Abstract
Interferon regulatory factor 6 (IRF6) is a novel and unique member of the IRF family of transcription factors. IRF6 has not been linked to the regulatory pathways or functions associated with other IRF family members, and the regulation and function of IRF6 remain unknown. We recently identified a protein interaction between IRF6 and the tumor suppressor maspin. To gain insight into the biological significance of the maspin-IRF6 interaction, we examined the regulation and function of IRF6 in relation to maspin in normal mammary epithelial cells. Our results demonstrate that in quiescent cells, IRF6 exists primarily in a nonphosphorylated state. However, cellular proliferation leads to rapid IRF6 phosphorylation, resulting in proteasome-dependent IRF6 degradation. These data are supported in situ by the increased expression of IRF6 in quiescent, differentiated lobuloalveolar cells of the lactating mammary gland compared to its expression in proliferating ductal and glandular epithelial cells during pregnancy. Furthermore, the reexpression of IRF6 in breast cancer cells results in cell cycle arrest, and the presence of maspin augments this response. These data support a model in which IRF6, in collaboration with maspin, promotes mammary epithelial cell differentiation by facilitating entry into the G0 phase of the cell cycle.
Interferon regulatory factor 6 (IRF6) is a poorly understood member of the IRF family of transcription factors. Currently with nine members, the IRF family plays a critical role in the innate immune response and has been implicated in numerous other cellular processes, including tumor suppression, cell cycle regulation, and apoptosis (1, 3, 9, 11, 15, 21, 31). However, IRF6 has not been implicated in any of the pathways regulating the activation and function of other IRFs. Although a role for IRF6 during embryonic development has recently been identified, its function and regulation remain unknown (12, 14, 26).
We recently reported a protein interaction between IRF6 and maspin (mammary serine protease inhibitor, serpin B5), based on the yeast two-hybrid system, in human mammary epithelial cells (2). We demonstrated that, similar to maspin, IRF6 expression is reduced or absent in breast carcinomas, suggesting that IRF6 may also possess tumor-suppressive properties (2). However, a role for IRF6 in the pathogenesis of breast cancer has not yet been established.
Phosphorylation plays an important role in the regulation and activation of IRFs (18, 20, 24). We have previously demonstrated that IRF6 exists in phosphorylated and nonphosphorylated states within mammary epithelial cells. However, the dual existence of both phosphorylated and nonphosphorylated forms within the cytoplasm is contrary to the established paradigm of IRF regulation and function. We also demonstrated that the phosphorylation of IRF6 facilitates its interaction with maspin, although the significance of the maspin-IRF6 interaction remains unknown (2).
Interestingly, phosphorylation of some IRFs not only renders them active by allowing nuclear translocation but also serves as a signal for proteasomal degradation (22, 27, 33). The proteasome is a multisubunit organelle that participates in the degradation of many cellular proteins, and proteasomal activity is known to regulate multiple key cellular processes, including the cell cycle (4). Recently, pharmacological inhibition of the proteasome has proven to be a promising therapy for certain types of cancer (19, 29). Natural proteasome inhibitors found in fruits and vegetables have also been reported, and several, including apigenin, have tumor-suppressive properties (5, 6). Apigenin is also a kinase inhibitor, with known inhibitory activity against phophatidylinositol 3-kinase-Akt, CK2, and mitogen-activated protein kinase signaling pathways (10, 16, 32).
In this article, we report that the treatment of mammary epithelial cells with proteasome inhibitors, including apigenin, results in the phosphorylation of IRF6, while maspin expression remains unchanged. The evidence suggests that the phosphorylation of IRF6 leads to decreased protein stability and is a signal for proteasome-dependent degradation. We further demonstrate that cellular proliferation, driven by mitogenic stimuli, is a key inducer of IRF6 phosphorylation and that cell cycle arrest reduces IRF6 phosphorylation while simultaneously increasing IRF6 protein levels. Notably, this reflects the differential expression we observe in IRF6 during mammary gland differentiation. Lactation represents the functional differentiation of mammary epithelial cells and is characterized by the presence of quiescent, secretory lobuloalveolar cells. In comparison to IRF6 expression in proliferating mammary epithelial cells during pregnancy, our data indicate that IRF6 expression during lactation is more than twofold above that seen during pregnancy in a murine model. Maspin protein levels also appear to increase, which is consistent with previous findings (34). Furthermore, when reexpressed in breast cancer cells which have lost IRF6 expression, IRF6 induces cell cycle arrest, and the simultaneous reexpression of maspin synergistically enhances this effect. These data highlight novel functions of IRF6 and suggest an intimate, cooperative role for maspin and IRF6 as key mediators of cellular proliferation and differentiation in mammary epithelial cells.
MATERIALS AND METHODS
Cell culture.
MCF-10A cells were maintained in complete growth medium consisting of Ham F-12 and Dulbecco's modified Eagle's medium (1:1; Gibco) supplemented with 5% horse serum (Invitrogen), epidermal growth factor (EGF) (20 ng/ml; Peprotech), hydrocortisone (0.5 μg/ml; Sigma), insulin (10 μg/ml; Sigma), and cholera toxin (100 ng/ml; Sigma) with penicillin and streptomycin (8). Primary human mammary epithelial cells were purchased from Cell Applications, Inc., and maintained in defined mammary epithelial cell basal medium provided by the company. The MLC8891 immortalized human prostate epithelial cell line was a kind gift from the laboratory of Ruth Sager and Arthur Pardee of the Dana-Farber Cancer Institute and was maintained in a serum-free keratinocyte growth medium supplemented with bovine pituitary extract (Invitrogen). The human breast cancer cell lines MCF-7 and MDA-MB-231 were maintained in RPMI medium supplemented with 10% fetal calf serum and gentamicin. MDA-MB-231 cells stably transfected with green fluorescent protein-maspin have been described previously (23). The cell cultures were determined to be free of mycoplasma contamination by using a GenProbe rapid detection system.
Adenoviral transfection and cell proliferation assay.
Cells were seeded at a density of 500,000 cells per 25 cm2. IRF6 transfection was performed by using the pacAd5CMV adenovirus construct developed at the University of Iowa DNA vector core (supplied at 10e11 particles per milliliter) at various multiplicities of infection ranging from 250 to 500. For viral infection, cells were treated with virus in antibiotic-free RPMI medium containing 1% serum for 2 h, after which RPMI complete medium was added to the cells. The transfection efficiency was verified by real-time PCR and Western blot analysis. All experiments were performed at 6 days postinfection. Proliferation was determined by cell counting using standard techniques. The experiments were performed and analyzed in triplicate.
Real-time PCR.
