Abstract
The transcription repressor BCL6 plays an essential role in the formation and function of germinal centers (GCs). While normal B cells promptly shut off BCL6 when they exit the GC, many GC-derived B-cell lymphomas sustain BCL6 expression through chromosomal translocations and activating mutations. We have previously shown that a common effect of lymphoma-associated BCL6 gene alterations is to bypass a negative autoregulatory loop that controls its transcription. In this study, we report that BCL6 autoregulation is independent of several known corepressor complexes including silencing mediator for retinoid and thyroid hormone receptors, nuclear receptor coreceptor, BCL6 corepressor, and MTA3/NuRD. Furthermore, we show that BCL6 can interact with the CtBP (C-terminal binding protein) corepressor both in vitro and in vivo and that CtBP is recruited by BCL6 to its 5′ regulatory region. In lymphoma cell lines carrying BCL6 translocations, small interfering RNA-mediated CtBP knock-down selectively relieved the previously silenced wild-type BCL6 allele but not the translocated alleles, which are driven by heterologous promoters. These results demonstrate that CtBP is a novel BCL6 corepressor and suggest that a unique corepressor requirement for BCL6 autoregulation may allow GC B cells to differentially control the expression of BCL6 and other BCL6 target genes in response to environmental stimuli during the GC stage of B cell development.
BCL6 is a sequence-specific transcription repressor that is required for the formation of germinal centers (GC), and its deregulated expression underlies development of many GC-derived B-cell lymphomas (12, 44, 47). Expression of BCL6 is developmentally regulated such that in the B-cell lineage, high levels of BCL6 are restricted to the GC stage. GC are dynamic and specialized structures in the secondary lymphoid organs within which B cells undergo immunoglobulin class switch recombination and somatic hypermutation to produce diverse, high-affinity antibodies (17, 26). Widely considered to be the master regulator of the GC stage of B-cell development, BCL6 maintains the GC-specific gene expression program by silencing genes involved in B-cell activation (CD69, CD80, and NF-κB1), response to DNA damage (TP53 and ATR), cell-cycle regulation (CCND2, CDKN1B, and CDKN1A), and plasma cell differentiation (PRDM1) (25, 29, 34, 37, 38, 40, 41). Thus, neither the memory nor the plasma cell differentiation program can be initiated until expression and activity of BCL6 are extinguished by GC exit signals.
BCL6 is a 95-kDa phosphoprotein with six Krüppel-type zinc fingers (ZF) at the C terminus and an N-terminal POZ/BTB domain. Our earlier work demonstrated that the maximum repression activity of BCL6 requires the entire POZ/BTB domain as well as a separate middle region, repression domain II (RDII) (7). To date, a variety of BCL6 corepressors have been described. Among the corepressors with well-documented in vivo functions, nuclear receptor coreceptor (NCoR), silencing mediator for retinoid and thyroid hormone receptors (SMRT), and BCL6 corepressor (BCoR) are recruited through the POZ/BTB domain while the central RDII region can interact with histone deacetylase 2 (HDAC2) and MTA3, which is a cell-type-specific component of the Mi-2/NuRD complex (8, 13-15, 19, 20, 24, 27, 49). Structural studies revealed that SMRT, NCoR, and BCoR all bind to a lateral groove in the POZ domain through a stretch of 17 amino acids (1). Using a cell-permeable BCL6 peptide inhibitor, BPI, designed to block POZ lateral groove corepressor recruitment, we have shown that SMRT/NCoR/BCoR control cell proliferation and survival in GC-derived B cells by mediating BCL6-dependent repression of genes such as ATR, TP53, and CDKN1A (32, 35, 37). The RDII corepressors, HDAC2 and MTA3/NuRD, bind to BCL6 in an acetylation-sensitive manner and are dissociated from BCL6 when the KKYK motif within RDII is acetylated (4, 15). MTA3/NuRD-dependent BCL6 target genes include PRDM1, and MTA3 is indispensable for the ability of BCL6 to inhibit plasma cell differentiation (15, 32). Unlike the lateral groove corepressors, MTA3 does not contribute to BCL6-mediated repression of ATR, TP53, and CDKN1A, and MTA3 depletion induces differentiation but not cell death in GC-derived B cell lines (32). These findings allude to functional separation of BCL6 repression mechanisms, particularly, differential utilization of corepressor complexes for target genes involved in different biological processes.
Aside from its important roles in the normal immune system, BCL6 is also the most frequently altered proto-oncogene in non-Hodgkin's lymphomas, the majority of which derive from normal GC B cells (22). The most common form of non-Hodgkin's B cell lymphomas is called diffuse large B cell lymphoma (DLBCL). In nearly half of DLBCLs, BCL6 expression is deregulated by chromosomal translocations and “activating” point mutations that target the 5′ regulatory region of this gene (33, 42, 46). We along with others have previously demonstrated that the BCL6 protein uses clustered BCL6 binding sites in the noncoding exon 1 to repress its own transcription and that both chromosomal translocations and activating mutations allow lymphoma cells to bypass this negative autoregulation mechanism (33, 42). A causative role for BCL6 in the pathogenesis of DLBCL has been subsequently demonstrated using mouse models that recapitulate the translocated BCL6 gene found in human DLBCL patients (3, 6). Despite its apparent functional importance, the mechanistic details including corepressor requirement for BCL6 autoregulation are not known.
In this study, we identify CtBP as a novel corepressor for BCL6. The CtBP family of proteins consists of evolutionarily conserved transcriptional corepressors (9). An increasing number of POZ- and ZF-containing transcription factors have been reported to use CtBP as their corepressors, including the human HIC1 and Drosophila Tramtrack 69 (Ttk69) proteins (11, 43). We show that BCL6 can directly bind to human CtBP1 and recruit it to a number of BCL6 transcriptional targets including the promoter region of BCL6. Our data indicate that neither the lateral groove corepressors nor MTA3/NuRD contributes to BCL6 negative autoregulation. In contrast, small interfering RNA (siRNA)-mediated knock-down experiments revealed that CtBP1 alone among known BCL6 corepressors is required for BCL6 autoregulation and that CtBP1 can also contribute to repression of several other BCL6 targets. Thus, our findings also provide a striking example of promoter-specific corepressor usage by BCL6.
MATERIALS AND METHODS
Cell culture.
293T cells were maintained in Dulbecco's modified Eagle medium supplemented with 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin (Gibco). The pre-B human leukemia cell line 697 was maintained in RPMI 1640 medium plus 10% FBS and 1% penicillin-streptomycin. All other human B cell lines were maintained in Iscove's modified Dulbecco's medium plus 10% FBS and 1% penicillin-streptomycin.
Plasmids.
The luciferase reporters are pLA/s5 for BCL6 (provided by Riccardo Dalla-Favera), HBM-Luc for c-Myc (provided by Linda Penn), pGL3-hBlimp-1 for PRDM1 (provided by Alexander Dent), and pGL2-hMCP1(−2910) for MCP1 (provided by Anthony J. Valente). To construct the pGL3-hCyclinD2 reporter, a 1.4-kb region of CCND2 was PCR amplified from human placental DNA using the forward primer 5′-CTAGCTAGCGGTCTCTCCCCTTCCTCCT-3′ and reverse primer 5′-GGAAGATCTGGTCCTCCCCTTAAAACTGG-3′ and cloned into the NheI and BglII sites of the pGL3 vector after restriction digestion. The synthetic BCL6 reporter, B6BS-tk-Luc (where tk is thymidine kinase and Luc is luciferase), and the expression plasmids pMT2T-HA-BCL6, pMT2T-HA-ΔPOZ, pMT2T-HA-ΔZF, pMT2T-HA-ZF, pMT2T-HA-ΔPEST, GST-BCL6, CMV-SMRT, and pBCL6FL_dbQC (where HA is hemagglutinin, GST is glutathione S-transferase, and CMV is cytomegalovirus) were previously described (7, 30). The other BCL6 deletion constructs used in Fig. 4 were generated by PCR-based approaches and verified by sequencing. pME18S_FlagHDAC2 and pMT2T-HA-QQYQ were also gifts from Riccardo Dalla-Favera. pCMV5-Flag-CtBP was generated from pCMV5-Flag-CtBP(K428R), a gift from David Wotton, using a QuikChange Site-Directed Mutagenesis Kit (Stratagene). Both pCS2(6X)MycCtBP1 and CtBP2 were provided by Douglas Dean. pMT2T-HA-BCL6cmpd and pcDNA3-Flag-BCL6Cmpd were generated from pBCL6FL_dbQC and pMT2T-HA-QQYQ using restriction-based subcloning.
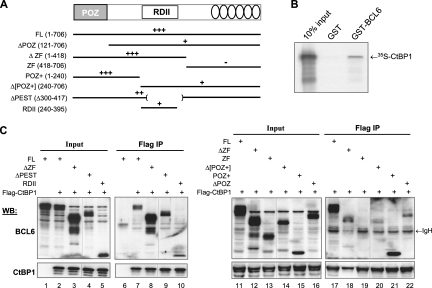
FIG. 4.
