Abstract
Centrosomes nucleate and organize interphase microtubules and are instrumental in mitotic bipolar spindle assembly, ensuring orderly cell cycle progression with accurate chromosome segregation. We report that the multifunctional structural protein 4.1R localizes at centrosomes to distal/subdistal regions of mature centrioles in a cell cycle-dependent pattern. Significantly, 4.1R-specific depletion mediated by RNA interference perturbs subdistal appendage proteins ninein and outer dense fiber 2/cenexin at mature centrosomes and concomitantly reduces interphase microtubule anchoring and organization. 4.1R depletion causes G1 accumulation in p53-proficient cells, similar to depletion of many other proteins that compromise centrosome integrity. In p53-deficient cells, 4.1R depletion delays S phase, but aberrant ninein distribution is not dependent on the S-phase delay. In 4.1R-depleted mitotic cells, efficient centrosome separation is reduced, resulting in monopolar spindle formation. Multipolar spindles and bipolar spindles with misaligned chromatin are also induced by 4.1R depletion. Notably, all types of defective spindles have mislocalized NuMA (nuclear mitotic apparatus protein), a 4.1R binding partner essential for spindle pole focusing. These disruptions contribute to lagging chromosomes and aberrant microtubule bridges during anaphase/telophase. Our data provide functional evidence that 4.1R makes crucial contributions to the structural integrity of centrosomes and mitotic spindles which normally enable mitosis and anaphase to proceed with the coordinated precision required to avoid pathological events.
Centrosomes nucleate and organize interphase microtubules and direct assembly of mitotic bipolar spindles responsible for accurate chromosome segregation in somatic vertebrate cells. Precise distribution of duplicated chromosomes to daughter cells is of paramount importance, since aberrant cell division is associated with genetic diseases and aneuploidy is a hallmark of malignant tumors (33). Centrosomes consist of a cylindrical tubulin-rich centriole pair, the more mature (or mother) centriole having “appendages” at its distal end appearing to anchor microtubules (42). Several proteins such as EB1, p150/glued, and centriolin as well as distal appendage proteins, including ninein, outer dense fiber 2 (ODF2)/cenexin, and ɛ-tubulin differentiate mature from immature centrioles (2, 3, 15, 32, 44). Recently, it was shown in Chlamydomonas that the mature centriole can position the daughter centriole and orient the nucleus (11) and that in Drosophila asymmetric cell division, the mature centrosome is always inherited by the stem cell (68). Surrounding centrioles is a fibrogranular matrix-like structure termed the pericentriolar material (PCM) containing many protein complexes. For example, γ-tubulin ring complexes (γ-TURCs) act as templates for nucleating microtubule growth (43, 59, 71) and pericentrin scaffolds appear to contact γ-TURCs for microtubule nucleation and anchorage (7a). Other proteins such as PCM-1 likely mediate microtubule anchoring via recruitment and assembly of subsets of centrosomal proteins (4).
Centrosomes are very dynamic organelles duplicating, maturing, and separating in stages coupled with cell cycle progression. During G1, the centrosome contains a mature mother and an immature daughter centriole. During S phase, each centriole spawns a procentriole such that by S/G2, cells have two centrosomes, each of which contains a mother-daughter pair. In G2/M, centrosomes separate and form the spindle poles during mitosis. In many cases, loss of centrosome integrity, induced by depleting a centrosome protein, causes a p53-dependent G1/S arrest, sometimes even leading to increased senescence (41, 61, 63). Although recently much has been learned about centrosomes, centrosomal structural components and functions remain only partially characterized, and mechanisms controlling centrosome duplication and interrelationships with cell cycle progression are not yet completely understood (47, 62).
The multifunctional structural protein 4.1 was recently discovered to be an important new structural component of cell division machinery, detected in centrosomes and mitotic spindles as well as the midbody and the nucleus (5, 7, 29, 30, 36, 37). Protein 4.1 formerly had been identified only as a crucial red cell adaptor protein, stabilizing spectrin-actin lattices and anchoring them to plasma membrane proteins. However, as adaptor proteins in nucleated cells, 4.1 isoforms could serve to integrate centrosomal structural components and thus be critical for the fidelity of centrosomal functions, such as mitotic spindle assembly, cytokinesis, and microtubule organization, regulating cell shape, polarity, motility, and intracellular transport (20, 27, 28, 53).
Previously, we reported that protein 4.1 is an integral or “core” centrosome component, present at centrosomes independent of microtubules (29). We also detected 4.1 epitopes in the basal bodies of murine, porcine, and Xenopus sperm (31). By confocal microscopy in mammalian cells, 4.1 colocalizes with centriolar tubulin and was detected in the surrounding PCM and on fibers connecting centriolar pairs (29). At higher resolution of whole-mount electron microscopy, 4.1 epitopes have a polar distribution on centrioles and decorate fibrous structures extending into the PCM (29). We used the cell-free Xenopus egg extract system to probe 4.1 functions and showed by depletion/add back that 4.1 is essential for spindle, centrosome aster, and even self-assembled microtubule aster assembly. Furthermore, dominant-negative peptides corresponding to 4.1 domains impaired microtubule dynamics and organization (31). Importantly, 4.1 has binding sites for microtubules (51), the spindle pole focusing protein NuMA (nuclear mitotic apparatus protein) (36, 37), and CPAP (centrosome protein 4.1-associated protein) (24), a regulator of microtubule dynamics.
The essential involvement of 4.1 in centrosome and spindle biogenesis is potentially very significant within a larger context of cell and tissue physiology, since defective centrosomes can affect S-phase entry and the accuracy of mitosis and cytokinesis (8, 9, 41, 60, 61). Recently, a 4.1 gene family was discovered, necessitating refinement in defining and distinguishing 4.1 family protein distributions, functions in subcellular structures, and possible effects on the cell cycle. The prototypical 4.1, now called 4.1R (red cell), is abundant both in erythroid tissues and nonerythroid cells. Other 4.1 genes have different mRNA distribution patterns: 4.1G is generally distributed (48, 65), 4.1N is predominantly neuronal (64), and 4.1B is detected mostly in the brain (50, 67). These genes have some conserved subdomains in common (49) as well as unique regions, and some tissues may express more than one 4.1 gene (7, 49, 58, 66). Thus, an important question is whether these 4.1 proteins have unique, redundant, or synergistic functions.
We report here that at centrosomes 4.1R is specifically a mature centriole protein, predominately localizing to distal/subdistal regions of mature centrioles in a cell cycle-dependent pattern. To selectively probe 4.1R function, we used RNA interference (RNAi)-mediated depletion without affecting 4.1G levels in cells expressing only these two 4.1 family members. By abrogating 4.1R function, we characterized a pleiotropic phenotype involving altered centrosomal and spindle structural and functional integrity, impaired anaphase/telophase, and an S-phase delay in transformed cells. Importantly, the effects of 4.1R depletion on centrosome integrity were not dependent on its effects on cell cycle progression. In untransformed cells with intact p53, 4.1R depletion induced G1 accumulation, similar to the effect reported for depletion of many other centrosomal proteins (41, 61, 63).
MATERIALS AND METHODS
Cells and media.
WI38, CaSki, and HeLa cells were obtained from American Type Culture Collection and grown at 37°C with 5% CO2. WI38 and HeLa cells were cultured in Dulbecco modified Eagle medium-H21 (Gibco BRL) and CaSki in RPMI 1640 (Cell Gro), both supplemented with 10% fetal calf serum and penicillin-streptomycin, as described previously (30). RPE-1 cells (Clontech) were cultured in Dulbecco modified Eagle medium-F-12. WI38 and HeLa cells were synchronized by double thymidine block using 2 mM thymidine. After an initial 16-h block, thymidine was washed out, and cells were released in S phase and then reblocked with thymidine for 15 h.
Immunofluorescence.
