Abstract
Cell proliferation and differentiation are governed by a finely controlled balance between repression and activation of gene expression. The vertebrate Ets transcriptional repressor Tel (ETV6) and its invertebrate orthologue Yan, play pivotal roles in cell fate determination although the precise mechanisms by which repression of gene expression by these factors is achieved are not clearly defined. Here, we report the identification and characterization of the primary site of sumoylation of Tel, lysine 11 (K11), which is highly conserved in vertebrates (except Danio rerio). We demonstrate that in cells PIAS3 binds to Tel and stimulates sumoylation of K11 in the nucleus. Both Tel monomers and oligomers are efficiently sumoylated on K11 in vitro; but in cells only Tel oligomers are found conjugated with SUMO, whereas sumoylation of Tel monomers is transitory and appears to sensitize them for proteasomal degradation. Mechanistically, sumoylation of K11 inhibits repression of gene expression by full-length Tel. In accordance with this observation, we found that sumoylation impedes Tel association with DNA. By contrast, a Tel isoform lacking K11 (TelM43) is strongly repressive. This isoform results from translation from an alternative initiation codon (M43) that is common to all Tel proteins that also contain the K11 sumoylation consensus site. We find that PIAS3 may have a dual, context-dependent influence on Tel; it mediates Tel sumoylation, but it also augments Tel's repressive function in a sumoylation-independent fashion. Our data support a model that suggests that PIAS-mediated sumoylation of K11 and the emergence of TelM43 in early vertebrates are linked and that this serves to refine spatiotemporal control of gene expression by Tel by establishing a pool of Tel molecules that are available either to be recycled to reinforce repression of gene expression or are degraded in a regulated fashion.
Genetic analyses of Tel (11, 39, 40) and its Drosophila orthologue Yan (14, 19, 23, 25) have yielded compelling evidence that these proteins are unique Ets repressors (14, 17, 19, 23) that are crucial regulators of progenitor cell differentiation. Moreover, perturbation of normal Tel function can lead to the development of cancers, especially leukemias (6, 7, 8), in which at least 22 translocations involving Tel have been reported. A model is emerging that suggests that monomers of Tel directly associate via their conserved SAM (sterile alpha motif) domains and that the resulting DNA-bound oligomers (currently of indeterminate length) act as a physical barrier to the transcription-activating apparatus (reviewed in references 22, 34, and 38). However, the exact nature of repression by Tel/Yan is incompletely defined. In Drosophila, a protein named Mae (modulator of the activity of Ets) orchestrates Yan derepression by binding to Yan, thereby disrupting Yan self-association and binding to DNA, and sensitizing it to mitogen-activated protein kinase-dependent down-regulation (1, 21, 31, 33). Hitherto, a similar mechanism of regulation of Tel has not been uncovered in vertebrates.
A number of recent reports have highlighted the importance of sumoylation as a generic regulator of protein function (reviewed in reference 10), including Tel (4, 42). Currently, an important focus is the role of PIAS (protein inhibitor of activated STAT) proteins, which have an intrinsic SUMO E3-ligase capacity that catalyzes covalent conjugation of SUMO (small ubiquitin-like modifier) proteins to target substrates (29). A variety of functions have been ascribed to PIAS proteins that serve to moderate transcriptional activity (30), including that of some ETS transcription factor family members such as Elk-1 (44, 45) and Fli-1 (36). Mammals have four separate PIAS genes named PIAS1, PIAS2 (also referred to as PIASx, of which there are two variants, PIASxα and PIASxβ), PIAS3, and PIAS4 (also referred to as PIASγ or PIASy). Loss-of-function studies in mice of either PIAS1 (16), PIAS2 (26), or PIASy (41) revealed relatively minor phenotypes, and mice homozygous for the loss of these alleles were viable, perhaps the result of genetic redundancy. Drosophila has a single PIAS gene [Su(Var)2-10] that is indispensable for embryo development and viability (3, 9) and that encodes potentially nine different polypeptides (see the FlyBase database [http://flybase.bio.indiana.edu]).
Deciphering the precise mechanisms of action of Yan/Tel is important for understanding the control of progenitor cell differentiation and tissue patterning. Here, we report the identification and characterization of a new, conserved Tel sumoylation site. We find that sumoylation of Tel is PIAS-mediated and serves to limit Tel's repressive function. Whereas in Drosophila, derepression by Yan is orchestrated by Mae, in vertebrates Tel function is regulated posttranslationally through sumoylation of K11 and further modulated posttranscriptionally, yielding a highly repressive nonsumoylated isoform of Tel.
METHODS AND MATERIALS
MS.
Protein bands were excised from sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels, reduced, alkylated, and in-gel digested using trypsin (modified, sequencing grade; Promega) as previously described (32). For matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) analysis, tryptic digestions were desalted using C18 ZipTips (Millipore). Peptides were eluted using approximately 1 μl of 10 mg/ml dihydroxybenzoic acid in 50% acetonitrile-0.1% trifluoroacetic acid directly on a stainless steel MALDI target plate (Bruker Daltonics, Bremen, Germany). MALDI-TOF analyses were performed on an Ultraflex II time-of-flight mass spectrometer (Bruker Daltonics, Bremen, Germany) controlled by the Flexcontrol, version 2.0, software package. For liquid chromatography-mass spectrometry (LC-MS) analysis, samples were injected onto a capillary high-performance LC system (Ultimate; Dionex, Amsterdam, The Netherlands) equipped with a peptide trap column (Pepmap 100; 0.3-mm internal diameter by 1 mm; Dionex, Amsterdam, The Netherlands) and an analytical column (Pepmap; 0.075 by 150 mm; Dionex, Amsterdam, The Netherlands). The mobile phases consisted of 0.04% formic acid-0.4% acetonitrile (phase A) and 0.04% formic acid-90% acetonitrile (phase B). A 45-min linear gradient from 0 to 60% mobile phase B was used at a flow rate of 0.2 μl/min. The outlet of the high-performance LC system was coupled to an HCT IonTrap (Bruker Daltonics, Bremen, Germany) using a nanoelectrospray ionization source. The spray voltage was set at 1.2 kV, and the temperature of the heated capillary was set to 165°C. Eluting peptides were analyzed using the data-dependent tandem MS (MS/MS) mode over an m/z range of 400 to 1,600. The five most abundant fragments in an MS spectrum were selected for MS/MS analysis by collision-induced dissociation using helium as the collision gas.
In vitro sumoylation assays.
Glutathione S-transferase (GST)-tagged Tel mutants were sumoylated in Escherichia coli BL21(DE3) by essentially following the published procedure (35). In vitro translated proteins were sumoylated according to methods previously described (37).
Cell-based sumoylation assays.
Sumoylation assays were adapted from the established methods (24) with the following modifications. His-Sumo pull-downs were performed with 50 μl of Ni-nitrilotriacetic acid beads (Qiagen) for 3 h at room temperature in 6 ml of 6 M guanidinium-HCl, 0.1 M Na2HPO4·NaH2PO4, and 0.01 M Tris-HCl (pH 8.0) plus 20 mM imidazole and 10 mM β-mercaptoethanol (buffer A). The beads were successively washed twice with 1 ml of each of the following buffers: buffer A plus 0.2% Triton X-100, 8 M urea, 0.1 M Na2HPO4·NaH2PO4, and 0.01 M Tris-HCl (pH 8.0) plus 20 mM imidazole, 10 mM β-mercaptoethanol, and 0.2% Triton X-100 (buffer B); and a buffer containing 8 M urea, 0.1 M Na2HPO4·NaH2PO4, and 0.01 M Tris-HCl (pH 6.3) plus 20 mM imidazole, 10 mM β-mercaptoethanol, and 0.2% Triton X-100 (buffer C). Sumoylated proteins were eluted in 60 μl of urea sample buffer: 37.5% buffer C, 39.3% Laemmli buffer (3×), 20 mM imidazol, and 3.2% β-mercaptoethanol. The samples were boiled and analyzed by Western blot analysis.
In vivo 35S labeling: pulse-chase experiments.
Cells were washed free of medium and seeded into 6-cm tissue culture dishes (Gibco) for each time point, in methionine-free Dulbecco's modified Eagle's medium (DMEM; Gibco). Cells were routinely incubated for 3 h, and then the medium was supplemented with 50 μCi of 35S-labeled methionine. After 3 h of labeling, cells were washed free of label and then incubated in DMEM containing 10% fetal calf serum for the times indicated in Fig. 1G. Labeled hemagglutinin (HA) epitope-tagged Tel proteins were immunoprecipitated from the cell lysates as described below.
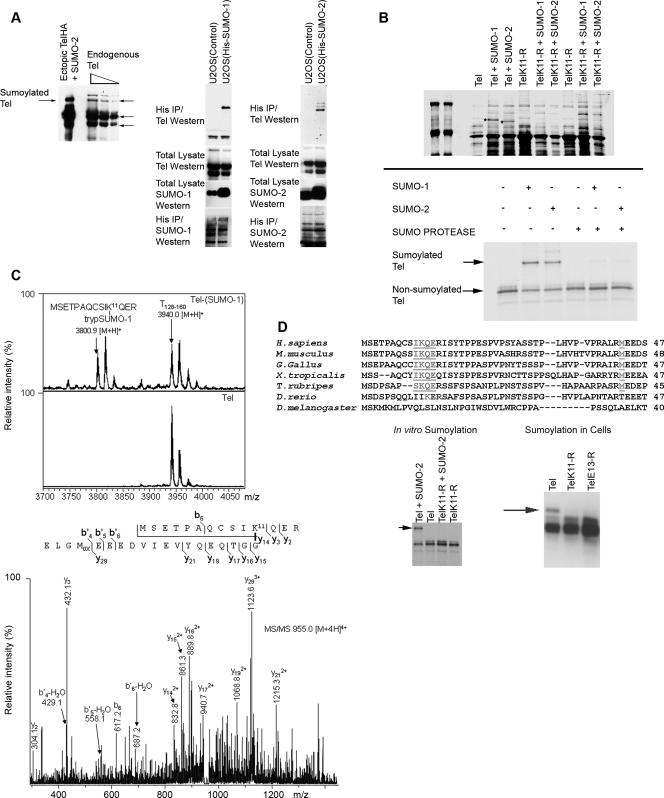
FIG. 1.
