenScript), MYC (catalog no. A00172; GenScript), E-cadherin monoclonal antibody (MAb) (catalog no. 610405; BD), N-cadherin MAb (catalog no. 610920; BD), vimentin MAb (catalog no. 550513, BD), occludin (catalog nos. H-279 and SC-5562; Santa Cruz Biotechnology), fibronectin MAb (catalog no. F0916; Sigma), pYAP(S127) (catalog no. 4911S; Cell Signaling), and beta-actin (13E5 [catalog no. 4970]; Cell Signaling).
For immunoprecipitation experiments, 500 μg of cell lysate was incubated for 12 h at 4°C with anti-FLAG M2-agarose (catalog no. A2220; Sigma), c-MYC (9E10) agarose conjugate beads (catalog no. sc-40 AC; Santa Cruz Biotechnology) or HA antibody (F-7 [catalog no. sc-7392]; Santa Cruz Biotechnology) plus 25 μg protein A agarose beads (catalog no. 16-125; Upstate). Beads were washed three times with lysis buffer and centrifuged for 5 min at 5,000 × g between each wash. Protein was eluted from beads with 50 μl of Laemmli sample buffer (Bio-Rad). Lysates were resolved on 8 to 10% SDS-PAGE gels and transferred onto nitrocellulose (Bio-Rad) for Western blotting.
Wound-healing assay.
Monolayer cells were wounded with a sterile plastic tip. Cell migration was observed 16 h later by microscopy.
Matrigel assay.
A Matrigel assay was performed as reported previously, with minor modifications (5). Briefly, the well of a chamber was precoated with 100% Matrigel (BD Pharmingen). Cells (5,000) were plated onto the Matrigel bed and cultured with mammary epithelial cell growth medium supplemented with hydrocortisone, insulin, epidermal growth factor, fetal bovine serum, and 2% Matrigel. Acinar formation was monitored by microscopy. For immunostaining, the acini were fixed with formalin, paraffin embedded, and stained with antibodies to PH3, a polyclonal antibody against mitosis-specific phosphorylated histone H3 (Upstate Biotechnology).
RESULTS
Transcription activity of TAZ is inhibited by the Hippo pathway.
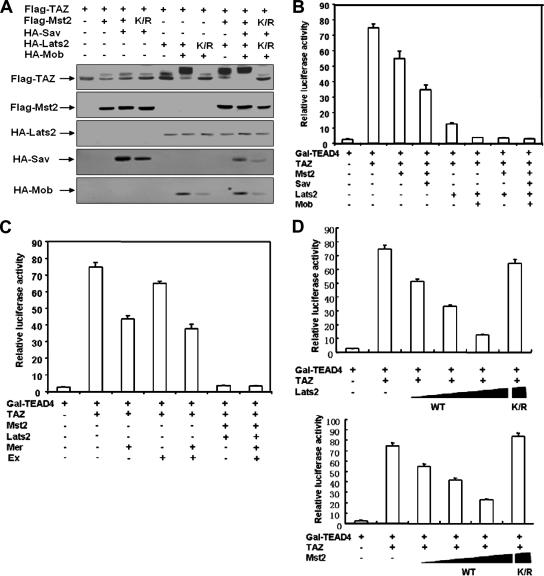
The TAZ transcription coactivator is closely related to YAP, which is the mammalian ortholog of the Drosophila Yki, a key component in the Hippo pathway (12). However, whether TAZ is regulated by the Hippo pathway has not been reported. TAZ has been implicated in the differentiation of mesenchymal tissue to bone and organ development. However, the molecular regulation of TAZ is largely unknown. We investigated whether the Hippo pathway regulates TAZ and found that the expression of Mst2 or Lats2 caused an obvious TAZ mobility shift (Fig. 1A), indicating that Mst2 and Lats2 may promote TAZ phosphorylation. Sav and Mob are regulatory subunits for the Mst2 and Lats2 kinases, respectively. Interestingly, the coexpression of Sav and Mob enhanced the effect of Mst2 and Lats2 on the mobility shift of TAZ (Fig. 1A). Moreover, the coexpression of both Mst2 and Lats2 led to an even more dramatic mobility shift of TAZ (Fig. 1A). The coexpression of kinase-dead mutants of Mst2 and Lats2 failed to induce a TAZ mobility shift, suggesting kinase activity is required (Fig. 1A). The mobility of TAZ induced by Mst2 and Lats2 can be reversed by treatment with lambda phosphatase (Fig. 2B). These results show that Mst2 and Lats2 induce TAZ phosphorylation.
FIG. 1.
The transcription coactivator activity of TAZ is inhibited by the Hippo pathway. (A) The cotransfection of Mst2 and Lats2 decreases TAZ electrophoretic mobility. Flag-TAZ was cotransfected with the indicated plasmids into BOSC cells (a cell line derived from HEK 293), and Western blotting was employed to examine TAZ mobility. (B) TAZ activity is repressed by Mst2 and Lats2. The 5× GAL4 UAS-luciferase reporter and GAL4-TEAD4 were transfected into BOCS cells, with the indicated plasmids. Renilla luciferase plasmid was also cotransfected as an internal control. Firefly luciferase activity was measured and normalized to Renilla luciferase activity. Data are representative of three independent experiments. The coexpression of the Hippo pathway components Mst2, Sav, Lats2, and Mob inhibited TAZ activity. (C) TAZ activity is inhibited by human Ex and Mer. The experiments shown are similar to those shown in panel B. Both Ex and Mer cause a significant inhibition of the TAZ reporter. (D) Kinase activity is required for Mst2 and Lats2 to inhibit TAZ. Increasing amounts of wild-type Lats2 (upper panel) or Mst2 (lower panel) were cotransfected with the TAZ reporter. Both Lats2 and Mst2 show a dose-dependent inhibition of the TAZ reporter. In contrast, the kinase-inactive mutants (K/R) of both Lats2 and Mst2 cannot inhibit the TAZ reporter.
FIG. 2.