Real-time PCR was performed using standard protocols (13). Total RNA was reverse transcribed using an Advantage PCR kit according to the manufacturer's instructions (Clontech). Real-time PCR was performed on a 7500 real-time PCR system (Applied Biosystems, Foster City, CA) using TaqMan gene expression human primer/probe sets for the following genes: IRF6 (Hs00196213_m1), Ki-67 (Hs00606991_m1), and maspin (Hs00184728_m1) (Applied Biosystems). Briefly, 5 μl cDNA, 1.25 μl 20× gene expression assay mix, and 12.5 μl 2× TaqMan universal PCR master mix in a total volume of 25 μl were amplified with the following thermocycler protocol: 1 cycle at 50°C for 2 min, 1 cycle at 95°C for 10 min, and 40 cycles at 95°C for 15 seconds and at 60°C for 1 min. All data were analyzed with Sequence Detection software (version 1.2.3; Applied Biosystems). The expression of each target gene was normalized to the expression of an endogenous control gene (glyceraldehyde-3-phosphate dehydrogenase [GAPDH], 4333764F; or RPLPO, 4333761).
Cellular fractionation and Western blot analysis.
Whole-cell lysate was acquired by using standard lysis techniques. Briefly, cells were washed in cold phosphate-buffered saline (PBS) and lysed in buffer A (10 mM HEPES [pH 7.9], 1.5 mM MgCl2, 10 mM KCl, 0.5 mM dithiothreitol, 0.1% NP-40 with 2 mM Na-ortho-vanadate, 2 mM NaF, plus protease inhibitor). The cells were scraped, and the slurry was briefly sonicated. The cells were then centrifuged at 12,000 × g for 20 min at 4°C to obtain the whole-cell lysate. Equal amounts of cellular protein (25 μg) were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (10% resolving gel) and Western blot analysis using specific antibody to maspin (1:5,000) or IRF6 (1:2,000) as described previously (2). Other antibodies and dilutions are as follows: beta-actin (1:10,000; Chemicon International), cyclin D3 (DCS22, 1:2,000; Cell Signaling Technology), and proliferating cell nuclear antigen (PCNA) (PC10, 1:1,000; Santa Cruz Biotechnology). All densitometry analysis was performed using Scion Image software (Scion Corporation).
Immunofluorescence and microscopy.
MCF-10A cells were seeded onto glass coverslips for growth and treatment. The cells were fixed and stained as described previously (2). Briefly, the cells were washed 3× in PBS and fixed in 100% ice-cold methanol for 5 min. Following fixation, the cells were again washed 3× in PBS and blocked with 10% goat serum in PBS for 1 h. Primary antibody was then added and incubated for 1 h. Following incubation, the cells were washed 3× in PBS and incubated with the secondary antibody for 1 h. The cells were then washed 3× in PBS and mounted onto glass slides with gelvatol. The primary antibody for IRF6 was used at 1:100 and for E-cadherin at 1:500. Alexa Fluor 663-conjugated goat anti-rabbit secondary antibody and Alexa Fluor 488-conjugated donkey anti-mouse secondary antibody (Molecular Probes) were used at 1:500. All immunofluorescence images were acquired by using a Zeiss 510 META laser scanning confocal microscope.
Pulse-chase and coimmunoprecipitataion assays.
For the ubiquitin coimmunoprecipitation assay, 500 μg total protein from whole-cell extracts from MCF-10A cells was incubated with 2 μg antiubiquitin antibody (monoclonal antibody clone 403B; Santa Cruz Biotechnology, Santa Cruz, CA) overnight at 4°C with gentle rotation. Antibody complexes were eluted with anti-mouse antibody immunoprecipitation beads (eBioscience, San Diego, CA) for 1 h at room temperature according to the manufacturer's instructions. Complexed beads were washed three times in PBS, and the complexes were eluted by boiling beads in 2× sample buffer with SDS for 10 min. The elution was evaluated by SDS-PAGE using a 10% gel and subsequent Western blot analysis for IRF6.
The pulse-chase assay was performed using MCF-10A cells grown to either 75% or 100% confluence. The cells were then maintained in methionine-free RPMI 1640 (Gibco) plus 5% dialyzed fetal calf serum, EGF (20 ng/ml), hydrocortisone (0.5 μg/ml), insulin (10 μg/ml), and cholera toxin (100 ng/ml) for 24 h. The cells were then exposed to 75 mCi Easytag l-[35S]methionine (Perkin Elmer) overnight, followed by a chase with nonlabeled methionine in excess (2 mM). Cell lysis and immunoprecipitation were performed as described above. Five microliters IRF6 MaxPab polyclonal antibody (Abnova) was used for the immunoprecipitation assay.
Animal husbandry and immunohistochemistry.
C57/Black6 female mice from Harlan (Indianapolis, IN) were kept according to International Animal Care and Use Committee (IACUC) standards and practices. Mice were euthanized at mid-pregnancy and 7 days postparturition following anesthetization with 10% ketamine-xylazine in PBS (ketamine, 100 mg/kg of body weight, and xylazine, 10 mg/kg). Pectoral and inguinal groups of mammary glands were removed, fixed in 10% neutral buffered formalin for 19 h, and embedded in paraffin. Tissue samples were sectioned at 4 μm, deparaffinized, and subjected to a water bath antigen-recovery protocol using citrate buffer (pH 6.0). Maspin and IRF6 expression were determined on a Dako Autostainer using a Vectastain universal elite ABC peroxidase kit (Vector Laboratories, Inc.). Antibody to maspin was used at a 1:200 dilution. Antibody to IRF6 was used at a 1:1,200 dilution, as previously described (2). Diaminobenzidine (DAB) was used to visualize antigen immunoreactivity. Slides were counterstained with Mayer's hematoxylin for 5 min, followed by dehydration in ethanol and xylene, and cover-slipped with permanent mounting medium. Slides, including the negative controls, represent serial sections. The relative staining intensity of DAB was quantified using a cyan-magenta-yellow-black (CMYK) color model adapted from a model by Pham et al. (25). Briefly, the chromogen intensity, based on a 0 to 255 scale, was determined from the yellow channel of a CMYK color image. Using Adobe Photoshop CS software, stained cells were selected by inversely selecting highlighted (white) areas. The mean intensity of the selected area was then measured for the yellow channel of the CMYK image. The inverse of the intensity was normalized and graphed using Microsoft Excel software. Images were captured with a Leica DM 4000B microscope mated to a Leica DFC480 5.0 megapixel charge-coupled-device camera.