The highly conserved lysine residue (K11) is the primary substrate for SUMO conjugation to Tel. (A) Endogenous Tel is sumoylated. The left panel shows a Western blot of different amounts of a cell lysate that were prepared from U2OS cells. Tel proteins were detected with a Tel antibody directed against the C terminus of Tel (highlighted with arrows) (20). Endogenous Tel proteins were compared with ectopically expressed Tel sumoylated with SUMO-2 as a control. We established U2OS cell lines stably expressing His epitope-tagged SUMO-1 or SUMO-2. Sumoylated endogenous Tel was recovered from cells lysed in guanidinium, by nickel bead purification (right panel) (sumoylation assay). (B) In vitro sumoylation assay. Both SUMO-1 and SUMO-2 are efficiently, covalently conjugated to Tel almost exclusively on K11 by one of two methods. Fusions between GST and either full-length wild-type Tel or full-length Tel in which lysine at position 11 was mutated to an arginine residue (TelK11-R) were coexpressed in E. coli along with a SUMO E1-ligase (Aos1 or Uba2) and a SUMO E2-ligase (Ubc9) either for SUMO-1 or for SUMO-2 conjugation; proteins were then purified onto glutathione-Sepharose beads. A Coomassie blue-stained gel shows sumoylated Tel, which is absent from TelK11-R preparations (highlighted with asterisks); the results were confirmed by Western blotting (data not shown). A complementary study shows in vitro [35S]methionine-translated Tel proteins that were sumoylated in vitro (37) and then incubated with or without the active site of a SUMO-protease (Lifesensors). (C) MS reveals Tel to be sumoylated on K11. In vitro sumoylated and nonsumoylated Tel were separated by SDS-PAGE and subjected to in-gel digestion with trypsin. MALDI-TOF MS demonstrated an m/z value of 3,800.9 [M+H]+ within the tryptic digest of Tel-SUMO-1 which was absent in unsumoylated Tel. This peptide corresponds to the tryptic peptide of SUMO-1 (trypSUMO-1, ELGMEEEDVIEVYQEQTGG) conjugated to K11 within the N-terminal tryptic peptide of Tel. As a control a nonsumoylated tryptic peptide (T128-160; m/z 3,940.0 [M+H]+) of Tel is shown. The SUMO-1-conjugated peptide was also identified by LC-iontrap MS (m/z 955.0 [M + 4H]4+), and its sequence was subsequently confirmed by MS/MS. (D) K11 forms part of a classic sumoylation consensus site (ψKXE; specifically IKQE, underlined) that is highly conserved in all sequenced vertebrates with the exception of D. rerio. An alternative initiation codon (M43; underlined) is present in all Tel proteins also harboring a K11 sumoylation site (and is absent in D. rerio). Western blotting of cell lysates revealed an additional Tel protein (indicated with an arrow), approximately 11 kDa larger than unmodified Tel that is lost following mutation of the sumoylation consensus site, TelK11-R or TelE13-A. Likewise, mutation of K11 abolishes in vitro sumoylation of Tel. (E to G) In cells, K11 is necessary and sufficient for full (detectable) sumoylation of Tel. HA epitope-tagged versions of wild-type Tel or Tel expressing mutations that disrupt the sumoylation consensus site, TelK11-R or TelE13-A, were cotransfected into 293T cells along with His epitope-tagged SUMO-1 or SUMO-2 (E). Also assayed are two other Tel mutants: TelK99-R, in which the lysine residue at position 99 has been replaced with an arginine residue, and TelD101-A, in which the aspartic acid residue at position 101 has been changed to an alanine residue. A schematic representation of the mutants tested is included. Sumoylated Tel was recovered from cells lysed in guanidinium by nickel bead purification. Conditions for immunoprecipitating sumoylated Tel from cells were optimized (F) to allow in vivo labeling of Tel with [35S]methionine in order to monitor the stability of the pool of sumoylated Tel (G). IP, immunoprecipitation.
Immunofluorescence.
Cells were grown on glass coverslips and transfected using Fugene-6 (Roche). Cells were fixed after 24 h with 4% paraformaldehyde for 15 min at room temperature (RT) (all the following steps were done at RT) and permeabilized in 0.2% Triton X-100-phosphate buffered saline for 5 min. Cells were washed with phosphate buffered saline and blocked with 5% goat serum for 1 h, incubated with primary antibodies for 1 h, washed, and incubated with secondary antibodies for 30 min. Following extensive washing, cells were mounted, and immunostaining was visualized with a Leica DM5500 B microscope.
Luciferase reporter.
Cells were seeded in 24-well plates and transfected with 0.75 μg of Tel, PIAS, or SUMO plasmids along with 2 μg of pGL2-TK-ETS luciferase reporter (where TK is thymidine kinase) (1) and 0.5 μg of lacZ reporter. Cells were lysed 24 h posttransfection, and luciferase activity was measured using a luciferase assay substrate (Promega). Luciferase activity was normalized by measuring β-galactosidase activity.
Analysis of stromelysin-1 mRNA in stable cell lines.
U20S cells were seeded at 40 to 60% confluence in 10-cm tissue culture dishes and transfected with 5 μg of Tel mutant plasmid and 0.5 μg of pCDNA3.1 using Fugene (Roche). After 48 h medium was replaced by medium containing 200 μg/ml G418.
After 3 weeks of selection, expression of Tel mutants was confirmed by Western blotting and immunofluorescence. Total RNA from stable cell lines was prepared using Trizol (Invitrogen) following the manufacturer's protocol. First-strand cDNA synthesis was derived from 1 μg of total RNA using a TaqMan kit (Roche) according to the manufacturer's protocol. Expression levels of stromelysin-1 and GAPDH (the glyceraldehyde-3-phosphate dehydrogenase gene) were determined by quantitative real-time PCR using the following primer sets: 5′-CAAAACATATTTCTTTGTAGAGGACAA and 3′-TTCAGCTATTTGCTTGGGAAA for Stromelysin-1 and 5′-TGCCATGTAGACCCCTTGAAG and 3′-ATGGTACATGACAAGGTGCGG for GAPDH. Real-Time PCRs were performed in 10-μl reaction volumes using a 7900 HT Fast Real-Time PCR System detection apparatus (Applied Biosystems) and Sybr Green PCR Mastermix (Applied Biosystems) according to the manufacturer's protocol.
Protein-DNA interaction assays.
A total of 50 pmol of biotinylated double-stranded oligonucleotides harboring three consecutive Ets DNA-binding sites (Invitrogen) was coupled to My One Streptavidin C1 beads (Invitrogen) according to the manufacturer (Invitrogen). Interaction with Tel proteins was assessed in binding buffer: 20 mM HEPES, 30 mM KCl, 0.1 mH EDTA (pH 8.0), 4 mM MgCl, 0.8 mM Na2HPO4, 20% glycerol, protease inhibitors, 1 μg of dI-dC competitor, and 4 mM spermidine. Either 100 ng of GST fusion proteins or 10% of an in vitro translated protein was used. Reactions were performed for 30 min at RT. The beads were successively washed four times with binding buffer using a magnetic holder. Associated proteins were eluted in 3× Laemmli buffer and analyzed by Western blotting following SDS-PAGE.
Cell culture, biochemistry, and molecular biology.
Cell lines were cultured in DMEM supplemented with 10% fetal bovine serum and antibiotics. Cells were transfected with Fugene-6 (Roche) according to the manufacturer's protocol. In general, 1 μg of each construct was transfected into cells seeded at 40 to 60% confluence in a 6-cm dish, and cells were lysed at 24 to 48 h posttransfection.
Tel-HA and Tel-Flag constructs were fused in frame with either an HA or Flag epitope tag and cloned into the pCS2 expression vector. GST Tel was cloned in frame with GST in pGEX-2TK vector. Mutants were generated by PCR. For immunoprecipitations cells were lysed in 1 ml of either ice-cold radioimmunoprecipitation assay buffer-SDS (1 mM EDTA, 50 mM Tris, pH 8.0, 150 mM NaCl, 10% glycerol, 1% Triton X-100) or ice-cold protein lysis buffer (50 mM Tris, pH 7.5, 1% NP-40, 0.1% SDS, 0.5% Na-deoxycholate, 150 mM NaCl) with protease inhibitors (phenylmethylsulfonyl fluoride, trypsin, pepstatin A, leucine, and aprotinin) and NaF. Cell lysates were needle treated and centrifuged. Immunoprecipitations were carried out with 0.75 μl of anti-HA (rabbit polyclonal antibody to HA tag from Abcam) or anti-Flag (mouse M2 from Sigma Aldrich) antibody. Generally, lysates were preincubated with the antibody for 1 h, following which suitable beads (protein G-Sepharose 4 fast flow [Amersham Pharmacia Biotech] for mouse Flag immunoprecipitations and protein A-Sepharose [Sigma-Aldrich] for rabbit HA immunoprecipitations) were added for a further 2-h incubation. Associated proteins were analyzed by Western blotting. For pull-down assays, Tel proteins were labeled with [35S]methionine using a TNT-coupled reticulocyte in vitro translation system (Promega) and incubated with GST Tel fusions that were immobilized on glutathione-Sepharose beads as previously described (1).
Antibodies and drugs.
The following antibodies were used: anti-Flag mouse M2 monoclonal (Sigma-Aldrich), anti-HA.11 mouse monoclonal (Covance), anti-GST rabbit (My Probe), anti-HA rabbit polyclonal (Abcam), anti-Tel rabbit polyclonal (kindly provided by Ruud Delwel and Olivier Bernard), anti-His (His1; Sigma), anti-SUMO1 (21C7; Zymed), anti-SUMO-2/3 (AV-SM23-0100; Eurogentec) (37), and anti-PIAS3 (sc-14017; Santa Cruz) antibodies. For MG132 experiments cells were incubated with 3 μM MG132 (Calbiochem) for 6 h prior to lysis.