Lats2 inhibits the function of TAZ via phosphorylation. (A) TAZ contains four HXRXXS motifs as the putative Lats phosphorylation sites. The yeast Dbf2 optimal target sequence was aligned with the four HXRXXS motifs of human TAZ. (B) The putative HXRXXS motifs in TAZ are required for a mobility shift by Lats2 and Mst2. The wild type (WT) or the mutant Flag-TAZ was cotransfected with HA-Mst2 and HA-Last2, as indicated. The TAZ mobility shift was examined by Western blotting. The 4SA mutation denotes the quadruple mutation of all four putative phosphorylation sites. As indicated, treatment with lambda phosphatase abolished the mobility shift by Lats2 and Mst2 cotransfection, indicating that the mobility shift is due to phosphorylation. (C) The S89A and S4A TAZ phosphorylation mutants are resistant to inhibition by Mst2 and Lats2. The S89A mutation and the phosphorylation mimic S89D mutant made no difference in the inhibition by Mst2 and Lats2. The reporter assay was performed similar to that shown in panel A. The inhibition change (n-fold) for each mutant is shown at the top of this panel. (D) The coexpression of Mst2 and Lats2 increases TAZ Ser89 phosphorylation. Lats2K/R dramatically decreases TAZ Ser89 phosphorylation. Flag-TAZ was cotransfected with Lats2, Mst2, or kinase-dead Lats2K/R into 293T cells, as indicated. Flag-TAZ was immunoprecipitated, and phosphorylation of Ser89 was detected by pYAP(Ser127) antibody, which can recognize the Ser89 phosphorylation site because of sequence conservation. Lambda phosphatase treatment and the TAZS89A mutant abolished the recognition of phosphoTAZ by the anti-phosphoYAP antibody. This result confirms the specificity of the phosphoYAP antibody. (E) In vitro phosphorylation of TAZ by Lats2. HA-Lats2 or Flag-Mst2 was immunoprecipitated from transfected 293T cells. An in vitro kinase assay was performed using purified His-TAZ as a substrate. Phosphorylation of TAZ was detected by pYAP(S127) antibody. His-TAZ input was shown by Coomassie blue staining. (F) A knockdown of Lats decreases TAZ Ser89 phosphorylation. HeLa cells cotransfected HA-TAZ with small interfering (siRNA) for Lats1 and Lats2, as indicated. Phosphorylation and protein levels of TAZ were determined by Western blotting. A knockdown of Lats was verified by quantitative PCR (data not shown).
Next, we examined the functional effect of the Hippo pathway components on TAZ, which has been reported to interact with and stimulate several transcription factors, including the TEAD family transcription factor. We utilized a TAZ transcription reporter system in which a luciferase was under the control of five Gal4 binding elements. The expression of Gal4-TEAD4 provided basal luciferase activity; however, the coexpression of TAZ dramatically increased the reporter activity (Fig. 1B). We examined the effect that the Hippo pathway components had on TAZ reporter activity. We found that the individual expression of Mst2 and of Lats2 inhibited TAZ activity and that the combination of both resulted in a further decrease of TAZ activity (Fig. 1B). In accord with the observation of TAZ phosphorylation, the coexpression of Sav or Mob with Mst2 and Lats2 further inhibited TAZ activity. An even more dramatic inhibition was observed by the combination of all five Hippo pathway components (Fig. 1B). These results indicate that the Hippo pathway inhibits TAZ and links the phosphorylation of TAZ by Mst2 and Lats2 with its functional inhibition.
We also assessed the effect of Mer and Ex on TAZ activity. It has been suggested that both Mer and Ex function upstream of Mst2. Cotransfection of Mer or Ex resulted in a modest but reproducible repression of TAZ activity (Fig. 1C). The weak effect of Mer and Ex could be due to the fact that components acting between Mer/Ex and Lats is limiting in this cell line. We tested the requirement of Mst2 and Lats2 kinase activity for their inhibition of TAZ. Both wild-type Mst2 and Lats2 displayed a dose-dependent inhibition of the TAZ reporter, while the kinase-dead mutants were completely inactive and had no inhibitory effect on TAZ reporter (Fig. 1D). Together, our data demonstrate that TAZ is inhibited by the Hippo pathway, probably via a phosphorylation-dependent mechanism.
Lats2 inhibits TAZ activity by phosphorylating the HXRXXS motif.
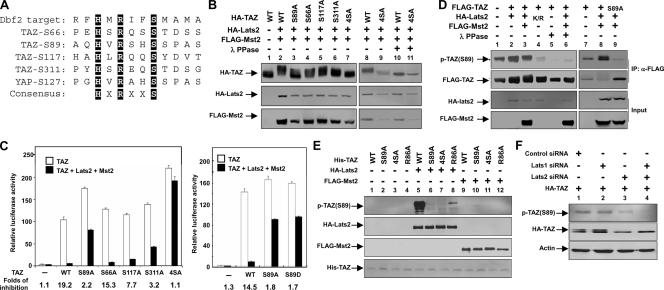
Lats2 and Wts belong to the nuclear Dbf2-related family of protein kinases. Biochemical studies of the Saccharomyces cerevisiae Dbf2 kinase indicate that it recognizes the RXXS motif in its substrates (15). In search of such consensus, we noticed that TAZ contains four HXRXXS motifs (Fig. 2A). It is worth noting that the His residue at the −5 position was also presented in the synthetic peptide used by Mah et al. (15). Therefore, we determined whether the four HXRXXS motifs in TAZ represent the putative Lats2 phosphorylation sites.
We generated a series of TAZ mutants by replacing individual serine residues in the HXRXXS motifs with alanine. Mst2 and Lats2 cotransfection could induce a mobility shift of all single-point TAZ mutants (TAZS89A, TAZS66A, TAZS117A, and TAZS311A), indicating that Lats2 phosphorylates on multiple sites (Fig. 2B). Lambda phosphatase treatment abolished the mobility shift of TAZ caused by Mst2 and Lats2, confirming that this mobility shift was indeed due to phosphorylation (Fig. 2B). In contrast, TAZ4SA, which had all four serine residues in the HXRXXS motif replaced with alanine, was resistant to the mobility shift caused by Mst2 and Lats2. These results indicate that some or all of the four serine sites in the HXRXXS motifs were phosphorylated by Lats2.