Interaction with CtBP requires both the POZ and the RDII domains of BCL6. (A) Schematic representation of BCL6 domain structure and the BCL6 deletion mutants used in the mapping studies. Relative strength of the interaction is depicted above the line structures: +++, strong, nearly wt level of binding; ++, moderate binding; +, weak binding; −, no binding. (B) Interaction between BCL6 and CtBP was studied in GST pull-down assays where immobilized GST or GST-BCL6 proteins were incubated with 35S-labeled CtBP1. (C) Co-IP assays were performed in 293T cells that were transiently transfected to express Flag-CtBP1 together with the indicated BCL6 deletion mutants. Full-length BCL6 was also expressed alone to serve as negative control for Co-IP (lanes 1 and 6). Whole-cell lysates were immunoprecipitated with anti-Flag beads and analyzed by Western blotting (WB) using either polyclonal anti-BCL6 (sc-858 plus sc-368) or CtBP antibodies. IgH, cross-reactive signals from the Ig heavy chain of the IP antibody. They are more pronounced in the right panel. Within each panel, vertical lines have been inserted to indicate a repositioned gel lane. FL, full-length BCL6.
BMM and retroviral infection.
Methods for in vitro differentiation of bone marrow-derived macrophages (BMM) and retrovirus-mediated transfection were described previously (48). In brief, the mouse stem cell virus-based retroviruses were packaged in 293T cells, and the resulting supernatants were used to infect day 6 BMM. Two days after infection, green fluorescent protein-positive (GFP+) cells were sorted by FACSVantage SE (BD Biosciences) and lysed in Trizol (Invitrogen) for total RNA preparation.
Transient transfection-mediated reporter assays.
Transient transfections were performed using Superfect reagent (Qiagen) for 293T and Mutu III cells according to the manufacturer's instructions with the following modification: 3 μl of Superfect reagent was used for each 12-well plate of 293T cells, and 8 μl of Superfect reagent was used for each 12-well plate of Mutu III cells. Two micrograms of luciferase reporter and 2 μg of a CMV-β-galactosidase plasmid were transfected with various amounts of effectors ranging from 0.02 to 0.1 pm/plate (see Fig. 2A) and 0.1 to 0.5 pm/plate (see Fig. 2B). All transfections were performed in duplicates and harvested 48 h later. Luciferase activities were measured using a luciferase assay system (Promega) and normalized by activities from the cotransfected control β-galactosidase plasmid.
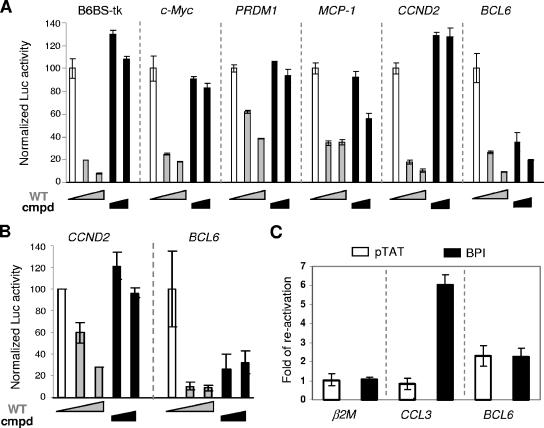
FIG. 2.
BCL6 autoregulation has a distinct corepressor requirement. (A) BCL6Cmpd potently repressed transcription from the BCL6 reporter but had limited activity on a number of other target gene reporters. Reporter assays were performed 293T cells with the indicated reporter constructs and increasing amounts of either wt BCL6 (wt) or BCL6Cmpd (Cmpd) expression plasmids. Activity of all reporters in the absence of BCL6 was set to 100. (B) Reporter assays were performed similar to those described in panel A but in the BCL6-negative Mutu III cells. (C) BPI treatment of SUDHL6 cells derepressed the expression of CCL3 but not BCL6. qRT-PCR was performed using primers specific for the three indicated genes. β2M, a gene not known to be regulated by BCL6, was examined as a specificity control. For each gene analyzed, the increase is relative to untreated cells. pTAT is a control peptide with only the carrier sequence. All graphs in this figure represent means and standard deviations of triplicate tests.
Treatment with BPI.
Cell-permeable peptides (BPI and its control pTAT) were synthesized by Bio-Synthesis, Inc., and stored at −20°C until reconstituted with sterile water immediately before use. Purity of the peptides was determined by high performance liquid chromatography-mass spectrometry to be 98% or higher. DLBCL cells were exposed to 4 μM BPI or pTAT for 8 h before being harvested for Trizol-based total RNA extraction.
RNA interference-mediated knockdown.
All the siRNA oligonucleotides were synthesized by Dharmacon. CtBP siRNA 1 is the Dharmacon SMART pool for human CtBP1 (catalog number M-008609-01). The target sequence for custom-designed CtBP siRNA 2 is GGAGGACCUGGAGAAGUUdTdG (50). Scramble II Duplex was used as a control. For each transfection, 8 million log phase growth cells were transfected with 10 μg of siRNA oligonucleotides using Amaxa Nucleofector solution T and program G16 (Amaxa Biosystems). The transfection efficiency, based on the use of an AlexaFluor 488-labeled control oligonucleotide, is typically between 90 to 95%.
Western blotting.
Whole-cell lysates were prepared using NP-40 buffer (50 mM Tris, pH 7.5, 100 mM NaCl, 0.25% Igepal CA630) or radioimmunoprecipitation assay (RIPA) buffer (4.55 mM Na2HPO4, 0.85 mM NaH2PO4, 50 mM Tris-HCl, pH 8, 150 mM NaCl, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate [SDS], 1% Igepal CA630) supplemented with protease inhibitor cocktail (Roche) and analyzed by Western blotting according to standard procedures. Primary antibodies used in this study include anti-BCL6 (sc-858 and sc-368; Santa Cruz), anti-CtBP (sc-11390; Santa Cruz), anti-CtBP1 (612042; BD Transduction Labs), anti-glyceraldehyde-3-phosphate dehydrogenase (anti-GAPDH) (sc-25778; Santa Cruz), and anti-Flag (F1804; Sigma).
RT-PCR and real-time qRT-PCR.
Total RNA samples were prepared using the Trizol reagent (Invitrogen). cDNA was synthesized using random primers and the Superscript II reverse transcriptase (Invitrogen). All PCRs were performed using Platinum Taq polymerase (Invitrogen) with serially diluted cDNA samples. Reverse transcription-PCR (RT-PCR) primers were the following: for mouse β-actin, 5′-TTGAGACCTTCAACACCCC-3′ (forward [F]) and 5′-CAGTAATCTCCTTCTGCATCC-3′ (reverse [R]); for mouse CCL3, 5′-AGATTCCACGCCAATTCAT-3′ (F) and 5′-GTGAACAACTGGGAGGGA-3′ (R). For quantitative RT-PCR (qRT-PCR), the total RNA was purified with an RNeasy Mini Kit (Qiagen). The levels of transcripts were detected by Sybr Green (Applied Biosystems) on an Opticon 2 thermal cycler (MJ Research). We normalized gene expression to hypoxanthine phosphoribosyltransferase and expressed values relative to control using the ΔΔCT (where CT is threshold cycle) method. The primers are the following: for wild-type (wt) BCL6 transcripts only, 5′-TCTAGGAAAGGCCGGACAC-3′ (F) and 5′-AATGCCTTGCTTCACAGTCC-3′ (R);for the BCL6 open reading frame, 5′-GACTCTGAAGAGCCACCTGC-3′ (F) and 5′-CTGGCTTTTGTGACGGAAAT-3′ (R); for CCL3, 5′-GGTCTCCACTGCTGCCCTTGC-3′ (F) and 5′-GGAATCTGCCGGGAGGTGTAGC-3′ (R); for CD69, 5′-AGCCCAAAATGCTTGTTCTG-3′ (F) and 5′-TTCCTCTCTACCTGCGTATCG-3′ (R); for CCND2 5′-CCGGACCTAATCCCTCACTC-3′ (F) and 5′-CACACCGATGCAGCTTTCTA-3′ (R); for TP53, 5′-CTTTGAGGTGCGTGTTTGTG-3′ (F) and 5′-TCTTGCGGAGATTCTCTTCC-3′ (R); for PRDM1, 5′-GTACACACGGGAGAAAAGCC-3′ (F) and 5′-TCTTGAGATTGCTGGTGCTG-3′ (R); for HPRT, 5′-AAAGGAACCCCACGAAGTGTT-3′ (F) and 5′-TCAAGGGCATATCCTACAACAA-3′ (R).
GST-pull down and Co-IP assays.