RESULTS
Tel is sumoylated on K11.
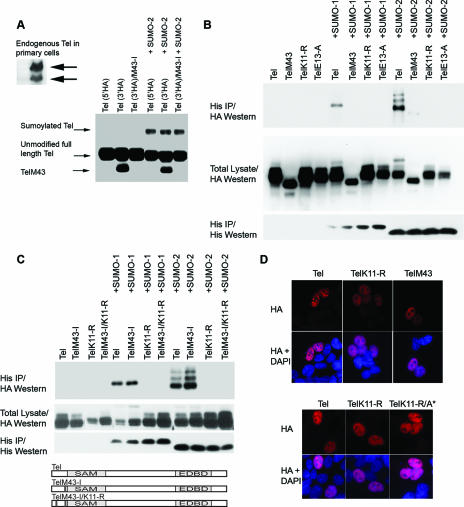
To study how the transcriptional repressor Tel is regulated, we monitored its posttranslational modifications. Figure 1A shows that in cells, a fraction of endogenous Tel is covalently conjugated with SUMO proteins. Tel-specific antibodies detect a species of endogenous Tel whose gel migration is indistinguishable from sumoylated (ectopically expressed) Tel. To confirm that this protein was indeed sumoylated Tel, we established cell lines that stably express relatively low levels of His epitope-tagged SUMO-1 or SUMO-2. This enabled the purification of cellular proteins that are covalently conjugated with SUMO following denaturing lysis with guanidinium solution. By this means we found that endogenous Tel was covalently conjugated with both SUMO-1 and SUMO-2 (Fig. 1A). These findings are consistent with previous analyses that identified Tel as a substrate for sumoylation (4, 42). To unambiguously identify the site(s) of sumoylation, we exploited the fact that both SUMO-1 and SUMO-2 are efficiently, covalently conjugated to Tel in vitro (Fig. 1B) and performed an MS analysis of the sumoylated and nonsumoylated fractions of Tel. While K99 of the Tel SAM domain was previously implicated as a site of sumoylation (4, 42), we found Tel to be predominantly sumoylated on the N-terminal lysine 11 (K11) (Fig. 1C) (this was also determined for SUMO-2 [P. J. Hensbergen and D. A. Baker, unpublished data]) but not on K99. Importantly K11 forms part of a “classic” sumoylation motif, ψKXE (where X is any residue and ψ is a large hydrophobic amino acid) (24), specifically IKQE, that is highly conserved in all sequenced vertebrates with the notable exception of Danio rerio (Fig. 1D). Significantly, a putative alternative initiation codon (M43) (20) is present in all known vertebrate Tel proteins also harboring a K11 sumoylation site (and is absent in D. rerio). We confirmed that K11 forms part of a bona fide sumoylation site and is a target for sumoylation both in vitro and in tissue culture cells. Figure 1B to E show that whereas wild-type Tel was efficiently sumoylated, Tel mutants that either lack K11 (TelK11-R) or carry a point mutation in the sumoylation consensus motif (TelE13-R) were almost completely resistant to SUMO conjugation both in vitro and in cells. Similar results were obtained using HeLa, U20S, MCF7, and U937 cells. Consistent with these findings, pulse-chase experiments in cells revealed a pool of Tel that is sumoylated on K11 that appears to be at least as stable as nonsumoylated Tel (Fig. 1F and G). In contrast to previous reports (4, 42), we found that mutation of K99 of the SAM domain to arginine or alanine did not abrogate Tel sumoylation (either monomeric or oligomeric Tel) either in cells (Fig. 1E) or in vitro (M. G. Roukens, M. Alloul-Ramdhani, D. A. Baker, and P. J. Hensbergen, unpublished data). Likewise, disruption of the putative consensus sumoylation site surrounding K99 as a result of replacing aspartic acid 101 with alanine (also arginine) (data not shown) had no significant effect on SUMO-1 and SUMO-2 conjugation (Fig. 1E). Collectively, these data argue that Tel is efficiently sumoylated and that the primary target of sumoylation is the highly conserved K11 residue.
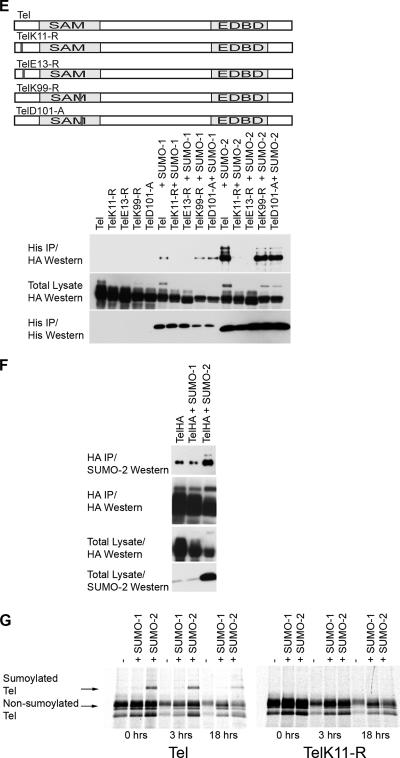
K11 of Tel oligomers, but not Tel monomers, is found conjugated with SUMO in cells.
There is strong molecular evidence that monomers of Tel form homotypic oligomers via the SAM domain (13). A current model suggests that once bound to DNA, these oligomers repress gene expression (13, 21, 31, 33). We initiated our analyses of the molecular mechanisms of Tel sumoylation by first determining whether sumoylation is common to both Tel monomers and oligomers. To this end we generated Tel mutants that are unable to self-associate. Figure 2A shows that whereas wild-type Tel efficiently self-associates, Tel proteins harboring mutations that inhibit SAM domain oligomerization (13, 21, 31) fail to associate with one another in cells (Fig. 2A) and in vitro (data not shown). Figure 2B and C show that in cells, monomeric Tel proteins (containing mutations of helices 2, 3, 4, and 5 but not helix 1 of the SAM domain) (Fig. 2A) (M. G. Roukens and D. A. Baker unpublished data) are only very weakly sumoylated (42). Rather than reflecting inefficient sumoylation, the more likely explanation is that sumoylation of monomers in cells is transient. Strong evidence for this comes from the fact that monomers are efficiently sumoylated on K11 in vitro (Fig. 2D). Moreover, a pool of Tel monomers that are efficiently sumoylated on K11 were readily detected in cells following treatment with proteosome inhibitors, suggesting that the sumoylating machinery does recognize Tel monomers (Fig. 2E). These results suggest that in common with Tel oligomers, monomers of Tel can be efficiently sumoylated but that in cells monomer sumoylation is transitory and appears to sensitize Tel for proteasomal degradation.
FIG. 2.
K11 of Tel polymers, but not Tel monomers, is found conjugated with SUMO in cells. (A) Disruption of the SAM domain prevents Tel self-association. HA epitope-tagged and Flag epitope-tagged fusions of wild-type Tel or Tel expressing mutations that disrupt the SAM domain (A*, which contains an arginine residue in place of an alanine residue; deletion of the SAM domain is shown as ΔSAM) were expressed in 293T cells in the indicated combinations. A schematic representation of the mutants tested is included. (B and C) Monomeric forms of Tel exhibit low levels of sumoylation in cells. (B) HA epitope-tagged versions of wild-type Tel or Tel expressing mutations that disrupt the SAM domain (described above) were cotransfected into 293T cells along with His epitope-tagged SUMO-1 or SUMO-2, and a sumoylation assay was performed. (C) The experiment in panel B was performed using Tel mutants in which each of the five helices that comprise the SAM domain were individually deleted. A schematic representation of the mutants tested is included. (D) Tel monomers are efficiently sumoylated in vitro on K11. Fusions between GST and either full-length wild-type Tel or full-length Tel harboring mutations that disrupt the SAM domain (see above) were sumoylated in E. coli as described in the legend of Fig. 1A. Sumoylated forms of Tel are highlighted (*) and were confirmed by Western blotting and also MS (data not shown). Shown also is an HA Western blot to detect Tel following an in vitro sumoylation assay (37) using the indicated Tel proteins that were made by in vitro translation in the presence of 1 mM unlabeled methionine. Sumoylated proteins that are absent from TelA* proteins containing a mutation of K11 to an arginine (TelA*/K11-R) are highlighted with arrows. (E) Inhibiting proteasome function stabilizes the pool of Tel monomers sumoylated on K11 in cells. Cells were cotransfected with the indicated Tel constructs along with His epitope-tagged SUMO-2. Following incubation with or without MG132, a sumoylation assay was performed. IP, immunoprecipitation.