We examined the functional significance of TAZ phosphorylation by using the TAZ reporter assay described in the legend to Fig. 1B. As shown, the wild-type TAZ was efficiently inhibited by the coexpression of Mst2 and Lats2. Among all single-point mutations, however, the TAZS89A mutant showed a significant resistance to inhibition by Mst2 and Lats2, while TAZS311A also displayed partial resistance to inhibition (Fig. 2C). Furthermore, the TAZ4SA mutant not only was resistant to inhibition by Mst2 and Lats2 but also displayed an elevated basal activity, presumably resulting from an increased resistance to the endogenous TAZ kinases (Fig. 2C). We also generated an S89D mutant and compared its activity with that of the wild type and the S89A mutant. We found that the S89D mutant is active and resistant to inhibition by Lats and Mst, similar to the S89A mutant (Fig. 2C). These data indicate that the mutation of Ser89 to aspartate does not mimic the effect of phosphorylation. Instead, the S89D mutant acts similarly to the nonphosphorylatable mutant. These data demonstrated that Lats2 phosphorylates the HXRXXS motifs in TAZ, especially Ser89 and Ser311, and this phosphorylation inhibits the transcriptional activity of TAZ.
To determine the function of Lats2 in TAZ phosphorylation in vivo, we examined TAZ Ser89 phosphorylation in culture cells. Ser89 in TAZ corresponds to Ser127 in YAP (Fig. 2A). We found that the phosphoYAP(Ser127) antibody could recognize TAZ. We observed that the coexpression of Lats2 or/and Mst2 enhanced TAZ Ser89 phosphorylation (Fig. 2D). To verify the specificity of the phosphoYAP(Ser127) antibody, we treated the immunoprecipitated TAZ with lambda phosphatase and included the expression of the TAZS89A mutant. Our data showed that the phosphatase treatment completely abolished the recognition by the phosphoYAP(Ser127) antibody and the expression of the TAZS89A mutant leads to the diminishment of p-TAZ(S89) (Fig. 2D). Interestingly, the Lats2 kinase-dead mutant (Lats2K/R) coexpression decreased the TAZ basal phosphorylation significantly below that of the control, indicating that Lats2K/R functions as a dominant negative and also suggesting the functional role for endogenous Lats2 in TAZ phosphorylation.
In order to address whether Lats2 or Mst2 directly phosphorylates TAZ, we performed an in vitro kinase assay using purified His-TAZ protein from bacteria and immunoprecipitated Lats2 or Mst2. We used p-YAP(S127) antibody, which can recognize the TAZ Ser89 phosphorylation site because of sequence conservation, to detect Ser89 phosphorylation by Lats2. As shown in Fig. 2E, Lats2 phosphorylated TAZ wild type but not TAZS89A or TAZ4SA. In contrast, Mst2 could not phosphorylate His-TAZ in vitro (Fig. 2E). These data demonstrate that Lats2 directly phosphorylates TAZ, while Mst2 stimulates TAZ phosphorylation indirectly in vivo, perhaps by activating Lats2. Therefore, our data suggest a linear model: Mst > Lats > TAZ. We generated an R86A mutant to determine the importance of the arginine in TAZ phosphorylation by Lats2. In vitro kinase assays showed that the R86A mutant dramatically decreased TAZ Ser89 phosphorylation (Fig. 2E), indicating that Arg86 was important for TAZ Ser89 phosphorylation by Lats. We further examined the functional role of endogenous Lats in TAZ phosphorylation by down-regulating Lats. Lats1 and Lats2 were down-regulated by RNA interference, and the phosphorylation of TAZ was determined. The knockdown of Lats2 caused a significant decrease in the Ser89 phosphorylation of transfected HA-TAZ, while the knockdown of both Lats1 and Lats2 almost abolished its phosphorylation (Fig. 2F). These results support the functional importance of endogenous Lats in TAZ phosphorylation.
Phosphorylation regulates TAZ localization and interaction with 14-3-3.
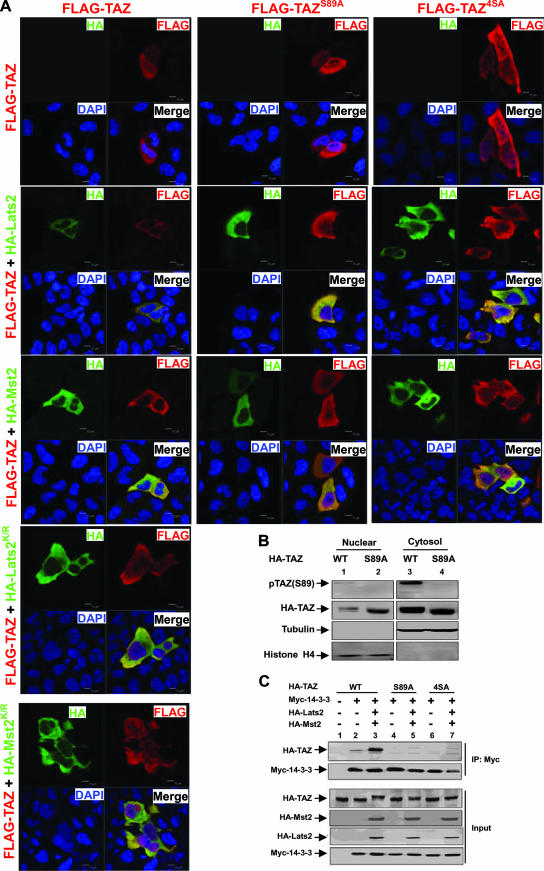
It was previously reported that TAZ phosphorylation promotes its cytoplasmic localization (13). We wondered if this phosphorylation is important for TAZ localization. Furthermore, Ser89 has been implicated in 14-3-3 binding, although the responsible kinase was unknown (10). Our studies have shown that Lats likely phosphorylates TAZ on Ser89. We wanted to test whether Mst2 and Lats2 might also regulate TAZ subcellular localization. We found that the transfected TAZ was distributed throughout the cell. However, the coexpression of Lats2 or Mst2 significantly decreased nuclear TAZ, resulting in a clear nuclear exclusion of TAZ (Fig. 3A). These effects depend on the kinase activity, as neither Lats2K/R nor Mst2K/R caused TAZ nuclear exclusion, indicating that the effects of Mst2 and Lats2 on TAZ localization is not nonspecific (Fig. 3A). We further tested the phosphorylation-defective TAZ mutations S89A and 4SA. Our results showed that Mst2 and Lats2 failed to promote the nuclear exclusion of the TAZ phosphorylation mutant. We also performed subcellular fractionation and found that more of the TAZS89A mutant protein was found in the nuclear fraction than in that of the wild-type protein (Fig. 3B). These results strongly suggest that the phosphorylation of TAZ retains it in the cytoplasm. These data are consistent with the TAZ transcription reporter assay, where Mst2 and Lats2 inhibit TAZ activity.