Cells grown on coverslips were fixed for 10 min in −20°C methanol and stained by indirect immunofluorescence as described previously (30). Anticentrin (20H5) was the kind gift of J. Salisbury (Mayo Clinic Foundation, Rochester, MN), mouse antininein was provided by G. Chan (Alberta Cancer Board, Alberta, Canada), rabbit anti-C-Nap1 was from E. Nigg (Max-Planck Institute, Martinsreid, Germany), anti-ODF2 MAb101 was from S. Tsukita (Kyoto University, Japan), and CREST serum was from A. von Hooser (University of California-Berkeley, Berkeley, CA). Affinity-purified rabbit antibodies against 4.1R and N were described previously (30, 58, 70). Chicken anti-4.1G was raised against a six-His peptide encoding the U1 region of human 4.1G (EP41L2) containing 217 amino acids following the AUG1 start site (48). Affinity-purified immune immunoglobulin Y was analyzed to confirm no cross-reaction with recombinant 4.1R, 4.1B, or 4.1N or with red blood cell 80-kDa 4.1R or an irrelevant six-His peptide. Commercial antibodies were rat antitubulin YL1/2 (Abcam), monoclonal GTU88 against γ-tubulin (Sigma), monoclonal anti-p150, monoclonal anti-C-Nap1 (BD Transduction Labs), rabbit antipericentrin (Covance), rabbit anti-Ki67 (BD Biosciences), and rabbit anti-phosphohistone 3 (Upstate). Secondary antibodies with minimal species cross-reactivity were from Molecular Probes or Jackson ImmunoResearch. Parallel samples probed with equal amounts of control nonimmune immunoglobulin G (IgG) or without primary antibody showed no fluorescent patterns under conditions used for experimental samples. DNA was stained with 4,6-diamidino-2-phenylindole (DAPI) (Vectorshield; VectoLabs). Images were acquired with a Nikon Eclipse 2000 using a 60× lens objective with a numerical aperture of 1.4 equipped with a Retiga Ex camera and ImagePro or, where indicated as deconvolved, with a Deltavision microscope on an Olympus IX70 platform with a 100× lens objective with a numerical aperture of 1.35. Images were processed using Adobe Photoshop, and quantification of immunofluorescence signals was done using ImageJ (http://rsb.info.nih.gov/ij/). Ninein immunofluorescence was scored as abnormal not only when the signals were dispersed relative to the circumscribed pattern in control cells but also when the pixels and area exceeded at least twice or were less than half that of the averages measured in 15 representative immunostained control cells.
4.1R RNAi.
Small interfering RNAs (siRNAs) against human 4.1R from Dharmacon were as follows: Duplex1, GAAAGUCUGUGUAGAACAUUU; Duplex 2, UGACACAGUUUAUGAAUGUUU; and Duplex 3, GGAUCCAAAUUUCGAUACAUU. Each duplex alone as well as a pool of all three specifically downregulated 4.1R expression. Control cells were transfected with duplex GCUAAGAAAUUAUGGAAAGUU, or with Lipofectamine 2000 alone. Each 35-mm well containing 2 × 105 cells was transfected with 100 nM RNA duplex complexed to Lipofectamine 2000 in the absence of antibiotics per the manufacturer's instructions. The transfection efficiency was generally about 85%. Downregulation of 4.1R expression by siRNA was detected by Western blotting, normalizing to actin staining, beginning at 48 h and was maximal 72 to 120 h. At 96 h in HeLa cells treated with 4.1R RNAi and immunostained for 4.1R, >80% of the cells showed 4.1R depletion ranging from 100% to 80% of controls. To determine the relative growth rates, cells were trypsinized and counted.
Microtubule regrowth.
siRNA-transfected and control HeLa or CaSki cells were incubated for 30 min at 4°C and then with 33 μM nocodazole for 30 min at 37°C to completely depolymerize microtubules. After washing out nocodazole with fresh media, regrowth was induced in cells incubated for indicated times at 37°C in prewarmed media. Samples were washed in phosphate-buffered saline, pH 7.4, containing 2.7 mM KCl, 1.5 mM KH2PO4, 1 mM MgCl2, 1 mM EGTA, 137 mM NaCl, and 8.1 mM NaHPO4 and fixed in −20°C methanol for immunostaining.
Flow cytometry.
Cells were either stained with propidium iodide alone or pulse-labeled for 30 min with 10 μg/ml bromodeoxyuridine (BrdU) (Sigma), fixed in methanol, and stained with propidium iodine and anti-BrdU (Becton Dickinson). Ten thousand cells were collected for each sample on a FACSCalibur (Becton Dickinson) and analyzed using Cellquest Pro (Becton Dickinson). Cells labeled with propidium iodide alone were analyzed using a Guava EasyCyte (Guava Technologies).
RESULTS
Protein 4.1R preferentially localizes to mature (“mother”) centrioles in the distal/subdistal region.
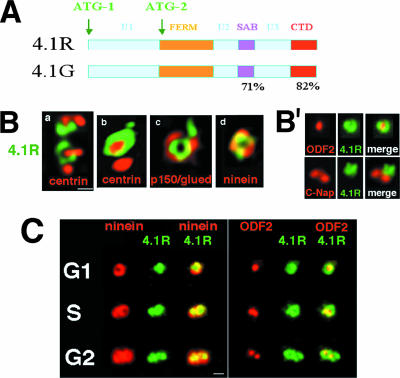
4.1R and 4.1G contain highly homologous spectrin/actin binding and C-terminal domains, two domains we reported as critical for microtubule organization and dynamics in a cell extract system (31) (Fig. 1A). To discriminate between these two 4.1 family members, we used antibodies tested to be specific for 4.1R and detected focal 4.1R immunofluorescent signals at centrosomes in a variety of cultured mammalian cells. Using deconvolution analysis, in asynchronous populations, 4.1R localized close to centrioles labeled with anticentrin (Fig. 1B, panels a and b).
FIG. 1.
Protein 4.1R epitopes are at mature centrioles. (A) Schematic map of 4.1R and 4.1G indicating three unique regions (U1 to U3) and three homologous domains: the highly homologous membrane-binding 30-kDa/FERM domain, the spectrin/actin binding domain (SAB), and the C-terminal domain (CTD). The percentages below the schematic map indicate the degree of homology, and arrows indicate translation initiation sites. (B) Deconvolution sections of WI38 human fibroblast centrosomes stained for indicated proteins. Similar results were obtained using HeLa and CaSki cells. (B′) Localization of 4.1R predominately at the distal/subdistal region of maternal centrioles of CaSki cells by indirect immunofluorescence. Proximal centriole regions were labeled with anti-C-Nap1, and distal/subdistal regions were labeled with anti-ODF2. Staining of HeLa cells gave similar patterns. (C) Distribution of 4.1R epitopes during cell cycle progression. WI38 human fibroblasts were synchronized using double thymidine block. By indirect immunofluorescence, 4.1R localizes exclusively to centrioles also containing ninein or ODF2 epitopes. Similar results were obtained using HeLa and CaSki cells. Bars in panel Ba and panel C = 0.5 μm; Bb to d, 2× magnification.
To more precisely investigate these centrosomal distributions, we compared 4.1R distribution to several key centrosomal proteins. Significantly, 4.1R localized at one of two or two of four centrioles labeled by centrin staining in several different cell types (Fig. 1B, panels a and b) or at a subset of supernumerary centrioles, as observed in human osteosarcoma U-2 OS cells (unpublished data). The 4.1R epitopes were consistently at centrioles bearing mature centriole marker p150/glued, part of the dynein/dynactin microtubule motor complex (Fig. 1B, panel c) (35, 56, 57) and colocalized extensively with ninein (Fig. 1B, panel d), a signature component of subdistal appendages of mature centrioles. Furthermore, 4.1R epitopes were in close proximity to another distal/subdistal appendage protein ODF2/cenexin but did not colocalize extensively with C-Nap1, a protein known to localize to proximal ends of centrioles (Fig. 1B′) (12, 22, 25, 32, 44). Taken together, these experiments define 4.1R as a newly recognized component of the distal/subdistal area of mature centrioles as observed by deconvolution microscopy and localization relative to centrosomal markers.
Distribution of centrosomal 4.1R during progression through interphase.
As a protein component of mature centrioles, 4.1R would be expected to associate with the second maturing centriole during completion of G2 phase. To test this prediction, we analyzed the distributions of 4.1R epitopes in synchronized diploid human fibroblasts using the microtubule anchoring protein ninein as a mature centriole marker (46). 4.1R colocalized with ninein at a single centriole in a ring-shaped distribution in G1, extended to a second ninein-labeled maturing centriole during S phase (one and a half rings), and colocalized with ninein at both mature centrioles (two-ring pattern) during G2 (Fig. 1C) (46). We also observed that 4.1R had a similar cell cycle-dependent distribution relative to ODF2 epitopes (44) (Fig. 1C). Therefore, centrosomal 4.1R localization varies during cell cycle progression, accumulating at centrioles as they mature as marked by ninein and ODF2.
4.1R RNAi affects microtubule organization during interphase.
Because 4.1R localization at mature and newly maturing centrioles appeared to be specifically in the region of distal/subdistal appendages, we reasoned that centriole morphogenesis and microtubule organization would be affected in cells depleted of 4.1R. To test this hypothesis, we first surveyed 4.1 family members expressed in various mammalian cells using gene-specific antibodies to identify cells that expressed only 4.1R. No cell line tested expressed exclusively 4.1R. However, CaSki cells and human diploid fibroblasts contained only 4.1R and 4.1G, a combination of 4.1 proteins that therefore must be sufficient for cell division. HeLa and U-2 OS cells additionally contain minor amounts of 4.1N, while thymocytes expressed 4.1R, 4.1G, and 4.1B (data not shown).
