Abstract
microRNAs in the miR-106b family are overexpressed in multiple tumor types and are correlated with the expression of genes that regulate the cell cycle. Consistent with these observations, miR-106b family gain of function promotes cell cycle progression, whereas loss of function reverses this phenotype. Microarray profiling uncovers multiple targets of the family, including the cyclin-dependent kinase inhibitor p21/CDKN1A. We show that p21 is a direct target of miR-106b and that its silencing plays a key role in miR-106b-induced cell cycle phenotypes. We also show that miR-106b overrides a doxorubicin-induced DNA damage checkpoint. Thus, miR-106b family members contribute to tumor cell proliferation in part by regulating cell cycle progression and by modulating checkpoint functions.
Changes in microRNA expression levels are correlated with and contribute to cancer development (7, 25, 33), but the cellular mechanisms by which microRNAs influence aberrant cell growth are poorly understood. Recent evidence suggests that microRNAs can act as either tumor suppressors or oncogenes. For example, microRNAs in the let-7 family may act as tumor suppressors by repressing certain oncogenes, including those encoding Ras family members and HMGA2, thereby inhibiting tumor growth (22, 28, 34). The miR-16 family also shows antiproliferative activity by negatively regulating cell cycle progression (32) and inducing apoptosis as a result of BCL2 silencing (12, 32). Moreover, we and others have shown that the miR-34 family plays a role in TP53 tumor suppressor function and causes cell cycle arrest and apoptosis (4, 9, 18, 37, 46, 47). Consistent with a tumor-suppressive role, let-7, miR-16, and miR-34 families of microRNAs are often deleted or down-regulated in cancers (6). microRNAs, including miR-21, miR-155/BIC, miR-372, and miR-373, may also have oncogenic properties. These microRNAs are often overexpressed or amplified in cancer and drive tumor progression in mouse models (14, 23, 41, 48, 51). Other oncogenic microRNAs include the miR-17-92 and miR-106a-363 polycistronic clusters. Ectopic expression of the miR-17-92 cluster, which is amplified in B-cell lymphomas, accelerates tumor growth in mice (19). The miR-106a-363 cluster is a site of retroviral insertion in a mouse T-cell lymphoma (24). These microRNA clusters contain multiple members of a microRNA family with seed region homology that we refer to here as the miR-106b family. Because they share a seed region (nucleotides 1 to 8), which largely dictates target mRNA recognition (30), these family members likely promote tumor growth through related, though poorly understood cellular mechanisms.
microRNAs regulate gene expression by inhibiting protein translation or by triggering cleavage of the target mRNAs. Each microRNA has the potential to target hundreds of genes with seed region complementary sites in their 3′ untranslated regions (UTRs) (31). For many microRNAs, a single key target has been put forth as the underlying cause of the observed phenotypes (20, 35, 44). Although individual targets responsible for the phenotypes elicited by miR-16 and miR-34a have been proposed, it is likely that these microRNAs function through cooperative down-regulation of multiple targets (8, 18, 32).
To better understand how the miR-106b family regulates oncogenesis, we investigated the functional properties of these microRNAs. We show that, consistent with a role in cancer, expression of the miR-106b family is correlated with expression of genes involved in cell proliferation, and our phenotypic studies indicate a role for this family in regulating the G1-to-S cell cycle transition.
MATERIALS AND METHODS
Functional annotation of microRNAs and microRNA levels in tumor samples.
microRNA levels in a collection of five different solid tumors (breast, colon, kidney, gastric, and lung) and adjacent noninvolved tissues obtained from 29 tumor and 28 normal adjacent samples were measured. The tissue blocks were from Genomics Collaborative, Inc. Each sample was pulverized and split into two tubes. One tube was used for RNA extraction with the RNEasy kit (Qiagen). Purified total RNA was profiled on 25,000-transcript human Agilent microarrays. Samples were hybridized against pooled normal samples from the same tissue. The other tube was used in Trizol RNA extraction protocols that preserve small RNAs. microRNA levels were measured as described previously (38). We looked at microRNAs in the signatures of 10 or more tumor samples and annotated sets of ≥100 mRNAs correlating with microRNAs at r values >0.4 or <−0.4. There was no cutoff for signature size.
Microarray analysis.
HCT116 Dicerex5 cells were transfected with microRNAs in six-well plates, and RNA was isolated 10 h after transfection. Microarray analysis was performed as described previously (21). Genes identified by expression arrays were validated using quantitative reverse transcription PCR (qRT-PCR). Transcripts containing the miR-106b family hexamers in their 3′ UTRs were identified as previously described (32). 3′ UTRs were analyzed because miRNA binding sites are easier to distinguish from background in the 3′ UTR (see, for instance, reference 31), although a small number of targets probably have coding region sites.
mRNA and protein levels.
mRNA levels were measured by qRT-PCR on an Applied Biosystems instrument. Protein levels were measured with antibodies against p21 (Cell Signaling) in an immunohistochemistry analysis in accordance with the manufacturer's instructions. Anti-HSP70 antibodies (Santa Cruz Biotechnology) were used to test for equal loading.
Cell cycle analysis.
Human mammary epithelial cells (HMECs) immortalized by stable integration of human telomerase (42) were obtained from J. Roberts (Fred Hutchinson Cancer Research Center) and were used in all experiments unless indicated otherwise. Tumor-derived human color cancer cell lines HCT116 p21+/+ and HCT116 p21−/− were obtained from B. Vogelstein (Johns Hopkins University School of Medicine). HCT116 Dicerex5 cells were previously described (13a). A549 lung epithelial cells were obtained from ATCC.