FIG. 3.
Phosphorylation increases TAZ cytoplasmic localization and 14-3-3 association. (A) Phosphorylation is important for TAZ nuclear exclusion caused by Lats2 and Mst2. Flag-TAZ (wild type and S89A and 4SA mutants) was cotransfected with HA-Lats2 or HA-Mst2, as indicated. Immunofluorescence using anti-Flag antibody (red) and HA antibody (green) was performed. DNA was stained with 4′,6′-diamidino-2-phenylindole (DAPI) (blue). The merged figures are shown in the lower right corner of each panel. (B) The mutation of Ser89 increases nuclear TAZ. HA-TAZ and the S89A mutant were cotransfected with Mst2 and Lats2 into HEK293 cells. Cytoplasmic and nuclear fractions were separated and analyzed by Western blotting. WT, wild type. (C) Flag-TAZ was cotransfected with Myc 14-3-3 and other indicated plasmids into 293T cells. Myc 14-3-3 was immunoprecipitated and coimmunoprecipitated. Flag-TAZ was probed with Flag antibody.
TAZ was initially isolated as a 14-3-3 binding protein (13). The 14-3-3 binding is a mechanism used commonly to promote the cytoplasmic retention of nuclear proteins, such as Foxo1 (39). We found that the coexpression of both Lats2 and Mst2 strongly increased the interaction between TAZ and 14-3-3ɛ, based on coimmunoprecipitation experiments. In contrast, the TAZS89A and TAZ4SA mutants showed little interaction with 14-3-3ɛ, and the coexpression of Mst2 and Lats2 did not enhance the interaction between the TAZ mutant and the 14-3-3ɛ protein (Fig. 3C). Our data indicate that Lats2 promotes TAZ cytoplasmic localization primarily by stimulating Ser89 phosphorylation and its binding with 14-3-3ɛ. Together, our data provide a possible biochemical mechanism, promoting TAZ's binding with 14-3-3 and thus cytoplasmic retention, for the inhibition of TAZ by the Hippo pathway.
TAZ stimulates cell proliferation.
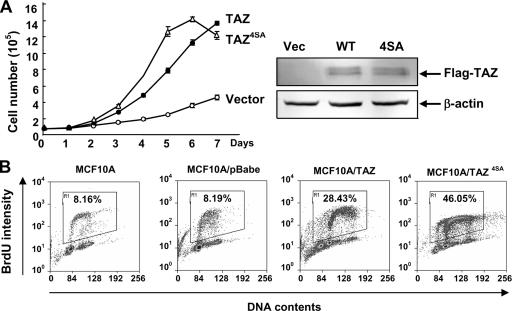
YAP is a candidate human oncogene in the amplicon 11q22 (23). Many components in the Hippo pathway, including Mer, Lats, and Sav, have been implicated as tumor suppressor genes (14, 19, 28, 29, 31). To explore the potential function of TAZ in oncogenes, we first examined the effect of TAZ overexpression on cell proliferation. The normal immortalized human breast epithelial cell line MCF10A was transduced with retroviruses expressing TAZ or TAZ4SA mutants. Stable pools were established and used for measuring cell proliferation. Both the MCF10A/TAZ and MCF10A/TAZ4SA cells proliferated significantly faster than the vector control cells (Fig. 4A). Furthermore, both the MCF10A/TAZ and MCF10A/TAZ4SA cells continued proliferating even after they reached confluence. To confirm this result, we pulse-labeled different pools of cells with BrdU for 30 min and analyzed their cell cycle distribution by flow cytometry. The parental MCF10A and vector control cells displayed similar cell cycle distributions, with 8.16% and 8.19% BrdU-positive cells, respectively. In contrast, the ectopic expression of either TAZ or TAZ4SA dramatically increased the S-phase cell population, in which 28.43% of MCF10A/TAZ cells and 46.05% MCF10A/TAZ4SA cells were BrdU positive, respectively (Fig. 4B). Together, these data indicate that an increased TAZ activity promotes cell proliferation and the cell proliferation activity of TAZ is negatively regulated by Mst2- and Lats2-mediated phosphorylation.
FIG. 4.
TAZ stimulates cell proliferation. (A) TAZ promotes cell growth. Growth curves of MCF10A cells stably expressing pBabe-vector, -TAZ, and -TAZ4SA were determined. (B) TAZ increases the proliferation of MCF10A cells. A synchronized parental MCF10A and pBabe-vector, -TAZ, and -TAZ4SA overexpressing MCF10A cells were labeled with BrdU (15 μM for 30 min), followed by staining with anti-BrdU and propidium iodide for flow cytometric analysis.
TAZ promotes epithelial-mesenchymal transition.
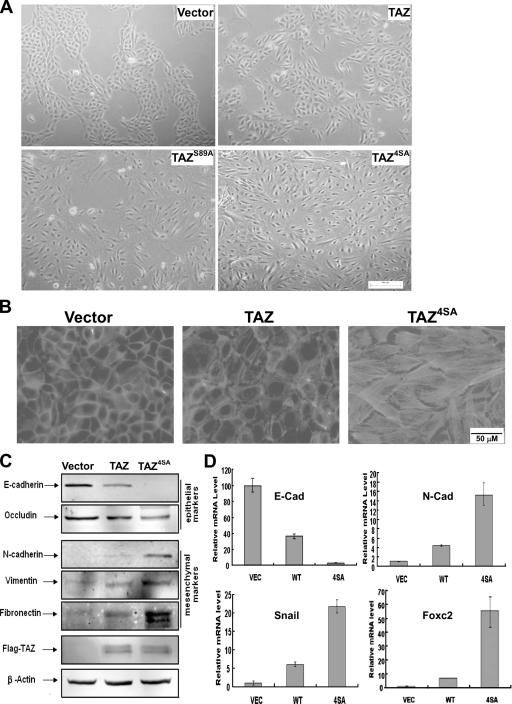
TAZ has been reported previously to modulate mesenchymal stem cell differentiation (9). We noticed that the TAZ-expressing cells, especially the constitutively active TAZ4SA-expressing cells, showed a dramatic alteration in cell morphology from that of the vector control cells (Fig. 5A). MCF10A vector control cells grew on monolayer cultures and displayed epithelium-type glands. TAZ-transduced MCF10A cells, on the other hand, showed an evident loss of epithelium-type characteristics. Both TAZ4SA-transduced and TAZS89A-transduced MCF10A cells displayed even stronger alterations in the morphology of spindle shape (Fig. 5A). The morphological alterations were also confirmed by phalloidin staining for F-actin, which is another hallmark of EMT, indicating disorganization of adherens junctions (22). The TAZ4SA-expressing cells displayed a dramatic increase in stress fibers (Fig. 5B).