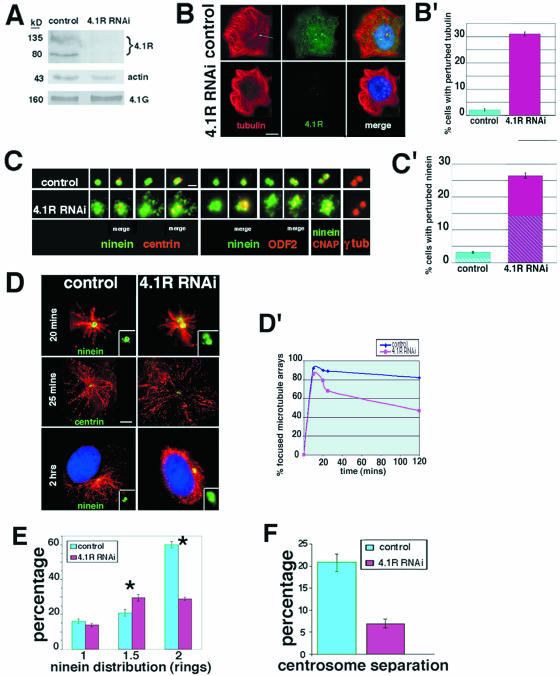
To explore 4.1R functions in intact cells, RNAi-mediated protein depletion experiments were performed. After transfection with a mixture of three RNA duplexes targeting 4.1R mRNA, Western blot analysis of a time course experiment showed that 4.1R protein levels were significantly reduced (70 to 85%) at 72 to 120 h (Fig. 2A). No significant effects on protein levels were detected at 24 h, and only partial inhibition (∼50%) was evident at 48 h (data not shown). It was crucial to evaluate whether 4.1R depletion also decreased 4.1G, since this family member contains regions that are highly homologous. 4.1G expression did not decline in RNAi-treated cells relative to controls; thus, silencing was specific for 4.1R expression (Fig. 2A). Transfection with individual RNAi duplexes also specifically targeted 4.1R, providing an additional control for off-target effects. A fourth control duplex with a variant sequence or mock transfection (Lipofectamine 2000 without duplex) did not affect 4.1R expression.
FIG. 2.
After downregulation of 4.1R expression, ninein is perturbed and microtubules become disorganized. (A) Representative Western blot of 4.1R and 4.1G after treatment for 96 h with 4.1R RNAi. Expression of 4.1R decreased 70 to 85% in HeLa cells and 50 to 70% in CaSki cells when normalized to actin as a loading control, while 4.1G expression was not markedly altered. The positions of molecular mass markers (in kilodaltons) are shown to the left of the blots. (B) Immunofluorescent staining of tubulin and 4.1R. In 4.1R RNAi-treated CaSki cells with no detectable 4.1R signals over background, microtubules appear whorled in contrast to controls with microtubules radiating from a focal point (arrow). Bar = 10 μm. (B′) Tubulin is perturbed in 31% of CaSki cells (n = 1,339) after 4.1R RNAi compared with 2.1% of control cells (n = 1,016) (average ± standard error [error bar] of three experiments). This was also observed in HeLa cells. (C) Ninein distribution at CaSki centrioles with depleted 4.1R is abnormal. Examples of irregular, dispersed, and larger immunostaining patterns relative to single or double circular ninein patterns in controls. Double-label experiments show centrin, ODF2, C-Nap1, and γ-tubulin distributions. Bar = 1 μm. (C′) Ninein is perturbed in 26.4% of CaSki cells (n = 803) relative to 3.1% of control cells (n = 607) (average ± standard error [error bar] of three independent experiments). Hatched areas indicate perturbed ODF2 at centrosomes with aberrant ninein. Similar results were obtained using HeLa cells. (D) Microtubule nucleation in 4.1R RNAi-treated CaSki cells is similar to controls up to 20 min after nocodozole exposure/washout. Centrioles are imaged using antibodies against centrin or ninein (enlarged inset), and microtubule growth was monitored by anti-β-tubulin staining at 2, 5, 10, 15, and 20 min and 2 h at 37°C. At later times, microtubules in 4.1R RNAi-treated cells become distanced from centrin-stained centrioles (25 min) and disorganized (2 h). DNA is imaged using DAPI. Bar = 3 μm. (D′) Data quantitating decreases in focused microtubule radial arrays (n = 300 per time point). Additional experiments at 20°C also showed that regrowth was not significantly different in 4.1R-depleted cells but subsequently became disarrayed. (E) 4.1R RNAi-treated CaSki cells with apparently normal ninein patterns show similar percentages of cells in G1 (1 ninein circle) as in controls but relatively more cells with ninein staining characteristic of S phase (1.5 circles) and fewer cells with G2 patterns (2 circles). Error bars show the standard errors from three experiments in which 2.9% controls (n = 442 cells) and 28% RNAi-treated cells (n = 462) had abnormal ninein staining. Values that were significantly different (P ≤ 0.05) are indicated by an asterisk. (F) In CaSki cells exposed to 4.1R RNAi, 6.9% of those with two ninein-stained centrosomes had separated centrosomes, while 21% of controls showed separation.
Using CaSki or HeLa cells, we specifically downregulated 4.1R expression and immunostained asynchronous 4.1R RNAi-treated cells. Tubulin staining revealed 31% of 4.1R-depleted cells with disorganized whorls of cytoplasmic microtubules, in contrast to well-organized radial arrays in 98% of control cells (Fig. 2B and B′). We also observed 26% of cells with abnormally dispersed ninein staining in 4.1R RNAi-treated cells relative to its characteristic circumscribed distribution in 97% of controls (Fig. 2C and C′). In 4.1R RNAi-treated cells simultaneously probed for ninein and tubulin, 79% of cells with disorganized microtubules also had perturbed ninein. In additional double-label experiments in which 50 cells with perturbed ninein were examined, 78% had normal C-Nap1 staining, while 53% had aberrant ODF2 patterns (Fig. 2C and C′). However, centrosomes with dispersed ninein had normal numbers of centrin-stained centrioles (either two or four), and γ-tubulin staining in the PCM was not affected (Fig. 2C). These results suggest that 4.1R depletion at centrosomes affects distal/subdistal appendage structural proteins ninein and ODF2 with little apparent effect on proximal protein C-Nap1, centriolar protein centrin, or γ-tubulin-containing microtubule nucleating complexes.
To further dissect the mechanism underlying the microtubule whorling phenotype, we tested whether microtubule nucleation and/or regrowth were impaired in 4.1R-depleted cells. Following microtubule depolymerization by cold and nocodazole exposure/washout, we did not find detectable differences in microtubule nucleation and regrowth between control and 4.1R RNAi-treated cells up to 20 min even when ninein distribution was abnormal (Fig. 2D, top). However, at 25 min, we observed asters in 4.1R RNAi-treated cells in which radial microtubules are distinctly separated from the centrin-stained centrosome (Fig. 2D, middle). At longer times of regrowth, there was an increased frequency of microtubule disorganization and microtubules not emanating from a focus of either centrin or perturbed ninein (Fig. 2D, bottom, and D′). Thus, 4.1R is required for microtubule anchoring at centrosomes but not required for microtubule nucleation from centrosomes. These results are consistent with our observations that in response to 4.1R depletion, ninein and, to a lesser extent, ODF2 localization are perturbed, while γ-tubulin staining is normal.
4.1R RNAi affects progression through the centrosome cycle.
In addition to quantitating centrosomal ninein perturbation after 4.1R RNAi treatment, we analyzed 4.1R-depleted cells with normal ninein patterns to assess centrosome maturation. Compared to controls, 4.1R RNAi-treated populations contained more cells with one and a half ninein rings (S phase) and fewer cells with two ninein rings (G2), indicating that progression toward centriole maturation during S phase was altered (Fig. 2E and see Fig. 3 below). Furthermore, in cells with two ninein rings, we quantitated those in which two centrosomes were separated versus juxtaposed. Normally in prophase, mature centrosomes separate and migrate around the nucleus as nuclear envelope breakdown proceeds, culminating in bipolar spindle pole formation. We observed that in asynchronous 4.1R RNAi-treated cultures, cells with two centrosomal ninein rings had about a threefold-lower frequency of separation of the two mature centrioles (Fig. 2F). This result indicates that even when there are two mature centrosomes with apparently normal ninein distribution in RNAi-treated cells, 4.1R downregulation impedes their capacity to efficiently separate in order to establish a bipolar spindle capable of accurate chromosome segregation. Thus, 4.1R is required for efficient centrosome separation independent of disrupting ninein.
