Abstract
The adipocyte integrates crucial information about metabolic needs in order to balance energy intake, storage, and expenditure. Whereas white adipose tissue stores energy, brown adipose tissue is a major site of energy dissipation through adaptive thermogenesis mediated by uncoupling protein 1 (UCP1) in mammals. In both white and brown adipose tissue, nuclear receptors and their coregulators, such as peroxisome proliferator-activated receptor γ (PPARγ) and PPARγ coactivator 1α (PGC-1α), play key roles in regulating their development and metabolic functions. Here we show the unexpected role of liver X receptor α (LXRα) as a direct transcriptional inhibitor of β-adrenergic receptor-mediated, cyclic AMP-dependent Ucp1 gene expression through its binding to the critical enhancer region of the Ucp1 promoter. The mechanism of inhibition involves the differential recruitment of the corepressor RIP140 to an LXRα binding site that overlaps with the PPARγ/PGC-1α response element, resulting in the dismissal of PPARγ. The ability of LXRα to dampen energy expenditure in this way provides another mechanism for maintaining a balance between energy storage and utilization.
Adipose tissue is the site of fat storage and oxidation and plays an important role in maintaining energy balance (45, 46). Two types of adipose tissue, namely, white adipose tissue (WAT) and brown adipose tissue (BAT), share many common features, such as the ability to take up glucose, synthesize and store triglycerides, and catabolically mobilize those triglycerides to free fatty acids for respiration and ATP production. However, they play largely opposing roles in mammalian physiology. WAT serves as the prime reservoir for storing excess caloric energy, in addition to its more recently recognized role as a source of endocrine hormones such as leptin, adiponectin, and resistin. In contrast, BAT is the major site of adaptive thermogenesis for the purpose of generating heat to maintain body temperature, particularly in response to cold environments (see review in reference 4). As such, it is a net consumer of caloric energy and is also a site of diet-induced thermogenesis (50).
In both WAT and BAT, catabolism is driven by the sympathetic nervous system (SNS) through the β-adrenergic receptor (βAR) signaling cascade. In BAT, the ability to mobilize and oxidize fuel is for the purpose of thermogenesis. In brown adipocytes, this thermogenic capability is due to the high density of mitochondria within this cell type and, most importantly, the presence of a brown adipocyte-specific uncoupling protein 1 (UCP1). UCP1 resides in the mitochondrial inner membrane, where it allows a regulated proton leak to uncouple respiration from ATP production (17, 54). Its role in diet-induced thermogenesis is related to the presence of brown adipocytes within various WAT depots, particularly in intra-abdominal or intrathoracic regions (10), where their appearance and function can be regulated by endogenous catecholamine stimulation or synthetic adrenergic agonists (11). Indeed, the inherent genetic propensity for this phenomenon is strongly associated with resistance to diet-induced obesity and metabolic syndrome (1, 12, 18). Although the existence of BAT in newborn humans is well accepted, brown fat has largely been considered irrelevant either as a source of heat or as a metabolic energy sink in adult humans. However, recent evidence from many clinical studies in the positron emission tomography imaging field, coupled with histological analyses, now challenge this notion and provide intriguing evidence for its existence (see reference 38), although the net impact on temperature or body weight and composition awaits further work.
The control of UCP1 expression in brown adipocytes by βARs and cyclic AMP (cAMP) has been known for many years, but other required components of the process have more recently come to light, one of which is the p38 mitogen-activated protein kinase (MAPK) pathway (6, 44). The brown fat-specific expression of the Ucp1 gene and the response to βAR activation resides predominantly in a small 220-bp enhancer region of the promoter of the Ucp1 gene, which is highly conserved among mammalian species (7, 15, 28). This region harbors a number of important sequence elements (7, 28) that have been identified as binding sites for key transcription factors, including peroxisome proliferator-activated receptors (PPARs) and activating transcription factor 2 (ATF-2) family members (2, 6, 43, 52). Coordinating these transcriptional regulatory activities is the family of associated nuclear coregulators (47). Of these, PPARγ coactivator 1α (PGC-1α) was discovered in a differential screen of transcriptional regulators in brown versus white adipocytes (41). PGC-1α has been shown to coordinate gene expression that stimulates mitochondrial biogenesis and oxidative metabolism in several cell types with relevance to metabolic fuel utilization (19). In BAT, the expression of PGC-1α is, like that of UCP1, increased in response to βAR stimulation, and its coregulator activity in this setting is also dependent on p38 MAPK phosphorylation (6).