FIG. 5.
TAZ induces the EMT of MCF10A cells. (A) TAZ induces a morphology change in MCF10A cells. Phase-contrast images of MCF10A cells expressing vector, TAZ, TAZS89A, and TAZ4SA are shown. WT, wild type. (B) TAZ alters the actin organization. Cells were stained with rhodamine-conjugated phalloidin. (C) TAZ causes EMT in MCF10A cells. Cell lysates from MCF10A cells expressing vector, TAZ, and TAZ4SA were separated and probed with antibodies for epithelial markers and mesenchymal markers as indicated. (D) TAZ decreases E-cadherin and increases N-cadherin expression through the activation of EMT transcription. Total RNA was extracted from MCF10A-vector, -TAZ, and -TAZ4SA cells, and quantitative PCR was performed to determine E-cadherin and N-cadherin expression (upper panel) and Foxc2 and Snail expression (lower panel). All data are normalized to GAPDH.
To test whether TAZ expression caused the EMT of MCF10A cells, Western blotting was performed to examine the expression of several well-characterized EMT markers. Epithelial markers such as E-cadherin and occludin were down-regulated in MCF10A/TAZ cells, and this downregulation was more dramatic with TAZ4SA cells (Fig. 5C). Furthermore, the mesenchymal markers N-cadherin, vimentin, and fibronectin were up-regulated in TAZ-overexpressing MCF10A cells and to a higher level in TAZ4SA cells (Fig. 5C). Together, these data indicate that TAZ activation induces EMT in MCF10A cells.
To gain insight into the molecular mechanism underlying TAZ-induced EMT, we next examined the mRNA levels of E-cadherin and N-cadherin, markers for epithelial and mesenchymal cells, respectively. Quantitative reverse transcription (RT)-PCR confirmed the Western blotting results indicating that the expression of TAZ decreased E-cadherin expression while increasing N-cadherin expression (Fig. 5D). We further examined the mRNA levels of two transcription factors, Foxc2 and Snail, which have been shown to participate in EMT control. We found that the expression of TAZ, and to a stronger degree, that of TAZ4SA, increased the levels of both Foxc2 and Snail mRNA (Fig. 5D), indicating that TAZ regulates the transcription program for EMT.
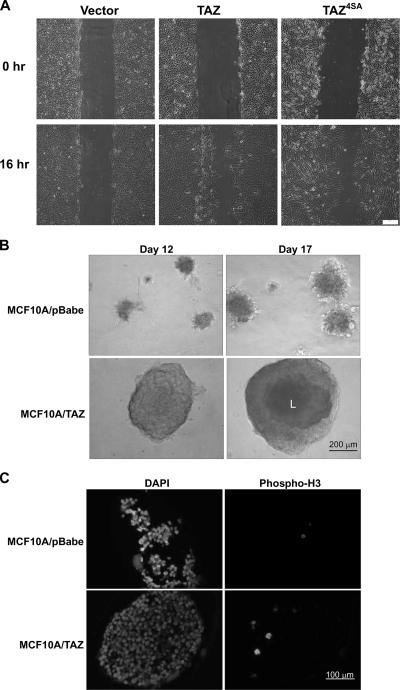
We then performed a wound-healing experiment to examine the activity of TAZ and TAZ4SA mutants in promoting cell migration and invasion. The motility of TAZ- and TAZ4SA-expressing MCF10A cells was dramatically increased compared with that of the parental MCF10A cells which exhibited minimal, if any, wound closure (Fig. 6A). Finally, to further examine the activity of TAZ in promoting cell migration and invasion, we performed a 3D Matrigel assay. An equal number of cells (5,000) were plated onto Matrigel, and acinar formation was followed at different times after cells were cultured and examined microscopically. MCF10A/TAZ cells formed acini as early as 3 to 4 days, while visible acini were seen in MCF10A control or MCF10A/pBabe cells only after 6 or 7 days (data not shown). At day 12, MCF10A/TAZ acini were much larger than those of the the control (Fig. 6B). At this stage, no acinus in either the MCF10A or the MCF10A/pBabe plate was larger than 200 μm in diameter, whereas more than five acini in each chamber of the MCF10A/TAZ plate were bigger than 300 μm in diameter (Fig. 6B). Lumen formation started to be seen in MCF10A/TAZ acini under phase-contrast microscopy (Fig. 6, dark-colored centers are noted in the MCF10A/TAZ acini). When acini were cultured further, at day 17, TAZ acini were bigger than 400 μM and lumen were observed clearly. Consistent with the previous result indicating that increased TAZ activity promoted cell proliferation, the mitotic index, as determined by the immunostaining of anti-phospho-histone H3 (PH3), was low in the MCF10A/pBabe control acini but was increased in the MCF10A/TAZ acini (day 17) (Fig. 6C). Collectively, these data show that TAZ promotes the EMT of MCF10A cells, possibly by regulating the expression of key transcription factors involved in EMT.
FIG. 6.
TAZ enhances cell migration and acinar growth in MCF10A cells. (A) TAZ overexpression promotes cell migration. MCF10A cells expressing vector, TAZ wild type (WT), and TAZ4SA (4SA) were analyzed for migration by a wound-healing assay. (B) TAZ stimulates acinar growth. MCF10A and derivative cells were cultured in the Matrigel. The acini formed at day 12 and 17 are shown. Note the large acini with lumen (L) formed by MCF-10A/TAZ. (C) TAZ promotes MCF10A proliferation in Matrigel culture. MCF10A and derivative cells were cultured in Matrigel and acini formed at day 17, fixed, paraffin embedded, and immunostained with antibody to PH3.
DISCUSSION
The TAZ transcription coactivator has been implicated in cell differentiation and organ development (10). The physiological regulation of the TAZ protein is not clear. Our study demonstrates that TAZ is regulated by the Hippo pathway, which is a novel tumor suppressor pathway initially defined by Drosophila genetic studies. Our data also indicate a role for TAZ in cell proliferation and EMT.