RESULTS
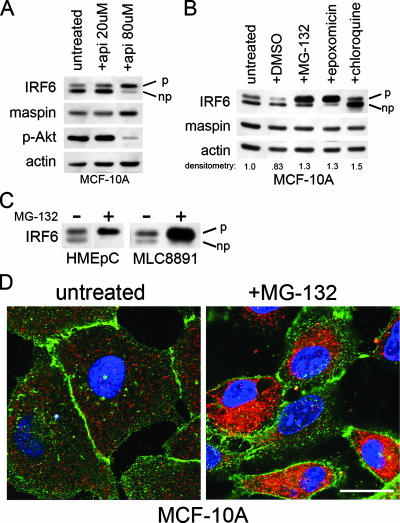
Because phosphorylation is an important step in the activation and function of IRFs, we employed the natural kinase inhibitor apigenin to evaluate its effects on IRF6 phosphorylation. Surprisingly, treatment of the mammary epithelial cell line MCF-10A with apigenin resulted in an increase in IRF6 phosphorylation (denoted by the upper band of a doublet in Western blotting) (2), suggesting an alternate mechanism apart from apigenin's kinase-inhibitory properties (Fig. 1A). Importantly, maspin expression appeared unchanged. As an internal control, a decrease in phospho-specific Akt levels verified apigenin activity.
FIG. 1.
Proteasomal inhibition induces IRF6 phosphorylation. (A) Western blot analysis of MCF-10A cells treated with two different concentrations of apigenin (api) for 18 h. The IRF6 doublet represents phosphorylated (upper band, denoted by “p”) and nonphosphorylated (lower band, denoted by “np”) forms. Phospho-Akt (p-Akt) levels verify apigenin function and act as an internal control. (B) Western blot analysis of MCF-10A cells treated with proteasome inhibitors MG-132 (0.5 μM) and epoxomicin (0.5 μM), the lysosomotropic agent chloroquine (50 μM), and the vehicle dimethyl sulfoxide (DMSO). All treatments were performed for 18 h. Densitometry represents the ratio of IRF6 to actin. (C) Western blot analysis demonstrating similar effects of proteasomal inhibition on IRF6 expression and phosphorylation in primary human mammary epithelial cells (HMEpC) and the nontransformed human prostate cell line MLC8891. +, present; −, absent. (D) Immunofluorescence analysis of MCF-10A cells grown on glass coverslips and treated with MG-132 (0.5 μM, 18 h). IRF6 is shown in red. E-cadherin (green) was used to highlight cellular architecture. 4′,6′-Diamidino-2-phenylindole was used to counterstain the nuclei (blue). Images were acquired on a Zeiss 510 META confocal microscope with a total optical magnification of ×630 with an additional 2.5× scan zoom. Bar represents 10 μm.
Based on apigenin's reported proteasome inhibitory properties, we tested whether the increase in IRF6 phosphorylation could be replicated following proteasomal inhibition. MCF-10A cells were treated with two well-characterized proteasome inhibitors (MG-132 and epoxomicin) for 18 h, and IRF6 phosphorylation was assayed by Western blotting (Fig. 1B). Similar to the effects of apigenin, proteasomal inhibition resulted in the phosphorylation of nearly the entire cellular pool of IRF6, whereas maspin again appeared to be unaffected by the treatment. Densitometric analysis further revealed a moderate increase in total IRF6 protein levels following proteasomal inhibition. Notably, an increase in IRF6 phosphorylation was not observed following treatment with chloroquine, a lysosomotropic agent which disrupts normal lysosomal function, despite a similar increase in IRF6 protein levels. Proteasomal inhibition induced a similar effect on IRF6 in primary human mammary epithelial cells (HMEpC) and the normal prostate cell line MLC8891, demonstrating that these effects are not cell type or tissue specific (Fig. 1C). The increase in IRF6 protein levels induced by MG-132 was confirmed by immunofluorescence microscopy, which also demonstrates the cytoplasmic localization of phosphorylated IRF6 following proteasomal inhibition (Fig. 1D).
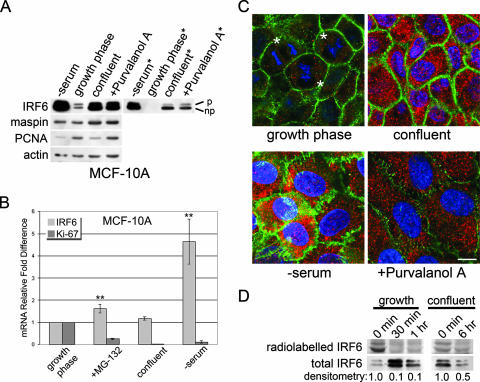
The proteasome plays a critical role in cell cycle regulation, and therefore, we tested whether cellular proliferation altered the phosphorylation status of IRF6. Following the induction of cell cycle arrest in MCF-10A cells by serum deprivation, pharmacological cell cycle inhibition, or cell contact-dependent cell cycle arrest, the status of IRF6 phosphorylation was assessed by Western blotting (Fig. 2A). Interestingly, quiescent cells predominantly expressed nonphosphorylated IRF6. Furthermore, total IRF6 protein levels were notably increased in arrested cells compared to their levels in asynchronous cells in growth phase (50% confluent), indicating that cell proliferation directly regulates IRF6 protein expression and phosphorylation and suggesting that the phosphorylation of IRF6 may lead to decreased protein stability and/or targeted protein degradation. We employed real-time PCR to ascertain whether increased IRF6 mRNA expression might account for a portion of the total increase in IRF6 protein levels (Fig. 2B). Importantly, only the serum-starved cells demonstrated a substantial increase in IRF6 mRNA, suggesting that the increase in IRF6 protein in confluent or MG-132-treated cells results from increased protein stabilization and/or a decrease in targeted degradation. The increase in IRF6 protein levels was verified by immunofluorescence microscopy (Fig. 2C).
FIG. 2.
Cell cycle arrest causes an increase in IRF6 protein levels and a reduction in IRF6 phosphorylation. (A) Western blot analysis of MCF-10A cells comparing proliferating cells (growth phase; 50% confluent) to quiescent cells arrested by serum starvation (−serum; 48 h), cell-cell contact inhibition (confluent; 96 h), or pharmacological cell cycle inhibition following treatment with the cyclin-dependent kinase inhibitor purvalanol A (+Purvalonol A; 10 μM, 18 h). Expression of PCNA was used as an indicator of quiescence. *, reduced exposure time demonstrating the prominence of the nonphosphorylated form of IRF6. p, phosphorylated; np, nonphosphorylated. (B) Real-time PCR results comparing IRF6 mRNA levels of proliferating versus quiescent cells. mRNA expression of the proliferation marker Ki-67 was used to verify cell cycle arrest. Treatment of cells was identical to that described for panel A. **, P ≤ 0.01 based on Student's t test comparing IRF6 mRNA expression to that in proliferating cells (growth phase). Error bars show standard deviations. (C) Immunofluorescence microscopy comparing proliferating MCF-10A cells (growth phase) with nonproliferating cells arrested by serum starvation (−serum; 48 h), contact inhibition (confluent; 96 h) or pharmacological inhibition (+Purvalanol A; 18 h). IRF6 is shown in red. E-cadherin (green) was used to highlight cellular architecture. 4′,6′-Diamidino-2-phenylindole was used to counterstain the nuclei (blue). *, actively dividing cells. Bar represents 10 μm. (D) Pulse-chase analysis of IRF6 protein stability. MCF-10A cells were pulsed with l-[35S]methionine followed by a cold methionine chase for the indicated time points. Radiolabeled IRF6 was immunoprecipitated and evaluated by SDS-PAGE. The presence of IRF6 protein (total IRF6) was confirmed by subsequent Western blot analysis. Densitometry represents radiolabeled IRF6 at 30 min, 1 h, or 6 h compared to that at 0 min.