Understanding the factors that promote the expression of UCP1 and the brown adipocyte phenotype shall undoubtedly help to unravel the molecular decisions that distinguish these two developmental programs. However, identifying factors that might actively repress or otherwise inhibit expression of UCP1 and the program of mitochondriogenesis are arguably no less important and are likely to prove equally informative. In this regard, there are several targeted gene deletions in rodents that result in a phenotype of “obesity resistance,” most commonly characterized at the molecular level by increased expression of UCP1 in WAT depots. Some of these genes include transcription factors and cofactors (8, 26, 30, 32). We were particularly interested in the phenotype of liver X receptor (LXR)-null mice for several reasons. Mice lacking both LXRα and -β are lean and show remarkable increases in UCP1 expression in WAT and muscle. Both subtypes of LXR are expressed in adipose tissue: LXRβ is ubiquitously expressed in every tissue, but LXRα is most abundant in liver and spleen BAT and WAT. Since activated LXRα stimulates glucose uptake (29) and promotes triglyceride synthesis and lipogenesis in adipose tissue (42, 53), coupled with the loss in adipose tissue mass observed for LXR knockout mice (26), this suggests that LXRα in particular is an important regulator of fat storage in adipose tissue (25). Because the role of LXRα in brown adipocytes is unknown, it was intriguing to speculate that LXRα may play a role in controlling energy expenditure in this tissue.
In this study, we show that LXRα can function as a transcriptional repressor in a gene-specific manner. In addition to the increases in basal expression of UCP1 in adipocytes from LXRα−/− mice, there is also a remarkable increase in mitochondrial biogenesis and oxygen consumption. Consistent with this increased capacity for thermogenesis, LXRα−/− mice possess a higher basal body temperature. Mechanistically, we show that LXRα antagonizes Ucp1 gene transcription by directly binding to a DR-4 (direct repeat spaced by 4) element in the Ucp1 enhancer in an LXR agonist-dependent manner. The nuclear corepressor RIP140 is recruited, and it serves together with LXRα to interfere with the cAMP-dependent binding of PPARγ to its response element at an adjacent cis-acting regulatory element in the Ucp1 enhancer. Collectively, our findings show that LXRα can function as a ligand-activated repressor and that in adipose tissue it serves as a negative break on the expression of UCP1 and the manifestation of the brown adipocyte phenotype.
MATERIALS AND METHODS
Animal experiments.
LXR+/+ (wild-type [WT]), LXRα−/−, and LXRαβ−/− mice (8 to 10 weeks old) were generated in a mixed-strain background (C57BL/6-129Sv/Ev) as described previously (26). They were housed in a 25°C environment with 12-h light/dark cycles on standard rodent chow diet for 3 days and then randomly divided into two groups for each genotype. One group for each genotype was transferred to a 5°C environment for 7 h; the other group for each genotype was kept in room temperature. Core body temperature was measured using a RET-3 microprobe (Physitemp). Following euthanasia, the interscapular BAT and gonadal WAT were rapidly collected. A portion of each tissue was frozen in liquid nitrogen and to recover RNA for measurement by real-time PCR (RT-PCR) using TaqMan or Sybr green probes as indicated in specific experiments. The other portion was fixed in 10% buffered formalin and used for histology. All animal experiments were approved by the Institutional Animal Care and Use Committees of The Hamner Institutes for Health Sciences and the University of Texas Southwestern Medical Center in accordance with National Institutes of Health guidelines for the care and use of laboratory animals.
Chemicals and plasmids.
T0901317 (T09) was from Cayman Chemical. 22R-hydroxycholesterol (22R-OH) and forskolin (FSK) were from Sigma (St. Louis, MO). Antibody against mouse LXRα was a very generous gift from Jean-Marc A. Lobaccaro (58). Antibodies against PPARγ (sc-7196x) and RIP140 (sc-8997x) were from Santa Cruz Biotechnology. The plasmid EN-Tk-CAT containing the mouse Ucp1 enhancer (−2530 to −2310) and the β-actin-luciferase (β-actin-luc) control vector for transfection were constructed as previously described (5). The plasmid pCMV5-hRXRα was purchased from Open Biosystems Company. The plasmid pCMX-mLXRα was described previously (24). The DR-4 mutation in the Ucp1 enhancer [Ucp1-En-DR-4 deletion tk-CAT, named Ucp1-En-tk-CAT(DR-4Δ)] was generated by removing the fold base spacer from the WT sequence, GCGTCACAGAGGGTCA (the 4-base spacer is underlined), to make GCGTCA----GGGTCA (where dashes indicate bases deleted) (see Fig. 7B). The integrity of all new plasmid constructs was verified by DNA sequencing (MWG).