The procedure for GST-pull down was previously described (8). For coimmunoprecipitation (Co-IP), transfected 293T cells were lysed in NP-40 buffer supplemented with 0.6 mM N-ethylmalemide and protease inhibitors. Approximately 1 mg of lysate was diluted 1:10 with NP-40 buffer without N-ethylmalemide before it was precleared with protein G-agarose beads (Sigma) for 30 min at 4°C. The lysates were then incubated with 40 μl of anti-Flag M2 affinity gel for 3 h at 4°C. Immunocomplexes were recovered by brief centrifugation and washed three to four times with NP-40 buffer supplemented with protease inhibitors. Immunocomplexes were eluted from the beads by adding 100 μl of NP-40 buffer containing Flag peptide at 150 μg/ml and incubating at 4°C for 30 min. The recovered supernatants were boiled in 1× SDS loading buffer before being used for Western blotting. For endogenous Co-IP, 40 million Ly7 or Mutu III cells were lysed in 500 μl of cold RIPA buffer supplemented with protease inhibitor cocktail (Roche) and 2 mM fresh dithiothreitol, 5 mM sodium orthovanadate, 50 mM sodium fluoride, and 10 mM β-glycerophosphate. After a preclearing step as described above, the lysates were incubated with 2 μg of anti-BCL6 (sc-858; Santa Cruz), anti-STAT1 (G16930; Transduction Labs), or anti-BCL3 (sc-185; Santa Cruz) antibodies for 3 h at 4°C followed by the addition of 50 μl of protein A agarose beads (RepliGen) and another 1-h incubation at 4°C. The beads were collected by brief centrifugation and washed four times with cold RIPA buffer supplemented as above. Washed beads were boiled in 50 μl of 1× SDS loading buffer and used for Western blotting.
ChIP and ChIP-on-chip assays.
Chromatin IP (ChIP) was carried out as previously described using a ChIP assay kit (Upstate biotechnology) (42). A total of 20 million cells were used for each reaction; chromatin was sheared to an average length of 600 bp; 2 μg of the anti-BCL6 N3 antibody, anti-CtBP antibody, normal rabbit immunoglobulin G (all from Santa Cruz), or anti-MTA3 (a gift from Paul Wade) was used. DNA sample recovered from each ChIP reaction was used for PCR with the following primers to amplify BCL6 exon 1 region: BCL6-e1, 5′-GGGTTCTTAGAAGTGGTGATGC-3′; and BCL6-i2, 5′-TGGGACTAATCTTCGGCATT-3′. To trace the allelic source, PCR products were subjected to EcoNI digestion prior to loading on 1.5% agarose gels. Gel images were analyzed by the ImageQuant software. For ChIP-on-chip analysis, BCL6, CtBP, or β-actin, ChIP products and their respective input genomic fragments were amplified by ligation-mediated PCR (31). Real-time PCR was performed again at this stage for selected positive control loci to verify that the enrichment ratios were retained. The genomic products of two biological ChIP replicates were labeled with Cy5 (for ChIP products) and Cy3 (for input) and cohybridized on custom-designed genomic tiling arrays generated by NimbleGen Systems. These high-density tiling arrays contain 50-residue oligonucleotides with an average overlap of 25 bases, omitting repetitive elements. Included in the arrays were genomic regions containing BCL6 (chr3: 188921082 to 188954417; size, 33,336 bp), TP53 (chr17: 7510832 to 7546927; size, 36,096 bp), CD69 (chr12: 9792142 to 9813738; size, 21,597 bp), CCL3 (chr17: 31438320 to 31445602; size, 7,283 bp), CCND2 (chr12: 4241461 to 4290594; size, 49,134 bp), PRDM1 (chr6: 106632981 to 106670412; size, 37,432 bp), GAPDH (chr12: 6513004 to 6518751; size, 5,748 bp) according to the human genome May 2004 assembly. After hybridization, the relative enrichment for each probe was calculated as the signal ratio of ChIP to input. Peaks of enrichment for each antibody relative to input were captured with a five-probe sliding window, and the results were uploaded as custom tracks into the University of California Santa Cruz genome browser and graphically represented as histograms. The cutoff threshold, represented as opaque horizontal masking windows in Fig. 6B, is defined as 2.5 times the standard deviation above the average relative enrichment on the entire array. Peaks involving five or more oligonucleotide probes above this threshold were considered positive hits.
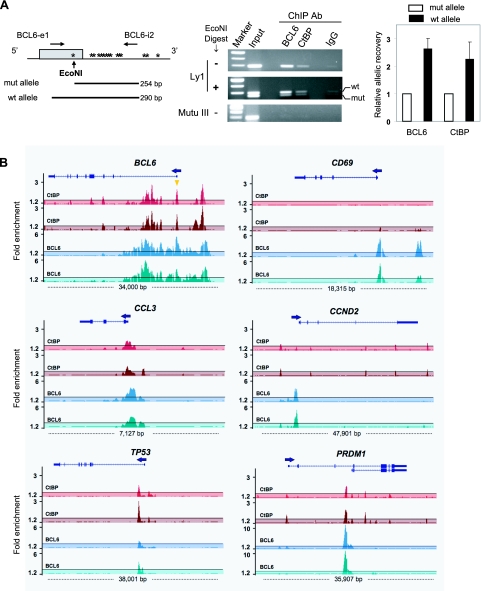
FIG. 6.
Recruitment of CtBP to the BCL6 exon 1 region and other BCL6 target loci. (A) CtBP is preferentially targeted to the wt BCL6 promoter in Ly1 cells. (Left) Shown is a schematic of the BCL6 exon 1 region in Ly1 cells which carry an “activating mutation” in the mutant allele (mut) that generates an EcoNI site. Other mutations (asterisks) are also present but do not affect BCL6 transcription (33). The mutant and wt alleles can be distinguished by EcoN1 digestion, which enables allelic analysis of amplified DNA fragments from ChIP. (Middle) Agarose gel images from a ChIP experiment show the DNA fragments before and after EcoN1 digestion. A ChIP assay was also carried out in the Mutu III cells to provide antibody specificity control. (Right) Following quantitation of the band intensity by the ImageQuant program, allelic enrichment was calculated as follows: (wt IP/wt input)/(mut IP/mut input). The bar graph shows the means and standard deviations of two independent ChIP experiments. (B) Occupancy of BCL6 and CtBP at selected BCL6 target loci was examined by ChIP-on-chip analysis using a custom-designed high-density tiling array. A graphic view of BCL6 and CtBP binding was generated in the University of California Santa Cruz web browser with the structure of the genes shown at the top of the histograms. Arrows indicate transcriptional orientation. In the BCL6 locus, the orange triangle indicates the exon 1 BCL6 binding sites studied in panel A. Opaque masking windows represent the cutoff defined as 2.5 times the standard deviation above the average relative enrichment on the entire array.
RESULTS
A BCL6 POZ/RDII compound mutant fails to recruit corepressors or repress a BCL6 target gene.
In order to uncover novel aspects of BCL6 repression with respect to corepressor usage, we constructed a mutant form of BCL6 which is incapable of interacting with several known corepressors (Fig. 1A). This construct, BCL6Cmpd, is a composite of two mutants that have been previously described. Specifically, it carries two point mutations in the POZ domain, N21K/H116A, which abrogate binding to SMRT/NCoR/BCoR (1) and a mutated acetylation motif, QQYQ, that significantly decreases binding to HDAC2 and the MTA3 subunit of the NuRD complex (4, 15). To verify whether BCL6Cmpd is in fact deficient in its ability to recruit known corepressors, we performed Co-IP assays with transiently transfected 293T cells. SMRT and HDAC2 were chosen as representative corepressors whose binding to BCL6 should be impaired by the mutations carried in the POZ and RDII domains, respectively. As expected, when SMRT was coexpressed with either Flag-BCL6, Flag-BCL6Cmpd, or empty control vector, anti-Flag antibody only immunoprecipitated SMRT in the presence of wt Flag-BCL6 but not Flag-BCL6Cmpd or of SMRT alone (Fig. 1B, compare lane 4 versus lanes 5 and 6). Similarly, when Flag-HDAC2 was coexpressed with HA-tagged BCL6 expression vectors, HA-BCL6Cmpd showed significantly reduced binding to HDAC2 compared to HA-BCL6 (Fig. 1B, compare lanes 11 and l2). We have also determined by gel shift assays that BCL6Cmpd possesses intact DNA binding activity (data not shown).
FIG. 1.