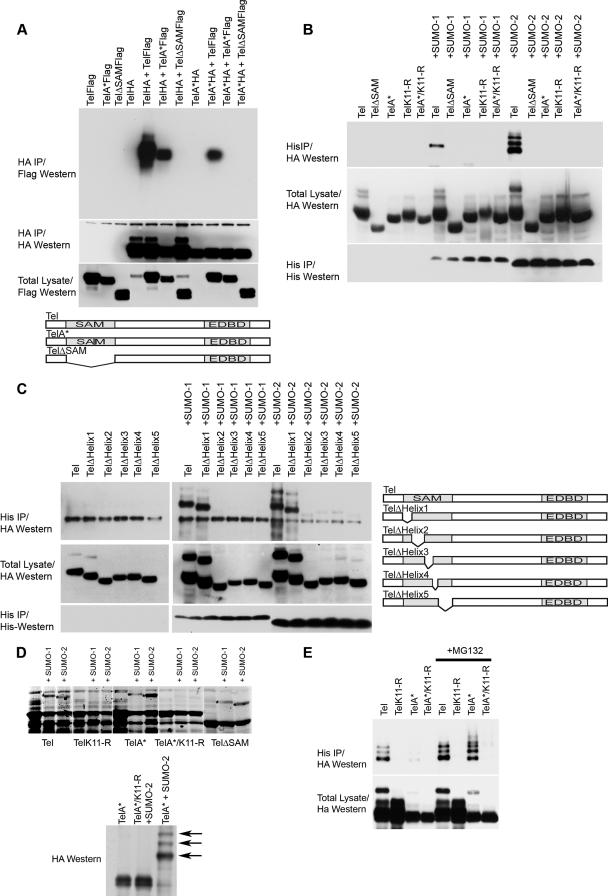
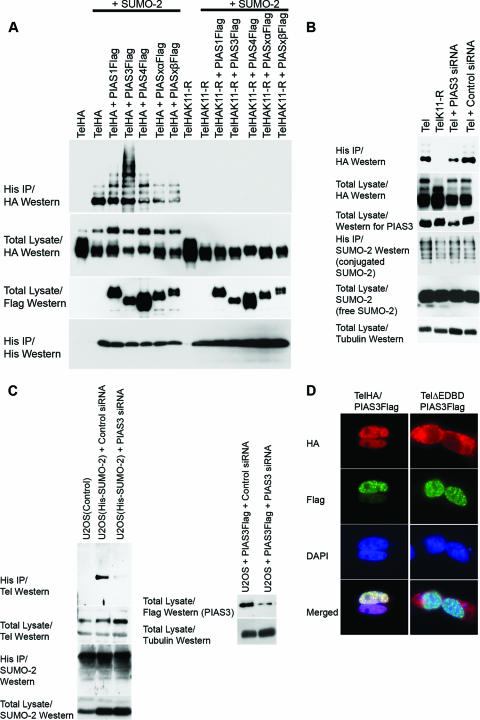
PIAS3 interacts with Tel and promotes sumoylation of K11.
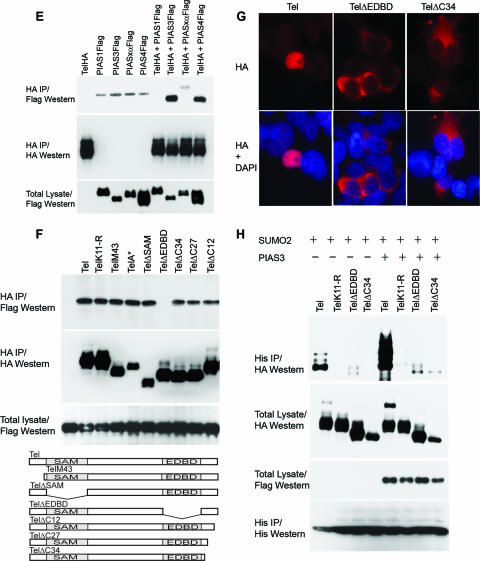
Next, we investigated the mechanisms mediating Tel sumoylation. PIAS proteins function as E3 ligases, which catalyze the covalent attachment of SUMO to numerous transcription factors, thereby modulating their activity (30). Therefore, we assessed the role of PIAS proteins in Tel sumoylation. Several lines of evidence indicate that PIAS3 acts as a SUMO E3 ligase for Tel. First, in cells coexpression of PIAS3 strongly stimulates Tel sumoylation (Fig. 3A) (similar results were obtained with SUMO-1) (data not shown). Coexpression of other PIAS family members had either only very modest effects on Tel sumoylation levels or no detectable impact (Fig. 3A). We further demonstrated that PIAS3 specifically stimulates SUMO conjugation on lysine K11 since mutation of this residue abolishes PIAS3-enhanced Tel sumoylation. Second, small interfering RNA (siRNA)-mediated knockdown of endogenous PIAS3 significantly reduced the levels of sumoylation of both ectopically expressed Tel (Fig. 3B) and most significantly also endogenous Tel (Fig. 3C), strongly implying that sumoylation of endogenous Tel is dependent on PIAS3. Third, immunohistochemistry (Fig. 3D) and live-cell imaging of cyan fluorescent protein-Tel and yellow fluorescent protein-PIAS3 fusion proteins (data not shown) showed that these proteins colocalize in cells (Fig. 3D). Finally, Tel efficiently coimmunoprecipitates both endogenous (data not shown) and exogenous PIAS3 from cellular extracts, indicating that the two proteins physically interact (Fig. 3E). Tel also interacted with PIAS4; however, Fig. 3A shows that PIAS4 had relatively little influence on Tel sumoylation even when expressed at very high levels. To map the region of Tel that is required for PIAS3 binding, we generated a number of Tel deletion mutants. While wild-type Tel readily bound to PIAS3, TelΔEDBD, which lacks the Ets DNA-binding domain (EDBD), failed to associate with PIAS3 (Fig. 3F). This interaction assay was performed in relatively stringent lysis buffer that disrupts cellular membranes and thereby allows proteins that are normally localized in the cytoplasm to interact with proteins that are normally localized in the nucleus. Two of the described mutations, TelΔEDBD and TelΔC34, which lacks the C-terminal 34 amino acids, are both mislocalized from the nucleus to the cytoplasm (Fig. 3G). The lack of PIAS3 binding is not due to mislocalization of TelΔEDBD since TelΔC34, which is similarly mislocalized, efficiently bound to PIAS3 under the same conditions (Fig. 3F). We noticed that TelΔC34 sumoylation levels are strongly attenuated, and in contrast to wild-type Tel, ectopic expression of PIAS3 fails to appreciably augment its sumoylation (Fig. 3H) even though TelΔC34 can efficiently associate with PIAS3 (Fig. 3E). Coupled to the fact that PIAS3 is predominantly a nuclear protein (Fig. 3D), this suggests that PIAS3-mediated sumoylation of Tel occurs in the nucleus. In sum, these results suggest that sumoylation of K11 of Tel occurs in the nucleus and is mediated by PIAS3 via binding to the Tel EDBD.
FIG. 3.
PIAS3 stimulates sumoylation of Tel on K11. (A) Ectopic expression of PIAS3 strongly stimulates sumoylation of K11 of Tel. Tel or TelK11-R was transfected into 293T cells with or without His epitope-tagged SUMO-2, either alone or together with the indicated PIAS constructs. Sumoylated Tel was recovered from cells by a sumoylation assay. (B) Endogenous PIAS3 is essential for normal Tel sumoylation. Cells were transfected with the indicated constructs and a sumoylation assay was performed 2 days later. In the absence of an effective antibody specific for human PIAS3, the efficiency of PIAS3 knockdown was assessed by targeting Flag epitope-tagged PIAS3 expressed in U2OS cells. A nonspecific siRNA was used as a control. (C) Endogenous PIAS3 is required for sumoylation of endogenous Tel. We established U2OS cell lines stably expressing either His epitope-tagged SUMO-1 or SUMO-2. The indicated cell lines were transfected with either a control siRNA (directed against GFP) or siRNAs directed against PIAS3, and a sumoylation assay was performed 2 days later. The efficiency of PIAS3 knockdown was assessed by targeting Flag epitope-tagged PIAS3 expressed in U2OS cells. A nonspecific siRNA was used as a control (right panel). (D) Colocalization of Tel and PIAS3 in the nucleus. Cells were transfected with the indicated constructs and proteins were detected with the indicated antibodies. TelΔEDBD is the same as wild-type Tel except it lacks the EDBD. (E) Tel interacts with PIAS3. 293T cells were transfected with the indicated constructs. Tel complexes were immunopurified from cell lysates, made using radioimmunoprecipitation assay-SDS lysis buffer, using an antibody directed against the HA epitope, and associated PIAS protein was detected using an antibody directed against the Flag epitope. (F) Tel binding to PIAS3 requires its EDBD in cells. The indicated HA epitope-tagged Tel proteins were coexpressed with or without PIAS3 (not shown since the background was clear). Tel complexes were immunopurified from cell lysates using an antibody directed against the HA epitope, and associated PIAS3 protein was detected using an antibody directed against the Flag epitope. Tel M43 lacks the N-terminal 42 amino acids; Tel A* contains an amino substitution (arginine residue in place of an alanine residue) at position 93; TelΔSAM lacks the SAM domain; and Tel ΔC34, TelΔC27, and TelΔC12 have deletions of the C terminus of the indicated lengths. A schematic representation of the mutants tested is included. (G) Mutations that disrupt the EDBD or the C terminus of Tel lead to mislocalization of Tel to the cytoplasm. Cells were transfected with the indicated constructs, and immunohistochemistry was performed with the antibodies shown. (H) Mislocalization strongly abrogates sumoylation. Cells were cotransfected with the indicated constructs, and a sumoylation assay was performed. DAPI, 4′,6′-diamidino-2-phenylindole; IP, immunoprecipitation.
Posttranscriptional regulation of Tel sumoylation.