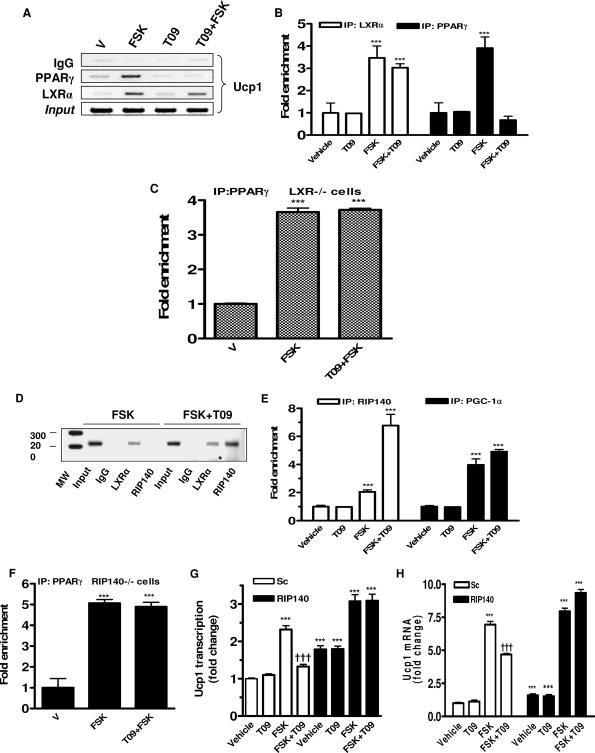
FIG. 7.
Dismissal of PPARγ from the Ucp1 enhancer by activated LXRα and role of RIP140 in suppressing of Ucp1 gene transcription. Differentiated HIB-1B cells (A, B, D, and E) and adipocytes derived from LXR−/− (C) or RIP140−/− (F) MEFs were treated with FSK (10 μM) or FSK plus T09 (5 μM) for 6 h and collected for immunoprecipitations with anti-RIP140, anti-LXRα, and anti-PPARγ sera as illustrated and as described in Materials and Methods. Occupancy of the Ucp1 enhancer by LXRα (A and B) and PPARγ (A, B, C, and F) and PGC-1α and RIP140 (D and E) was detected by ChIP. (G and H) HIB-1B cells were transfected with siRNA targeting RIP140 as described in Materials and Methods and subsequently treated (G) or not (H) with delivery of vector-containing Ucp1 enhancer (Ucp1-En-tk-CAT) and β-actin-luc as an internal control. Six hours prior to collection for assays, cells were treated with the following: T09 (5 μM), FSK (10 μM), or FSK plus T09. Ucp1 enhancer activity and transcripts were measured by CAT and luciferase enzyme activity assay and by RT-PCR using TaqMan probes with GAPDH as an internal control, respectively. Immunoprecipitated DNA was analyzed by agarose gel imaging (A and D) and quantitative PCR using specific primers for Ucp1 enhancer (B, C, E, and F). Bar graphs are presented as means ± SEM. V, vehicle; MW, molecular weight. Symbols: ***, P < 0.001 versus vehicle or vehicle of scrambled sequence; †††, P < 0.001 versus FSK of scrambled sequence (one-way ANOVA).
Histology.
Tissues were fixed in buffered formalin and embedded in paraffin. Five-micrometer sections were incubated with 1:500-diluted UCP1 antisera (kind gift of T. Gettys and L. Kozak) overnight at 4°C. After being washed with phosphate-buffered saline (PBS), the sections were incubated with 1:200-diluted biotinylated anti-sheep immunoglobulin G (IgG) (BA-6086; Vector Labs, Burlingame, CA) for 30 min, washed, and incubated with 1:200-diluted streptavidin horseradish peroxidase (43-4323; Zymed Labs, San Francisco, CA) for 30 min, all at room temperature. Following staining with AEC chromogen (00-2007; Zymed Labs, San Francisco, CA) for 5 min at room temperature, the sections were dehydrated and mounted for evaluation and photography.