During the course of this study, it was reported that YAP is phosphorylated and inhibited by the Hippo pathway (6). TAZ shares approximately 50% sequence identity with YAP. We have determined that serine 89 in the HXRXXS motif is the major Lats2 phosphorylation site in TAZ, while serine 311 is also a functionally important site. Interestingly, the major Lats-dependent phosphorylation site in TAZ (Ser89) identified in this study corresponds to the major Lats-dependent phosphorylation site in YAP (Ser127), reported by Dong et al. (6). We also have supporting data indicating that YAP Ser127 is the major Lats phosphorylation site.
Several lines of evidence support the phosphorylation of TAZ by Lats2. First, TAZ contains four conserved HXRXXS motifs, which likely represent the Lats recognition consensus. Second, the coexpression of Lats2 induces a mobility shift of TAZ, and this mobility shift can be completely reversed by lambda phosphatase treatment. Third, Lats2 coexpression increases the phosphorylation of TAZ Ser89, as indicated by Western blotting with the phosphoYAP(Ser127) antibody. Fourth, Lats2 directly phosphorylates TAZ in an in vitro kinase assay. Fifth, a knockdown of endogenous Lats significantly decreases TAZ Ser89 phosphorylation. Sixth, Lats2 promotes cytoplasmic localization of the wild-type TAZ but not the S89A mutant. Furthermore, the mutation of Ser89 or Ser311 resulted in a TAZ mutant that is partially resistant to inhibition by Lats2.
It has been reported that Akt/PKB was identified as a kinase that phosphorylated YAP at its 14-3-3 binding site and inhibited its transactivation function (1). However, the reported YAP inhibition by Akt-dependent phosphorylation was inconsistent with the findings in our studies and those of Dong et al. that Lats2 was responsible for YAP2 Ser127 phosphorylation but not that of AKT (6, 38). This raises the question of whether Akt also phosphorylates TAZ at its 14-3-3 binding site (Ser89). Interestingly, Hong et al. have also reported that Akt does not phosphorylate TAZ at its 14-3-3 binding site (Ser89) (10). Therefore, we propose that TAZ is phosphorylated and inhibited by Lats but not by AKT. Matallanas et al. have recently reported that RASSF1A alleviated YAP1 cytoplasmic retention, thereby culminating in p73-mediated apoptosis, indicating that Yap1 might play a critical role in tumor suppression (18). These observations are inconsistent with the prevailing model in which YAP functions as an oncogene and is inhibited by the Hippo tumor suppressor pathway. Future studies are required to fully understand the cellular function of YAP and TAZ.
TAZ shares approximately 50% sequence identity with YAP1 and YAP2, which are alternative splicing products of the YAP gene. Both YAP and TAZ are phosphorylated and inhibited by the Lats kinase. YAP controls organ size and has been implicated in cancer development. Consistently we found that TAZ also promotes cell growth. Furthermore, Yap has been reported to induce EMT, while our study also indicates a role for TAZ in promoting EMT. Therefore, YAP and TAZ likely will have overlapping and distinct functions.
TAZ knockout mice have been independently reported by two groups (9, 33). Hossain et al. showed that Wwtr1-null mice have only minor skeletal defects but develop renal cysts (11), and Makita et al. reported that TAZ inactivation in mice results in pathological changes in the kidney and lung, resembling polycystic kidney disease and pulmonary emphysema, respectively (17). Whether the TAZ knockout animals are more resistant to tumor development has not been investigated.
We propose a model of TAZ regulation by the Hippo pathway via the following sequence. Lats phosphorylates TAZ on Ser89. This phosphorylation creates a 14-3-3 binding site. The phosphorylated TAZ then binds to 14-3-3 and is retained in cytoplasm, resulting in the depletion of TAZ from the nucleus. Therefore, TAZ fails to bind nuclear transcription factors and cannot activate target genes when it is phosphorylated by Lats. This model suggests that TAZ inactivation is an important downstream signaling output of the Hippo pathway.
Our study indicates a function for TAZ in cell proliferation and EMT, which are both important steps for cancer development. The expression of TAZ in MCF10A cells significantly increases the growth rate of the cells. Furthermore, the TAZ-expressing cells continue to proliferate even when cells reach confluence. This finding is supported by a high fraction of TAZ-expressing cells that continue to synthesize DNA in a confluent culture. Our study suggests the growth-stimulating activity of TAZ.
Another interesting finding from this study is that TAZ promotes EMT. The expression of TAZ, especially the phosphorylation-defective S89A and 4SA mutants, alters the morphology of MCF10A cells. Moreover, the TAZ-expressing cells showed a dramatic decrease in epithelial markers with a concomitant increase in mesenchymal markers. Therefore, TAZ may play a role in cancer development by promoting proliferation and EMT. This notion is consistent with the fact that TAZ is inhibited by the Hippo tumor suppressor pathway. More interestingly, the expression of the TAZ phosphorylation-defective 4SA mutant not only significantly promotes cell proliferation but also dramatically alters the morphology of MCF10A cells, indicating that the functional role of TAZ in cancer development may result from the dysregulation of the Hippo pathway. Dysregulation of the upstream region of the Hippo pathway such as NF2, Mst/Sav, and Lats/Mob would lead to the inhibition of the Hippo pathway and subsequent activation of TAZ by the loss of inhibitory phosphorylation. Mer is a tumor suppressor, and Sav and Mob mutation have also been reported in tumor cell lines (14, 31). Therefore, the mutation or dysregulation of the components of the Hippo pathway may result in promoting cell proliferation and inducing EMT via activation of TAZ.
Studies of Drosophila have shown that the Hippo pathway inhibits cell proliferation and induces apoptosis (12, 26). Dysregulation of the Hippo pathway results in organ and tissue hypertrophy (7, 23, 26). Consistently, mutations of the tumor suppressor NF2 gene coding for Mer provide direct evidence supporting the tumor suppressor function of this pathway in humans (19, 25, 35). Our discovery that TAZ promotes proliferation and EMT provides a molecular basis for and additional support to the function of the Hippo pathway in tumor suppression. We speculate that the phosphorylation and inactivation of TAZ and YAP may represent the major downstream effects of the Hippo pathway in tumor suppression. TAZ and YAP may be the key physiological substrates of Lats, which is also a candidate of a tumor suppressor gene. In summary, our study has connected TAZ to the Hippo pathway and suggests a potential function for TAZ in tumorigenesis.