The BCL6Cmpd mutant is defective in its corepressor interactions and transcription repression activity. (A) Schematic representation of mutations carried by BCL6Cmpd and affected corepressors. (B) BCL6Cmpd has significantly decreased binding to HDAC2 and SMRT. 293T cells were transiently transfected to express SMRT together with Flag-BCL6 or Flag-BCL6Cmpd. Alternatively, Flag-HDAC2 was expressed with either HA-BCL6 or HA-BCL6Cmpd. Whole-cell lysates were immunoprecipitated with Flag beads and analyzed by Western blotting (WB) using the indicated antibodies. Vertical lines are inserted to indicate a repositioned gel lane. (C) BCL6Cmpd was incapable of restoring aberrantly expressed CCL3 in BCL6−/− BMM. BCL6+/+ and BCL6−/− BMM were infected with either a control murine stem cell virus retrovirus or viral constructs expressing BCL6 or BCL6Cmpd. Semiquantitative RT-PCR was performed using twofold serially diluted cDNA prepared from GFP+ cells sorted by fluorescence-activated cell sorting. β-Actin was used as a control for normalization. Ctrl, control.
To make an initial inquiry as to whether BCL6Cmpd is indeed compromised in its ability to repress BCL6 target genes in vivo, we turned to BMM. There are a number of well-characterized BCL6 target genes in BMM, and these cells can be derived from BCL6 knockout mice whereas GC B cells are completely absent in these animals. Through retrovirus-mediated reconstitution, we expressed either wt BCL6 or BCL6Cmpd in BCL6−/− BMM and compared their ability to suppress the BCL6 target gene, CCL3. As a control, wt BMM were similarly infected. Semiquantitative RT-PCR was used to assess CCL3 expression levels in GFP+ cells, purified by fluorescence-activated cell sorting. As shown in Fig. 1C, CCL3 was expressed at a very low level in wt BMM but markedly upregulated in BCL6−/− BMM. Reconstitution of BCL6−/− cells with wt BCL6 readily silenced CCL3; reconstitution with BCL6Cmpd, on the other hand, only led to slightly reduced CCL3 expression. These observations indicate that BCL6Cmpd is, in fact, deficient in repression of an endogenous target gene as predicted by its known biochemical properties.
BCL6 autoregulation demonstrates distinct corepressor requirement compared to other targets.
To further characterize the repression activity of BCL6Cmpd, we compared and contrasted its ability to repress a series of natural target promoters using 293T-based reporter assays. A synthetic BCL6 reporter, B6BS-tk-Luc, which contains a consensus BCL6 binding site linked to the tk200 promoter was also tested. These assays demonstrated that transcription from the B6BS-tk-Luc, c-Myc, PRDM-1, MCP-1, CCND2, and BCL6 promoters was significantly inhibited by wt BCL6, resulting in a 3- to 10-fold loss of activity (Fig. 2A). In comparison, BCL6Cmpd was almost completely inactive on four of the reporters (i.e., B6BS-tk-Luc, c-Myc, PRDM-1, and CCND2) and caused only weak repression on the MCP-1 promoter. The only exception was the BCL6 promoter construct, which in marked contrast to other reporters was repressed by BCL6Cmpd to a similar level as wt BCL6, suggesting that BCL6Cmpd might still retain the ability to mediate negative autoregulation. These results cannot be attributed to aberrant protein expression of BCL6Cmpd because this mutant and the wt constructs were expressed at similar levels in 293T cells (compare lanes 1 and 2 in Fig. 1 and Fig. 3). A number of additional BCL6 target gene reporters including mouse interleukin-6 were also tested, all of which failed to be repressed by BCL6Cmpd (data not shown).
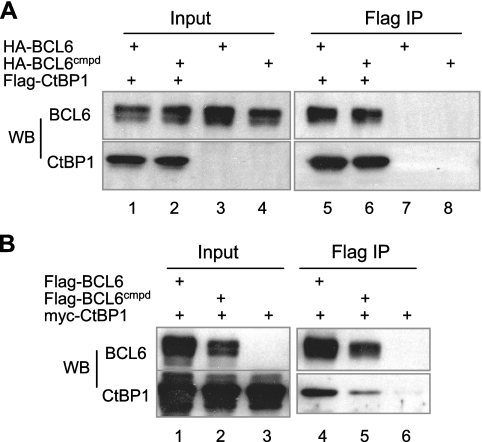
FIG. 3.
CtBP1 corepressor interacts with both wt BCL6 and BCL6Cmpd. 293T cells were transiently transfected to express Flag-CtBP1 together with either HA-BCL6 or HA-BCL6Cmpd (A). Alternatively, myc-CtBP1 was expressed with either Flag-BCL6 or Flag-BCL6Cmpd (B). Whole-cell lysates were immunoprecipitated with Flag beads and analyzed by Western blotting using the indicated antibodies. WB, Western blotting.
To validate our observations made in 293T-based reporter assays, we turned to mature B cells, a cellular context relevant to GC biology and lymphomagenesis. Two experimental strategies were employed to evaluate the contributions of lateral groove corepressors or HDAC2/MTA3 to BCL6 autoregulation. First, we compared the activity of BCL6 and BCL6Cmpd in reporter assays performed in the BCL6-negative Mutu III B cells (Fig. 2B). Our results showed that on the BCL6 reporter, both wt BCL6 and BCL6Cmpd exhibited substantial inhibitory activity although the latter is slightly compromised compared to wt BCL6. On the CCND2 reporter, however, BCL6Cmpd showed little repression activity whereas wt BCL6 repressed this reporter by 70%. Thus, the reporter assays in Mutu III cells produced results identical to those obtained in 293T cells.
Transiently transfected plasmids may not adopt the same chromatin structure as endogenous genes, which could limit the significance of reporter assay results. To address this issue, we evaluated the response of the endogenous BCL6 gene to the POZ lateral groove corepressor blocking peptide, BPI. We have previously shown that BPI treatment can derepress a number of BCL6 target genes in mature B cell lines (35). BCL6-positive SUDHL6 cells were treated with BPI and assayed for changes in BCL6 expression by qRT-PCR. As shown in Fig. 2C, CCL3 mRNA was up-regulated by sixfold following BPI treatment while the BCL6 mRNA level remained unchanged, as was the expression of the negative control gene, β2M. Similarly, it was previously shown that the level of BCL6 mRNA is insensitive to MTA3 siRNA (15). Thus, BCL6 autoregulation appears to require neither the lateral groove corepressors nor MTA3/NuRD. One possible explanation is that BCL6 autoregulation works through a competitive mechanism in which mere occupancy of the exon 1 BCL6 binding sites by the BCL6 protein is sufficient to preclude binding by transcription activators such as the STAT proteins (2). However, this model is not supported by our results showing that the BCL6 DNA binding domain alone is insufficient to mediate autoregulation in reporter assays (data not shown). Taken together, these observations suggest that BCL6 autoregulation requires a novel corepressor.
CtBP interacts with wt BCL6 and BCL6Cmpd.
In search for novel BCL6 corepressors, we turned our attention to a previous study of the Drosophila POZ-containing protein, Ttk69, which genetically interacts with Drosophila CtBP to specify cell fate in the developing Drosophila eye (43). Because the POZ domain of Ttk69 physically interacts with Drosophila CtBP and this domain can be functionally replaced by that from human BCL6, we asked whether mammalian CtBP1 could be a novel BCL6 corepressor fulfilling the unique corepressor requirement for BCL6 autoregulation. First, we performed Co-IP experiments in transiently transfected 293T cells to determine whether the two proteins associate in vivo. As shown in Fig. 3A, HA-BCL6 was efficiently coprecipitated with Flag-CtBP1 (lane 5). Likewise, Myc-CtBP1 also coprecipitated with Flag-BCL6 (Fig. 3B, lane 4). Of note, BCL6Cmpd demonstrated a comparable ability to complex with CtBP1 as does its wt counterpart in these assays (Fig. 3A, lane 6, and B, lane 5). CtBP1 also interacts efficiently with a BCL6-RDII KKYR mutant (data not shown), suggesting that the status of the KKYK motif does not influence the interaction between BCL6 and CtBP1 as it does that of BCL6 and HDAC2/MTA3. The fact that CtBP1 is able to bind to BCL6Cmpd makes it an attractive candidate to support BCL6 autoregulation.
To further characterize the interaction between BCL6 and CtBP1, we used both GST pull-down and 293T-based Co-IP assays. Our results are schematically summarized in Fig. 4A. In GST pull-down assays, interaction was consistently detected between full-length BCL6 and CtBP1, indicating that the two proteins are capable of direct binding (Fig. 4B). We consider this interaction to be specific because it persisted in high-salt wash solution (400 mM NaCl), and no interaction was ever detected between CtBP1 and GST alone. Next, we performed Co-IP experiments in 293T cells using a large panel of BCL6 deletion mutants to delineate the BCL6 domain(s) involved in binding to CtBP1. Our results indicated that the BCL6 ZF domain was dispensable for this interaction because the ZF mutant showed no detectable binding to CtBP1 (Fig. 4C, lanes 13 and 19) while the ΔZF mutant retained a binding ability comparable to that of the full-length BCL6 (compare lane 2 to lane 3 in the input panel and lane 7 to lane 8 in the IP panel; lanes 12 and 18 are another ΔZF experiment, but the CtBP1 pull-down was suboptimal). As to the role played by the N terminus POZ domain, both Δ[POZ+] (lanes 14 and 20) and ΔPOZ (lanes 16 and 22) had weaker interactions than the full-length BCL6 (lanes 11 and 17), while the POZ+ construct showed robust binding (lanes 15 and 21). As to the contribution by the RDII region, binding of ΔPEST to CtBP1 was very comparable to that of the full-length BCL6 (compare lane 2 to 4 in the input panel and lane 7 to 9 in the IP panel) while the RDII domain on its own showed a rather weak interaction (lanes 5 and 10). Taken together, these results suggest that the BCL6 POZ domain serves as the primary interaction interface between BCL6 and CtBP1 while the RDII domain makes a minor contribution. Not only is this interpretation in line with the previous Drosophila work (43), it may also explain our observation that CtBP1 binding to BCL6 is unaffected by an acetylated RDII (Fig. 3).