Cell culture and transfections.
HIB-1B brown preadipocytes (48) were maintained in Dulbecco's modified Eagle medium (DMEM) with 10% fetal bovine serum (FBS). Upon reaching confluence, they were differentiated into adipocytes by the addition of rosiglitazone (1 μM) for 5 days (5). Primary brown adipocytes from C57BL/6J mice were prepared from BAT as described previously (5, 6, 36, 37, 44) and cultured in DMEM supplemented with 10% FBS, 0.14 μg/ml amphotericin B (Fungizone), 17 μM d-pantothenic acid, 33 μM d-biotin, 100 μM ascorbic acid, and 4 nM insulin. Differentiation of the cells was performed in the presence of 50 nM insulin, 50 nM thyroid hormone (T3), and 1 μM rosiglitazone for 10 days (5, 6, 44). Mouse embryo fibroblasts (MEFs) were isolated from WT and LXRα−/− and RIP140−/− mouse embryos. The cells were cultured in DMEM supplemented with 10% heat-inactivated FBS. When they approached 100% confluence, they were differentiated with dexamethasone (20 μg/ml), isobutylmethylxanthine (110 μg/ml), rosiglitazone (2.5 μM), and insulin (10 μg/ml) for 6 to 8 days. Where indicated, fully differentiated primary brown adipocytes, HIB-1B cells, or MEFs were treated for 6 h with vehicle (dimethyl sulfoxide [DMSO]; final concentration, ≤0.1%), FSK (10 μM), T09 (5 μM), or FSK and T09 together (T09 added 30 min prior to FSK) in DMEM supplemented with 10% dextran-coated charcoal (Sigma)-stripped FBS.
Transfections of HIB-1B cells were performed in six-well plates (Falcon) with a total amount of plasmid DNA equal to 2.2 μg/well and 5 μl Lipofectamine 2000 (Life Technologies). As indicated in particular experiments, these DNA mixtures included WT EN-tk-CAT or mutant EN-tk-CAT(DR-4Δ) (1.0 μg), pCMX-mLXRα (0.5 μg), pCMV5-hRXRα (0.5 μg), and β-actin-luc (0.2 μg). The PPARγ agonist rosiglitazone (1 μM) was added to the cells 6 h after transfection. Where indicated, during the final 6 h of the transfection, the cells were treated with FSK (10 μM) or T09 (5 μM) or with both together as described above. Forty-eight hours after transfection, cells were collected for chloramphenicol acetyltransferase (CAT) assays (Roche Molecular Biochemicals) and luciferase assays (Promega) as described previously (33).
Mitochondrial mass.
Primary MEFs (WT and LXRα−/−) were differentiated into adipocytes by use of the protocol described above. We used green fluorescent MitoTracker green FM (M7514; molecular probe from Invitrogen) to measure the mitochondrial DNA mass according to the product manual. When the cells fully differentiated into adipocytes, we removed the medium from the dish and added the prewarmed (37°C) growth medium containing the MitoTracker (final concentration, 75 nM) and incubated the cells for 30 min. After staining, we removed the medium and washed the cells with prewarmed medium two times, and then the cells were digested by trypsin, transferred to 1 ml cold PBS, and kept on ice for running-flow cytometry. In this experiment, a no-dye (without adding MitoTracker green FM) negative control was used to remove the background.
Oxygen consumption.
Oxygen consumption was measured for adipocytes derived from MEFs by using a Clark-type oxygen electrode (Hansatech Instruments Ltd., Norfolk, England). Each sample was analyzed by incubating 1 × 106 cells with 1 ml DMEM containing 25 mM HEPES and 2% fatty acid-free bovine serum albumin (pH 7.4) in a magnetically stirred chamber at 37°C. In each sample, after the basal respiration was recorded, 2.0 μg/ml oligomycin (F1Fo ATPase inhibitor) was added to determine the uncoupled respiration following the addition of 8 μM carbonyl cyanide p-(trifluoromethoxy)phenylhydrazone (FCCP; uncoupler) to determine the maximum respiration. The results are presented as the means ± standard errors of the mean (SEM) of three independent experiments.
RIP140 gene silencing in HIB-1B and WT MEFs.