Acknowledgments
We thank the members of the Fudan MCB laboratory for discussions throughout this study and the Institutes of Biomedical Sciences for their support.
This work was supported by the 985 Program from the Chinese Ministry of Education and the Shanghai Leading Academic Discipline Project, project number B110.
Footnotes
Published ahead of print on 28 January 2008.
REFERENCES
- 1.Basu, S., N. F. Totty, M. S. Irwin, M. Sudol, and J. Downward. 2003. Akt phosphorylates the Yes-associated protein, YAP, to induce interaction with 14-3-3 and attenuation of p73-mediated apoptosis. Mol. Cell 1111-23. [DOI] [PubMed] [Google Scholar]
- 2.Cho, E., Y. Feng, C. Rauskolb, S. Maitra, R. Fehon, and K. D. Irvine. 2006. Delineation of a Fat tumor suppressor pathway. Nat. Genet. 381142-1150. [DOI] [PubMed] [Google Scholar]
- 3.Cui, C. B., L. F. Cooper, X. Yang, G. Karsenty, and I. Aukhil. 2003. Transcriptional coactivation of bone-specific transcription factor Cbfa1 by TAZ. Mol. Cell. Biol. 231004-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dan, I., N. M. Watanabe, and A. Kusumi. 2001. The Ste20 group kinases as regulators of MAP kinase cascades. Trends Cell Biol. 11220-230. [DOI] [PubMed] [Google Scholar]
- 5.Debnath, J., S. K. Muthuswamy, and J. S. Brugge. 2003. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods 30256-268. [DOI] [PubMed] [Google Scholar]
- 6.Dong, J., G. Feldmann, J. Huang, S. Wu, N. Zhang, S. A. Comerford, M. F. Gayyed, R. A. Anders, A. Maitra, and D. Pan. 2007. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell 1301120-1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Edgar, B. A. 2006. From cell structure to transcription: Hippo forges a new path. Cell 124267-273. [DOI] [PubMed] [Google Scholar]
- 8.Hamaratoglu, F., M. Willecke, M. Kango-Singh, R. Nolo, E. Hyun, C. Tao, H. Jafar-Nejad, and G. Halder. 2006. The tumour-suppressor genes NF2/Merlin and Expanded act through Hippo signalling to regulate cell proliferation and apoptosis. Nat. Cell Biol. 827-36. [DOI] [PubMed] [Google Scholar]
- 9.Hong, J. H., E. S. Hwang, M. T. McManus, A. Amsterdam, Y. Tian, R. Kalmukova, E. Mueller, T. Benjamin, B. M. Spiegelman, P. A. Sharp, N. Hopkins, and M. B. Yaffe. 2005. TAZ, a transcriptional modulator of mesenchymal stem cell differentiation. Science 3091074-1078. [DOI] [PubMed] [Google Scholar]
- 10.Hong, J. H., and M. B. Yaffe. 2006. TAZ: a beta-catenin-like molecule that regulates mesenchymal stem cell differentiation. Cell Cycle 5176-179. [DOI] [PubMed] [Google Scholar]
- 11.Hossain, Z., S. M. Ali, H. L. Ko, J. Xu, C. P. Ng, K. Guo, Z. Qi, S. Ponniah, W. Hong, and W. Hunziker. 2007. Glomerulocystic kidney disease in mice with a targeted inactivation of Wwtr1. Proc. Natl. Acad. Sci. USA 1041631-1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang, J., S. Wu, J. Barrera, K. Matthews, and D. Pan. 2005. The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila homolog of YAP. Cell 122421-434. [DOI] [PubMed] [Google Scholar]
- 13.Kanai, F., P. A. Marignani, D. Sarbassova, R. Yagi, R. A. Hall, M. Donowitz, A. Hisaminato, T. Fujiwara, Y. Ito, L. C. Cantley, and M. B. Yaffe. 2000. TAZ: a novel transcriptional co-activator regulated by interactions with 14-3-3 and PDZ domain proteins. EMBO J. 196778-6791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lai, Z. C., X. Wei, T. Shimizu, E. Ramos, M. Rohrbaugh, N. Nikolaidis, L. L. Ho, and Y. Li. 2005. Control of cell proliferation and apoptosis by mob as tumor suppressor, mats. Cell 120675-685. [DOI] [PubMed] [Google Scholar]
- 15.Mah, A. S., A. E. Elia, G. Devgan, J. Ptacek, M. Schutkowski, M. Snyder, M. B. Yaffe, and R. J. Deshaies. 2005. Substrate specificity analysis of protein kinase complex Dbf2-Mob1 by peptide library and proteome array screening. BMC Biochem. 622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mahoney, W. M., Jr., J. H. Hong, M. B. Yaffe, and I. K. Farrance. 2005. The transcriptional co-activator TAZ interacts differentially with transcriptional enhancer factor-1 (TEF-1) family members. Biochem. J. 388217-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Makita, R., Y. Uchijima, K. Nishiyama, T. Amano, Q. Chen, T. Takeuchi, A. Mitani, T. Nagase, Y. Yatomi, H. Aburatani, O. Nakagawa, E. V. Small, P. Cobo-Stark, P. Igarashi, M. Murakami, J. Tominaga, T. Sato, T. Asano, Y. Kurihara, and H. Kurihara. 2008. Multiple renal cysts, urinary concentration defects, and pulmonary emphysematous changes in mice lacking TAZ. Am. J. Physiol. Renal Physiol. [Epub ahead of print] doi: 10.1152/ajprenal.00201.2007. [DOI] [PubMed]
- 18.Matallanas, D., D. Romano, K. Yee, K. Meissl, L. Kucerova, D. Piazzolla, M. Baccarini, J. K. Vass, W. Kolch, and E. O'Neill. 2007. RASSF1A elicits apoptosis through an MST2 pathway directing proapoptotic transcription by the p73 tumor suppressor protein. Mol. Cell 27962-975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McCartney, B. M., M. Kulikauskas, R. D. LaJeunesse, and R. G. Fehon. 2000. The neurofibromatosis-2 homologue, Merlin, and the tumor suppressor expanded function together in Drosophila to regulate cell proliferation and differentiation. Development 1271315-1324. [DOI] [PubMed] [Google Scholar]