CtBP physically interacts with BCL6 in GC-derived mature B cells.
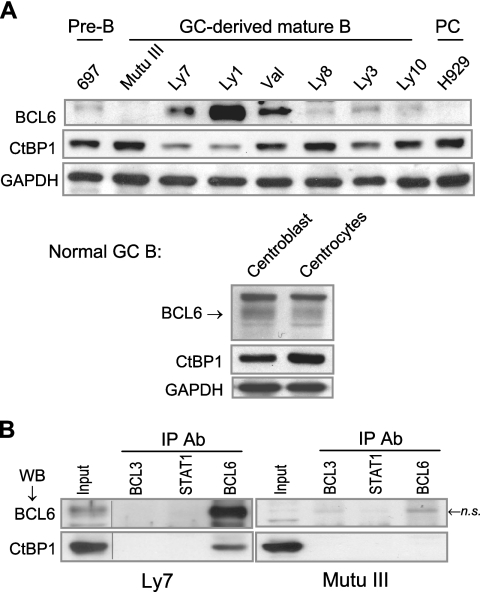
Because BCL6 autoregulation was initially characterized in DLBCL cell lines and its importance has been shown for the pathogenesis of DLBCL, we carried out a series of experiments to determine the role of CtBP in BCL6 negative autoregulation in DLBCL cells. First, we examined the expression pattern of CtBP using a panel of cell lines representing different stages of B-cell development (Fig. 5A). The pre-B and plasma cell stages are represented by 697 and H929, respectively; the other cell lines are GC-derived mature B-cell lines. With the exception of Mutu III, which is an Epstein-Barr virus-positive Burkitt's lymphoma cell line, the rest are DLBCL cell lines. Western blot analysis showed that the DLBCL cell lines vary in their levels of BCL6, largely reflecting the stage of B-cell development. The levels of CtBP also vary in these cells such that there appears to be a loose, inverse correlation between the two proteins. We have also determined that BCL6 and CtBP are coexpressed in two types of primary GC B cells, centroblasts and centrocytes, isolated from reactive human tonsils (Fig. 5A, bottom panel). To determine whether endogenous BCL6 and CtBP interact in B cells, Co-IP assays were performed in one of the DLBCL cell lines, Ly7. CtBP was only recovered in the precipitates using the anti-BCL6 antibody but not the control antibodies against two other transcription factors, BCL3 and STAT1 (Fig. 5B, top panel). To exclude the possibility that the BCL6 antibody cross-reacted with CtBP, a control experiment was carried out in parallel using the BCL6-negative Mutu III cells. As expected, although CtBP1 is abundantly expressed in Mutu III, it was not recovered by any of the antibodies used in this experiment (Fig. 5B, bottom panel). These results indicate that in GC-derived mature B cells, a fraction of endogenous CtBP and BCL6 is associated in a complex. Of note, because both CtBP1 and the highly homologous CtBP2 can interact with BCL6 in GST pull-down assays (Fig. 4B and data not shown) and because we cannot exclude the possibility that the anti-CtBP1 antibody cross-reacts with CtBP2, the BCL6 corepressor activity detected in this study can be potentially attributable to both CtBP proteins.
FIG. 5.
CtBP forms a complex with BCL6 in B cells. (A) Western blot analysis of BCL6 and CtBP expression in cell lines representing pre-B, GC-derived mature B, and plasma cells (PC). GAPDH was included as a loading control. Human centroblasts and centrocytes were purified by magnetic cell separation using the MidiMACS system (Miltenyi Biotechnology) following a published protocol (21). (B) CtBP1 is present in a complex with BCL6 in the BCL6-positive Ly7 cells. Co-IP assays in Ly7 and Mutu III cells were carried out using an antibody for BCL6 or two control antibodies (BCL3 and STAT1). The presence of BCL6 and CtBP1 was examined in the precipitates by Western blotting (WB). In the BCL6-negative Mutu III cells, neither BCL6 nor CtBP was immunoprecipitated by the panel of antibodies we used although a weak band with similar migration to BCL6 was nonspecifically pulled down by both the BCL6 and the BCL3 antibodies (n.s.). Vertical lines are inserted to indicate a repositioned gel lane. Ab, antibody.
CtBP is associated with BCL6 in the BCL6 promoter region and a subset of other BCL6 target loci.
In negative autoregulation, BCL6 docks to clustered BCL6 binding sites in its exon 1 to repress its own transcription. We have devised an assay to decipher autoregulation-specific DNA binding in Ly1 cells, where BCL6 preferentially binds to the wt allele with intact BCL6 sites over the mutant allele that harbors a point mutation in the high-affinity BCL6 site (42). Specifically, in a pool of DNA fragments obtained from a given ChIP assay, those derived from the mutated allele are shortened by EcoN1 digestion while those derived from the wt allele are spared (Fig. 6A, left panel). Using this allele-specific ChIP method, we found that not only is CtBP associated with the exon 1 region of the endogenous BCL6 gene, but, like the BCL6 protein, it also preferentially occupies the wt allele (Fig. 6A, middle and right panels). In further support of BCL6-dependent CtBP recruitment, we found that neither BCL6 nor CtBP is bound to the exon 1 region in the BCL6-negative Mutu III cells (Fig. 6A, middle panel).
Given that CtBP can interact with BCL6 in vivo and be recruited to the BCL6 promoter region, we expanded our CtBP ChIP analysis to additional BCL6 target genes. To this end, we hybridized ChIP-enriched DNA fragments to a densely tiled custom oligonucleotide microarray covering the genomic loci of a number of known BCL6 target genes and GAPDH (negative control). We performed duplicate ChIP experiments using antibodies for BCL6 and CtBP, hybridized the enriched DNA fragments to the arrays, and assessed the enrichment relative to input chromatin to identify sites of specific association. As expected, among the six target genes showing specific BCL6 enrichment is BCL6 (Fig. 6B). Interestingly, although there was very strong and colocalized BCL6 and CtBP enrichment over the exon 1/promoter region (Fig. 6B, orange triangle in the BCL6 panel), thus confirming the conventional ChIP data shown in Fig. 6A, a number of additional BCL6 and CtBP peaks were also present in the 5′ regulatory region as well as the first intron. This widespread pattern of BCL6 enrichment was also observed in the Ramos cell line that has a wt BCL6 gene (data not shown). Thus, although prior studies have indicated that only the clustered exon 1 BCL6 sites are critical for autoregulation, multiple sites throughout the BCL6 locus can be used to target binding of BCL6 and CtBP. Colocalized CtBP and BCL6 signals were also detected in three other BCL6 target genes, namely, CCL3, TP53, and PRDM1. In the PRDM1 locus, very robust BCL6 enrichment and a colocolized CtBP peak were detected in the third intron where BCL6 binding was shown previously (32, 40). There were additional, BCL6-independent CtBP signals in the PRDM1 promoter region and the fourth intron. We have confirmed binding of BCL6 and CtBP to the promoter regions of BCL6, CCL3, TP53, and the third intron of PRDM1 by single-locus quantitative ChIP (data not shown). There were also two genes that showed BCL6 occupancy without targeted CtBP recruitment. In the CD69 locus, although the BCL6 signals were strong, the CtBP enrichment did not reach the cutoff level. In the CCND2 locus, weak CtBP peaks were present at several positions; none of these, however, coincided with the BCL6 peak in the promoter region. Thus, CtBP is likely to be targeted to CCND2 by another sequence-specific transcription factor(s). Collectively, these ChIP studies demonstrate that, in addition to the BCL6 promoter region, CtBP is also recruited to a subset of other BCL6 target genes.
CtBP is required for BCL6 autoregulation in vivo.