RNA duplexes corresponding to mature microRNAs were designed as previously described (31). The mutant miR-106b duplex contains changes in nucleotides 2 and 3 of the miR-106b seed region. miRNAs (10 nM) and anti-miRs (50 nM; Exiqon) were transfected using Lipofectamine RNAiMax (Invitrogen). For small interfering RNA (siRNA)-mediated target knockdown, three siRNAs (obtained from Sigma-Proligo) designed with an algorithm that increases silencing efficiency and decreases off-target effects (21) were pooled and transfected at 25 nM of each duplex. Subsequently, the siRNA pools were deconvolved and each siRNA was transfected at 25 nM, unless otherwise indicated. The control siRNA used in all experiments targets the firefly luciferase gene, which is not present in these cells.
Transfections were performed as described previously (32). Cells were plated 24 h prior to transfection with microRNA mimics or siRNAs in six-well plates. Transfection efficiency for each cell line was optimized for greater than 85% efficiency using siRNAs against known genes. Where indicated (see the legends to Fig. 2C, 3A, 4D, and 5B), nocodazole (100 ng ml−1; Sigma Aldrich) was added 24 h after transfection for 16 to 48 h. Doxorubicin (500 nM) was added for 48 h. The supernatant from each well (floating cells) was combined with cells harvested by trypsinization (adherent cells), and cells were fixed with 95% ethanol at −20°C for 1 h. Cell cycle distributions were measured by staining with propidium iodide as described previously (32), followed by analysis on a FACSCalibur flow cytometer (Becton Dickinson). A total of 10,000 events were counted for each sample. Data were analyzed with FlowJo software (Tree Star). In each instance, flow cytometry was performed at least twice, and a representative experiment is shown in each figure.
FIG. 2.
Gain of function of the miR-106b family promotes cell cycle progression; knockdown reverses the phenotype. (A) miR-106b promotes cell division. A growth curve measuring cell numbers following transfection of a control duplex or miR-106b into HMECs shows that miR-106b promotes cell division. Error bars represent coefficients of variation. (B) miR-106b and miR-106a gain of function led to an increase in S-phase cells. HMECs were transfected with the indicated microRNA or control duplex (luciferase). BrdU incorporation was analyzed using flow cytometry. Shown are scatter plots of fluorescence intensities of BrdU incorporation (y axis) against DNA content (x axis). Magenta gates capture the S-phase populations (positive for BrdU incorporation), and the numbers depict percentages of cells in S phase. 7-AAD, 7-amino-actinomycin D. (C) The miR-106b family is required for the G1-to-S transition. HMECs were transfected with control duplex (luciferase, top left), microRNAs (middle), or anti-miRs (bottom) and treated with nocodazole for 16 h. “miR-106b mutant” has mutations in positions 2 and 3 of the seed region (top right). Cell cycle profiles were analyzed using flow cytometry. Shown are histograms of cell numbers (y axis) against DNA content (determined by measuring fluorescence intensity; x axis).
FIG. 3.
p21/CDKN1A silencing phenocopies the miR-106b family gain of function. (A) siRNA silencing of p21/CDKN1A led to reduction in G1-phase cells upon treatment with nocodazole as seen for miR-106b gain of function. Cell cycle profiles were analyzed using flow cytometry. Shown are histograms of cell numbers (y axis) against DNA content (determined by measuring fluorescence intensity; x axis). Numbers above the 2N peaks represent the percentages of G1 cells, as captured by the indicated gates. The experiment was repeated three times, and representative data are shown. (B) p21 knockdown results in an increase in S phase. HMECs were transfected with control duplex (luciferase; left), miR-106b (middle), or p21 (right) siRNAs. BrdU incorporation was analyzed using flow cytometry. Shown are scatter plots of fluorescence intensities of BrdU incorporation (y axis) against DNA content (x axis). Magenta gates capture the S-phase populations (positive for BrdU incorporation), and the numbers depict percentages of cells in S phase. 7-AAD, 7-amino-actinomycin D.
FIG. 4.
p21 is a direct, in vivo target of miR-106b. (A) Endogenous p21 mRNA levels were reduced by miR-106b gain of function. HMECs were treated with miR-106b or a luciferase control, and p21 mRNA levels were measured by qRT-PCR. Shown are relative levels normalized against the human β-glucuronidase gene (a housekeeping gene). (B) p21 protein levels were reduced by miR-106b overexpression and increased by anti-miR-106b. HMECs were treated with luciferase control, miR-106b, anti-miR-106b, or a p21 siRNA, and p21 protein levels were measured by immunoblotting. Shown is a Western blot with anti-p21 and anti-HSP70 antibodies. Numbers above lanes represent relative levels normalized against HSP70 (a housekeeping protein). (C) miR-106b family members modulate a p21 mRNA 3′ UTR reporter plasmid. The luciferase open reading frame was fused to the entire p21 mRNA 3′ UTR. Cotransfection of this construct with miR-106b family duplexes resulted in down-regulation of luciferase activity (gray bars), compared to a construct in which the miR-106b seed region complementary sites were mutated (black bars). Averages and standard deviations were calculated from two independent experiments. (D) p21 is required for the anti-miR-106b phenotype. HMECs were transfected with a control duplex (luciferase), anti-miR-106b, p21 siRNAs, or anti-miR-106b and p21 siRNAs and were treated with nocodazole for 24 h. Cell cycle profiles were analyzed using flow cytometry. Shown are histograms of cell numbers (y axis) against DNA content (determined by measuring fluorescence intensity; x axis). Numbers denote the percentages of cells in G1.
FIG. 5.