Sequence-specific RIP140 gene silencing by small interfering RNA (siRNA) was performed in HIB-1B cells and LXRα-null and WT MEFs. The sequence of siRNA duplexes were designed as described previously (40). HIB-1B cells were transfected with siRNA duplex by use of Lipofectamine 2000 in six-well plates (20 pmol siRNA in 1 × 105 cells). Twelve hours later, cells were transfected with plasmids Ucp1-EN-tk-CAT and β-actin-luc (as an internal control); 72 h after the siRNA transfection, cells were collected for CAT assays (Roche Molecular Biochemicals) and luciferase assays (Promega).
WT MEFs were transfected with siRNA duplexes by use of Lipofectamine 2000 in six-well plates (20 pmol siRNA in 1 × 105 cells). Eighteen hours later, the cells were transfected with the same amount of siRNA. Six hours after the second transfection, the cells were incubated with differentiation medium as described above. Ninety-six hours after the first transfection, cells were treated with FSK (10 μM) and/or T09 (5 μM) for 6 h as described above and then collected for RNA isolation and RT-PCR.
RNA isolation and analysis.
Total RNA was extracted from cultured cells by use of Trizol reagent (Sigma). Prior to RT-PCR, samples were treated with DNA-free DNase (Ambion) to remove contaminating genomic or plasmid DNA. RNA was quantified by NanoDrop. cDNA was generated using the high-capacity cDNA archive kit from Applied Biosystems in a reaction volume scaled down to a 50-μl total. RT-PCR was performed on an ABI Prism 7700 sequence detector (Perkin Elmer) using TaqMan probes or Sybr green reagent and specific primers (IDT). Expression levels for all genes were normalized to the mean value for internal control GAPDH. Primer and probe sequences are given elsewhere (see http://www.thehamner.org/docs/figures.pdf).
Electrophoretic mobility shift assay.
Nuclear extracts were prepared by use of a TransFactor extraction kit from BD Biosciences (Palo Alto, CA). Double-stranded gel shift probes were end labeled with [γ-32P]ATP and T4 polynucleotide kinase. The binding reaction mixtures (10 μl) contained 5 μg nuclear protein in a buffer composed of 25 mM HEPES (pH 7.9), 0.5 mM EDTA, 0.5 mM dithiothreitol, 1% Nonidet P-40, 5% glycerol, 50 mM NaCl, and 1 μg poly(dΙ-dC). Unlabeled competitor oligonucleotides were added to the binding reaction mixtures in molar excess as detailed in the figure legends prior to the addition of labeled oligonucleotides. Reaction mixtures were incubated at room temperature for 15 min, and DNA-protein complexes were resolved on a 6% DNA retardation gel for 40 min at 180 V. Specificities of binding interactions were determined by the addition of 1 μl of antiserum (either mouse anti-LXRα or mouse anti-IgG) to the binding reaction mixtures 10 min after the components were mixed, followed by an additional 5 min of incubation before loading into the gel. Gels were then dried and subjected to autoradiography. The nucleotide sequences of sense oligonucleotide probes are provided below (see Fig. 7A), and a consensus LXR response element (LXRE) from the mouse GLUT4 promoter was used as a positive control (13).
ChIP assays.
Chromatin immunoprecipitation (ChIP) assays were performed using a commercial reagent (Upstate) according to the manufacturer's protocol. Briefly, differentiated HIB-1B cells grown in 10-cm2 dishes were treated with vehicle or with FSK (10 μM), T09 (5 μM), or both together for 6 h. For the cross-linking step, an aliquot of formaldehyde (37% [vol/vol]) was added directly to the culture medium to achieve a final concentration of 1%, and incubation was done for 10 min. Cells were then rinsed twice with cold PBS, harvested with cell scrapers, and lysed in 10 ml of a sodium dodecyl sulfate (SDS) buffer (1% SDS, 10 mM EDTA, 50 mM Tris, pH 8.1). The lysates were sonicated to shear genomic DNAs to achieve lengths of between 200 and 1,000 base pairs. The sheared lysates were centrifuged at 16,000 × g for 10 min, supernatants were collected and diluted 10-fold with ChIP dilution buffer (24 ml of 0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris-HCl, pH 8.1, 167 mM NaCl) in the presence of protease inhibitors (1 mM phenylmethylsulfonyl fluoride, 1 μg/ml aprotin, and 1 μg/ml pepstatin A). An aliquot (75 μl) of a 50% slurry of salmon sperm DNA-protein A-agarose was added and incubated for 30 min. Agarose beads were precipitated by brief centrifugation, and the precleared supernatant was collected. Antibodies against LXRα or PPARγ and control antibody IgG were added to this 2-ml supernatant fraction and incubated overnight at 4°C with gentle mixing. Sixty microliters of the salmon sperm DNA-protein A agarose slurry was added and incubated for an additional 1 h at 4°C to collect the antibody/histone/DNA complex by gentle and brief spinning. The presence of target DNA fragments was detected by PCR after the DNA was de-cross-linked with proteins. The primers used for the Ucp1 enhancer were 5′-AGTGAAGCTTGCTGTCACTC-3′ and 5′-GTCTGAGGAAAGGGTTGACC-3′.