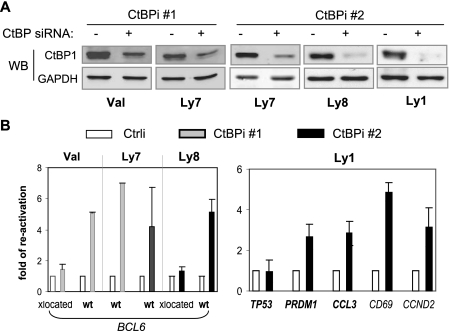
To directly address the functional contribution of CtBP to BCL6 autoregulation, we used siRNA to knock down endogenous CtBP in DLBCL cells and analyzed the resulting gene expression changes by qRT-PCR. Two of the four cell lines we used, Val and Ly8, carry BCL6 translocations such that the BCL6 coding region is fused to a heterologous promoter in one allele while the second allele remains in wt configuration (10, 45). Compared to cells treated with the control siRNA oligonucleotide (Ctrli), 40 h after treatment with either one of the two CtBP siRNA oligonucleotides (CtBPi 1 and CtBPi 2), the levels of CtBP1 protein decreased by 75% or more (Fig. 7A). As a result, expression of the wt BCL6 allele was upregulated five- to sevenfold in all three cell lines tested (Fig. 7B). In comparison, the translocated (Fig. 7B, xlocated) BCL6 alleles in Val and Ly8 were largely unaffected. Combined with our ChIP results shown in Fig. 6, these data indicate that while the wt BCL6 allele is controlled by a CtBP-mediated autoregulation mechanism, the translocated allele driven by a heterologous promoter is no longer subject to such regulation. To evaluate the role of CtBP in regulation of other BCL6 target genes, we also knocked down CtBP in Ly1 cells where the ChIP-on-chip study was conducted. Among the other three BCL6 target genes where CtBP is colocalized with BCL6, CtBP siRNA moderately upregulated PRDM1 and CCL3 (2.7- and 2.8-fold, respectively) but did not affect TP53 expression. As to the two genes showing no BCL6-directed CtBP recruitment, the mRNA changes caused by CtBP knock-down are likely to be CtBP related but BCL6 independent in the case of CCND2 and an indirect effect in the case of CD69.
FIG. 7.
CtBP is a required corepressor for BCL6 negative autoregulation. Both alleles of BCL6 in Ly7 are in germ line configuration, while in Val and Ly8 there is one translocated (xlocated) and one wt allele. (A) Effect of siRNA-mediated CtBP knockdown in 4 DLBCL cell lines. Western blot (WB) analysis was performed on cell lysates 40 h after transfection with CtBP siRNA oligonucleotides (CtBPi 1 and CtBPi 2) or a control oligonucleotide (−). (B) Gene expression changes caused by CtBP siRNA were analyzed by qRT-PCR. The ΔΔCT method was used to calculate the increase relative to control-treated cells defined as 1.0. Plotted in the graph are the means and standard deviations of two independent experiments. At the bottom of the graph, gene names appearing in bold indicate occupancy by both BCL6 and CtBP.
DISCUSSION
Negative autoregulation is a central mechanism governing the expression and function of BCL6 in normal B cells and oncogenesis. Although a number of corepressor complexes have been shown to participate in BCL6-mediated gene silencing, the specific corepressor requirement for BCL6 autoregulation has not been addressed. We demonstrate in this study that BCL6 autoregulation operates independently of several known BCL6 corepressors and, instead, requires a novel cofactor, CtBP. Several lines of findings support this conclusion. (i) In reporter assays, the BCL6Cmpd mutant, deficient in its ability to recruit the BBD corepressors as well as HDAC2 and MTA3/NuRD, retained nearly wt activity on the BCL6 promoter. (ii) Expression of the endogenous BCL6 gene was not affected by treatment with either the BPI peptide or, as reported by Fujita et al. (15), by MTA3 shRNA. (iii) CtBP can physically interact with BCL6 both in vitro and in vivo, and this interaction is not compromised by mutations in BCL6Cmpd. (iv) In BCL6-positive lymphoma B cells, CtBP wass recruited by BCL6 to the BCL6 exon 1/promoter region, and CtBP siRNA specifically derepressed the wt BCL6 allele but spared the translocated allele driven by heterologous promoters. Our results not only identify a novel corepressor for BCL6 but also substantiate a model in which corepressors are selectively used by BCL6 to regulate subsets of target genes and thus different cellular functions.
Although CtBP is uniquely required for BCL6 autoregulation, it is not the only corepressor recruited to the BCL6 promoter region. This region has been shown to be occupied by BCoR and even contains ubiquitylated histone H2A, indicative of the E3 ligase activity contained in the BCoR complex (16). Our unpublished data indicate that BCL6 can also recruit SMRT, NCoR, and MTA3 to this region (data not shown). It appears that corepressors can be targeted to a given promoter without playing an active role in the maintenance of gene expression status. These observations imply that BCL6 autoregulation requires unique chromatin remodeling activities that are conferred by only CtBP but not other known BCL6 corepressors. In contrast to this selective requirement for one corepressor in BCL6 autoregulation, several BCL6 corepressors contribute to BCL6-mediated repression of the chemokine gene CCL3. It was previously reported that silencing of CCL3 in B cells depends upon the lateral groove corepressors as well as the MTA3/NuRD complex (15, 35). Our CtBP siRNA experiment revealed that CtBP also contributes to repression of CCL3 (Fig. 7B). CtBP alone, however, is inadequate because the BCL6Cmpd mutant, with its intact capability to recruit CtBP, was unable to turn off CCL3 in BCL6−/− BMM (Fig. 1C). Thus, CCL3 appears to be an unusual BCL6 target in that its full inhibition requires multiple BCL6 corepressors, e.g., those binding to the POZ domain and RDII as well as CtBP. An example between these two extremes is the PRDM1 gene encoding Blimp-1, the master regulator of plasma cell differentiation. BCL6-mediated silencing of PRDM1 is insensitive to BPI treatment, critically dependent upon MTA3/NuRD (15, 32), and partially responsive to CtBP siRNA (Fig. 7B). Interestingly, in Ly1 cells, although CtBP was found to localize with BCL6 in the TP53 5′ regulatory region, TP53 mRNA levels were unchanged following CtBP siRNA. Thus, the phenomenon of corepressor recruitment without apparent regulatory consequences may occur quite frequently although probably in a cell-type- and locus-specific manner.
Taken together, our data suggest that BCL6 mediates transcriptional repression through multiple mechanisms even in the same cell type. These involve promoter context-dependent requirements for specific corepressors in the case of CtBP in BCL6 negative autoregulation and the lateral groove corepressors on TP53, promoter-specific combinatorial actions of corepressors such as CtBP and MTA3 on PRDM1, or all three on CCL3. Moreover, BCL6 can recruit additional corepressors such as ETO, which may contribute to repression of CCND2 (8). Our ChIP-on-chip experiments demonstrate that BCL6 binding is variable in pattern and intensity between loci, which may further contribute to the complexity of the BCL6-dependent transcriptional program in B cells. Nevertheless, as discussed above, BCL6 autoregulation and differentiation-coupled control of PRDM1 expression offer two excellent model systems in which mechanistic details of BCL6-mediated transcription inhibition can be investigated in the future. Structural studies of the interaction between BCL6 and the lateral groove corepressors suggest that each BCL6 dimer can potentially bind to two such corepressors (SMRT, NCoR, and BCoR) at a time (1). Since our data indicate that BCL6 binds to CtBP1 primarily through the POZ domain with a minor contribution from RDII, it raises the question whether a BCL6 dimer already in complex with CtBP can further engage one or more of the lateral groove corepressors (which are very large proteins) and/or the RDII corepressors (HDAC2 or MTA3/NuRD) and vice versa. Thus, one important issue that should be addressed in future studies is the stoichiometry and dynamics of assembled BCL6-corepressor complexes over a given target promoter where multiple corepressors are recruited, such as BCL6, CCL3, and PRDM1.
CtBP1 is unique among known BCL6 corepressors in that it appears to contact both the POZ domain and the RDII region of BCL6. Interestingly, BCL6 contains neither the typical PXDLS motif, which is present in many CtBP-binding transcription factors, nor the alternative RRT motif (RRTGXPPXL) found in a number of CtBP-interacting ZF proteins (28, 36). Although the results of our GST pull-down experiment suggest that BCL6 and CtBP are capable of direct binding (Fig. 4B), we cannot formally rule out the requirement for a third, bridging protein in vivo. Candidates for such a potential bridging molecule include HDAC2 and hPC2; both are core constituents of the CtBP1 complex (23, 39). HDAC2 was previously shown to bind to BCL6 RDII (4) (Fig. 1B), and we have found that BCL6 and hPC2 can readily interact in 293T cells (not shown). At the moment, the contribution of CtBP to BCL6-regulated immune functions is unknown. Knockout mouse models for the CtBP proteins have been described before (18). While germ line CtBP2 inactivation results in embryonic lethality, CtBP1 knockout mice are viable although smaller in size, and some die by postnatal day 20. The impact of CtBP deficiency on B-cell functions in these animal models has not been reported.