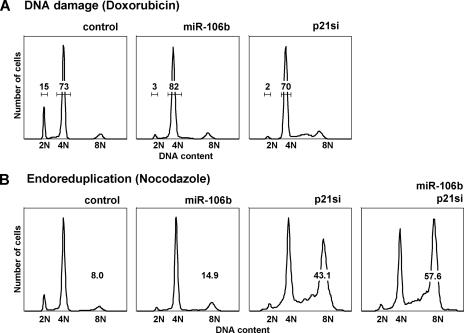
miR-106b modulates the p21-mediated checkpoint. (A) miR-106b overrides a DNA damage-induced G1 arrest. HMECs were transfected with a control duplex (luciferase), miR-106b, or p21 siRNAs and were treated with doxorubicin for 48 h. Cell cycle profiles were analyzed using flow cytometry. Shown are histograms of cell numbers (y axis) against DNA content (determined by measuring fluorescence intensity; x axis). Numbers denote the percentages of cells in G1 and G2/M, respectively. (B) miR-106b and p21 loss promote endoreduplication in nocodazole-blocked cells. HMECs were transfected with a control duplex (luciferase), miR-106b, p21 siRNAs, or miR-106b and p21 siRNAs and were treated with nocodazole for 48 h. Cell cycle profiles were analyzed using flow cytometry. Shown are histograms of cell numbers (y axis) against DNA content (determined by measuring fluorescence intensity; x axis). Numbers denote the percentages of cells with 8N DNA content.
For bromodeoxyuridine (BrdU) incorporation analysis, 48 h after transfection, HMECs were pulsed with BrdU for 1 h (BD Bioscience). Cells were fixed and stained with fluorescein isothiocyanate-conjugated anti-BrdU antibody and the DNA dye 7-amino-actinomycin D in accordance with the manufacturer's instructions (BD Biosciences).
Dual-luciferase assay.
The 3′ UTR from the human p21 gene was cloned into a vector containing the luciferase open reading frame (pSGG_3UTR; SwitchGear Genomics). Seed regions were mutated to remove complementarity to the miR-106b by using the QuikChange II XL mutagenesis kit (Stratagene). Specifically, two miR-106b-complementary sites, with sequence gcaCTtt, were changed to gcaGAtt (mutated nucleotides are capitalized) (nucleotides 1057 to 1063 and 1738 to 1744 in NM_000389). HCT116 Dicerex5 cells were cotransfected with the reporter construct and microRNAs using Lipofectamine 2000 (Invitrogen). Cells were lysed 24 h after transfection, and ratios between firefly luciferase and Renilla luciferase activity were measured with a dual-luciferase assay (Promega).
RESULTS
Expression of the miR-106b family of microRNAs is correlated with the expression of cell cycle genes.
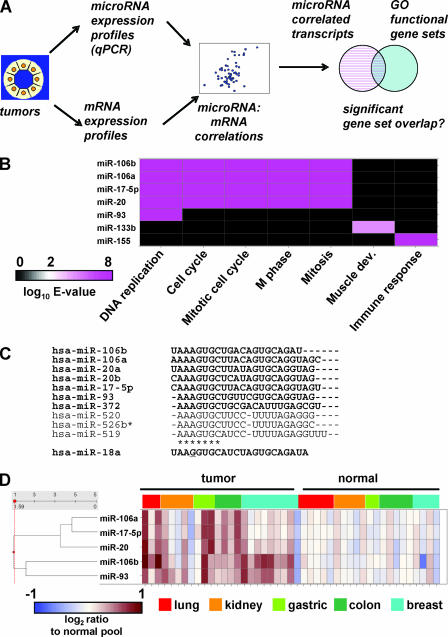
We classified microRNAs by correlating their levels with the expression of ∼40,000 transcripts (Fig. 1A) in human tumors and adjacent normal tissue samples. To functionally classify correlated transcripts, we annotated them with Gene Ontology (GO) biological process terms (1). Our rationale was that transcripts correlated with the expression of individual microRNAs and enriched for genes in known cellular pathways could indicate roles for the microRNAs in those pathways. Figure 1B depicts a heat map of the expectation (E value) of enrichment for GO biological process terms in sets of transcripts that were correlated with a subset of microRNAs. Several members of the miR-106b family (Fig. 1C) correlated with cell cycle-related transcripts (Fig. 1B). Specifically, miR-106b, miR-106a, miR-20, and miR-17-5p were correlated with transcripts involved in DNA replication and mitosis, whereas miR-93 was correlated with regulators of DNA replication. Several microRNAs with known functions were correlated with transcripts with the expected annotation. For example, miR-133b (as well as miR-1 and miR-133a [not shown]) levels correlated with transcripts annotated for muscle development (10). Also, consistent with previous studies (14, 23), miR-155 (B-cell integration cluster), a leukemogenic microRNA, correlated with transcripts regulating the immune response. Despite its association with tumorigenesis, miR-155 did not correlate significantly with cell cycle transcripts. Thus, the correlation of the miR-106b family members with cell cycle-related transcripts suggests a role for these microRNAs in cell cycle progression.
FIG. 1.
The miR-106b family. (A) A method for assigning functional annotation to microRNAs. RNA was extracted from a panel of tumor and adjacent normal tissues, and the endogenous expression levels of microRNAs and mRNA transcripts were profiled. For each microRNA, a set of transcripts whose expression levels correlated with the expression of the microRNA was identified. The correlated transcripts were annotated for GO biological process functional gene sets. A significant gene set overlap indicates that the microRNA-correlated transcripts were enriched for a particular biological process and is expressed as the E value for enrichment. (B) miR-106b family expression levels are correlated with cell cycle functional annotation in a human tumor atlas. microRNA levels measured in human tumor samples were correlated with mRNA levels in the same samples. Shown is a heat map depicting a two-dimensional cluster of E-value enrichment for the most common GO biological process terms (x axis) associated with microRNAs (y axis). The E value is a Bonferroni-corrected P value that takes into account that multiple sets were tested. (C) Sequence alignment of the miR-106b family. This is one of the largest families of microRNAs, with 18 members (not shown are four additional miR-520 and four additional miR-519 variants). *, seed region. miR-106b, miR-106a, miR-20a, miR-20b, and miR-17-5p share seed region identity. miR-93, miR-372, miR-520, miR-526*, and miR-519 have seed regions that are offset by one nucleotide, and miR-18 has a seed region with one divergent base (underlined). (D) The miR-106b family is overexpressed in tumors. microRNA levels in tumor and adjacent normal tissues from a tumor atlas were measured (38). Shown are log2 values for ratios of miR-106b family levels in tumor and normal samples to average levels of the same microRNAs in the corresponding normal tissues (normal pool).