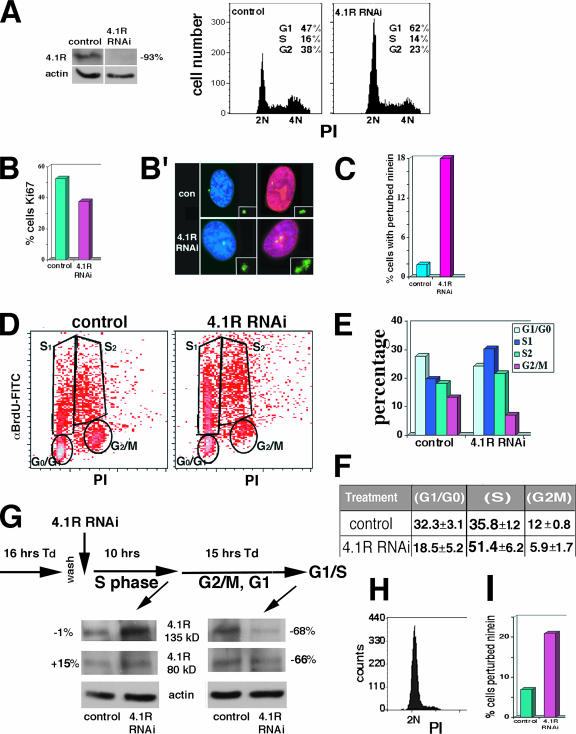
FIG. 3.
4.1R depletion affects cell cycle progression and can perturb centrosomal ninein distribution during G1. (A) (Right) Flow cytometric analysis of RPE-1 control cells and 4.1R RNAi-treated cells (96 h). The cells were labeled with propidium iodide (PI), and 10,000 events were scored. (Left) Western blot analysis of 4.1R depletion in 4.1R RNAi-treated RPE-1 cells relative to controls. Control RPE-1 cells contained a strong 80-kDa band (shown), but a 135-kDa 4.1R band was not detected. Actin was used as a loading control. (B) Percentage of RPE-1 cells stained with Ki67. For each condition, 1,040 cells were examined; 52% of controls were Ki67 positive relative to 38% for RNAi-treated cells. Cells were scored positive when there was strong staining throughout the nucleus as shown in the right panel of B′. (B′) Perturbed ninein in RPE-1 cells after 4.1R RNAi treatment for 96 h. By indirect immunofluorescence, aberrant ninein distribution at centrosomes was detected (enlarged images) in both Ki67-negative and -positive cells (Ki67 [red]; ninein [green]). Negative cell images were captured at the same exposure as positive cells. con, control. (C) Percentage of RPE-1 cells with perturbed ninein after 4.1R RNAi. In RNAi-treated cells, 18% had aberrant ninein distributions relative to 2% of controls (n = 300 for each condition). (D) Flow cytometric analysis of BrdU incorporation and propidium iodide (PI)-labeled DNA content of 10,000 4.1R RNAi-treated cells and 10,000 control CaSki cells in a representative experiment. αBrdU-FITC, fluorescein isothiocyanate-conjugated anti-BrdU antibody. (E) Percentages of cells in G1, early S (S1), late S (S2), and G2/M based on the cell distribution in the corresponding gated areas indicated in panel D. (F) Percentages of CaSki cells in cell cycle phases from four independent experiments (average ± standard error). HeLa cells treated with 4.1R RNAi and then analyzed in parallel had 1.7-fold-higher numbers of cells in S phase and 2.1-fold fewer cells in G2/M relative to control cells. (G) Scheme for double thymidine block with Western blot analysis of 4.1R levels at various cell cycle phases. 4.1R RNAi treatment did not deplete 4.1R after S-phase transition, but there was significant 4.1R depletion following the second thymidine (Td) block of cells in G1/S. Percent depletion (shown at the sides of the blots) is normalized to actin loading controls. The positions of 4.1R proteins and actin are shown between the blots. (H) Flow cytometry of 10,000 propidium iodide (PI)-labeled 4.1R RNAi-treated cells after the second thymidine treatment, confirming blockage of cells in G1 in a representative experiment. In five independent experiments, 85.5% ± 2.0% of cells were in G1. (I) Graph of the percentage of cells with perturbed ninein following the second thymidine block. For each condition, 300 cells were scored and 21% of RNAi-treated cells had abnormal ninein distribution compared with 7% in control cells.
4.1R RNAi-treated cells have increased G1 phase and decreased G2/M populations in cells with functional p53.
It is well established that the centrosome duplication/maturation cycle is intimately coordinated with the cell cycle in most cells (9, 19, 39). Since depleting 4.1R affected the distribution of the mature centriolar marker ninein, altered centrosome separation, and slowed the growth rate relative to controls (Fig. 2E and F; also data not shown), we tested whether cell cycle progression was altered. It was recently reported that centrosome defects could cause p53-dependent cell cycle alterations (41, 61, 63). Therefore, we analyzed effects of 4.1R depletion in telomerase-immortalized diploid human cells with normal p53 (RPE-1 cells). In 4.1R RNAi-treated RPE-1 cells, with 93% 4.1R depletion, there was an increased proportion of G1 cells shown both by analyzing cells stained with propidium iodide and by probing for the proliferation marker Ki67 (Fig. 3A, B, and B′). The 4.1R-depleted cells also had a significantly increased frequency of abnormal ninein distribution at centrosomes (Fig. 3C). These results show that, similar to depletion of many other centrosome proteins (41, 61, 63), 4.1R depletion affects G1 progression dependent on p53.
4.1R RNAi-treated cells have increased S-phase and decreased G2/M-phase populations in cells with compromised p53.
In order to test whether a cell cycle delay caused by 4.1R depletion can be overcome in cells without functional p53, we analyzed the effect of 4.1R depletion on cell cycle progression in CaSki and HeLa cells. Dual-color flow cytometry analyzing BrdU incorporation versus DNA content stained with propidium iodide showed that 4.1R RNAi-treated cells had increased numbers of cells in S phase and decreased numbers of cells in G2/M (Fig. 3D). Furthermore, we noted that more 4.1R RNAi-treated cells incorporating BrdU in S phase appeared to be in the earlier part of S phase relative to the control population (Fig. 3D and E). The finding that there is a larger proportion of cells in S phase after 4.1R RNAi exposure and a lesser proportion in G2/M (Fig. 3F) is consistent with our independent data evaluating ninein staining patterns showing decreased numbers of cells with characteristic G2/M ninein labeling (Fig. 2E and F). Thus, 4.1R depletion alters S-phase progression and ninein recruitment to maturing centrosomes with a corresponding decrease in G2/M in cells with compromised p53. To determine whether cells with perturbed ninein could enter mitosis, we immunostained 4.1R-depleted CaSki cells for phosphohistone H3 (a mitosis marker). We observed 4.1R RNAi-treated cells in prophase with irregular ninein patterns, indicating that an altered distribution of ninein does not prevent transitioning into prophase (data not shown).
4.1R RNAi perturbs ninein independent of S-phase delay.
To address whether the loss of centrosome structural integrity during 4.1R depletion is secondary to an S-phase delay, we used double thymidine synchronization to test for aberrant ninein centrosomal distribution under conditions where 4.1R is depleted prior to S-phase entry. After an initial 2 mM thymidine exposure, which blocks cells in all stages of S phase, thymidine was washed out to release cells from the S-phase block and cells were transfected with 4.1R RNAi duplexes. After S-phase transit, there was no detectable 4.1R depletion (Fig. 3G). After the second thymidine exposure, 87% of cells were blocked at the G1/S transition (Fig. 3H) with 66 to 68% depletion of 4.1R (Fig. 3G) indicating that 4.1R depletion occurred subsequent to S-phase transit and prior to reentry into S phase. Importantly, 21% of the G1/S population had aberrant ninein distributions (Fig. 3I). These data clearly show that 4.1R depletion can perturb ninein independent of its effects on S-phase progression. Thus, disruption of centrosome integrity is a primary effect of 4.1R depletion, rather than a secondary effect of S-phase delay.
Mitotic spindle defects in cells treated with 4.1R RNAi.
We previously showed that 4.1 has critical mitotic functions in spindle assembly and maintenance in Xenopus egg extracts using dominant-negative peptides corresponding to 4.1R domains (31). Since mammalian centrosomes (precursors of spindle poles) contain 4.1R, we wanted to dissect 4.1R-specific effects on mammalian mitotic spindle formation in vivo using 4.1R RNAi duplexes under conditions we knew did not decrease 4.1G levels. To gain functional insight into mitotic defects caused by 4.1R depletion, we used a variety of informative spindle and chromatin probes to characterize mitotic spindles in CaSki and HeLa cells by double-label immunofluorescence (Fig. 4).
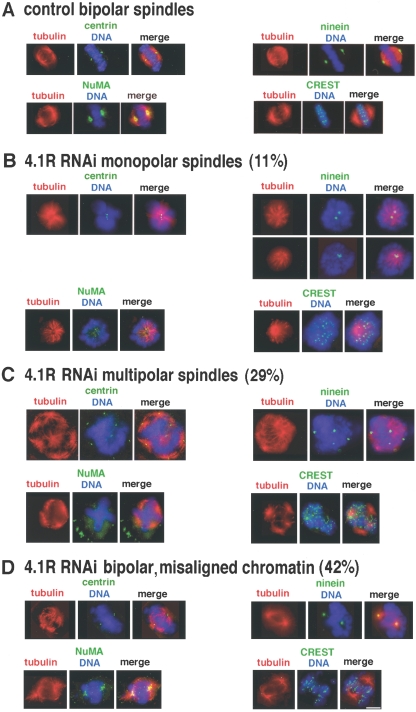
FIG. 4.