We next evaluated the relative stability of the IRF6 protein in proliferating versus quiescent MCF-10A cells using a pulse-chase assay. Our results demonstrate a substantial reduction of radiolabeled IRF6 in proliferating cells following a 30-min chase, whereas radiolabeled IRF6 remains detectable following a 6-h chase in confluent cells (Fig. 2D). Notably, the upper (phosphorylated) IRF6 band appears to diminish more quickly than the lower band, suggesting an increased stability of nonphosphorylated IRF6.
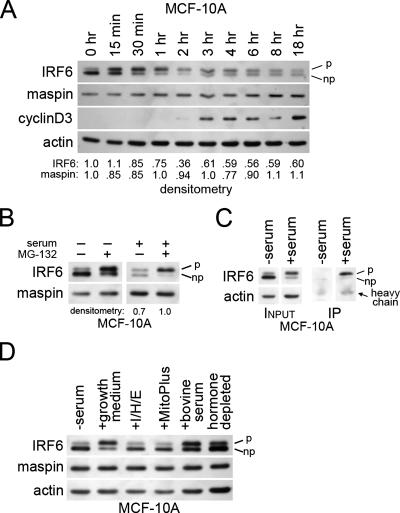
Since IRF6 expression and phosphorylation appear to be regulated by cell proliferation, we next investigated at what stage of the cell cycle IRF6 becomes phosphorylated. MCF-10A cells were synchronized by serum deprivation, and IRF6 phosphorylation was analyzed at multiple time points following the readdition of serum. The results shown in Fig. 3A demonstrate that IRF6 phosphorylation is an early event in cell cycle progression, with maximal IRF6 phosphorylation occurring 30 min to 1 h following the addition of serum. Importantly, by 2 h, total IRF6 protein levels appeared to steadily decline, as determined by densitometric analysis. This was achieved primarily through a decrease in phosphorylated IRF6 levels, whereas the level of nonphosphorylated IRF6 appeared to change very little following the initial decrease coincident with serum addition. Importantly, the decrease in IRF6 expression occurred concomitantly with the appearance of cyclin D3, an important marker of the G1 phase of the cell cycle. Maspin levels were not significantly altered over the 18 h following the serum addition. These data support the possibility that IRF6 phosphorylation may facilitate its degradation, which in turn may allow progression from G0 (quiescence) to the G1 phase of the cell cycle.
FIG. 3.
IRF6 protein expression and phosphorylation are regulated by the cell cycle in a proteasome-dependent manner. (A) Western blot analysis depicting changes in IRF6 phosphorylation and expression upon addition of serum to synchronized MCF-10A cells. Time 0 (0 h) represents 48 h of serum starvation, with each subsequent time point indicating time elapsed following serum addition. Densitometry represents the ratio of IRF6 or maspin to actin. (B) Western blot analysis demonstrating the effects of MG-132 on IRF6 expression and phosphorylation in the presence and absence of serum. (C) Coimmunoprecipitation of whole-cell lysate from MCF-10A cells either serum-starved for 48 h (−serum) or stimulated for 1 h with growth medium (+serum). Complexes were immunoprecipitated (IP) with antiubiquitin antibody, and the resulting elution was analyzed by SDS-PAGE and Western blot probed for IRF6. (D) Western blot analysis of IRF6 expression and phosphorylation following various mitogenic stimuli. Growth medium consisted of Dulbecco's modified Eagle's medium-F12 (1:1) supplemented with 5% horse serum, EGF (20 ng/ml), hydrocortisone (0.5 μg/ml), and insulin (10 μg/ml). “I/H/E” represents the addition of insulin, hydrocortisone, and EGF at the same concentrations used in the growth medium but in the absence of serum. The serum supplement MitoPlus was used at 1:1,000. Bovine serum and charcoal-dextran-stripped serum (hormone depleted) were used at 10%. p, phosphorylated; np, nonphosphorylated; +, present; −, absent.
To test whether IRF6 phosphorylation results in protein degradation via a proteasome-dependent pathway, serum was added to synchronized MCF-10A cells in the presence or absence of the proteasome inhibitor MG-132. Consistent with previous results, IRF6 protein levels noticeably decreased in the absence of MG-132 after 6 h following serum addition. However, in the presence of MG-132, total IRF6 protein levels were unchanged over 6 h, even though IRF6 was phosphorylated (Fig. 3B). Interestingly, in the absence of serum, MG-132 retained its ability to induce IRF6 phosphorylation. The exact mechanism for the phosphorylation of IRF6 following proteasomal inhibition in quiescent cells is not known.
Polyubiquitination of the target protein is the most common signal for proteasome-mediated degradation. To test whether IRF6 is ubiquitinated following phosphorylation, we performed coimmunoprecipitation analysis with an antiubiquitin antibody, in which we compared serum-starved cells (nonphosphorylated IRF6) with serum-stimulated cells (phosphorylated IRF6). Figure 3C demonstrates that ubiquitin conjugation specifically occurs on phosphorylated IRF6 in serum-stimulated cells, which supports the hypothesis that the phosphorylation of IRF6 is a signal for proteasome-mediated degradation.
We next sought to identify the serum component responsible for IRF6 phosphorylation in MCF-10A cells. Because IRF6 phosphorylation occurs quickly after serum stimulation, we tested whether the mitogenic signaling of EGF, insulin, or hydrocortisone affected IRF6 phosphorylation. When added in concentrations similar to those present in complete growth medium, these growth factors were not sufficient to induce rapid IRF6 phosphorylation (Fig. 3D). Furthermore, neither the serum supplement MitoPlus, which contains an assortment of growth factors and hormones, nor charcoal-dextran-stripped serum were able to efficiently induce IRF6 phosphorylation. Expectedly, maspin levels remained constant. We are currently working to identify the unknown serum component(s) responsible for the induction of IRF6 phosphorylation.