Figure 1D highlighted a highly conserved methionine residue (M43) common to all Tel proteins that also contain the K11 sumoylation consensus site. In agreement with previous reports we found that cells express different Tel isoforms (Fig. 4A), and it has been shown that one of these isoforms, TelM43, results from use of M43 as an alternative initiation codon (2, 20, 27, 28). Importantly, TelM43 lacks the N-terminal 42 amino acid residues including the K11 sumoylation site. As the results shown in Fig. 1B indicate, whereas full-length endogenous Tel appears to be detectably sumoylated, we were unable to detect a protein of a size that would be expected if TelM43 were sumoylated. We explored how this mechanism regulates Tel function. To that end we expressed a C-terminally tagged tel construct in cells that yielded two Tel proteins: full-length Tel and a shorter form of Tel (Fig. 4A). The shorter form of Tel results from initiation of translation from codon M43 because expression of a tel mutant that lacks this alternative start codon (TelM43-I) generates only full-length Tel. Figure 4B shows that, as expected, sumoylation of TelM43, like TelK11-R, is strongly abrogated. We found that the full-length TelM43-I protein, which is manufactured in cells without translation of the smaller TelM43 protein (Fig. 4A), is sumoylated on K11 as efficiently as wild-type full-length Tel (Fig. 4C), suggesting that sumoylation of full-length Tel K11 neither requires nor is inhibited by coexpressed TelM43. Collectively, these results show that from a single tel transcript, two different initiation codons can be used to generate a full-length Tel protein that is sumoylated and a smaller Tel isoform (TelM43) that is not sumoylated on K11 and, furthermore, does not influence sumoylation of full-length Tel K11. In cells, TelM43 and TelK11-R adopt a defined speckled pattern of nuclear distribution in contrast to wild-type full-length Tel that has a more uniform, less speckled nuclear distribution (Fig. 4D). This speckled pattern likely reflects the distribution of complexes containing Tel oligomers (Tel repressive conformation) since TelK11-R and TelM43 proteins that are unable to oligomerize (Fig. 3A, TelK11/A* and TelM43/A*) do not have a speckled nuclear distribution (Fig. 4D; also data not shown).
FIG. 4.
Posttranscriptional regulation of Tel sumoylation. (A and B) An alternative Tel isoform produced through use of an alternative initiation codon (M43) escapes sumoylation. TelM43 can be translated from the full-length Tel cDNA. 293T cells were separately transfected with three different Tel constructs (A). One expressed an HA epitope at the N terminus, another expressed the HA epitope at the C terminus enabling visualization of both Tel and TelM43 isoforms, and a final construct contained an HA epitope at the C terminus but expressed an isoleucine residue in place of the methionine at position 43 (TelM43-I). A separate panel highlights endogenous Tel proteins detected following Western blotting of lysates prepared from primary human hemopoietic blast cells. Sumoylation of the TelM43 isoform is strongly abrogated (B). Cells were cotransfected with the indicated constructs, and a sumoylation assay was performed. (C) Sumoylation of K11 is independent of the TelM43 isoform. Cells were cotransfected with the indicated constructs, and a sumoylation assay was performed. TelM43-I is described for panel A. TelM43-I/K11-R is identical to TelM43-I except that lysine at position 11 has been mutated into an arginine residue. A schematic representation of the mutants tested is included. (D) Nonsumoylated Tel isoforms adopt a more speckled subcellular distribution. Cells were transfected with the indicated constructs, and proteins were detected with the indicated antibodies.
Sumoylation of K11 inhibits Tel's repressive function.
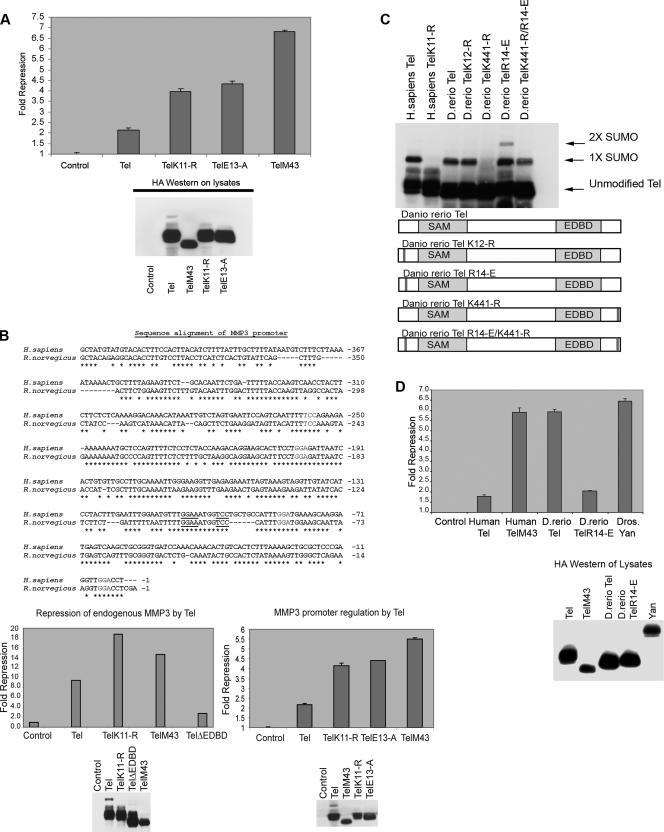
We next examined how sumoylation of K11 affects Tel's ability to repress gene expression. To determine the role of sumoylation in Tel's repressive function, we employed three different but complementary measures. First, we utilized a luciferase trans repression reporter assay in which an engineered minimal Tel regulatable promoter controls constitutive TK-driven luciferase expression (1). Figure 5A shows that ectopic expression of Tel represses luciferase activity. By contrast, we found that both TelM43 and TelK11-R, which are not sumoylated, exerted a significantly stronger repressive effect than wild-type Tel (Fig. 5A). To confirm that the enhanced repressiveness of TelK11-R and TelM43 results from the absence of sumoylation rather than from a different posttranslational modification of K11, we also assayed the repressive activity of TelE13-A, which disrupts sumoylation of K11 (Fig. 1E) while leaving the K11 residue accessible to other modifications. Figure 5A shows that comparable protein levels of TelE13-A repressed luciferase expression to the same degree as TelK11-R, suggesting that the enhanced repressive function of TelK11-R and TelM43 does indeed result from their resistance to repression-inhibiting covalent conjugation of SUMO to K11. Next, we assessed Tel's repressive function by monitoring an in vivo target of Tel. A previous report highlighted the rat stromelysin-1 (MMP3) promoter as a target of repression by Tel (5). Figure 5B shows that the stromelysin-1 promoter is highly conserved between human and rat, and, in particular, both share nearly identical consensus Tel DNA-binding sites. To determine if Tel controls expression of endogenous human stromelysin-1 and if sumoylation of Tel K11 regulates this process, we established a number of stable cell lines expressing either wild-type full-length Tel, TelM43, TelK11-R, or TelΔEDBD. By quantitative PCR we found that Tel strongly inhibited expression of endogenous stromelysin-1, which requires Tel DNA-binding, because TelΔEDBD failed to strongly inhibit stromelysin-1 expression. Significantly, comparable expression levels of both TelK11-R and TelM43 impeded stromelysin-1 expression far more potently than wild-type Tel. Indeed, both were at least twice as repressive as wild-type Tel (Fig. 5B) which almost exactly mirrors our findings with the artificial Tel promoter presented in Fig. 5A. In support of this, we performed chromatin immunoprecipitation assays and found that disruption of Tel sumoylation (TelK11-R and TelM43) leads to enhanced Tel association to endogenous stromelysin-1 (data not shown). To further corroborate these results, we placed expression of the luciferase reporter gene under the control of the human stromelysin-1 promoter and compared repression of gene expression by full-length wild-type Tel, which is sumoylated on K11, with TelM43, TelK11-R, and TelE13-A, which are not sumoylated on K11. The results using this system were in strict agreement with the prior assays (Fig. 5B); Tel that was not conjugated with SUMO on K11 was at least two times more repressive than wild-type Tel, which is sumoylated on K11. Collectively, these data suggest that sumoylation of K11 negatively regulates Tel's ability to suppress gene expression.
FIG. 5.
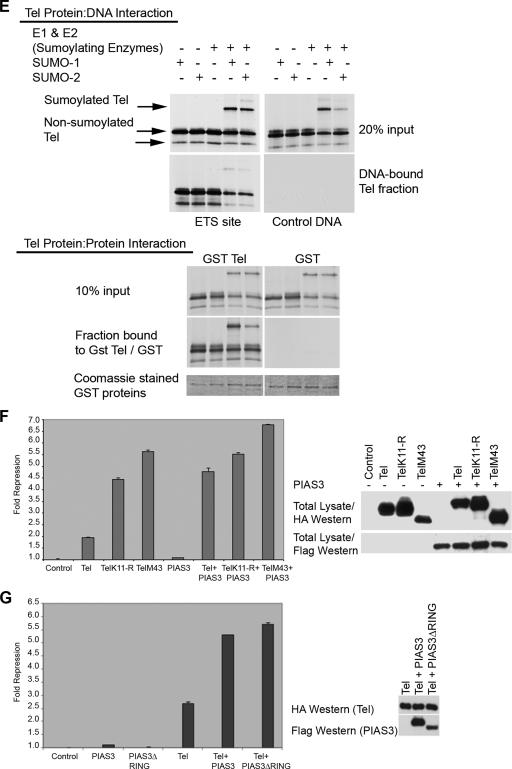
Sumoylation of K11 inhibits repression by Tel. (A) Either 293T, U2OS, or NIH 3T3 cells were cotransfected with the indicated Tel constructs along with a luciferase reporter in which an engineered minimal Tel/Yan regulatable promoter controls TK-driven luciferase expression. Equivalent transfection efficiencies of the indicated constructs were determined by a β-galactosidase assay for expression of a cotransfected LacZ reporter, and protein levels of ectopically expressed Tel constructs were determined by Western blotting of lysates. Shown is a representative experiment using 293T cells. Experiments were performed in triplicate. (B) Tel repression of expression of endogenous human stromelysin-1 (MMP3) is inhibited by sumoylation of K11. The upper panel shows that the promoters of rat and human stromelysin-1 share a number of conserved, potential Tel DNA binding sites. Highlighted in bold are previously characterized Tel binding sites (5). Additional highly conserved putative Tel binding sites are underlined. In the lower left panel, we established stable cell lines expressing the indicated Tel constructs. We prepared cDNA from these lines and performed quantitative PCR to assess the levels of expression of endogenous stromelysin-1. Shown graphically are the relative repression levels normalized against a control human gapdh gene. Shown in the right panel is a luciferase reporter assay. Luciferase expression was placed under the control of the human stromelysin promoter. Equivalent transfection efficiencies of the indicated constructs were determined by a β-galactosidase assay for expression of a cotransfected LacZ reporter, and protein levels of ectopically expressed Tel constructs were determined by Western blotting of lysates. Shown is a representative experiment using U2OS cells. Experiments were performed in triplicate. (C) D. rerio Tel does not express an N-terminal site of sumoylation but is sumoylated at position K441. Cells were transfected with the indicated constructs together with SUMO-2, and Tel sumoylation was assessed by Western blotting. D. rerio TelK12-R is the same as wild-type D. rerio Tel except that the lysine residue at position 12 has been replaced with an arginine residue. D. rerio TelR14-E is identical to wild-type D. rerio Tel except that the N-terminal sumoylation consensus site has been created by replacing the arginine residue with a glutamic acid residue at position 14. D. rerio TelK441-R is the same as wild-type D. rerio Tel except that the lysine residue at position 441 has been replaced with an arginine residue. D. rerio TelK441-R/R14-E is the same as D. rerio TelK441-R except that the arginine residue at position 14 has been replaced with a glutamic acid residue. A schematic representation of the mutants tested is included. (D) Cells were transfected with the indicated constructs, and a luciferase repression assay was performed as described in panel A. (E) Sumoylation of K11 impedes the ability of Tel to associate with DNA. Tel proteins were efficiently sumoylated following in vitro translation in reticulocyte lysates as previously described (37). Biotinylated Ets DNA-binding sites were coupled to streptavidin beads following the manufacturer's advice (Invitrogen) and incubated with the indicated proteins. Associated Tel proteins were recovered by SDS-PAGE. Tel-Tel interactions were assessed by incubating GST Tel fusion proteins or GST proteins alone, along with sumoylated and nonsumoylated in vitro translated forms of Tel. (F and G) PIAS3 enhancement of repression by Tel is sumoylation independent. The experiment shown in panel F is the same as that described for panel A except that cells were cotransfected with or without PIAS3. In the experiment in panel G, cells were transfected with the indicated constructs, and a luciferase reporter assay was performed as described for panel A. PIAS3ΔRING is the same as PIAS3 except that the RING domain has been deleted.