Correlation with cell cycle transcripts predicted that the miR-106b family expression levels may be elevated in highly proliferative tissues and cancer samples. microRNAs in the miR-17-92 locus are overexpressed in B-cell lymphomas with chromosomal amplifications of this locus (19) and in some lung cancer cell lines (16). We extended these findings and compared a panel of tumor samples from several tissues. We found that miR-106b family members were overexpressed in tumor samples from five tissues compared with expression in adjacent, normal samples (Fig. 1D). Overexpression of miR-106b family members in tumor samples suggests that high levels of these microRNAs may contribute to the highly proliferative nature of the tumors. The correlation with cell cycle terms and the high levels in proliferative tissues suggest that the miR-106b family may influence tumor growth by promoting cell proliferation.
miR-106b affects cell cycle progression.
To study the effects of the miR-106b family on cell cycle progression, we transfected synthetic RNA duplexes (to mimic the microRNAs) or anti-miRs (to inhibit the microRNAs) into asynchronously growing HMECs immortalized by stable integration of human telomerase (42). The miR-106b duplex promoted cell division compared with a control duplex (Fig. 2A), whereas anti-miR-106b had no effect (data not shown).
We also analyzed cell cycle effects by flow cytometry and measurement of BrdU incorporation. Gain of function of miR-106b and miR-106a following transfection of RNA duplexes resulted in an increase in the population of cells that are in S phase by 14 to 15% in three independent experiments. Figure 2B shows a representative experiment in which 17.7% of control-treated cells were in S phase, whereas miR-106b- and miR-106a-treated cultures had 31.8% and 31.0% S-phase cells, respectively. Comparable results were observed in A549 lung carcinoma cells (see Fig. S1 in the supplemental material). microRNA mimics with unrelated seed regions (miR-18, miR-19, and miR-92; data not shown) did not cause an increase in S phase. We also failed to observe an S-phase increase with miR-93 and miR-372 mimics, suggesting that these miR-106b family members have more-subtle effects. These results indicate that select miR-106b family microRNAs specifically affect cell cycle progression during either G1 or S phase.
To investigate effects on cell cycle progression that may be too subtle to observe in asynchronously cycling cultures, we treated cells with the microtubule-depolymerizing drug nocodazole, which blocks cell cycle progression at G2/M (Fig. 2C). Compared with control-treated cells (Fig. 2C, top left), miR-106b, miR-106a, miR-20b, and miR-17-5p mimics substantially and consistently reduced the G1 population by 15 to 17% in three independent experiments, indicating that they function as positive regulators of the G1-to-S transition (Fig. 2C, middle). This phenotype is dependent on seed region complementarity, as mutating nucleotides 2 and 3 in miR-106b abrogated the effect, indicating that the phenotype is dependent on target recognition by the microRNA (Fig. 2C, top right). As with the BrdU assay described above, miR-93 and miR-372 mimics had more-subtle effects (see Fig. S2 in the supplemental material). Therefore, we concentrated on the four microRNAs with the most robust phenotypes for further study.
Inhibition of miR-106b family members causes accumulation of cells in G1.
To address whether the cell cycle function of the miR-106b family reflects the intrinsic activity of the microRNAs or an ectopic gain of function of nonphysiological levels, we used locked-nucleic-acid-based anti-miRs to suppress the endogenous microRNAs. If the miR-106b family is required for progression from G1 to S, then a disruption of mature microRNA levels by anti-miRs should result in an accumulation of cells in G1. We found that anti-miR-106b, anti-miR-106a, anti-miR-20, and anti-miR-17-5p produced a greater percentage of G1 cells after treatment with nocodazole (Fig. 2C, bottom). Even after prolonged exposure to nocodazole (72 h), a subpopulation of cells remained blocked in G1, suggesting that the miR-106b family is required for the G1-to-S transition (see Fig. S3 in the supplemental material). This observation is consistent with the results of our gain-of-function analysis and indicates that, endogenously, these microRNAs function at the G1-to-S transition.
The miR-106b family targets cell cycle regulators.
The positive effects of the miR-106b family on the cell cycle likely result from down-regulation of a gene(s) that negatively regulates cell cycle progression. To identify targets of the miR-106b family, we performed mRNA expression profiling after transfection of microRNA mimics. By microarray analysis, 103 transcripts that contained miR-106b family complementary hexamers in their 3′ UTRs were down-regulated by miR-106b, miR-106a, miR-20b, and miR-17-5p within 10 h of transfection (see Table S1 in the supplemental material), indicating that these transcripts are likely direct targets of miR-106b. Previously identified single targets of the miR-106b family (20, 35, 44) were not regulated in our experimental system, whereas transcripts from the reported miR-106b signature (8, 32) were represented in this set (see Fig. S1 in the supplemental material).
To identify the targets that mediate the cell cycle function of the miR-106b family of microRNAs, we tested whether down-regulation of individual targets phenocopied miR-106b gain of function. A subset of 88 targets that included genes down-regulated in two cell lines were tested. The miR-106b family signature contained 14 genes annotated as belonging to the “cell cycle” category by GO biological processes (see Table S2 in the supplemental material). To test whether suppression of these genes can phenocopy miR-106b family gain of function, we targeted them with siRNAs. The gene encoding the cyclin-dependent kinase inhibitor p21/CDKN1A was the target whose silencing fully reflected the phenotype (Fig. 3A and B; see Fig. S4A in the supplemental material). In addition, partial silencing of p21 to levels comparable to silencing by miR-106b (see below) produced the same phenotype (see Fig. S5 in the supplemental material). Silencing of the additional 74 targets did not reveal any that strongly phenocopied miR-106b (data not shown). Given the robust miR106b-like phenotype generated by silencing of p21 along with the well-characterized role of p21 in G1-to-S progression (reviewed in reference 40), we explored further the relationship between p21 and miR-106b.
p21 is a direct target of miR-106b.