Protein kinase assays for PKA and p38 MAPK.
WT and LXRα−/− MEFs were differentiated into adipocytes as described above and were treated with 10 μM isoproterenol (Iso) for either 5 min (protein kinase A [PKA] activity) or 10 min (p38 activity). Cells were washed twice in PBS and lysed in 25 mM HEPES buffer, pH 7.4 (containing 150 mM NaCl, 5 mM β-glycerophosphate, 1 mM sodium orthovanadate, 5 mM sodium pyrophosphate, 5 mM EDTA, 10% glycerol, 0.9% Triton X-100, 0.1% Igepal, and 1 complete mini tablet of protease inhibitors per 10 ml) for 15 min, and supernatants were recovered as described above.
To measure PKA activity assay, 16 μl of cell lysate was incubated for 30 min at 37°C with 5 μl of the 5× PepTag PKA reaction buffer, 3 μl of PepTag A1 peptide (fluorescent kemptide), and 1 μl of the peptide protection solution (assay kit from Promega, Madison, WI) in a 25-μl total volume. The reaction was stopped by boiling the samples at 95°C for 10 min, and 4 μl of 50% glycerol solution was added to the samples. Samples (10 μl) were loaded and resolved on a 0.8% agarose gel. Images were acquired using a Typhoon 9410 variable-mode imager (GE Healthcare, Piscataway, NJ) and then quantified by densitometry analysis using ImageQuant 5.2 software (Molecular Dynamics, Piscataway, NJ).
To measure p38 MAPK activity, 10-μl cell lysates were incubated with 4 μg glutathione transferase-ATF-2(1-109) fusion substrate (New England Biolabs), 200 μM ATP, and 25 μl of 4× kinase reaction buffer (80 mM HEPES, pH 7.4, 80 mM MgCl2, 100 mM β-glycerophosphate, 8 mM dithiothreitol, and 0.4 mM sodium orthovanadate) in a 100-μl reaction volume at 37°C for 30 min. Glutathione-Sepharose 4B (60 μl) was used to recover the substrate. Proteins were resolved with 4 to 20% Tris-glycine gels and transferred to nitrocellulose membrane. Protein phosphorylation was visualized by immunoblotting with anti-phospho-ATF-2 (Thr71) antibody (Cell Signaling Technology, Beverly, MA). Image acquisition was performed on a Typhoon 9410 variable-mode imager.
Statistics.
Significant differences between groups were determined by either two-tailed t tests or one-way analysis of variance (ANOVA) as indicated for specific experiments in figure legends. A minimum P value of <0.05 was considered statistically significant.
RESULTS
Elevated core body temperature of LXRα-null mice.