For differentiation stage-specific transcription factors, like BCL6, coupling of their expression status to cell differentiation control is of critical importance. That the wt BCL6 allele is silent in lymphoma cells bearing BCL6 translocations or activating mutations convincingly demonstrates that the B-cell differentiation program is set up so that, in late-stage GC B cells, autoregulation is capable of completely shutting off BCL6. The timing of this event is intriguing as it coincides with upregulation of differentiation-related BCL6 targets. One possible approach to achieve this dichotomy of target gene regulation is to allow a graded response to the subsiding levels of BCL6 protein based on target gene avidity for BCL6 binding. In support of this notion, we have previously reported that BCL6 autoregulation is operative in many cell types across a wide range of BCL6 concentration (42). An alternative strategy is to use different corepressors or corepressor combinations. The subcellular localization and transcriptional activities of CtBP are known to be highly regulated at the levels of protein conformation as well as posttranslational modification by a range of stimuli including hypoxia, metabolic changes, and UV-induced DNA damage (5). It is conceivable that, due to its principle reliance on CtBP, BCL6 autoregulation will follow a distinct course of response compared to its other cellular targets in GC B cells that are exposed to dynamic extracellular and intracellular changes. The fact that CtBP can bind to an acetylated BCL6 protein suggests that when BCL6 acetylation triggers dissociation of MTA3/NuRD leading to activation of Blimp-1 and its associated plasma cell program, the CtBP-dependent BCL6 autoregulation will proceed unperturbed. Finally, insensitivity of autoregulation to BPI also suggests that in lymphoma cells subject to BPI-based therapies, reactivation of the wt BCL6 allele, which could compromise treatment success, is unlikely. Thus, our study not only uncovers new mechanistic insights of BCL6-mediated transcription repression but also has important implications for development of future lymphoma therapies.
Acknowledgments
We thank Arthur Skoultchi for his insightful suggestions, Fiona Pixley for preparation of primary BMM, Stella Maris Ranuncolo for isolating tonsilar human B-cell subsets, and Tania Dell'Oso for help with qRT-PCR analysis. We are also grateful to a number of colleagues whose generosity in sharing unique reagents has made this study possible.
This work was supported by grants from the National Institute of Health (RO1 CA85573) and the G & P Foundation for Cancer Research to B.H.Y., a grant from the National Institutes of Health (R01 CA104348) to A.M, and a grant from the Lauri Strauss Leukemia Foundation to B.B.D. J.M.P. is supported by a predoctoral fellowship from the National Cancer Center.
Footnotes
Published ahead of print on 22 January 2008.
REFERENCES
- 1.Ahmad, K. F., A. Melnick, S. Lax, D. Bouchard, J. Liu, C. L. Kiang, S. Mayer, S. Takahashi, J. D. Licht, and G. G. Prive. 2003. Mechanism of SMRT corepressor recruitment by the BCL6 BTB domain. Mol. Cell 121551-1564. [DOI] [PubMed] [Google Scholar]
- 2.Arguni, E., M. Arima, N. Tsuruoka, A. Sakamoto, M. Hatano, and T. Tokuhisa. 2006. JunD/AP-1 and STAT3 are the major enhancer molecules for high BCL6 expression in germinal center B cells. Int. Immunol. 181079-1089. [DOI] [PubMed] [Google Scholar]
- 3.Baron, B. W., J. Anastasi, A. Montag, D. Huo, R. M. Baron, T. Karrison, M. J. Thirman, S. K. Subudhi, R. K. Chin, D. W. Felsher, Y. X. Fu, T. W. McKeithan, and J. M. Baron. 2004. The human BCL6 transgene promotes the development of lymphomas in the mouse. Proc. Natl. Acad. Sci. USA 10114198-14203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bereshchenko, O. R., W. Gu, and R. Dalla-Favera. 2002. Acetylation inactivates the transcriptional repressor BCL6. Nat. Genet. 32606-613. [DOI] [PubMed] [Google Scholar]
- 5.Bergman, L. M., and J. P. Blaydes. 2006. C-terminal binding proteins: emerging roles in cell survival and tumorigenesis. Apoptosis 11879-888. [DOI] [PubMed] [Google Scholar]
- 6.Cattoretti, G., L. Pasqualucci, G. Ballon, W. Tam, S. V. Nandula, Q. Shen, T. Mo, V. V. Murty, and R. Dalla-Favera. 2005. Deregulated BCL6 expression recapitulates the pathogenesis of human diffuse large B cell lymphomas in mice. Cancer Cell 7445-455. [DOI] [PubMed] [Google Scholar]
- 7.Chang, C. C., B. H. Ye, R. S. Chaganti, and R. Dalla-Favera. 1996. BCL-6, a POZ/zinc-finger protein, is a sequence-specific transcriptional repressor. Proc. Natl. Acad. Sci. USA 936947-6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chevallier, N., C. M. Corcoran, C. Lennon, E. Hyjek, A. Chadburn, V. J. Bardwell, J. D. Licht, and A. Melnick. 2004. ETO protein of t(8;21) AML is a corepressor for Bcl-6 B-cell lymphoma oncoprotein. Blood 1031454-1463. [DOI] [PubMed] [Google Scholar]
- 9.Chinnadurai, G. 2002. CtBP, an unconventional transcriptional corepressor in development and oncogenesis. Mol. Cell 9213-224. [DOI] [PubMed] [Google Scholar]
- 10.Dallery, E., S. Galiegue-Zouitina, M. Collyn-d'Hooghe, S. Quief, C. Denis, M. P. Hildebrand, D. Lantoine, C. Deweindt, H. Tilly, C. Bastard, et al. 1995. TTF, a gene encoding a novel small G protein, fuses to the lymphoma-associated LAZ3 gene by t(3;4) chromosomal translocation. Oncogene 102171-2178. [PubMed] [Google Scholar]
- 11.Deltour, S., S. Pinte, C. Guerardel, B. Wasylyk, and D. Leprince. 2002. The human candidate tumor suppressor gene HIC1 recruits CtBP through a degenerate GLDLSKK motif. Mol. Cell. Biol. 224890-4901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dent., A. L., A. L. Shaffer, X. Yu, D. Allman, and L. M. Staudt. 1997. Control of inflammation, cytokine expression, and germinal center formation by BCL-6. Science 276589-592. [DOI] [PubMed] [Google Scholar]
- 13.Dhordain, P., O. Albagli, R. J. Lin, S. Ansieau, S. Quief, A. Leutz, J. P. Kerckaert, R. M. Evans, and D. Leprince. 1997. Corepressor SMRT binds the BTB/POZ repressing domain of the LAZ3/BCL6 oncoprotein. Proc. Natl. Acad. Sci. USA 9410762-10767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dhordain, P., R. J. Lin, S. Quief, D. Lantoine, J. P. Kerckaert, R. M. Evans, and O. Albagli. 1998. The LAZ3(BCL-6) oncoprotein recruits a SMRT/mSIN3A/histone deacetylase containing complex to mediate transcriptional repression. Nucleic Acids Res. 264645-4651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fujita, N., D. L. Jaye, C. Geigerman, A. Akyildiz, M. R. Mooney, J. M. Boss, and P. A. Wade. 2004. MTA3 and the Mi-2/NuRD complex regulate cell fate during B lymphocyte differentiation. Cell 11975-86. [DOI] [PubMed] [Google Scholar]
- 16.Gearhart, M. D., C. M. Corcoran, J. A. Wamstad, and V. J. Bardwell. 2006. Polycomb group and SCF ubiquitin ligases are found in a novel BCoR complex that is recruited to BCL6 targets. Mol. Cell. Biol. 266880-6889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Han, S., B. Zheng, Y. Takahashi, and G. Kelsoe. 1997. Distinctive characteristics of germinal center B cells. Semin. Immunol. 9255-260. [DOI] [PubMed] [Google Scholar]
- 18.Hildebrand, J. D., and P. Soriano. 2002. Overlapping and unique roles for C-terminal binding protein 1 (CtBP1) and CtBP2 during mouse development. Mol. Cell. Biol. 225296-5307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huynh, K. D., and V. J. Bardwell. 1998. The BCL-6 POZ domain and other POZ domains interact with the co-repressors N-CoR and SMRT. Oncogene 172473-2484. [DOI] [PubMed] [Google Scholar]