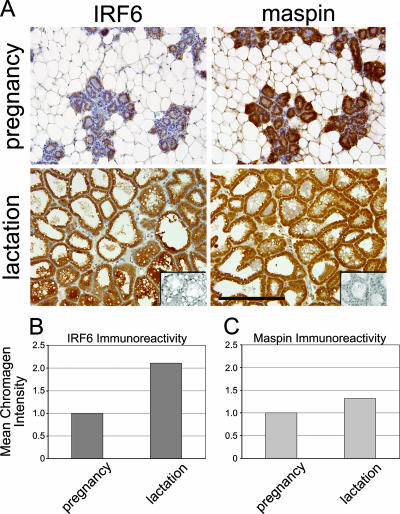
Mammary gland development and differentiation are characterized by heightened periods of proliferation (during pregnancy) prior to the functional differentiation of the secretory lobuloalveolar cells which comprise the lactating gland. We therefore sought to confirm our findings in situ using immunohistochemistry to compare IRF6 expression levels during pregnancy and lactation in mice. The results shown in Fig. 4A demonstrate a noticeable increase in IRF6 immunoreactivity in the lactating lobuloalveolar cells compared to the IRF6 immunoreactivity in ductal and glandular epithelial cells during pregnancy. A mild increase was also observed in maspin expression, an observation which coincides with the findings in a previous report showing maximal expression of maspin mRNA during lactation in the differentiating gland (34). Quantification of the relative mean intensity of the staining indicated a greater-than-twofold increase in IRF6 immunoreactivity, which is very similar to the increase observed as MCF-10A cells become confluent in culture (Fig. 4B and Fig. 2A and C, respectively).
FIG. 4.
IRF6 expression is maximal during lobuloalveolar differentiation (lactation). (A) Immunohistochemistry demonstrating increased IRF6 immunoreactivity during lactation compared to immunoreactivity during pregnancy. Wild-type C57/Black6 mice were harvested at mid-pregnancy and 7 days postparturition. Expression is visualized by the staining intensity of DAB. Slides are counterstained with Mayer's hematoxylin. The negative control inset represents staining with secondary antibody only. Slides, including the negative control, represent serial sections. Bar represents 200 μm. Total magnification is ×200. Pictures were acquired with a Leica DM 4000B microscope mated to a Leica DFC480 charge-coupled-device camera. (B and C) The relative mean intensity of DAB indicating IRF6 or maspin was quantified by using a CMYK color model adapted from a model by Pham et al. (25), based on the chromogen intensity determined from the yellow channel of a CMYK color image.
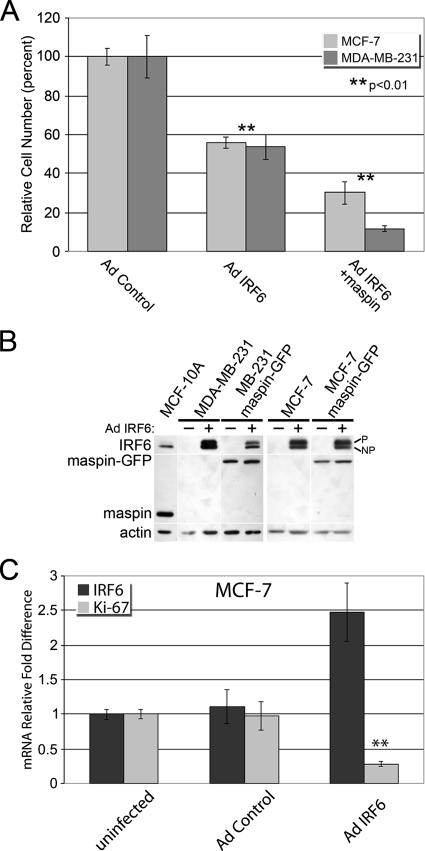
These findings led us to postulate that IRF6 is an important component of cell cycle regulation. Hence, we evaluated the effects of IRF6 reexpression on cell proliferation in breast cancer cells which no longer express readily detectable amounts of IRF6. Proliferation assays comparing IRF6-transfected breast cancer cells with control cells revealed a significant reduction in total cell number(s) following IRF6 transfection (Fig. 5A). In both poorly aggressive MCF-7 and highly aggressive MDA-MB-231 breast cancer cells, the reexpression of IRF6 reduced cellular proliferation by more than 40%. Furthermore, the reexpression of maspin in combination with IRF6 synergistically augmented the growth-inhibitory effects of IRF6, with the most-pronounced effect occurring in the highly aggressive cells, where the total cell number was reduced by nearly 90%. Importantly, MCF-7 and MDA-MB-231 cells stably transfected with maspin alone do not exhibit significantly altered doubling times. Real-time PCR analysis of IRF6-transfected MCF-7 cells indicated an 80% reduction in the cell proliferation marker Ki-67, suggesting an important role for IRF6 in promoting cell cycle arrest (Fig. 5C).
FIG. 5.
IRF6 reexpression promotes cell cycle arrest in cancer cells. (A) Proliferation assay of poorly invasive (MCF-7) or highly invasive (MDA-MB-231) breast cancer cells infected with IRF6-expressing adenovirus (Ad IRF6) in the presence or absence of maspin, compared to Ad controls. Cells were counted 6 days postinfection. (B) Western blot analysis depicting the relative expression of Ad-transfected IRF6 or stably transfected maspin-green fluorescent protein (GFP) in MCF-7 and MDA-MB-231 cells. +, present; −, absent; p, phosphorylated; np, nonphosphorylated. (C) Real-time PCR results demonstrating the inverse correlation between IRF6 reexpression in MCF-7 breast cancer cells and the cellular proliferation marker Ki-67. Error bars represent the standard deviations of the results of three independent experiments. **, P ≤ 0.01 based on Student's t test.
DISCUSSION
We recently reported a novel protein interaction between IRF6 and maspin, a tumor suppressor in the mammary gland. Both proteins are generally lost during the development of breast cancer, and an increased understanding of maspin and IRF6 function may yield novel diagnostic and therapeutic approaches. In this study, we present data which demonstrate that IRF6 protein expression and phosphorylation are regulated concurrently with the cell cycle in a proteasome-dependent manner, whereas maspin protein expression is not significantly altered. Furthermore, we demonstrate that the reexpression of IRF6 in breast cancer cells results in decreased proliferation, and this effect is augmented by the presence of maspin. When taken together, these findings suggest a novel role for IRF6, together with maspin, in regulating mammary epithelial cell proliferation.
The regulation of IRF6 protein expression by the proteasome is not unique among IRF family members, since proteasomal degradation, initiated by IRF phosphorylation, is employed to regulate the protein expression of several IRFs (18, 22, 27, 33). Our study has shown that the proteasome regulates IRF6 in at least two different ways. First, the proteasome appears to be directly involved in regulating IRF6 phosphorylation, since inhibition of the proteasome resulted in IRF6 phosphorylation even in the absence of a mitogenic stimulus. One possible explanation is that the kinase responsible for IRF6 phosphorylation is also regulated by the proteasome in a cell cycle-dependent manner. Thus, inhibition of the proteasome in quiescent cells would result in an increase in kinase expression, thereby inducing an increase in IRF6 phosphorylation. However, it is also possible that the phosphorylation of IRF6 following proteasomal inhibition is secondary to the induction of other stimuli initiated by the inhibition of the proteasome, such as apoptosis. Further studies are required to definitively identify the cause of IRF6 phosphorylation resulting from proteasomal inhibition.