Mitotic spindle abnormalities after 4.1R RNAi treatment for 96 h. (A) Control mitotic spindles with microtubules converging at two opposing poles each containing focal centrin, ninein, and NuMA. Microtubules extend from each pole toward chromatin which forms a bisecting metaphase plate and has paired centromeres stained by CREST antibody. (B to D) Mitotic cells with downregulated 4.1R expression have three main classes of mitotic spindle abnormalities. (B) Monopolar spindles with microtubules radiating toward chromatin organized in a rosette pattern and a central polar area containing centrioles (anticentrin) and ninein. Two examples of ninein staining are shown: a monopolar spindle with two centrosomes displaying circular ninein patterns and a monopolar spindle with perturbed ninein. NuMA is unfocused at the polar region, and centromeres (CREST staining) are unpaired in the surrounding chromatin. (C) Multipolar spindles with more than two poles marked by multiple centrin and ninein epitopes with weakly or strongly convergent microtubules, mislocalized NuMA, and relatively uncondensed chromatin with misaligned centromeres (CREST). (D) Bipolar spindles with misaligned, uncondensed chromatin and unpaired centromeres (CREST). NuMA is not focally organized at the poles. Bar = 5 μm. Quantitation of each class of abnormal mitotic spindles resulting from 4.1R depletion is indicated for CaSki cells in three experiments. There were 322 control cells examined and 316 4.1R RNAi-treated cells analyzed. For HeLa cells, downregulation of 4.1R expression produced 9% monopolar, 33% multipolar, and 36% bipolar spindles with misaligned chromatin in 216 mitotic cells scored.
Control mitotic cells have focused spindle poles with centrin and ninein and are capped by NuMA epitopes. Ninein has been detected at spindles as well as at centrosomes (2a, 46), although after using an antibody against the ninein CII region, it was reported to diminish during mitosis (3a). In these control cell spindles, bipolar microtubule arrays emanate toward a metaphase plate with regularly aligned kinetochore epitopes (stained by CREST antibody) on the condensed chromatin (Fig. 4A). In contrast, 4.1R depletion resulted in three major classes of mitotic spindle defects with a distribution of 11% monopolar, 29% multipolar, and 42% bipolar with decondensed, misaligned chromatin and dispersed CREST staining (Fig. 4B to D).
Monopolar spindles (Fig. 4B) have chromatin organized in a rosette pattern with a central polar area containing centrioles marked by centrin. Two categories of ninein staining were observed: either perturbed or two ninein rings in close proximity indicative of mature unseparated centrosomes. NuMA, a 4.1 binding partner, is diffuse in the polar region, and centromeres are unpaired (CREST staining).
Multipolar spindles observed after 4.1R downregulation (Fig. 4C) have multiple poles marked by paired centrin dots and ninein epitopes with weakly or strongly convergent microtubules. We noted that multipolar spindles can have both perturbed and unperturbed ninein. The multiple poles were also marked by γ-tubulin (data not shown). Strikingly, NuMA is markedly mislocalized, and poles are broadened. Chromatin is relatively uncondensed with misaligned centromeres.
The third class of perturbed spindles, bipolar spindles with misaligned, uncondensed chromatin (Fig. 4D), also had NuMA broadly distributed at poorly focused poles and mislocalized at nonpolar areas. Furthermore, spindle microtubules were disorganized, and centromeres were unpaired.
Using these multiple combinations of spindle and chromatin protein probes, quantitation of 4.1R RNAi-treated cells revealed a dramatic increase in mitotic abnormalities by 96 h (81% in CaSki cells and 78% in HeLa cells) compared with 8% in control cells. Mitotic defects were also observed after 4.1R RNAi treatment in RPE-1 cells (data not shown). Thus, in intact mammalian cells, 4.1R is required for the fidelity of proper spindle pole formation likely by impacting the integrity of precursor centrosomes and microtubule focusing at spindle poles.
Lagging chromosomes and cytokinesis defects are detected in 4.1R RNAi-treated cells.
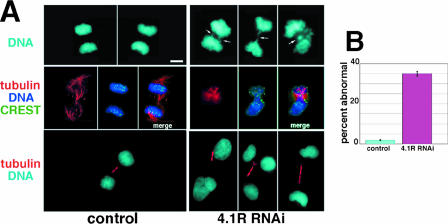
Since properly anchored and organized microtubules at centrosomes and spindles are prerequisites for proper chromosome segregation and cytokinesis, we predicted that knockdown of 4.1R expression might produce defects at these stages of mitosis. Indeed, 4.1R RNAi-treated anaphase/telophase cells often displayed lagging chromatin trapped between nascent daughters and inappropriately localized spindles (Fig. 5A, top and middle panels). Furthermore, while control cells at cytokinesis were connected by short bundled tubulin bridges, 4.1R knockdown cells had a variety of cytokinesis defects, including elongated tubulin connecting structures, wide intercellular bridges, broken bridges, and tubulin bridges contacting more than two cells (Fig. 5A, lower panels). Anaphase/telophase defects were apparent in 35% of 4.1R RNAi-treated cells (Fig. 5B). In sum, 4.1R depletion causes centrosomal and mitotic spindle abnormalities that likely contribute to improper chromosome segregation and aberrant intercellular bridges.
FIG. 5.
Anaphase/telophase defects after 4.1R RNAi. (A) Examples of anaphase cells with lagging chromosomes (right top panel, arrows), decondensed chromatin with dispersed CREST staining and misaligned spindles (tubulin, middle panel, right) relative to controls (left). The lower right panels show examples of abnormalities in tubulin bridges formed at cytokinesis in 4.1R RNAi-treated cells, including broken, tricellular, and elongated bridges. Bar = 10 μm. (B) Quantitation of anaphase/telophase defects. In 4.1R RNAi-treated cell populations, 35% anaphase cells (n = 300) were abnormal relative to 1.8% cells (n = 264) in control populations in three independent experiments. DNA is imaged using DAPI.
DISCUSSION
It was only recently recognized that protein 4.1 is not exclusively a red cell protein but in fact is abundant in most nonerythroid and erythroid cells. Additionally, earlier characterizations of “protein 4.1” in mammalian cells now have to be revisited in light of the discovery of a 4.1 gene family. Here we report evidence that at centrosomes 4.1R is a mature centriole protein.
To understand potential functions of 4.1R at mature centrioles, we first examined in detail its distribution relative to several centrosomal markers. We found that 4.1R surrounds one or two centrin-marked centrioles, was itself surrounded by mature centriole markers p150/glued, associates with mature centriole bearing ODF2 epitopes, and colocalizes extensively with the subdistal appendage maker ninein (42, 44). However, 4.1R epitopes are largely noncoincident with C-Nap1 at the proximal ends of centrioles. We also found that 4.1R epitopes were consistently targeted in tandem with ninein and ODF2 to newly maturing centrioles during centriole duplication, unlike some other proteins, such as Nudel, that arrive after ninein (18). 4.1R localization at centrosomes is not dependent on microtubules (29), and our data reported here suggest that 4.1R may be an integral component of centrioles bearing distal/subdistal appendages.
To directly explore 4.1R centrosome function, we specifically downregulated 4.1R but not 4.1G expression in cultured mammalian cells. After 4.1R depletion, distribution of centrosomal ninein and, to a lesser but significant extent, ODF2 was aberrant, demonstrating that 4.1R deficiency has a direct effect on mature centrosome structure and may modulate organization of several distal/subdistal proteins. Since localization of γ-tubulin, the defining member of the γ-TURC, was normal, we speculated that abrogating 4.1R expression may perturb ninein's microtubule anchoring functions but not γ-tubulin microtubule nucleating functions. In fact, it was reported that mutations in ninein can cause microtubule anchoring defects without perturbing γ-TURC localization (6). We tested our hypothesis by analyzing microtubule regrowth which was similar to that in controls. Interestingly, ectopic overexpression of a variant 4.1R isoform also does not impair microtubule nucleation/regrowth (52). We did find that 4.1R-depleted cells had a loss of focused microtubule arrays, thus confirming that 4.1R is involved in microtubule anchoring at centrosomes. Microtubule anchorage and radial organization depend on many proteins, and anchorage occurs both at mature centriolar subdistal appendages and in the PCM. Our data indicate that 4.1R functions specifically in microtubule anchorage at mature centrioles. Future experiments will test direct interactions of 4.1R with mature centriole microtubule anchoring proteins to uncover relevant molecular mechanisms. In sum, our data show that 4.1R is a component of mature centrioles, required for proper localization of other mature centriole proteins, such as ninein and ODF2, and contributes to the microtubule anchoring function of mature centrioles.