- 20.Huynh, K. D., W. Fischle, E. Verdin, and V. J. Bardwell. 2000. BCoR, a novel corepressor involved in BCL-6 repression. Genes Dev. 141810-1823. [PMC free article] [PubMed] [Google Scholar]
- 21.Klein, U., Y. Tu, G. A. Stolovitzky, J. L. Keller, J. Haddad, Jr., V. Miljkovic, G. Cattoretti, A. Califano, and R. Dalla-Favera. 2003. Transcriptional analysis of the B cell germinal center reaction. Proc. Natl. Acad. Sci. USA 1002639-2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuppers, R., U. Klein, M. L. Hansmann, and K. Rajewsky. 1999. Cellular origin of human B-cell lymphomas. N. Engl. J. Med. 3411520-1529. [DOI] [PubMed] [Google Scholar]
- 23.Kuppuswamy, M., S. Vijayalingam, L. J. Zhao, Y. Zhou, T. Subramaninan, J. Ryerse, and G. Chinnadurai. 2008. Role of the PLDLS-binding cleft region of CtBP1 in recruitment of core and auxiliary components of the corepressor complex. Mol. Cell. Biol. 28269-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lemercier, C., M. P. Brocard, F. Puvion-Dutilleul, H. Y. Kao, O. Albagli, and S. Khochbin. 2002. Class II histone deacetylases are directly recruited by BCL6 transcriptional repressor. J. Biol. Chem. 27722045-22052. [DOI] [PubMed] [Google Scholar]
- 25.Li, Z., X. Wang, R. Y. Yu, B. B. Ding, J. J. Yu, X. M. Dai, A. Naganuma, E. R. Stanley, and B. H. Ye. 2005. BCL-6 negatively regulates expression of the NF-κB1 p105/p50 subunit. J. Immunol. 174205-214. [DOI] [PubMed] [Google Scholar]
- 26.MacLennan, I. C. 1994. Somatic mutation. From the dark zone to the light. Curr. Biol. 470-72. [DOI] [PubMed] [Google Scholar]
- 27.Mascle, X., O. Albagli, and C. Lemercier. 2003. Point mutations in BCL6 DNA-binding domain reveal distinct roles for the six zinc fingers. Biochem. Biophys. Res. Commun. 300391-396. [DOI] [PubMed] [Google Scholar]
- 28.Molloy, D. P., P. M. Barral, K. H. Bremner, P. H. Gallimore, and R. J. Grand. 2001. Structural determinants outside the PXDLS sequence affect the interaction of adenovirus E1A, C-terminal interacting protein and Drosophila repressors with C-terminal binding protein. Biochim. Biophys. Acta 154655-70. [DOI] [PubMed] [Google Scholar]
- 29.Niu, H., G. Cattoretti, and R. Dalla-Favera. 2003. BCL6 controls the expression of the B7-1/CD80 costimulatory receptor in germinal center B cells. J. Exp. Med. 198211-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Niu, H., B. H. Ye, and R. Dalla-Favera. 1998. Antigen receptor signaling induces MAP kinase-mediated phosphorylation and degradation of the BCL-6 transcription factor. Genes Dev. 121953-1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oberley, M. J., J. Tsao, P. Yau, and P. J. Farnham. 2004. High-throughput screening of chromatin immunoprecipitates using CpG-island microarrays. Methods Enzymol. 376315-334. [DOI] [PubMed] [Google Scholar]
- 32.Parekh, S., J. M. Polo, R. Shaknovich, P. Juszczynski, P. Lev, S. M. Ranuncolo, Y. Yin, U. Klein, G. Cattoretti, R. Dalla Favera, M. A. Shipp, and A. Melnick. 2007. BCL6 programs lymphoma cells for survival and differentiation through distinct biochemical mechanisms. Blood 1102067-2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pasqualucci, L., A. Migliazza, K. Basso, J. Houldsworth, R. S. Chaganti, and R. Dalla-Favera. 2003. Mutations of the BCL6 proto-oncogene disrupt its negative autoregulation in diffuse large B-cell lymphoma. Blood 1012914-2923. [DOI] [PubMed] [Google Scholar]
- 34.Phan, R. T., and R. Dalla-Favera. 2004. The BCL6 proto-oncogene suppresses p53 expression in germinal-centre B cells. Nature 432635-639. [DOI] [PubMed] [Google Scholar]
- 35.Polo, J. M., T. Dell'Oso, S. M. Ranuncolo, L. Cerchietti, D. Beck, G. F. Da Silva, G. G. Prive, J. D. Licht, and A. Melnick. 2004. Specific peptide interference reveals BCL6 transcriptional and oncogenic mechanisms in B-cell lymphoma cells. Nat. Med. 101329-1335. [DOI] [PubMed] [Google Scholar]
- 36.Quinlan, K. G., M. Nardini, A. Verger, P. Francescato, P. Yaswen, D. Corda, M. Bolognesi, and M. Crossley. 2006. Specific recognition of ZNF217 and other zinc-finger proteins at a surface groove of C-terminal binding proteins. Mol. Cell. Biol. 268159-8172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ranuncolo, S. M., J. M. Polo, J. Dierov, M. Singer, T. Kuo, J. Greally, R. Green, M. Carroll, and A. Melnick. 2007. Bcl-6 mediates the germinal center B cell phenotype and lymphomagenesis through transcriptional repression of the DNA-damage sensor ATR. Nat. Immunol. 8705-714. [DOI] [PubMed] [Google Scholar]
- 38.Shaffer, A. L., A. Rosenwald, E. M. Hurt, J. M. Giltnane, L. T. Lam, O. K. Pickeral, and L. M. Staudt. 2001. Signatures of the immune response. Immunity 15375-385. [DOI] [PubMed] [Google Scholar]
- 39.Shi, Y., J. Sawada, G. Sui, B. Affar el, J. R. Whetstine, F. Lan, H. Ogawa, M. P. Luke, and Y. Nakatani. 2003. Coordinated histone modifications mediated by a CtBP co-repressor complex. Nature 422735-738. [DOI] [PubMed] [Google Scholar]
- 40.Tunyaplin, C., A. L. Shaffer, C. D. Angelin-Duclos, X. Yu, L. M. Staudt, and K. L. Calame. 2004. Direct repression of prdm1 by Bcl-6 inhibits plasmacytic differentiation. J. Immunol. 1731158-1165. [DOI] [PubMed] [Google Scholar]
- 41.Vasanwala, F. H., S. Kusam, L. M. Toney, and A. L. Dent. 2002. Repression of AP-1 function: a mechanism for the regulation of Blimp-1 expression and B lymphocyte differentiation by the B cell lymphoma-6 protooncogene. J. Immunol. 1691922-1929. [DOI] [PubMed] [Google Scholar]
- 42.Wang, X., Z. Li, A. Naganuma, and B. H. Ye. 2002. Negative autoregulation of BCL-6 is bypassed by genetic alterations in diffuse large B cell lymphomas. Proc. Natl. Acad. Sci. USA 9915018-15023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wen, Y., D. Nguyen, Y. Li, and Z. C. Lai. 2000. The N-terminal BTB/POZ domain and C-terminal sequences are essential for Tramtrack69 to specify cell fate in the developing Drosophila eye. Genetics 156195-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ye, B. H., G. Cattoretti, Q. Shen, J. Zhang, N. Hawe, R. de Waard, C. Leung, M. Nouri-Shirazi, A. Orazi, R. S. Chaganti, P. Rothman, A. M. Stall, P. P. Pandolfi, and R. Dalla-Favera. 1997. The BCL-6 proto-oncogene controls germinal-centre formation and Th2-type inflammation. Nat. Genet. 16161-170. [DOI] [PubMed] [Google Scholar]
- 45.Ye, B. H., S. Chaganti, C. C. Chang, H. Niu, P. Corradini, R. S. Chaganti, and R. Dalla-Favera. 1995. Chromosomal translocations cause deregulated BCL6 expression by promoter substitution in B cell lymphoma. EMBO J. 146209-6217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ye, B. H., F. Lista, F. Lo Coco, D. M. Knowles, K. Offit, R. S. Chaganti, and R. Dalla-Favera. 1993. Alterations of a zinc finger-encoding gene, BCL-6, in diffuse large-cell lymphoma. Science 262747-750. [DOI] [PubMed] [Google Scholar]
- 47.Ye, B. H., P. H. Rao, R. S. Chaganti, and R. Dalla-Favera. 1993. Cloning of BCL-6, the locus involved in chromosome translocations affecting band 3q27 in B-cell lymphoma. Cancer Res. 532732-2735. [PubMed] [Google Scholar]
- 48.Yu, R. Y., X. Wang, F. J. Pixley, J. J. Yu, A. L. Dent., H. E. Broxmeyer, E. R. Stanley, and B. H. Ye. 2005. BCL-6 negatively regulates macrophage proliferation by suppressing autocrine IL-6 production. Blood 1051777-1784. [DOI] [PubMed] [Google Scholar]
- 49.Zhang, H., S. Okada, M. Hatano, S. Okabe, and T. Tokuhisa. 2001. A new functional domain of Bcl6 family that recruits histone deacetylases. Biochim. Biophys. Acta 1540188-200. [DOI] [PubMed] [Google Scholar]
- 50.Zhang, Q., Y. Yoshimatsu, J. Hildebrand, S. M. Frisch, and R. H. Goodman. 2003. Homeodomain interacting protein kinase 2 promotes apoptosis by downregulating the transcriptional corepressor CtBP. Cell 115177-186. [DOI] [PubMed] [Google Scholar]