We tested whether endogenous p21 mRNA and protein levels fluctuate upon miR-106b gain of function and knockdown. miR-106b reduced p21 mRNA levels by 38% (Fig. 4A) and p21 protein levels by 46%, as usually observed for microRNA-mediated target down-regulation (2). Multiple miR-106b family members do not synergize to further decrease the levels of p21 (data not shown). Anti-miR-106b increased p21 protein levels by 53% (Fig. 4B), consistent with p21 being a direct target of miR-106b.
The p21 mRNA 3′ UTR contains two hexamers complementary to the miR-106b family seed region. To test for direct effects on the p21 transcript by miR-106b microRNAs, we used a luciferase reporter carrying the complete p21 mRNA 3′ UTR. As shown in Fig. 4C, mimics of multiple members of the miR-106b family down-regulated the reporter. This effect depends on the seed region hexamer complements in the p21 mRNA 3′ UTR, as mutations in these sequences rendered the reporter irresponsive to microRNA transfection (Fig. 4C). In contrast, the effects of miR-106b family members do not appear to result from indirect effects on p21 gene transcription as a p21 gene promoter reporter did not respond to miR-106b transfection (data not shown). Because several miR-106b family members behaved comparably in this assay, we concentrated on miR-106b as a representative for further studies.
To further establish a functional connection between miR-106b and p21, we tested whether p21 is required for the anti-miR-106b phenotype. If the anti-miR-106b phenotype (Fig. 2C) depends on increased levels of p21 (Fig. 4C), then the absence of p21 should abrogate the effect. When we silenced p21 with an siRNA, anti-miR-106b no longer elicited an accumulation in G1 (Fig. 4D). This phenotype is not due to competition between the p21 siRNA and anti-miR-106b, as similar results were obtained with HCT116 p21−/− cells (data not shown). These results show that p21 is required for the anti-miR-106b phenotype and that the observed increase in p21 protein levels in anti-miR-106b-treated cells is not a secondary consequence of increased numbers of cells in G1.
miR-106b modulates the p21-mediated checkpoint.
Cells respond to DNA damage by arresting at G1 and G2/M to allow repair or to initiate cell death if the damage cannot be repaired (50). Activation of TP53 and p21 mediates the G1 checkpoint block (49). To test whether miR-106b affects the checkpoint functions of p21, we induced DNA damage with doxorubicin after transfection of miR-106b mimics. As shown in Fig. 5A, miR-106b prevented a DNA damage-induced G1 block (middle panel), reflecting a p21 loss-of-function phenotype (right panel). These observations suggest that miR-106b gain of function is sufficient to override a DNA damage-induced checkpoint.
p21 and TP53 also function to prevent G2/M-arrested cells from entering an unscheduled S phase and endoreduplicating into polyploid cells (Fig. 5B) (13, 26, 43). miR-106b gain of function and p21 knockdown both led to an accumulation of polyploid (8N) cells upon prolonged nocodazole block in G2/M (Fig. 5B). The miR-106b phenotype (15% 8N) is less pronounced than that elicited by the p21 siRNA (43% 8N), likely because more p21 protein remains in the microRNA-treated cells than in siRNA-treated cells (Fig. 4B). In this assay, miR-106b gain of function and p21 silencing were additive (Fig. 5B, right), indicating that additional miR-106b targets contribute to the phenotype. We also observed endoreduplication in HCT116 cells treated with miR-106b and/or null for p21 (see Fig. S4B in the supplemental material) (5, 43). In summary, miR-106b gain of function overrides the cell cycle checkpoint established by the TP53-p21 pathway, presumably due to p21 silencing.
DISCUSSION
We demonstrate an efficient approach for determining microRNA function comprising functional annotation (i.e., correlation of microRNA expression and GO annotations), phenotypic analysis to identify cellular functions, and expression profiling to select putative targets relevant to the observed phenotypes. Thus, starting with the indication that expression of miR-106b family members is related to that of cell cycle genes, we formulated and tested the hypothesis that these microRNAs play a functional role in cell cycle progression. The results of our in vitro studies have shown that the miR-106b family promotes the G1-to-S transition and targets the CDK inhibitor p21.
MicroRNA phenotypes are a result of regulation of cellular programs (8, 32) or key targets (20, 22, 28, 34, 35, 44). Consistent with the first model, the miR-106b family down-regulates multiple targets (Fig. 3A). In addition, miR-106b gain-of-function and p21 silencing have an additive effect on endoreduplication (Fig. 5B), indicating that miR-106b down-regulation of additional targets contributes to this phenotype. Nevertheless, individual silencing of other targets did not phenocopy miR-106b gain of function, whereas silencing of p21 to levels reflecting miR-106b-mediated knockdown recapitulated several microRNA phenotypes (Fig. 3 and 5 and data not shown). Thus, p21 is a key target for the cell cycle function of the miR-106b family.
Our phenotypic analyses revealed that the miR-106b family promotes exit from G1 and entry into S phase. These data are consistent with a model in which the miR-106b family accelerates the G1-to-S transition, as has been observed in cells depleted of negative regulators of S-phase entry (e.g., Rb [39]).
Similar to our findings, the cyclin-dependent kinase inhibitor p27Kip1 is regulated by miR-221 and miR-222 (15, 29). Down-regulation of p27Kip1 by miR-221/222 promotes cancer cell proliferation (15, 29), whereas down-regulation of miR-221/222 by anti-miRs arrests the cell cycle, indicating a dependence on elevated miR-221/222 levels for cancer cell proliferation. Indeed, miR-221 is often upregulated in cancer cell lines and tumor samples (11, 17, 27, 36). Thus, an emerging common theme is that microRNAs that down-regulate negative regulators of cell cycle progression are overexpressed in tumors.