Depletion of centrosome proteins can alter cell cycle progression, and in cells with functional p53, loss of centrosome integrity can specifically prevent entry into S phase. For example, centrosomal PCM1 depletion induces a p53-dependent exit from the cell cycle in MRC-5 cells (61), and depleting various other centrosomal proteins in RPE-1 cells induces a p53-dependent G1-S arrest or partial arrest (41). We found that 4.1R depletion delays transitioning into S phase in untransformed RPE-1 cells, supporting the important role of 4.1R in centrosome structure. Although 4.1R depletion does not cause a robust G1 checkpoint arrest in RPE-1 cells, our findings are consistent with another study in untransformed human cells that has shown that centrosomal defects act synergistically with additional stresses to induce a G1 arrest (63). In cells with compromised p53 pathways, such as CaSki or HeLa cells, 4.1R depletion delays S-phase progression significantly. In order to test whether centrosomal ninein perturbation induced by 4.1R RNAi is a secondary effect caused by the S-phase delay in these cells, we depleted 4.1R expression in between two S phases. We found that cells that had not gone through an S-phase delay had significant ninein perturbation at centrosomes, providing strong evidence that 4.1R has a direct role in centrosomal structure organization. It is interesting that perturbing ninein, ODF2, and potentially other centrosomal proteins induced by 4.1R depletion differs from loss of centrosomal ninein alone, which does not cause a G1 arrest, even in p53-proficient cells (41). This result raises the intriguing possibility that as a scaffolding protein, 4.1R interactions may additionally affect functions of other proteins at the centrosome that regulate cell cycle progression or DNA replication/repair, such as the ORC protein family (1, 9, 34, 45, 55).
In addition to its functions at interphase centrosomes and in cell cycle regulation, 4.1R is required for formation of mitotic bipolar spindles and for proper cytokinesis. As previously reported (23), aberrant mitotic spindles were found after downregulation of 4.1R. Here we have analyzed three classes of mitotic defects which are likely linked to specific 4.1R functions at centrosomes and spindle poles. Some monopolar spindles had normal double-ring ninein patterns, resulting from decreased centrosome separation at prophase, as predicted by our evidence that 4.1R is required for efficient centrosome separation independent of ninein perturbation. Other monopolar spindles had centrosomes with perturbed ninein at the single pole, reflecting the fact that centrosomes with perturbed ninein were able to enter prophase as shown independently by phosphohistone H3 staining (unpublished observations) but that these centrosomes were unable to separate at G2/M. Additionally the 4.1 binding partner NuMA, a protein required for highly focused spindle poles (14, 40) was mislocalized in monopolar spindles. Monopolar spindles have been observed following injection of anti-NuMA into cells (69). Thus, perturbation of several centrosomal and spindle proteins in 4.1R-depleted cells likely contributes to monopolar spindle formation during mitosis.
Multipolar spindles resulting from 4.1R downregulation could be generated in several ways, such as centrosome overduplication, pole fragmentation, cytokinesis failure, and/or NuMA mislocalization (26). Since most all poles contained paired centrin-stained centrioles, γ-tubulin, and ninein, along with the fact that HeLa cells are not prone to centrosome overduplication even when stressed by hydroxyurea treatment (72), it appears that neither centrosome amplification nor pole fragmentation is a major source of the multipolarity we observed. Rather we attribute multipolar spindle formation mechanistically to cell division failure that was observed in 4.1R-depleted cells, similar to what has been observed for Aurora A overexpression (38). This is further supported by our observation that in 4.1R RNAi-treated cultures, only multinucleated or enormous mononucleated cells had more than two centrosomes. Of cells with four or more centrioles (by centrin or C-Nap1 staining), 92% were ninein positive, with 64% having four or more ninein staining rings and the remainder having perturbed ninein distribution at centrioles (S. Krauss, unpublished observations). Thus, these centrioles, potentially giving rise to multipolar spindles, had progressed into late G2/M to acquire a maturation-specific protein. A similar diagnostic for centriole amplification due to failed cell division has been described for Cep170 labeling (17).
The final class of mitotic spindle defects resulting from 4.1R depletion was bipolar spindles with misaligned, uncondensed chromosomes. Microtubule interactions with kinetochores are a prerequisite for correct chromosome alignment and segregation (13, 54). Therefore, these spindles likely resulted from aberrant precursor centrosome structure and improper microtubule organization/anchorage leading to the failure to establish proper bipolar spindle architecture, poorly condensed misaligned chromatin, and dispersed kinetochores. Additionally, a chromosome missegregation phenotype has been reported after NuMA RNAi (10).
In sum, 4.1R depletion leads to structurally aberrant centrosomes during interphase and at the onset of mitosis, to insufficient centrosome separation (an effector of spindle bipolarity), and to decreased centrosome-anchored microtubules that could contribute not only to spindle defects but also to mechanisms responsible for subsequent chromosome missegregation, spindle dysmorphology, and improper tubulin bridge formation at anaphase/telophase. Several laboratories have reported that centrosomal function and the completion of cytokinesis are interrelated (15, 21, 28) and, more specifically, that the maternal centriole moves to the intercellular bridge before the completion of cytokinesis (53). However, we do not exclude possible direct roles of 4.1R in cytokinesis as was reported recently for the centrosome scaffolding protein centriolin (15, 16).
The spectrum of pleiotropic defects generated when 4.1R is depleted is strongly predictive of inaccurate chromosome segregation, especially in cells with defective p53. 4.1R at centrosomes or even elsewhere appears critical for molecular networks integrating centrosome maturation and microtubule anchorage, centrosome separation, and effective assembly of bipolar spindles capable of faithful chromosome distribution to daughter cells during cytokinesis. Our investigations highlight a role of 4.1R as a multifunctional structural protein component of mature centrioles ensuring the integrity of cell division and suggest that loss or decrements in 4.1R function may lead to as yet unrecognized pathological consequences.
Acknowledgments
We thank W. James Nelson, Rebecca Heald, Jeffrey Salisbury, Carolyn Larabell, Steve Yannone, Janice Pluth, and Andreas Merdes for valuable discussions. We are particularly grateful to G. Chan, J. Salisbury, E. Nigg, S. Tsukita, L. Walensky, and P. Gascard for sharing valuable antibody reagents. We thank Abby Dernberg for use of Deltavision microscopy.
This work was supported by NIH grants DK059079 and DOD grant BC032806 to S. W. Krauss.
S.W.K. dedicates this paper to the memory of Myra Berman Kurtz.
Footnotes
Published ahead of print on 22 January 2008.