This notion was further strengthened by our studies with D. rerio Tel. Although D. rerio Tel encodes a lysine at position 12 (K12) that is equivalent to Homo sapiens Tel K11, this lysine is not part of a typical sumoylation consensus motif, and, moreover, D. rerio Tel lacks an internal initiation codon M43 (Fig. 1D). However, D. rerio Tel is sumoylated in cells (Fig. 5C). There are three obvious possibilities: K12 is in fact sumoylated, K99 is sumoylated (the sequence surrounding D. rerio Tel K99 is identical to human Tel), or the C-terminal K441 is sumoylated since this forms part of a classic sumoylation consensus motif. Figure 5C shows that mutation of K441 to an arginine residue completely abolished D. rerio Tel sumoylation, whereas mutation of K12 or K99 (data not shown) had no detectable effect on D. rerio sumoylation levels. Furthermore, mutation of R14 to E (R14-E) created a sumoylation consensus site that allowed sumoylation of K12 (Fig. 5C). These data show that wild-type D. rerio Tel is not normally sumoylated on K12 but that the mutation R14-E establishes a new sumoylation site in a position that is equivalent to H. sapiens K11. We tested these constructs using our luciferase trans repression reporter assay. Figure 5D shows that wild-type D. rerio Tel efficiently repressed gene expression. The ability of D. rerio Tel to repress reporter gene expression was comparable to the levels of repression by human TelM43. In contrast, the D. rerio TelR14-E mutant, which is sumoylated on K12, displayed a significantly reduced repressive activity compared to wild-type D. rerio Tel. Repression by D. rerio TelR14-E was similar to repression by wild-type full-length human Tel (Fig. 5D). Together, these data showed that sumoylation of K11 limits the ability of Tel to repress gene expression while TelM43 is strongly repressive by comparison with full-length wild-type Tel because it escapes sumoylation of K11. In the assay systems described above, PIAS3 siRNA oligonucleotides had little effect; this result may reflect genetic redundancy, insufficient PIAS3 knockdown, or the fact that PIAS3 appears to exert a sumoylation-independent, stimulatory effect on repression by Tel (Fig. 5F) such that the effects of the PIAS3 siRNA oligonucleotides on this system are cancelled out. Disruption of other components of the sumoylation machinery such as the SUMO E1 ligase SAE2 or the SUMO E2 ligase Ubc9 by using siRNAs had significant impact in our reporter assays even in the absence of Tel (Roukens and Baker, unpublished), presumably because they are generic regulators of protein function. This precluded any direct, specific quantitative analysis of their roles in repression by Tel.
We next examined how sumoylation might inhibit repression. To that end we coupled biotinylated Ets DNA-binding sites to beads and compared the efficiency of binding of sumoylated and nonsumoylated Tel. Consistent with our reporter assay results, Fig. 5E shows that nonsumoylated Tel alone interacted efficiently with the Ets DNA-binding sites while sumoylation of K11 served to impair Tel binding to DNA of both sumoylated Tel and also nonsumoylated Tel in the same mixture. Under these experimental conditions, we found that both sumoylated and nonsumoylated Tel could efficiently associate with one another, as shown by the fact that a GST-Tel fusion protein was able to efficiently purify both sumoylated and nonsumoylated Tel from a mixture of the two (Fig. 5E). These results suggest that sumoylation does not inhibit Tel self-association but does impede Tel DNA-binding.
Interestingly, in the absence of added SUMO-1 or SUMO-2, although PIAS3 alone had little detectable influence on luciferase expression, it synergistically enhanced repression by Tel (Fig. 5F). SUMO-1 and SUMO-2 are rate limiting for stimulation of sumoylation by PIAS3, because in the absence of added SUMO-1 or SUMO-2 ectopic expression of PIAS3 fails to appreciably augment sumoylation (M. Alloul-Ramdhani and D. A. Baker, unpublished data). Consistent with the idea that the observed PIAS3 corepressor function may be independent of sumoylation, Fig. 5G shows that a PIAS3 protein lacking the RING domain synergistically enhanced Tel repression of gene expression to approximately the same degree as wild-type PIAS3. Furthermore, in the presence of ectopic SUMO-1 or SUMO-2, PIAS3 failed to promote repression of gene expression by Tel (Roukens and Baker, unpublished). These data suggest that as well as strongly stimulating Tel sumoylation that inhibits repression by Tel (Fig. 3), PIAS3 might also act as a SUMO-independent Tel corepressor.
DISCUSSION
Our study shows that PIAS3 mediates sumoylation of full-length Tel on K11 but not of TelM43, whose evolution appears to be linked to that of the K11 sumoylation site. Sumoylation of K11 inhibits the ability of Tel to repress gene expression. This establishes a mechanism that presumably allows finer spatiotemporal control of Tel function through the production of a nonsumoylated, repressive version of Tel and by promoting the formation of a pool of (sumoylated) Tel that can subsequently be recruited for repression or be degraded in a regulated fashion.
Tel K11 is the primary site of covalent conjugation of SUMO.
We establish here that endogenous Tel is sumoylated, and through a combination of biochemical and molecular analyses, including MS, we have determined that the N-terminal lysine at position 11 (K11) is the primary substrate for covalent conjugation of both SUMO-1 and SUMO-2 to Tel. K11 forms part of a bona fide sumoylation motif that is highly conserved in all vertebrates except D. rerio. Although this is the first report of sumoylation of Tel K11, sumoylation of Tel K99 has been described previously (4, 42). K99 is found in the Tel SAM domain that characterizes a subfamily of Ets transcription factors. Although it is embedded in a sequence that is not a perfect match to known sumoylation sites, nevertheless the sequence does resemble a consensus sumoylation site. Moreover, this sequence is not only highly conserved in all known vertebrate Tel proteins but also shared by invertebrate orthologues of Tel such as Bombyx mori Yan and, indeed, in the SAM domains of related Ets family members such as Tel2, Fli-1, and Erg. However, unlike our findings for K11, we failed to uncover K99 as a target for sumoylation in any of our analyses, leading us to conclude that K11 is the primary, regulatory site of Tel sumoylation. Interestingly, and perhaps significantly, the sequence immediately adjacent to K99 conforms to a recently described SUMO interaction motif (15), raising the possibility that this hydrophobic core (LLLL in all sequenced vertebrates including D. rerio) may serve as an interface for noncovalent docking of SUMO to Tel and may also modulate Tel function.
K11 of Tel oligomers and monomers can be sumoylated, but only a pool of Tel oligomers are found conjugated with SUMO in cells.
Tel can exist in two basic forms: as a monomer or as homotypic oligomers formed by self-association of monomers via their SAM domains (1, 13, 21, 31, 33). Our results presented in Fig. 2 strongly suggest that both Tel monomers and Tel polymers can be sumoylated but that in cells only a pool of oligomeric Tel is found covalently conjugated with SUMO, whereas the fraction of monomeric forms of Tel that are sumoylated is minimal by comparison. This latter observation was reported previously (42). These differences could be due to rapid desumoylation or inherent monomer instability. For example, unlike Tel monomers, Tel oligomers may adopt a conformation or subcellular localization that renders the sumoylated K11 site less accessible to desumoylating enzymes. Certainly, desumoylation will prove to play an important role in Tel function (M. G. Roukens, M. Alloul-Ramdhani, and D. A. Baker, unpublished data); however, we believe that the absence of a pool of sumoylated monomers reflects pronounced monomer instability. In support of this, Fig. 2E shows that inhibiting the proteosome leads to a dramatic stabilization of sumoylated forms of the monomer but not of sumoylated forms of Tel oligomers, suggesting that Tel monomers are intrinsically more unstable than Tel oligomers and that perhaps sumoylation sensitizes Tel monomers for proteasomal degradation. Consistent with this, in a complementary study we find that Tel monomers are especially sensitive to ubiquitin-mediated degradation (Roukens, Alloul-Ramdhani, and Baker, unpublished).
Sumoylation of Tel K11 is mediated by PIAS3.