The miR-106b family is emerging as an important component of the cellular networks relevant to oncology. The presence of high microRNA levels in tumor samples suggests a causal relationship between microRNA abundance and promotion of cell division. Two miR-106b family members, miR-17-5p and miR-20a, reside in an oncogenic microRNA polycistron (16, 19, 35, 45). Our findings suggest that regulation of the cell cycle by these microRNAs contributes in part to the mechanism by which they drive tumor progression. Thus, the oncogenic properties of the miR-106b family of microRNAs may stem from combined positive regulation of the cell cycle and additional functions.
Supplementary Material
Acknowledgments
We thank the Rosetta Inpharmatics Gene Expression Lab for microarray analysis, Rosetta Biosoftware for data analysis software, and members of the Cancer Biology Department for invaluable discussions and support. We thank James Roberts, Fred Hutchinson Cancer Research Center, for advice and critical reading of the manuscript.
Rosetta Inpharmatics LLC is a wholly owned subsidiary of Merck & Co., Inc.
Footnotes
Published ahead of print on 22 January 2008.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Ashburner, M., C. A. Ball, J. A. Blake, D. Botstein, H. Butler, J. M. Cherry, A. P. Davis, K. Dolinski, S. S. Dwight, J. T. Eppig, M. A. Harris, D. P. Hill, L. Issel-Tarver, A. Kasarskis, S. Lewis, J. C. Matese, J. E. Richardson, M. Ringwald, G. M. Rubin, G. Sherlock, et al. 2000. Gene ontology: tool for the unification of biology. Nat. Genet. 2525-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartel, D. P., and C. Z. Chen. 2004. Micromanagers of gene expression: the potentially widespread influence of metazoan microRNAs. Nat. Rev. Genet. 5396-400. [DOI] [PubMed] [Google Scholar]
- 3.Reference deleted.
- 4.Bommer, G. T., I. Gerin, Y. Feng, A. J. Kaczorowski, R. Kuick, R. E. Love, Y. Zhai, T. J. Giordano, Z. S. Qin, B. B. Moore, O. A. Macdougald, K. R. Cho, and E. R. Fearon. 2007. p53-mediated activation of miRNA34 candidate tumor-suppressor genes. Curr. Biol. 171298-1307. [DOI] [PubMed] [Google Scholar]
- 5.Bunz, F., A. Dutriaux, C. Lengauer, T. Waldman, S. Zhou, J. P. Brown, J. M. Sedivy, K. W. Kinzler, and B. Vogelstein. 1998. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science 2821497-1501. [DOI] [PubMed] [Google Scholar]
- 6.Calin, G. A., C. D. Dumitru, M. Shimizu, R. Bichi, S. Zupo, E. Noch, H. Aldler, S. Rattan, M. Keating, K. Rai, L. Rassenti, T. Kipps, M. Negrini, F. Bullrich, and C. M. Croce. 2002. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. USA 9915524-15529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Calin, G. A., C. G. Liu, C. Sevignani, M. Ferracin, N. Felli, C. D. Dumitru, M. Shimizu, A. Cimmino, S. Zupo, M. Dono, M. L. Dell'Aquila, H. Alder, L. Rassenti, T. J. Kipps, F. Bullrich, M. Negrini, and C. M. Croce. 2004. MicroRNA profiling reveals distinct signatures in B cell chronic lymphocytic leukemias. Proc. Natl. Acad. Sci. USA 10111755-11760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carleton, M., M. A. Cleary, and P. S. Linsley. 2007. MicroRNAs and cell cycle regulation. Cell Cycle 62127-2132. [DOI] [PubMed] [Google Scholar]
- 9.Chang, T. C., E. A. Wentzel, O. A. Kent, K. Ramachandran, M. Mullendore, K. H. Lee, G. Feldmann, M. Yamakuchi, M. Ferlito, C. J. Lowenstein, D. E. Arking, M. A. Beer, A. Maitra, and J. T. Mendell. 2007. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol. Cell 26745-752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, J. F., E. M. Mandel, J. M. Thomson, Q. Wu, T. E. Callis, S. M. Hammond, F. L. Conlon, and D. Z. Wang. 2006. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat. Genet. 38228-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ciafre, S. A., S. Galardi, A. Mangiola, M. Ferracin, C. G. Liu, G. Sabatino, M. Negrini, G. Maira, C. M. Croce, and M. G. Farace. 2005. Extensive modulation of a set of microRNAs in primary glioblastoma. Biochem. Biophys. Res. Commun. 3341351-1358. [DOI] [PubMed] [Google Scholar]
- 12.Cimmino, A., G. A. Calin, M. Fabbri, M. V. Iorio, M. Ferracin, M. Shimizu, S. E. Wojcik, R. I. Aqeilan, S. Zupo, M. Dono, L. Rassenti, H. Alder, S. Volinia, C. G. Liu, T. J. Kipps, M. Negrini, and C. M. Croce. 2005. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc. Natl. Acad. Sci. USA 10213944-13949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cross, S. M., C. A. Sanchez, C. A. Morgan, M. K. Schimke, S. Ramel, R. L. Idzerda, W. H. Raskind, and B. J. Reid. 1995. A p53-dependent mouse spindle checkpoint. Science 2671353-1356. [DOI] [PubMed] [Google Scholar]