REFERENCES
- 1.Andersen, J. S., C. J. Wilkinson, T. Mayor, P. Mortensen, E. A. Nigg, and M. Mann. 2003. Proteomic characterization of the human centrosome by protein correlation profiling. Nature 426570-574. [DOI] [PubMed] [Google Scholar]
- 2.Bornens, M. 2002. Centrosome composition and microtubule anchoring mechanisms. Curr. Opin. Cell Biol. 1425-34. [DOI] [PubMed] [Google Scholar]
- 2a.Bouckson-Castaing, V., M. Moudjou, D. J. Ferguson, S. Mucklow, Y. Belkaid, and G. Milon. 1996. Molecular characterisation of ninein, a new coiled-coil protein of the centrosome. J. Cell Sci. 109179-190. [DOI] [PubMed] [Google Scholar]
- 3.Chang, P., T. H. Giddings, Jr., M. Winey, and T. Stearns. 2003. Epsilon-tubulin is required for centriole duplication and microtubule organization. Nat. Cell Biol. 571-76. [DOI] [PubMed] [Google Scholar]
- 3a.Chen, C. H., S. L. Howng, T. S. Cheng, M. H. Chou, C. Y. Huang, and Y. R. Hong. 2003. Molecular characterization of human ninein protein: two distinct subdomains required for centrosomal targeting and regulating signals in cell cycle. Biochem. Biophys. Res. Commun. 308975-983. [DOI] [PubMed] [Google Scholar]
- 4.Dammermann, A., and A. Merdes. 2002. Assembly of centrosomal proteins and microtubule organization depends on PCM-1. J. Cell Biol. 159255-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Carcer, G., M. J. Lallena, and I. Correas. 1995. Protein 4.1 is a component of the nuclear matrix of mammalian cells. Biochem. J. 312871-877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Delgehyr, N., J. Sillibourne, and M. Bornens. 2005. Microtubule nucleation and anchoring at the centrosome are independent processes linked by ninein function. J. Cell Sci. 1181565-1575. [DOI] [PubMed] [Google Scholar]
- 7.Delhommeau, F., C. Vasseur-Godbillon, P. Leclerc, P. O. Schischmanoff, L. Croisille, P. Rince, M. Moriniere, E. J. Benz, Jr., G. Tchernia, G. Tamagnini, L. Ribeiro, J. Delaunay, and F. Baklouti. 2002. A splicing alteration of 4.1R pre-mRNA generates 2 protein isoforms with distinct assembly to spindle poles in mitotic cells. Blood 1002629-2636. [DOI] [PubMed] [Google Scholar]
- 7a.Dictenberg, J. B., W. Zimmerman, C. A. Sparks, A. Young, G. Vidair, Y. Zheng, W. Carrington, F. S. Fay, and S. J. Doxsey. 1998. Pericentrin and gamma-tubulin form a protein complex and are organized into a novel lattice at the centrosome. J. Cell Biol. 141163-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Doxsey, S., D. McCollum, and W. Theurkauf. 2005. Centrosomes in cellular regulation. Annu. Rev. Cell Dev. Biol. 21411-434. [DOI] [PubMed] [Google Scholar]
- 9.Doxsey, S., W. Zimmerman, and K. Mikule. 2005. Centrosome control of the cell cycle. Trends Cell Biol. 15303-311. [DOI] [PubMed] [Google Scholar]
- 10.Du, Q., and I. G. Macara. 2004. Mammalian Pins is a conformational switch that links NuMA to heterotrimeric G proteins. Cell 119503-516. [DOI] [PubMed] [Google Scholar]
- 11.Feldman, J. L., S. Geimer, and W. F. Marshall. 2007. The mother centriole plays an instructive role in defining cell geometry. PLoS Biol. 5e149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fry, A. M., T. Mayor, P. Meraldi, Y. D. Stierhof, K. Tanaka, and E. A. Nigg. 1998. C-Nap1, a novel centrosomal coiled-coil protein and candidate substrate of the cell cycle-regulated protein kinase Nek2. J. Cell Biol. 1411563-1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gadde, S., and R. Heald. 2004. Mechanisms and molecules of the mitotic spindle. Curr. Biol. 14R797-R805. [DOI] [PubMed] [Google Scholar]
- 14.Gaglio, T., A. Saredi, and D. A. Compton. 1995. NuMA is required for the organization of microtubules into aster-like mitotic arrays. J. Cell Biol. 131693-708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gromley, A., A. Jurczyk, J. Sillibourne, E. Halilovic, M. Mogensen, I. Groisman, M. Blomberg, and S. Doxsey. 2003. A novel human protein of the maternal centriole is required for the final stages of cytokinesis and entry into S phase. J. Cell Biol. 161535-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gromley, A., C. Yeaman, J. Rosa, S. Redick, C. T. Chen, S. Mirabelle, M. Guha, J. Sillibourne, and S. J. Doxsey. 2005. Centriolin anchoring of exocyst and SNARE complexes at the midbody is required for secretory-vesicle-mediated abscission. Cell 12375-87. [DOI] [PubMed] [Google Scholar]
- 17.Guarguaglini, G., P. I. Duncan, Y. D. Stierhof, T. Holmstrom, S. Duensing, and E. A. Nigg. 2005. The forkhead-associated domain protein Cep170 interacts with Polo-like kinase 1 and serves as a marker for mature centrioles. Mol. Biol. Cell 161095-1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guo, J., Z. Yang, W. Song, Q. Chen, F. Wang, Q. Zhang, and X. Zhu. 2006. Nudel contributes to microtubule anchoring at the mother centriole and is involved in both dynein-dependent and -independent centrosomal protein assembly. Mol. Biol. Cell 17680-689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hinchcliffe, E. H. 2003. Cell cycle: seeking permission from the mother centriole. Curr. Biol. 13R646-R648. [DOI] [PubMed] [Google Scholar]
- 20.Hinchcliffe, E. H., and G. Sluder. 2001. Centrosome duplication: three kinases come up a winner! Curr. Biol 11R698-R701. [DOI] [PubMed] [Google Scholar]
- 21.Hinchcliffe, E. H., and G. Sluder. 2001. Centrosome reproduction in Xenopus lysates. Methods Cell Biol. 67269-287. [DOI] [PubMed] [Google Scholar]
- 22.Hoyer-Fender, S., J. Neesen, J. Szpirer, and C. Szpirer. 2003. Genomic organisation and chromosomal assignment of ODF2 (outer dense fiber 2), encoding the main component of sperm tail outer dense fibers and a centrosomal scaffold protein. Cytogenet. Genome Res. 103122-127. [DOI] [PubMed] [Google Scholar]
- 23.Huang, S. C., R. Jagadeeswaran, E. S. Liu, and E. J. Benz, Jr. 2004. Protein 4.1R, a microtubule-associated protein involved in microtubule aster assembly in mammalian mitotic extract. J. Biol. Chem. 27934595-34602. [DOI] [PubMed] [Google Scholar]
- 24.Hung, L.-Y., C.-J. C. Tang, and T. K. Tang. 2000. Protein 4.1 R-135 interacts with a novel centrosomal protein (CPAP) which is associated with the γ-tubulin complex. Mol. Cell. Biol. 207813-7825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ishikawa, H., A. Kubo, and S. Tsukita. 2005. Odf2-deficient mother centrioles lack distal/subdistal appendages and the ability to generate primary cilia. Nat. Cell Biol. 7517-524. [DOI] [PubMed] [Google Scholar]
- 26.Kallajoki, M., K. Weber, and M. Osborn. 1991. A 210 kDa nuclear matrix protein is a functional part of the mitotic spindle: a microinjection study using SPN monoclonal antibodies. EMBO J. 103351-3362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kellogg, D. R., M. Moritz, and B. M. Alberts. 1994. The centrosome and cellular organization. Annu. Rev. Biochem. 63639-674. [DOI] [PubMed] [Google Scholar]
- 28.Khodjakov, A., and C. L. Rieder. 2001. Centrosomes enhance the fidelity of cytokinesis in vertebrates and are required for cell cycle progression. J. Cell Biol. 153237-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krauss, S. W., J. A. Chasis, C. Rogers, N. Mohandas, G. Krockmalnic, and S. Penman. 1997. Structural protein 4.1 is located in mammalian centrosomes. Proc. Natl. Acad. Sci. USA 947297-7302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krauss, S. W., C. A. Larabell, S. Lockett, P. Gascard, N. Mohandas, and J. A. Chasis. 1997. Structural protein 4.1 in the nucleus of human cells: dynamic rearrangements during cell division. J. Cell Biol. 137275-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krauss, S. W., G. Lee, J. A. Chasis, N. Mohandas, and R. Heald. 2004. Two protein 4.1 domains essential for mitotic spindle and aster microtubule dynamics and organization in vitro. J. Biol. Chem. 27927591-27598. [DOI] [PubMed] [Google Scholar]
- 32.Lange, B. M., and K. Gull. 1995. A molecular marker for centriole maturation in the mammalian cell cycle. J. Cell Biol. 130919-927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lengauer, C., K. W. Kinzler, and B. Vogelstein. 1998. Genetic instabilities in human cancers. Nature 396643-649. [DOI] [PubMed] [Google Scholar]
- 34.Lesca, C., M. Germanier, B. Raynaud-Messina, C. Pichereaux, C. Etievant, S. Emond, O. Burlet-Schiltz, B. Monsarrat, M. Wright, and M. Defais. 2005. DNA damage induce gamma-tubulin-RAD51 nuclear complexes in mammalian cells. Oncogene 245165-5172. [DOI] [PubMed] [Google Scholar]
- 35.Louie, R. K., S. Bahmanyar, K. A. Siemers, V. Votin, P. Chang, T. Stearns, W. J. Nelson, and A. I. Barth. 2004. Adenomatous polyposis coli and EB1 localize in close proximity of the mother centriole and EB1 is a functional component of centrosomes. J. Cell Sci. 1171117-1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mattagajasingh, S., S. Huang, J. Hartenstein, and E. J. Benz, Jr. 1998. Protein 4.1R associates with mitotic spindle pole organizing proteins NuMA, dynein and dynactin. Blood 925a. [Google Scholar]