Figure 3 describes our evidence that PIAS3 mediates Tel sumoylation. A number of reports and, in particular, loss-of-function studies in mice (16, 26, 41) suggest that PIAS proteins can perform overlapping roles and compensate for the lack of other PIAS proteins as a result of genetic redundancy. Importantly, we found that disruption of PIAS3 alone strongly inhibited Tel sumoylation, suggesting that it plays a key role in Tel function, although one cannot rule out the possibility that other PIAS proteins also regulate Tel perhaps in a cell-type-specific fashion. We mapped the PIAS3 binding site to the EDBD of Tel, and this finding resembles the requirement of the Fli-1 EDBD for interaction with PIASxα (36). This raises the possibility that the EDBD of other ETS proteins is also a site of interaction with PIAS protein family members and thus PIAS-ETS interactions are a defining feature of at least a subfamily of ETS proteins.
Sumoylation of Tel K11 suppresses repression of gene expression by Tel.
To date, many studies of transcriptional regulators have demonstrated that, by and large, sumoylation serves to reinforce repression of gene expression (44), for example, by facilitating the recruitment of histone deacetylases (43). By contrast, we find that sumoylation of Tel K11 strongly suppresses Tel's repressive function. We used three different but complementary means to examine repression of gene expression by Tel (Fig. 5), and the robustness of these mechanistic studies is revealed by the strict agreement in results between the various assays. Specifically, we find that forms of Tel that are not sumoylated on K11 are at least twice as repressive as versions of Tel that can be sumoylated on K11. The consensus sumoylation site containing K11 is a feature of every predicted vertebrate Tel protein with the exception of D. rerio Tel. This offered a naturally occurring “mutant” with which to corroborate our finding that sumoylation of Tel does indeed limit its ability to suppress gene expression. Our results with H. sapiens Tel predicted that wild-type D. rerio should act as a relatively efficient repressor, comparable to the highly repressive H. sapiens TelM43, which also lacks a K11 site (Fig. 5A and B), and that acquisition of a site equivalent to that of H. sapiens K11, which can be sumoylated (Fig. 5C), would result in a D. rerio Tel protein with a much reduced capacity for repression of gene expression. This is indeed what we found (Fig. 5D). The differences between D. rerio Tel and other vertebrate Tel proteins provide a means to explore how sumoylation of K11 has refined Tel function during the course of evolution. Strikingly, an internal initiation codon at position 43 (M43), which leads to the production of a particularly repressive version of Tel because it cannot be sumoylated at the N terminus since it lacks K11, appears to emerge along with K11 (and is absent in D. rerio). We suggest that these dual, early vertebrate innovations of tel and Tel have collectively refined Tel function: the capacity to yield, posttranscriptionally a nonsumoylated, strongly repressive version of Tel (TelM43) coupled to posttranslational regulation by sumoylation of K11, which acts to limit Tel repression. This mechanism may not be limited to Tel. The related Ets transcription factors Fli-1 and Erg are also sumoylated, at positions K68 and K67, respectively (36; also M. G. Roukens, A. Anvarian, and D. A. Baker, unpublished data). These proteins also express an upstream sequence (Fli-1, MDEKN; Erg, MEEKH) that resembles the sequence in which M43 of Tel is embedded (MEEDS). It will be of interest to determine if Fli-1 and Erg are regulated similarly to Tel.
We found that both TelM43 and full-length Tel lacking K11 (TelK11-R) adopted a more speckled (repressive) subnuclear distribution by comparison with wild-type full-length Tel, suggesting that sumoylation of Tel influences its cellular localization. Also consistent with our reporter data, we found that sumoylation of Tel impeded its binding to DNA, suggesting that sumoylation of K11 inhibits Tel repressive function by regulating DNA occupancy by Tel. Previously, it has been reported that Tel sumoylation regulates its subcellular localization (42). Furthermore, the oncogene v-SRC stimulated kinase-dependent mislocalization from the nucleus to the cytoplasm of full-length Tel but not TelM43, suggesting that the N-terminal 43 amino acids of Tel encode a “signal” required for SRC-dependent nuclear export (18). Together, these findings suggest that sumoylation of K11 generates a repository of Tel molecules that are not tethered to DNA and either are available to be recycled, perhaps through desumoylation, to reinforce gene expression repression or are degraded in a regulated, context-dependent fashion. Tel sumoylation provides a framework for deciphering the precise mechanisms that determine the cellular status of Tel, which should prove to be of fundamental importance to understanding differentiation of early progenitor cells that express Tel.
A potential role for PIAS3 as a sumoylation-independent corepressor of Tel.
Interestingly, and perhaps paradoxically, under conditions that do not favor Tel sumoylation we found that PIAS3 synergistically enhanced Tel's repressive function, suggesting that PIAS3 might be a context-dependent modulator of Tel function: on the one hand, it strongly promotes Tel sumoylation, thus inhibiting Tel's repressive function, but on the other hand, it can act as a sumoylation-independent Tel corepressor. It will be important to ascertain whether PIAS3 has such a corepressor function in vivo and whether PIAS has a more ancient role as a corepressor of the invertebrate Tel orthologue, Yan, and D. rerio Tel that is independent of its sumoylating function. Drosophila Yan does not express an equivalent K11 residue (or M43 residue) and is not detectably sumoylated in insect Schneider cells (Roukens, Anvarian, and Baker, unpublished). It is worth noting that lowered levels of PIAS in Drosophila lead to a failure of normal photoreceptor and blood cell differentiation (9), which tellingly resembles Yan loss-of-function mutations in Drosophila (abnormal eye development phenotype) and Tel loss-of-function mutations in mice (abnormal blood cell differentiation phenotype).
Finally, the N terminus of Tel, which encodes K11, is indispensable for activating fusion proteins that result from chromosomal translocations found in various leukemias (6, 12). It will be instructive to determine a potential role for K11 sumoylation in the activity of these fusion proteins.
Acknowledgments
We are indebted to members of the Department of Molecular and Cellular Biology for invaluable technical assistance, especially Joost Schimmel, Martijn van Hagen, and Yolanda Ramos. We also thank Peter ten Dijke and A. G. Jochemsen for invaluable advice during the course of the work. We are most grateful to Pascal Meier for critically reading the manuscript and to Simon Baker for editorial advice and assistance. We thank those who very kindly provided us with materials: R. T. Hay (Dundee, United Kingdom) for SUMO cDNAs and plasmids for in vitro sumoylation, J. J. Palvimo (Kuopio, Finland) for PIAS constructs, G. Grosveld (Memphis, TN) for TEL cDNA, H. Saitoh (Kumamoto, Japan) for the bacterial sumoylation system, and Olivier Bernard and Ruud Delwel for Tel antibodies.
Footnotes
Published ahead of print on 22 January 2008.
REFERENCES
- 1.Baker, D. A, B. Mille-Baker, S. M. Wainwright, D. Ish-Horowicz, and N. J. Dibb. 2001. Mae mediates MAP kinase phosphorylation of Ets transcription factors in Drosophila. Nature 411330-334. [DOI] [PubMed] [Google Scholar]
- 2.Barjesteh van Waalwijk van Doorn-Khosrovani, S. B., D. Spensberger, Y. de Knegt, M. Tang, B. Löwenberg, and R. Delwel. 2005. Somatic heterozygous mutations in ETV6 (TEL) and frequent absence of ETV6 protein in acute myeloid leukemia. Oncogene 244129-4137. [DOI] [PubMed] [Google Scholar]
- 3.Betz, A, N Lampen, S. Martinek, M. W. Young, and J. E. Darnell, Jr. 2001. A Drosophila PIAS homologue negatively regulates stat92E. Proc. Natl. Acad. Sci. USA 989563-9568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chakrabarti, S. R., R. Sood, S. Nandi, and G. Nucifora. 2000. Posttranslational modification of TEL and TELyAML1 by SUMO-1 and cell-cycle-dependent assembly into nuclear bodies. Proc. Acad. Natl. Sci. USA 9713281-13285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fenrick, R., L. Wang, J. Nip, J. M. Amann, R. J. Rooney, J. Walker-Daniels, H. C. Crawford, D. L. Hulboy, M. S. Kinch, L. M. Matrisian, and S. W. Hiebert. 2000. TEL, a putative tumor suppressor, modulates cell growth and cell morphology of Ras-transformed cells while repressing the transcription of stromelysin-1. Mol. Cell. Biol. 205828-5839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Golub, T. R., G. F. Barker, M. Lovett, and G. D. Gilliland. 1994. Fusion of PDGF receptor beta to a novel ets-like gene, tel, in chronic myelomonocytic leukemia with t(5;12) chromosomal translocation. Cell 77307-316. [DOI] [PubMed] [Google Scholar]