Second, proteasomal inhibition results in an increase in IRF6 protein levels similar to that seen in serum-starved, confluent, and purvalanol A-treated samples. Whereas the increase in nonphosphorylated IRF6 resulting from quiescence or cell cycle arrest is probably due to decreased protein turnover and may be augmented in some cases by increased IRF6 mRNA, the increase in phosphorylated IRF6 in MG-132-treated cells likely results from direct blocking of the proteasomal degradation of phosphorylated IRF6. We hypothesize that the regulation of IRF6 expression occurs primarily at the protein level, and our data demonstrate that the phosphorylation of IRF6 is a key step toward IRF6 ubiquitination and suggest that IRF6 phosphorylation is a signal for its degradation. Furthermore, the expression of IRF6 appears to be regulated by the growth status of the cell. Both cell cycle arrest (induced by purvalanol A, a selective inhibitor of cdk1 and cdk2) and cellular quiescence (induced by serum starvation or cell contact-mediated arrest) were sufficient to cause a buildup of IRF6 protein levels. The high PCNA expression in purvalanol A-treated cells demonstrates that these cells have not entered the G0 phase but are arrested in the cell cycle, since PCNA synthesis occurs early in the G1 phase, while cdk1 and cdk2 (targets of purvalanol A) act primarily at the transition stages of G1 to S and G2 to M, respectively (28). These findings suggest that active cycling through all phases of the cell cycle facilitates IRF6 degradation, whereas cell cycle arrest (natural or pharmacologically induced) is sufficient to increase IRF6 protein stability.
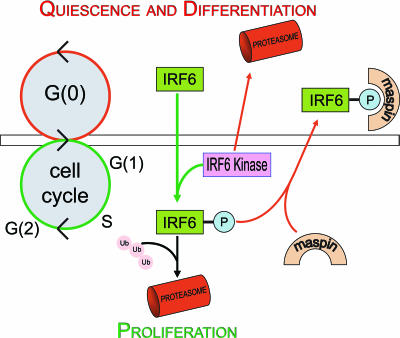
Cell cycle regulation by other IRFs establishes precedence for the involvement of IRF6 in this process. Specifically, IRF5 has been shown to induce G2/M cell cycle arrest concomitant with the induction of proapoptotic genes in B-cell lymphomas, and IRF2 is involved in aspects of the G1/S phase transition in HeLa cells (1, 3). Our data support a model (depicted in Fig. 6) in which the buildup of nonphosphorylated IRF6 promotes quiescence (an important step toward functional differentiation) through entry into the G0 phase of the cell cycle. Upon transition from G0 to G1 following a mitogenic stimulus, the degradation of the unknown IRF6 kinase is mitigated, leading to an increase in kinase activity and the resultant increase in IRF6 phosphorylation, thus targeting IRF6 for proteasome-mediated degradation. This is followed by an increase in cyclin D3 (Fig. 3A) and progression through the G1 phase.
FIG. 6.
Model depicting the function and regulation of IRF6 protein expression and phosphorylation. Nonphosphorylated IRF6 promotes differentiation and entry into the G0 stage. During this stage, the unknown IRF6 kinase is targeted for degradation by the proteasome. Upon receipt of a proliferation signal, degradation of the IRF6 kinase is ablated and expression of the kinase increases, leading to IRF6 phosphorylation (P). Phosphorylated IRF6 is either targeted for proteasomal degradation or is sequestered by maspin, which may regulate IRF6 function and preserve IRF6 expression. Arrows depicting differentiation signals are shown in red, while those depicting cell proliferation signals are shown in green. Ub, ubiquitin.
Coordination between proliferation and differentiation is critical for proper mammary gland development and function, with the two processes often antagonistic. By initiating entry into the G0 phase of the cell cycle, IRF6 may promote cellular differentiation in mammary epithelial cells. We report the differential expression of IRF6 in murine mammary epithelial cells during pregnancy and lactation. IRF6 is maximally expressed in lobuloalveolar cells during lactation, which represents the pinnacle of functional differentiation for mammary epithelial cells. Expectedly, IRF6 is only weakly expressed in ductal and glandular epithelial cells during pregnancy, which is characterized by heightened proliferation and ductal outgrowth. Maspin expression follows a similar, albeit less-dramatic trend. These in situ findings support a putative role for IRF6, likely in cooperation with maspin, in regulating cell growth and promoting the differentiation of mammary epithelial cells. This hypothesis is consistent with two recent reports which demonstrate that the Irf6 knockout mutation in mice is perinatally lethal, resulting in a severe skin phenotype characterized by abnormal keratinocyte differentiation and unchecked proliferation (12, 26). This is also in line with the probable role for IRF6 in palatal fusion, during which the terminal differentiation of the medial edge epithelium may be critical for proper palatal fusion (14, 30).
This study further enhances our understanding of the relationship between IRF6 and maspin, although a precise role for maspin remains somewhat unclear. Interestingly, a limited number of reports have previously implicated maspin in proteasome regulation and function (7, 17). Chen and colleagues demonstrated that maspin reexpression in breast carcinoma cells down-regulated the β5 subunit of the proteasome, which effectively decreased chymotrypsin-like proteasomal activity in these cells. Maspin's ability to regulate proteasome function may provide an alternate mechanism for its regulation of IRF6. Furthermore, because maspin is known to promote epithelial differentiation, it is possible that the maspin-IRF6 interaction may serve to regulate the proliferation-differentiation exchange (23). The fact that maspin synergizes IRF6-induced growth-regulatory effects on breast cancer cells suggests a cooperative role for maspin and IRF6 in regulating cell proliferation. Paradoxically, we have previously demonstrated that the reexpression of maspin in invasive MDA-MB-231 breast cancer cells mitigates the genotypic effects induced by IRF6 reexpression, suggesting a complex and coordinated regulation of various cellular functions (2). Because maspin preferentially interacts with phosphorylated IRF6, it is possible that the interaction serves to sequester and protect a portion of IRF6 from proteasomal degradation, which could rapidly promote cell cycle arrest (as depicted by the model in Fig. 6). This would allow maspin to oversee proliferation and differentiation in mammary epithelial cells through the regulation of IRF6 expression. Further work is necessary to ascertain the function of the maspin-IRF6 protein interaction and the effects induced by the loss of these two proteins in breast cancer.