- 13a.Cummins, J. M., et al. 2006. The colorectal microRNAome. Proc. Natl. Acad. Sci. USA 1033687-3692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eis, P. S., W. Tam, L. Sun, A. Chadburn, Z. Li, M. F. Gomez, E. Lund, and J. E. Dahlberg. 2005. Accumulation of miR-155 and BIC RNA in human B cell lymphomas. Proc. Natl. Acad. Sci. USA 1023627-3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gillies, J. K., and I. A. Lorimer. 2007. Regulation of p27Kip1 by miRNA 221/222 in glioblastoma. Cell Cycle 62005-2009. [DOI] [PubMed] [Google Scholar]
- 16.Hayashita, Y., H. Osada, Y. Tatematsu, H. Yamada, K. Yanagisawa, S. Tomida, Y. Yatabe, K. Kawahara, Y. Sekido, and T. Takahashi. 2005. A polycistronic microRNA cluster, miR-17-92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res. 659628-9632. [DOI] [PubMed] [Google Scholar]
- 17.He, H., K. Jazdzewski, W. Li, S. Liyanarachchi, R. Nagy, S. Volinia, G. A. Calin, C. G. Liu, K. Franssila, S. Suster, R. T. Kloos, C. M. Croce, and A. de la Chapelle. 2005. The role of microRNA genes in papillary thyroid carcinoma. Proc. Natl. Acad. Sci. USA 10219075-19080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He, L., X. He, L. P. Lim, E. de Stanchina, Z. Xuan, Y. Liang, W. Xue, L. Zender, J. Magnus, D. Ridzon, A. L. Jackson, P. S. Linsley, C. Chen, S. W. Lowe, M. A. Cleary, and G. J. Hannon. 2007. A microRNA component of the p53 tumour suppressor network. Nature 4471130-1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He, L., J. M. Thomson, M. T. Hemann, E. Hernando-Monge, D. Mu, S. Goodson, S. Powers, C. Cordon-Cardo, S. W. Lowe, G. J. Hannon, and S. M. Hammond. 2005. A microRNA polycistron as a potential human oncogene. Nature 435828-833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hossain, A., M. T. Kuo, and G. F. Saunders. 2006. Mir-17-5p regulates breast cancer cell proliferation by inhibiting translation of AIB1 mRNA. Mol. Cell. Biol. 268191-8201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jackson, A. L., S. R. Bartz, J. Schelter, S. V. Kobayashi, J. Burchard, M. Mao, B. Li, G. Cavet, and P. S. Linsley. 2003. Expression profiling reveals off-target gene regulation by RNAi. Nat. Biotechnol. 21635-637. [DOI] [PubMed] [Google Scholar]
- 22.Johnson, S. M., H. Grosshans, J. Shingara, M. Byrom, R. Jarvis, A. Cheng, E. Labourier, K. L. Reinert, D. Brown, and F. J. Slack. 2005. RAS is regulated by the let-7 microRNA family. Cell 120635-647. [DOI] [PubMed] [Google Scholar]
- 23.Kluiver, J., S. Poppema, D. de Jong, T. Blokzijl, G. Harms, S. Jacobs, B. J. Kroesen, and A. van den Berg. 2005. BIC and miR-155 are highly expressed in Hodgkin, primary mediastinal and diffuse large B cell lymphomas. J. Pathol. 207243-249. [DOI] [PubMed] [Google Scholar]
- 24.Landais, S., S. Landry, P. Legault, and E. Rassart. 2007. Oncogenic potential of the miR-106-363 cluster and its implication in human T-cell leukemia. Cancer Res. 675699-5707. [DOI] [PubMed] [Google Scholar]
- 25.Landgraf, P., M. Rusu, R. Sheridan, A. Sewer, N. Iovino, A. Aravin, S. Pfeffer, A. Rice, A. O. Kamphorst, M. Landthaler, C. Lin, N. D. Socci, L. Hermida, V. Fulci, S. Chiaretti, R. Foa, J. Schliwka, U. Fuchs, A. Novosel, R. U. Muller, B. Schermer, U. Bissels, J. Inman, Q. Phan, M. Chien, D. B. Weir, R. Choksi, G. De Vita, D. Frezzetti, H. I. Trompeter, V. Hornung, G. Teng, G. Hartmann, M. Palkovits, R. Di Lauro, P. Wernet, G. Macino, C. E. Rogler, J. W. Nagle, J. Ju, F. N. Papavasiliou, T. Benzing, P. Lichter, W. Tam, M. J. Brownstein, A. Bosio, A. Borkhardt, J. J. Russo, C. Sander, M. Zavolan, and T. Tuschl. 2007. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 1291401-1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lanni, J. S., and T. Jacks. 1998. Characterization of the p53-dependent postmitotic checkpoint following spindle disruption. Mol. Cell. Biol. 181055-1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee, E. J., Y. Gusev, J. Jiang, G. J. Nuovo, M. R. Lerner, W. L. Frankel, D. L. Morgan, R. G. Postier, D. J. Brackett, and T. D. Schmittgen. 2007. Expression profiling identifies microRNA signature in pancreatic cancer. Int J. Cancer 1201046-1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee, Y. S., and A. Dutta. 2007. The tumor suppressor microRNA let-7 represses the HMGA2 oncogene. Genes Dev. 211025-1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.le Sage, C., R. Nagel, D. A. Egan, M. Schrier, E. Mesman, A. Mangiola, C. Anile, G. Maira, N. Mercatelli, S. A. Ciafre, M. G. Farace, and R. Agami. 2007. Regulation of the p27(Kip1) tumor suppressor by miR-221 and miR-222 promotes cancer cell proliferation. EMBO J. 263699-3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lewis, B. P., I. H. Shih, M. W. Jones-Rhoades, D. P. Bartel, and C. B. Burge. 2003. Prediction of mammalian microRNA targets. Cell 115787-798. [DOI] [PubMed] [Google Scholar]
- 31.Lim, L. P., N. C. Lau, P. Garrett-Engele, A. Grimson, J. M. Schelter, J. Castle, D. P. Bartel, P. S. Linsley, and J. M. Johnson. 2005. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 433769-773. [DOI] [PubMed] [Google Scholar]