- 7.Golub, T. R. 1997. TEL gene rearrangements in myeloid malignancy. Hematol. Onclo. Clin. N. Am. 111207-1220. [DOI] [PubMed] [Google Scholar]
- 8.Golub, T. R., G. F. Barker, K. Stegmaier, and G. D. Gilliland. 1997. The TEL gene contributes to the pathogenesis of myeloid and lymphoid leukemias by diverse molecular genetic mechanisms. Curr. Top. Microbiol. Immunol. 22067-79. [DOI] [PubMed] [Google Scholar]
- 9.Hari, K. L., K. R. Cook, and G. H. Karpen. 2001. The Drosophila Su(var)2-10 locus regulates chromosome structure and function and encodes a member of the PIAS protein family. Genes Dev. 151334-1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hay, R. T. 2005. SUMO: a history of modification. Mol. Cell 181-12. [DOI] [PubMed] [Google Scholar]
- 11.Hock, H., E. Meade, S. Medeiros, J. W. Schindler, P. J. M. Valk, Y. Fujiwara, and S. H. Orkin. 2004. Tel/Etv6 is an essential and selective regulator of adult hematopoietic stem cell survival. Genes Dev. 182336-2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jousset, C., C. Carron, A. Boureux, C. T. Quang, C. Oury, I. Dusanter-Fourt, M. Charon, L. Levin, O. Bernard, and J. Ghysdael. 1997. A domain of TEL conserved in a subset of ETS proteins defines a specific oligomerization interface essential to the mitogenic properties of the TEL-PDGFR beta oncoprotein. EMBO J. 1669-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim, C. A., M. L. Phillips, W. Kim, M. Gingery, H. H. Tran, M. A. Robinson, S. Faham, and J. U. Bowie. 2001. Polymerization of the SAM domain of TEL in leukemogenesis and transcriptional repression. EMBO J. 204173-4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lai, Z. C., and G. M Rubin. 1992. Negative control of photoreceptor development in Drosophila by the product of the Yan gene, an ETS domain protein. Cell 70609-620. [DOI] [PubMed] [Google Scholar]
- 15.Lin, D.-Y., Y.-S. Huang, J.-C. Jeng, H.-Y. Kuo, C.-C. Chang, T.-T. Chao, C.-C. Ho, Y.-C. Chen, T.-P. Lin, H.-I. Fang, C.-C. Hung, C.-S. Suen, M.-J. Hwang, K.-S. Chang, G. G. Maul, and H.-M. Shih. 2006. Role of SUMO-interacting motif in Daxx SUMO modification, subnuclear localization, and repression of sumoylated transcription factors. Mol. Cell 24341-354. [DOI] [PubMed] [Google Scholar]
- 16.Liu, B., S. Mink, K. A. Wong, N. Stein, C. Getman, P. W. Dempsey, H. Wu, and K. Shuai. 2004. PIAS1 selectively inhibits interferon-inducible genes and is important in innate immunity. Nat. Immunol. 5891-898. [DOI] [PubMed] [Google Scholar]
- 17.Lopez, R. G., C. Carron, C. Oury, P. Gardellin, O. Bernard, and J. Ghysdael. 1999. TEL is a sequence-specific transcriptional repressor. J. Biol. Chem. 27430132-30138. [DOI] [PubMed] [Google Scholar]
- 18.Lopez, R. G., C. Carron, and J. Ghysdael. 2003. v-SRC specifically regulates the nucleo-cytoplasmic delocalization of the major isoform of TEL (ETV6). J. Biol. Chem. 27841316-41325. [DOI] [PubMed] [Google Scholar]
- 19.O'Neill, E. M., I. Rebay, R. Tjian, and G. M. Rubin. 1994. The activities of two Ets-related transcription factors required for Drosophila eye development are modulated by the Ras/MAPK pathway. Cell 78137-147. [DOI] [PubMed] [Google Scholar]
- 20.Poirel, H., C. Oury, C. Carron, E. Duprez, Y. Laabi, A. Tsapis, S. P. Romana, M. Mauchauffe, M. Le Coniat, R. Berger, J. Ghysdael, and O. A. Bernard. 1997. The TEL gene products: nuclear phosphoproteins with DNA binding properties. Oncogene 14349-357. [DOI] [PubMed] [Google Scholar]
- 21.Qiao, F., H. Song, C. A. Kim, M. R. Sawaya, J. B. Hunter, M. Gingery, I. Rebay, A. J. Courey, and J. U. Bowie. 2004. Derepression by depolymerization: structural insights into the regulation of Yan by Mae. Cell 118163-173. [DOI] [PubMed] [Google Scholar]
- 22.Qiao, F., and J. U. Bowie. 31 May 2005. The many faces of SAM. Sci. STKE 2005re7. doi: 10.1126/stke.2862005re7. [DOI] [PubMed] [Google Scholar]
- 23.Rebay, I., and G. M. Rubin. 1995. Yan functions as a general inhibitor of differentiation and is negatively regulated by activation of the Ras1/MAPK pathway. Cell 80857-866. [DOI] [PubMed] [Google Scholar]
- 24.Rodriguez, M. S., C. Dargemont, and R. T. Hay. 2001. SUMO-1 conjugation in vivo requires both a consensus modification motif and nuclear targeting. J. Biol. Chem. 27612654-12659. [DOI] [PubMed] [Google Scholar]
- 25.Rogge, R., P. J. Green, J. Urano, S. Horn-Saban, M. Mlodzik, B.-Z. Shilo, V. Hartenstein, and U. Banerjee. 1995. The role of Yan in mediating the choice between cell division and differentiation. Development 1213947-3958. [DOI] [PubMed] [Google Scholar]
- 26.Santti, H., L. Mikkonen, A. Anand, S. Hirvonen-Santti, J. Toppari, M. Panhuysen, F. Vauti, M. Perera, G. Corte, W. Wurst, O. A. Jänne, and J. J. Palvimo. 2005. Disruption of the murine PIASx gene results in reduced testis weight. J. Mol. Endocrinol. 34645-654. [DOI] [PubMed] [Google Scholar]
- 27.Sasaki, K., Y. Nakamura, K. Maki, K. Waga, F. Nakamura, H. Arai, Y. Imai, H. Hirai, and K. Mitani. 2004. Functional analysis of a dominant-negative DETS TEL/ETV6 isoform. Biochem. Biophys. Res. Comm. 3171128-1137. [DOI] [PubMed] [Google Scholar]
- 28.Schick, N., E. J. Oakeley, N. E. Hynes, and A. Badachi. 2004. TEL/ETV6 is a signal transducer and activator of transcription 3 (Stat3)-induced repressor of Stat3 activity. J. Biol. Chem. 27938787-38796. [DOI] [PubMed] [Google Scholar]
- 29.Schmidt, D., and S. Müller. 2003. PIAS/SUMO: new partners in transcriptional regulation. Cell Mol. Life Sci. 602561-2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sharrocks, A. D. 2006. PIAS proteins and transcriptional regulation—more than just SUMO E3 ligases? Genes Dev. 20754-758. [DOI] [PubMed] [Google Scholar]
- 31.Song, H., M. Nie, F. Qiao, J. U Bowie, and A. J. Courey. 2005. Antagonistic regulation of Yan nuclear export by Mae and Crm1 may increase the stringency of the Ras response. Genes Dev. 191767-1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Steen, H., A. Pandey, J. S. Andersen, and M. Mann. 2002. Analysis of tyrosine phosphorylation sites in signaling molecules by a phosphotyrosine-specific immonium ion scanning method. Sci. STKE 2002PL16. doi: 10.1126/stke.2002.154.pl16. [DOI] [PubMed] [Google Scholar]
- 33.Tootle, T. L., P. S. Lee, and I. Rebay. 2003. CRM1-mediated nuclear export and regulated activity of the receptor tyrosine kinase antagonist YAN require specific interactions with MAE. Development 130845-857. [DOI] [PubMed] [Google Scholar]
- 34.Tootle, T. L., and I. Rebay. 2005. Post-translational modifications influence transcription factor activity: a view from the ETS superfamily. Bioessays 27285-298. [DOI] [PubMed] [Google Scholar]
- 35.Uchimura, Y., M. Nakamura, K. Sugasawa, M. Nakao, and H. Saitoh. 2004. Overproduction of eukaryotic SUMO-1 and SUMO-2-conjugated proteins in Escherichia coli. Anal. Biochem. 331204-206. [DOI] [PubMed] [Google Scholar]
- 36.van den Akker, E., S. Ano, H.-M. Shih, L.-C. Wang, M. Pironin, J. J. Palvimo, N. Kotaja, O. Kirsh, A. Dejean, and J. Ghysdael. 2005. FLI-1 functionally interacts with PIASxα, a member of the PIAS E3 SUMO ligase family. J. Biol. Chem. 28038035-38046. [DOI] [PubMed] [Google Scholar]
- 37.Vertegaal, A. C. O., S. C. Ogg, E. Jaffray, M. S. Rodriguez, R. T. Hay, J. S. Andersen, M. Mann, and A. Lamond. 2004. A proteomic study of SUMO-2 target proteins. J. Biol. Chem. 27933791-33798. [DOI] [PubMed] [Google Scholar]
- 38.Vivekanand, P., and I. Rebay. 2006. Intersection of signal transduction pathways and development. Annu. Rev. Genet. 40139-157. [DOI] [PubMed] [Google Scholar]
- 39.Wang, L. C., F. Kuo, Y. Fujiwara, D. G. Gilliland, T. R. Golub, and S. H. Orkin. 1997. Yolk sac angiogenic defect and intra-embryonic apoptosis in mice lacking the Ets-related factor TEL. EMBO J. 164374-4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang, L. C., W. Swat, Y. Fujiwara, L. Davidson, J. Visvader, F. Kuo, F. W. Alt, D. G. Gilliland, T. R. Golub, and S. H. Orkin. 1998. The TEL/ETV6 gene is required specifically for hematopoiesis in the bone marrow. Genes Dev. 122392-2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wong, K. A., R. Kim, H. Christofk, J. Gao, G. Lawson, and H. Wu. 2004. Protein inhibitor of activated STAT Y (PIASy) and a splice variant lacking exon 6 enhance sumoylation but are not essential for embryogenesis and adult life. Mol. Cell. Biol. 245577-5586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wood, L. D., B. J. Irvin, G. Nucifora, K. S. Luce, and S. W. Hiebert. 2003. Small ubiquitin-like modifier conjugation regulates nuclear export of TEL, a putative tumor suppressor. Proc. Acad. Natl. Sci. USA 1003257-3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang, S.-H., and A. D. Sharrocks. 2004. SUMO promotes HDAC-mediated transcriptional repression. Mol. Cell 13611-617. [DOI] [PubMed] [Google Scholar]
- 44.Yang, S.-H., and A. D. Sharrocks. 2005. PIASx acts as an Elk-1 coactivator by facilitating derepression. EMBO J. 242161-2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang, S.-H., and A. D. Sharrocks. 2006. PIASxa differentially regulates the amplitudes of transcriptional responses following activation of the ERK and p38 MAPK pathways. Mol. Cell 22477-487. [DOI] [PubMed] [Google Scholar]