There remain several important questions regarding IRF6 regulation and function in mammary epithelial cells. First, the exact stimulus to signal IRF6 phosphorylation remains unknown. The inability of specific growth factors, such as EGF or the serum supplement MitoPlus, to elicit a rapid effect on IRF6 phosphorylation invites the possibility that other serum components may be responsible. However, the inability of charcoal-dextran-stripped serum to induce IRF6 phosphorylation suggests that serum hormones may play a role. The most-pronounced effect was observed following the addition of complete growth medium, suggesting that a combination of factors which efficiently induce proliferation may be necessary for IRF6 phosphorylation. It is important to note that, while treatment with EGF alone strongly induces mitogen-activated protein kinase signaling through extracellular signal-regulated kinases 1 and 2, this signal is not sufficient to efficiently induce cell proliferation in MCF-10A cells, compared to the effects of complete growth medium. Second, the link between IRF6 and the cell cycle is not clear. Following the paradigm established by other IRFs, the phosphorylation of IRF6 would result in activation prior to degradation. Because IRF6 phosphorylation occurs early during the transition from G0 to G1, it is possible that phosphorylated IRF6 may act to facilitate exit from G0 prior to its degradation. However, the buildup of IRF6 protein during quiescence and the ability of reexpressed IRF6 to dramatically reduce cell proliferation in poorly aggressive (MCF-7) and highly invasive (MDA-MB-231) breast cancer cells suggest that IRF6 may actively promote cell cycle arrest, perhaps by prompting entry into the G0 phase. Third, the cytoplasmic localization of IRF6 following phosphorylation remains a conundrum. It is known that mutations in amino acid residues within the IRF6 DNA binding domain which are predicted to directly interact with DNA result in a more-severe syndrome (popliteal pterygium syndrome) than other mutations in the Irf6 gene that cause Van der Woude syndrome (14). This would suggest that IRF6-DNA interactions are important, yet IRF6 is not readily observed in the nucleus at any stage of the cell cycle or during quiescence. We are currently working to address these important issues.
In summary, we present data which suggest a novel and important role for IRF6 in regulating the proliferation and differentiation of mammary epithelial cells. These data highlight important new findings regarding mammary epithelial cell differentiation and support the hypothesis that IRF6 acts in collaboration with maspin as a regulator of cellular proliferation. Increased understanding of these growth-regulatory pathways will further our knowledge of cancer development and progression and may lead to novel therapeutic and/or diagnostic tools for breast cancer.
Acknowledgments
This work was supported by National Institutes of Health grant CA 75681.
Footnotes
Published ahead of print on 22 January 2008.
REFERENCES
- 1.Aziz, F., A. J. van Wijnen, J. L. Stein, and G. S. Stein. 1998. HiNF-D (CDP-cut/CDC2/cyclin A/pRB-complex) influences the timing of IRF-2-dependent cell cycle activation of human histone H4 gene transcription at the G1/S phase transition. J. Cell. Physiol. 177453-464. [DOI] [PubMed] [Google Scholar]
- 2.Bailey, C. M., Z. Khalkhali-Ellis, S. Kondo, N. V. Margaryan, R. E. Seftor, W. W. Wheaton, S. Amir, M. R. Pins, B. C. Schutte, and M. J. Hendrix. 2005. Mammary serine protease inhibitor (Maspin) binds directly to interferon regulatory factor 6: identification of a novel serpin partnership. J. Biol. Chem. 28034210-34217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barnes, B. J., M. J. Kellum, K. E. Pinder, J. A. Frisancho, and P. M. Pitha. 2003. Interferon regulatory factor 5, a novel mediator of cell cycle arrest and cell death. Cancer Res. 636424-6431. [PubMed] [Google Scholar]
- 4.Brandeis, M., and T. Hunt. 1996. The proteolysis of mitotic cyclins in mammalian cells persists from the end of mitosis until the onset of S phase. EMBO J. 155280-5289. [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, D., M. S. Chen, Q. C. Cui, H. Yang, and Q. P. Dou. 2007. Structure-proteasome-inhibitory activity relationships of dietary flavonoids in human cancer cells. Front. Biosci. 121935-1945. [DOI] [PubMed] [Google Scholar]
- 6.Chen, D., K. G. Daniel, M. S. Chen, D. J. Kuhn, K. R. Landis-Piwowar, and Q. P. Dou. 2005. Dietary flavonoids as proteasome inhibitors and apoptosis inducers in human leukemia cells. Biochem. Pharmacol. 691421-1432. [DOI] [PubMed] [Google Scholar]
- 7.Chen, E. I., L. Florens, F. T. Axelrod, E. Monosov, C. F. Barbas III, J. R. Yates III, B. Felding-Habermann, and J. W. Smith. 2005. Maspin alters the carcinoma proteome. FASEB J. 191123-1124. [DOI] [PubMed] [Google Scholar]
- 8.Debnath, J., S. K. Muthuswamy, and J. S. Brugge. 2003. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods 30256-268. [DOI] [PubMed] [Google Scholar]
- 9.Duguay, D., F. Mercier, J. Stagg, D. Martineau, J. Bramson, M. Servant, R. Lin, J. Galipeau, and J. Hiscott. 2002. In vivo interferon regulatory factor 3 tumor suppressor activity in B16 melanoma tumors. Cancer Res. 625148-5152. [PubMed] [Google Scholar]
- 10.Ford, H. L., E. Landesman-Bollag, C. S. Dacwag, P. T. Stukenberg, A. B. Pardee, and D. C. Seldin. 2000. Cell cycle-regulated phosphorylation of the human SIX1 homeodomain protein. J. Biol. Chem. 27522245-22254. [DOI] [PubMed] [Google Scholar]
- 11.Hu, G., M. E. Mancl, and B. J. Barnes. 2005. Signaling through IFN regulatory factor-5 sensitizes p53-deficient tumors to DNA damage-induced apoptosis and cell death. Cancer Res. 657403-7412. [DOI] [PubMed] [Google Scholar]
- 12.Ingraham, C. R., A. Kinoshita, S. Kondo, B. Yang, S. Sajan, K. J. Trout, M. I. Malik, M. Dunnwald, S. L. Goudy, M. Lovett, J. C. Murray, and B. C. Schutte. 2006. Abnormal skin, limb and craniofacial morphogenesis in mice deficient for interferon regulatory factor 6 (Irf6). Nat. Genet. 381335-1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kirschmann, D. A., E. A. Seftor, S. F. Fong, D. R. Nieva, C. M. Sullivan, E. M. Edwards, P. Sommer, K. Csiszar, and M. J. Hendrix. 2002. A molecular role for lysyl oxidase in breast cancer invasion. Cancer Res. 624478-4483. [PubMed] [Google Scholar]