- 37.Mattagajasingh, S. N., S. C. Huang, J. S. Hartenstein, M. Snyder, V. T. Marchesi, and E. J. Benz, Jr. 1999. A nonerythroid isoform of protein 4.1R interacts with the nuclear mitotic apparatus (NuMA) protein. J. Cell Biol. 14529-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meraldi, P., R. Honda, and E. A. Nigg. 2002. Aurora-A overexpression reveals tetraploidization as a major route to centrosome amplification in p53−/− cells. EMBO J. 21483-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Meraldi, P., and E. A. Nigg. 2002. The centrosome cycle. FEBS Lett. 5219-13. [DOI] [PubMed] [Google Scholar]
- 40.Merdes, A., R. Heald, K. Samejima, W. C. Earnshaw, and D. W. Cleveland. 2000. Formation of spindle poles by dynein/dynactin-dependent transport of NuMA. J. Cell Biol. 149851-862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mikule, K., B. Delaval, P. Kaldis, A. Jurcyzk, P. Hergert, and S. Doxsey. 2007. Loss of centrosome integrity induces p38-p53-p21-dependent G1-S arrest. Nat. Cell Biol. 9160-170. [DOI] [PubMed] [Google Scholar]
- 42.Mogensen, M. M., A. Malik, M. Piel, V. Bouckson-Castaing, and M. Bornens. 2000. Microtubule minus-end anchorage at centrosomal and non-centrosomal sites: the role of ninein. J. Cell Sci. 1133013-3023. [DOI] [PubMed] [Google Scholar]
- 43.Moritz, M., M. B. Braunfeld, J. W. Sedat, B. Alberts, and D. A. Agard. 1995. Microtubule nucleation by gamma-tubulin-containing rings in the centrosome. Nature 378638-640. [DOI] [PubMed] [Google Scholar]
- 44.Nakagawa, Y., Y. Yamane, T. Okanoue, and S. Tsukita. 2001. Outer dense fiber 2 is a widespread centrosome scaffold component preferentially associated with mother centrioles: its identification from isolated centrosomes. Mol. Biol. Cell 121687-1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Okano, S., L. Lan, A. E. Tomkinson, and A. Yasui. 2005. Translocation of XRCC1 and DNA ligase IIIalpha from centrosomes to chromosomes in response to DNA damage in mitotic human cells. Nucleic Acids Res. 33422-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ou, Y. Y., G. J. Mack, M. Zhang, and J. B. Rattner. 2002. CEP110 and ninein are located in a specific domain of the centrosome associated with centrosome maturation. J. Cell Sci. 1151825-1835. [DOI] [PubMed] [Google Scholar]
- 47.Paoletti, A., and M. Bornens. 1997. Organisation and functional regulation of the centrosome in animal cells. Prog. Cell Cycle Res. 3285-299. [DOI] [PubMed] [Google Scholar]
- 48.Parra, M., P. Gascard, L. D. Walensky, S. H. Snyder, N. Mohandas, and J. G. Conboy. 1998. Cloning and characterization of 4.1G (EPB41L2), a new member of the skeletal protein 4.1 (EPB41) gene family. Genomics 49298-306. [DOI] [PubMed] [Google Scholar]
- 49.Parra, M., S. Gee, N. Chan, D. Ryaboy, I. Dubchak, N. Mohandas, P. Gascard, and J. Conboy. 2004. Differential domain evolution and complex RNA processing in a family of paralogous EPB41 (protein 4.1) genes facilitates expression of diverse tissue-specific isoforms. Genomics 84637-646. [DOI] [PubMed] [Google Scholar]
- 50.Parra, M., L. Walensky, N. Chan, S. Snyder, N. Mohandas, and J. Conboy. 1998. Characterization of protein 4.1B, a new gene in the protein 4.1 family with high level focal expression in brain. Mol. Biol. Cell 9265a. [DOI] [PubMed] [Google Scholar]
- 51.Perez-Ferreiro, C. M., C. M. Luque, and I. Correas. 2001. 4.1R proteins associate with interphase microtubules in human T cells: a 4.1R constitutive region is involved in tubulin binding. J. Biol. Chem. 27644785-44791. [DOI] [PubMed] [Google Scholar]
- 52.Perez-Ferreiro, C. M., I. Vernos, and I. Correas. 2004. Protein 4.1R regulates interphase microtubule organization at the centrosome. J. Cell Sci. 1176197-6206. [DOI] [PubMed] [Google Scholar]
- 53.Piel, M., J. Nordberg, U. Euteneuer, and M. Bornens. 2001. Centrosome-dependent exit of cytokinesis in animal cells. Science 2911550-1553. [DOI] [PubMed] [Google Scholar]
- 54.Pinsky, B. A., and S. Biggins. 2005. The spindle checkpoint: tension versus attachment. Trends Cell Biol. 15486-493. [DOI] [PubMed] [Google Scholar]
- 55.Prasanth, S. G., K. V. Prasanth, K. Siddiqui, D. L. Spector, and B. Stillman. 2004. Human Orc2 localizes to centrosomes, centromeres and heterochromatin during chromosome inheritance. EMBO J. 232651-2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Quintyne, N. J., S. R. Gill, D. M. Eckley, C. L. Crego, D. A. Compton, and T. A. Schroer. 1999. Dynactin is required for microtubule anchoring at centrosomes. J. Cell Biol. 147321-334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Quintyne, N. J., and T. A. Schroer. 2002. Distinct cell cycle-dependent roles for dynactin and dynein at centrosomes. J. Cell Biol. 159245-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ramez, M., M. Blot-Chabaud, F. Cluzeaud, S. Chanan, M. Patterson, L. D. Walensky, S. Marfatia, A. J. Baines, J. A. Chasis, J. G. Conboy, N. Mohandas, and P. Gascard. 2003. Distinct distribution of specific members of protein 4.1 gene family in the mouse nephron. Kidney Int. 631321-1337. [DOI] [PubMed] [Google Scholar]
- 59.Schnackenberg, B. J., A. Khodjakov, C. L. Rieder, and R. E. Palazzo. 1998. The disassembly and reassembly of functional centrosomes in vitro. Proc. Natl. Acad. Sci. USA 959295-9300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sluder, G. 2005. Two-way traffic: centrosomes and the cell cycle. Nat. Rev. Mol. Cell Biol. 6743-748. [DOI] [PubMed] [Google Scholar]
- 61.Srsen, V., N. Gnadt, A. Dammermann, and A. Merdes. 2006. Inhibition of centrosome protein assembly leads to p53-dependent exit from the cell cycle. J. Cell Biol. 174625-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stearns, T. 2001. Centrosome duplication. A centriolar pas de deux. Cell 105417-420. [DOI] [PubMed] [Google Scholar]
- 63.Uetake, Y., J. Loncarek, J. J. Nordberg, C. N. English, S. La Terra, A. Khodjakov, and G. Sluder. 2007. Cell cycle progression and de novo centriole assembly after centrosomal removal in untransformed human cells. J. Cell Biol. 176173-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Walensky, L. D., S. Blackshaw, D. Liao, C. C. Watkins, H. U. Weier, M. Parra, R. L. Huganir, J. G. Conboy, N. Mohandas, and S. H. Snyder. 1999. A novel neuron-enriched homolog of the erythrocyte membrane cytoskeletal protein 4.1. J. Neurosci. 196457-6467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Walensky, L. D., P. Gascard, M. E. Fields, S. Blackshaw, J. G. Conboy, N. Mohandas, and S. H. Snyder. 1998. The 13-kD FK506 binding protein, FKBP13, interacts with a novel homologue of the erythrocyte membrane cytoskeletal protein 4.1. J. Cell Biol. 141143-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Walensky, L. D., Z. T. Shi, S. Blackshaw, A. C. DeVries, G. E. Demas, P. Gascard, R. J. Nelson, J. G. Conboy, E. M. Rubin, S. H. Snyder, and N. Mohandas. 1998. Neurobehavioral deficits in mice lacking the erythrocyte membrane cytoskeletal protein 4.1. Curr. Biol. 81269-1272. [DOI] [PubMed] [Google Scholar]
- 67.Yamakawa, H., R. Ohara, D. Nakajima, M. Nakayama, and O. Ohara. 1999. Molecular characterization of a new member of the protein 4.1 family (brain 4.1) in rat brain. Brain Res. Mol. Brain Res. 70197-209. [DOI] [PubMed] [Google Scholar]
- 68.Yamashita, Y. M., A. P. Mahowald, J. R. Perlin, and M. T. Fuller. 2007. Asymmetric inheritance of mother versus daughter centrosome in stem cell division. Science 315518-521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yang, C. H., and M. Snyder. 1992. The nuclear-mitotic apparatus protein is important in the establishment and maintenance of the bipolar mitotic spindle apparatus. Mol. Biol. Cell 31259-1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ye, K., D. A. Compton, M. M. Lai, L. D. Walensky, and S. H. Snyder. 1999. Protein 4.1N binding to nuclear mitotic apparatus protein in PC12 cells mediates the antiproliferative actions of nerve growth factor. J. Neurosci. 1910747-10756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zheng, Y., M. L. Wong, B. Alberts, and T. Mitchison. 1995. Nucleation of microtubule assembly by a gamma-tubulin-containing ring complex. Nature 378578-583. [DOI] [PubMed] [Google Scholar]
- 72.Zou, C., J. Li, Y. Bai, W. T. Gunning, D. E. Wazer, V. Band, and Q. Gao. 2005. Centrobin: a novel daughter centriole-associated protein that is required for centriole duplication. J. Cell Biol. 171437-445. [DOI] [PMC free article] [PubMed] [Google Scholar]