- 20.Murakami, M., M. Nakagawa, E. N. Olson, and O. Nakagawa. 2005. A WW domain protein TAZ is a critical coactivator for TBX5, a transcription factor implicated in Holt-Oram syndrome. Proc. Natl. Acad. Sci. USA 10218034-18039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Murakami, M., J. Tominaga, R. Makita, Y. Uchijima, Y. Kurihara, O. Nakagawa, T. Asano, and H. Kurihara. 2006. Transcriptional activity of Pax3 is co-activated by TAZ. Biochem. Biophys. Res. Commun. 339533-539. [DOI] [PubMed] [Google Scholar]
- 22.Overholtzer, M., J. Zhang, G. A. Smolen, B. Muir, W. Li, D. C. Sgroi, C. X. Deng, J. S. Brugge, and D. A. Haber. 2006. Transforming properties of YAP, a candidate oncogene on the chromosome 11q22 amplicon. Proc. Natl. Acad. Sci. USA 10312405-12410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pan, D. 2007. Hippo signaling in organ size control. Genes Dev. 21886-897. [DOI] [PubMed] [Google Scholar]
- 24.Park, K. S., J. A. Whitsett, T. Di Palma, J. H. Hong, M. B. Yaffe, and M. Zannini. 2004. TAZ interacts with TTF-1 and regulates expression of surfactant protein-C. J. Biol. Chem. 27917384-17390. [DOI] [PubMed] [Google Scholar]
- 25.Ruttledge, M. H., J. Sarrazin, S. Rangaratnam, C. M. Phelan, E. Twist, P. Merel, O. Delattre, G. Thomas, M. Nordenskjöld, V. P. Collins, J. P. Dumanski, and G. A. Rouleau. 1994. Evidence for the complete inactivation of the NF2 gene in the majority of sporadic meningiomas. Nat. Genet. 6180-184. [DOI] [PubMed] [Google Scholar]
- 26.Saucedo, L. J., and B. A. Edgar. 2007. Filling out the Hippo pathway. Nat. Rev. Mol. Cell Biol. 8613-621. [DOI] [PubMed] [Google Scholar]
- 27.Silva, E., Y. Tsatskis, L. Gardano, N. Tapon, and H. McNeill. 2006. The tumor-suppressor gene fat controls tissue growth upstream of expanded in the hippo signaling pathway. Curr. Biol. 162081-2089. [DOI] [PubMed] [Google Scholar]
- 28.St John, M. A., W. Tao, X. Fei, R. Fukumoto, M. L. Carcangiu, D. G. Brownstein, A. F. Parlow, J. McGrath, and T. Xu. 1999. Mice deficient of Lats1 develop soft-tissue sarcomas, ovarian tumours and pituitary dysfunction. Nat. Genet. 21182-186. [DOI] [PubMed] [Google Scholar]
- 29.Takahashi, Y., Y. Miyoshi, C. Takahata, N. Irahara, T. Taguchi, Y. Tamaki, and S. Noguchi. 2005. Down-regulation of LATS1 and LATS2 mRNA expression by promoter hypermethylation and its association with biologically aggressive phenotype in human breast cancers Clin. Cancer Res. 111380-1385. [DOI] [PubMed] [Google Scholar]
- 30.Tamaskovic, R., S. J. Bichsel, and A. Hemmings. 2003. NDR family of AGC kinases—essential regulators of the cell cycle and morphogenesis. FEBS Lett. 54673-80. [DOI] [PubMed] [Google Scholar]
- 31.Tapon, N., K. F. Harvey, D. W. Bell, D. C. Wahrer, T. A. Schiripo, D. A. Haber, and I. K. Hariharan. 2002. Salvador promotes both cell cycle exit and apoptosis in Drosophila and is mutated in human cancer cell lines. Cell 110467-478. [DOI] [PubMed] [Google Scholar]
- 32.Thompson, B. J., and S. M. Cohen. 2006. The Hippo pathway regulates the bantam microRNA to control cell proliferation and apoptosis in Drosophila. Cell 126767-774. [DOI] [PubMed] [Google Scholar]
- 33.Tian, Y., R. Kolb, J.-H. Hong, J. Carroll, D. Li, J. You, R. Bronson, M. B. Yaffe, J. Zhou, and T. Benjamin. 2007. TAZ promotes PC2 degradation through a SCFβ-Trcp E3 ligase complex. Mol. Cell Biol. 276383-6395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tian, Y., D. Li, J. Dahl, J. You, and T. Benjamin. 2004. Identification of TAZ as a binding partner of the polyomavirus T antigens. J. Virol. 7812657-12664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Trofatter, J. A., M. M. Rutter, J. MacCollin, J. L. Murrell, R. Duyao, M. P. M. D. Parry, R. Eldridge, N. Kley, A. G. Menon, K. Pulaski, V. H. Haase, C. M. Ambrose, D. Munroe, C. Bove, J. L. Haines, R. L. Martuza, M. E. MacDonald, B. R. Seizinger, M. P. Short, A. J. Buckler, and J. F. Gusella. 1993. A novel moesin-, ezrin-, radixin-like gene is a candidate for the neurofibromatosis 2 tumor suppressor. Cell 72791-800. [DOI] [PubMed] [Google Scholar]
- 36.Wei, X., T. Shimizu, and Z. C. Lai. 2007. Mob as tumor suppressor is activated by Hippo kinase for growth inhibition in Drosophila. EMBO J. 261772-1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Willecke, M., F. Hamaratoglu, M. Kango-Singh, R. Udan, C. L. Chen, C. Tao, X. Zhang, and G. Halder. 2006. The fat cadherin acts through the hippo tumor-suppressor pathway to regulate tissue size. Curr. Biol. 162090-2100. [DOI] [PubMed] [Google Scholar]
- 38.Zhao, B., X. Wei, W. Li, R. S. Udan, Q. Yang, J. Kim, J. Xie, T. Ikenoue, J. Yu, L. Li, P. Zheng, K. Ye, A. Chinnaiyan, G. Halder, Z. C. Lai, and K. L. Guan. 2007. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 212747-2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao, X., L. Gan, H. Pan, D. Kan, M. Majeski, S. A. Adam, and T. G. Unterman. 2004. Multiple elements regulate nuclear/cytoplasmic shuttling of FOXO1: characterization of phosphorylation- and 14-3-3-dependent and -independent mechanisms. Biochem. J. 378839-849. [DOI] [PMC free article] [PubMed] [Google Scholar]