- 14.Kondo, S., B. C. Schutte, R. J. Richardson, B. C. Bjork, A. S. Knight, Y. Watanabe, E. Howard, R. L. de Lima, S. Daack-Hirsch, A. Sander, D. M. McDonald-McGinn, E. H. Zackai, E. J. Lammer, A. S. Aylsworth, H. H. Ardinger, A. C. Lidral, B. R. Pober, L. Moreno, M. Arcos-Burgos, C. Valencia, C. Houdayer, M. Bahuau, D. Moretti-Ferreira, A. Richieri-Costa, M. J. Dixon, and J. C. Murray. 2002. Mutations in IRF6 cause Van der Woude and popliteal pterygium syndromes. Nat. Genet. 32285-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kroger, A., A. Dallugge, S. Kirchhoff, and H. Hauser. 2003. IRF-1 reverts the transformed phenotype of oncogenically transformed cells in vitro and in vivo. Oncogene 221045-1056. [DOI] [PubMed] [Google Scholar]
- 16.Kuo, M. L., and N. C. Yang. 1995. Reversion of v-H-ras-transformed NIH 3T3 cells by apigenin through inhibiting mitogen activated protein kinase and its downstream oncogenes. Biochem. Biophys. Res. Commun. 212767-775. [DOI] [PubMed] [Google Scholar]
- 17.Li, X., D. Chen, S. Yin, Y. Meng, H. Yang, K. R. Landis-Piwowar, Y. Li, F. H. Sarkar, G. P. Reddy, Q. P. Dou, and S. Sheng. 2007. Maspin augments proteasome inhibitor-induced apoptosis in prostate cancer cells. J. Cell. Physiol. 212298-306. [DOI] [PubMed] [Google Scholar]
- 18.Lin, R., C. Heylbroeck, P. M. Pitha, and J. Hiscott. 1998. Virus-dependent phosphorylation of the IRF-3 transcription factor regulates nuclear translocation, transactivation potential, and proteasome-mediated degradation. Mol. Cell. Biol. 182986-2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ma, C., S. J. Mandrekar, S. R. Alberts, G. A. Croghan, A. Jatoi, J. M. Reid, L. J. Hanson, L. Bruzek, A. D. Tan, H. C. Pitot, C. Erlichman, J. J. Wright, and A. A. Adjei. 2007. A phase I and pharmacologic study of sequences of the proteasome inhibitor, bortezomib (PS-341, Velcade), in combination with paclitaxel and carboplatin in patients with advanced malignancies. Cancer Chemother. Pharmacol. 59207-215. [DOI] [PubMed] [Google Scholar]
- 20.Marie, I., E. Smith, A. Prakash, and D. E. Levy. 2000. Phosphorylation-induced dimerization of interferon regulatory factor 7 unmasks DNA binding and a bipartite transactivation domain. Mol. Cell. Biol. 208803-8814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moriyama, Y., S. Nishiguchi, A. Tamori, N. Koh, Y. Yano, S. Kubo, K. Hirohashi, and S. Otani. 2001. Tumor-suppressor effect of interferon regulatory factor-1 in human hepatocellular carcinoma. Clin. Cancer Res. 71293-1298. [PubMed] [Google Scholar]
- 22.Nakagawa, K., and H. Yokosawa. 2000. Degradation of transcription factor IRF-1 by the ubiquitin-proteasome pathway. The C-terminal region governs the protein stability. Eur. J. Biochem. 2671680-1686. [DOI] [PubMed] [Google Scholar]
- 23.Odero-Marah, V. A., Z. Khalkhali-Ellis, J. Chunthapong, S. Amir, R. E. Seftor, E. A. Seftor, and M. J. Hendrix. 2003. Maspin regulates different signaling pathways for motility and adhesion in aggressive breast cancer cells. Cancer Biol. Ther. 2398-403. [DOI] [PubMed] [Google Scholar]
- 24.Paz, S., Q. Sun, P. Nakhaei, R. Romieu-Mourez, D. Goubau, I. Julkunen, R. Lin, and J. Hiscott. 2006. Induction of IRF-3 and IRF-7 phosphorylation following activation of the RIG-I pathway. Cell. Mol. Biol. (Noisy-le-Grand) 5217-28. [PubMed] [Google Scholar]
- 25.Pham, N. A., A. Morrison, J. Schwock, S. Aviel-Ronen, V. Iakovlev, M. S. Tsao, J. Ho, and D. W. Hedley. 2007. Quantitative image analysis of immunohistochemical stains using a CMYK color model. Diagn. Pathol. 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Richardson, R. J., J. Dixon, S. Malhotra, M. J. Hardman, L. Knowles, R. P. Boot-Handford, P. Shore, A. Whitmarsh, and M. J. Dixon. 2006. Irf6 is a key determinant of the keratinocyte proliferation-differentiation switch. Nat. Genet. 381329-1334. [DOI] [PubMed] [Google Scholar]
- 27.Saitoh, T., A. Tun-Kyi, A. Ryo, M. Yamamoto, G. Finn, T. Fujita, S. Akira, N. Yamamoto, K. P. Lu, and S. Yamaoka. 2006. Negative regulation of interferon-regulatory factor 3-dependent innate antiviral response by the prolyl isomerase Pin1. Nat. Immunol. 7598-605. [DOI] [PubMed] [Google Scholar]
- 28.Senderowicz, A. M., and E. A. Sausville. 2000. Preclinical and clinical development of cyclin-dependent kinase modulators. J. Natl. Cancer Inst. 92376-387. [DOI] [PubMed] [Google Scholar]
- 29.Stanford, B. L., and S. D. Zondor. 2003. Bortezomib treatment for multiple myeloma. Ann. Pharmacother. 371825-1830. [DOI] [PubMed] [Google Scholar]
- 30.Takigawa, T., and K. Shiota. 2004. Terminal differentiation of palatal medial edge epithelial cells in vitro is not necessarily dependent on palatal shelf contact and midline epithelial seam formation. Int. J. Dev. Biol. 48307-317. [DOI] [PubMed] [Google Scholar]
- 31.Tanaka, N., M. Ishihara, M. Kitagawa, H. Harada, T. Kimura, T. Matsuyama, M. S. Lamphier, S. Aizawa, T. W. Mak, and T. Taniguchi. 1994. Cellular commitment to oncogene-induced transformation or apoptosis is dependent on the transcription factor IRF-1. Cell 77829-839. [DOI] [PubMed] [Google Scholar]
- 32.Way, T. D., M. C. Kao, and J. K. Lin. 2004. Apigenin induces apoptosis through proteasomal degradation of HER2/neu in HER2/neu-overexpressing breast cancer cells via the phosphatidylinositol 3-kinase/Akt-dependent pathway. J. Biol. Chem. 2794479-4489. [DOI] [PubMed] [Google Scholar]
- 33.Xiong, H., H. Li, H. J. Kong, Y. Chen, J. Zhao, S. Xiong, B. Huang, H. Gu, L. Mayer, K. Ozato, and J. C. Unkeless. 2005. Ubiquitin-dependent degradation of interferon regulatory factor-8 mediated by Cbl down-regulates interleukin-12 expression. J. Biol. Chem. 28023531-23539. [DOI] [PubMed] [Google Scholar]
- 34.Zhang, M., D. Magit, F. Botteri, H. Y. Shi, K. He, M. Li, P. Furth, and R. Sager. 1999. Maspin plays an important role in mammary gland development. Dev. Biol. 215278-287. [DOI] [PubMed] [Google Scholar]