- 32.Linsley, P. S., J. Schelter, J. Burchard, M. Kibukawa, M. M. Martin, S. R. Bartz, J. M. Johnson, J. M. Cummins, C. K. Raymond, H. Dai, N. Chau, M. Cleary, A. L. Jackson, M. Carleton, and L. Lim. 2007. Transcripts targeted by the microRNA-16 family cooperatively regulate cell cycle progression. Mol. Cell. Biol. 272240-2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu, J., G. Getz, E. A. Miska, E. Alvarez-Saavedra, J. Lamb, D. Peck, A. Sweet-Cordero, B. L. Ebert, R. H. Mak, A. A. Ferrando, J. R. Downing, T. Jacks, H. R. Horvitz, and T. R. Golub. 2005. MicroRNA expression profiles classify human cancers. Nature 435834-838. [DOI] [PubMed] [Google Scholar]
- 34.Mayr, C., M. T. Hemann, and D. P. Bartel. 2007. Disrupting the pairing between let-7 and Hmga2 enhances oncogenic transformation. Science 3151576-1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.O'Donnell, K. A., E. A. Wentzel, K. I. Zeller, C. V. Dang, and J. T. Mendell. 2005. c-Myc-regulated microRNAs modulate E2F1 expression. Nature 435839-843. [DOI] [PubMed] [Google Scholar]
- 36.Pallante, P., R. Visone, M. Ferracin, A. Ferraro, M. T. Berlingieri, G. Troncone, G. Chiappetta, C. G. Liu, M. Santoro, M. Negrini, C. M. Croce, and A. Fusco. 2006. MicroRNA deregulation in human thyroid papillary carcinomas. Endocr. Relat. Cancer 13497-508. [DOI] [PubMed] [Google Scholar]
- 37.Raver-Shapira, N., E. Marciano, E. Meiri, Y. Spector, N. Rosenfeld, N. Moskovits, Z. Bentwich, and M. Oren. 2007. Transcriptional activation of miR-34a contributes to p53-mediated apoptosis. Mol. Cell 26731-743. [DOI] [PubMed] [Google Scholar]
- 38.Raymond, C. K., B. S. Roberts, P. Garrett-Engele, L. P. Lim, and J. M. Johnson. 2005. Simple, quantitative primer-extension PCR assay for direct monitoring of microRNAs and short-interfering RNAs. RNA 111737-1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sage, J., G. J. Mulligan, L. D. Attardi, A. Miller, S. Chen, B. Williams, E. Theodorou, and T. Jacks. 2000. Targeted disruption of the three Rb-related genes leads to loss of G1 control and immortalization. Genes Dev. 143037-3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sherr, C. J., and J. M. Roberts. 1999. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev. 131501-1512. [DOI] [PubMed] [Google Scholar]
- 41.Si, M. L., S. Zhu, H. Wu, Z. Lu, F. Wu, and Y. Y. Mo. 2007. miR-21-mediated tumor growth. Oncogene 262799-2803. [DOI] [PubMed] [Google Scholar]
- 42.Smith, L. L., H. A. Coller, and J. M. Roberts. 2003. Telomerase modulates expression of growth-controlling genes and enhances cell proliferation. Nat. Cell Biol. 5474-479. [DOI] [PubMed] [Google Scholar]
- 43.Stewart, Z. A., S. D. Leach, and J. A. Pietenpol. 1999. p21Waf1/Cip1 inhibition of cyclin E/Cdk2 activity prevents endoreduplication after mitotic spindle disruption. Mol. Cell Biol. 19205-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sylvestre, Y., V. De Guire, E. Querido, U. K. Mukhopadhyay, V. Bourdeau, F. Major, G. Ferbeyre, and P. Chartrand. 2007. An E2F/miR-20a autoregulatory feedback loop. J. Biol. Chem. 2822135-2143. [DOI] [PubMed] [Google Scholar]
- 45.Tagawa, H., K. Karube, S. Tsuzuki, K. Ohshima, and M. Seto. 2007. Synergistic action of the microRNA-17 polycistron and Myc in aggressive cancer development. Cancer Sci. 981482-1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tarasov, V., P. Jung, B. Verdoodt, D. Lodygin, A. Epanchintsev, A. Menssen, G. Meister, and H. Hermeking. 2007. Differential Regulation of microRNAs by p53 revealed by massively parallel sequencing: miR-34a is a p53 target that induces apoptosis and G1-arrest. Cell Cycle 61586-1593. [DOI] [PubMed] [Google Scholar]
- 47.Tazawa, H., N. Tsuchiya, M. Izumiya, and H. Nakagama. 2007. Tumor-suppressive miR-34a induces senescence-like growth arrest through modulation of the E2F pathway in human colon cancer cells. Proc. Natl. Acad. Sci. USA 10415472-15477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Voorhoeve, P. M., C. le Sage, M. Schrier, A. J. Gillis, H. Stoop, R. Nagel, Y. P. Liu, J. van Duijse, J. Drost, A. Griekspoor, E. Zlotorynski, N. Yabuta, G. De Vita, H. Nojima, L. H. Looijenga, and R. Agami. 2006. A genetic screen implicates miRNA-372 and miRNA-373 as oncogenes in testicular germ cell tumors. Cell 1241169-1181. [DOI] [PubMed] [Google Scholar]
- 49.Waldman, T., K. W. Kinzler, and B. Vogelstein. 1995. p21 is necessary for the p53-mediated G1 arrest in human cancer cells. Cancer Res. 555187-5190. [PubMed] [Google Scholar]
- 50.Zhou, B. B., and S. J. Elledge. 2000. The DNA damage response: putting checkpoints in perspective. Nature 408433-439. [DOI] [PubMed] [Google Scholar]
- 51.Zhu, S., M. L. Si, H. Wu, and Y. Y. Mo. 2007. MicroRNA-21 targets the tumor suppressor gene tropomyosin 1 (TPM1). J. Biol. Chem. 28214328-14336. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.