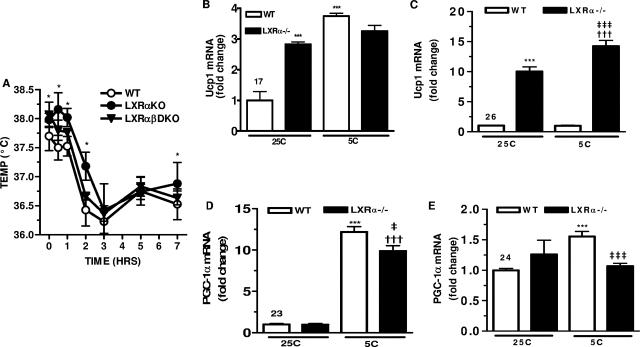
When placed in a cold environment, the SNS activates β-ARs in BAT to quickly mobilize free fatty acids for stimulating UCP1-dependent uncoupling and nonshivering thermogenesis. Since previous work showed that LXRα−/− LXRβ−/− mice had increased UCP1 expression, we examined the potential for increased nonshivering thermogenesis in these animals. WT, LXRα−/−, and LXRα−/− LXRβ−/− mice (6- to 8-week-old males) were placed at 5°C for up to 7 h. As shown in Fig. 1A, at 25°C the body temperature of LXRα−/− mice was significantly higher (0.25°C) than that of WT mice, and they maintained this elevated temperature difference during the cold challenge. The temperature of LXRα−/− LXRβ−/− mice tended to be intermediate. The fact that mice lacking LXRs have a higher body temperature prior to the cold challenge, which tends to persist through the 5°C period, suggests that the machinery of nonshivering thermogenesis involving Ucp1 and PGC-1α can be constitutively elevated in LXR-null mice. Gene expression was measured in BAT and WAT of these animals at room temperature and following cold challenge. At room temperature, Ucp1 mRNA levels in LXRα−/− mice were threefold higher than in WT mice (Fig. 1B). Following 7 h of cold exposure, there was a fourfold increase in Ucp1 expression in WT mice, with only a modest further increase in LXRα−/− mice. As observed previously (26), the difference in Ucp1 expression levels between the genotypes was more dramatic in WAT (Fig. 1C). Compared with what was seen for WT mice, there was a 10-fold increase in Ucp1 mRNA for LXRα−/− mice in gonadal WAT even at 25°C (26), and there was a further significant elevation after 7 h at 5°C. In WT mice, Ucp1 expression in WAT was unchanged in response to the cold (Fig. 1C). Interestingly, in BAT there was no significant difference between WT and LXRα−/− mice in the expression of PGC-1α at room temperature or in the ability to robustly induce PGC-1α mRNA in response to cold exposure (Fig. 1D). In WAT of WT mice, there was a 1.5-fold increase in PGC-1α mRNA levels following the 5°C challenge, as observed previously (6, 41). However, in WAT of LXRα−/− mice, PGC-1α gene expression was only slightly higher than that seen for WT mice at 25°C, with no further increase in response to cold (Fig. 1E). Taken together, these data reveal a significant inverse relationship between expression of LXRα and Ucp1 and cold sensitivity, but not with PGC-1α gene expression.
FIG. 1.
Higher core body temperature and expression of Ucp1 in BAT and WAT of LXRα−/− mice. (A) Core body temperature in WT (○), LXRα−/− (•), and LXRα−/− LXRβ−/− (▾) mice before and after cold (5°C) challenge. *, P < 0.05 versus WT (two-tailed t test). (B to E) RT-PCR analysis. Ucp1 gene in BAT (B) and WAT (C) of WT and LXRα−/− mice at 25°C and 5°C. Symbols: ***, P < 0.001 versus WT at 25°C; †††, P < 0.001 versus LXRα−/− at 25°C; ‡‡‡, P < 0.001versus WT at 5°C (one-way ANOVA). PGC-1α mRNA in BAT (D) and WAT (E) of WT and LXRα−/− mice at 25°C and 5°C. Symbols: *, P < 0.05; ***, P < 0.001 versus WT at 25°C; ‡, P < 0.05; ‡‡‡, P < 0.001 versus WT at 5°C (one-way ANOVA). All results are means ± SEM for five mice for each genotype.
Adipocytes from LXRα−/− mice exhibit multiple characteristics of the brown adipocyte phenotype.
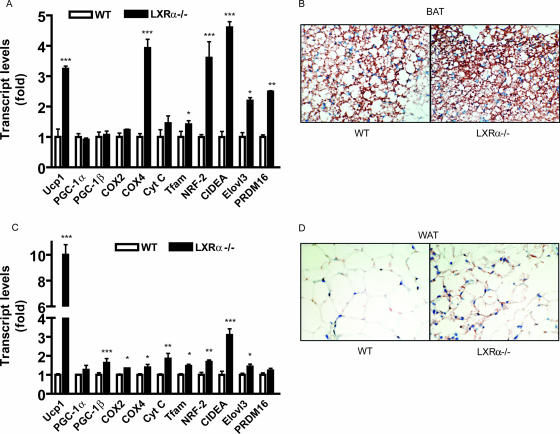
Given the increased basal body temperature and expression of UCP1 in adipose tissue of LXRα−/− mice, we also investigated the possibility that the adipocytes in these mice have higher thermogenic activity due to a higher density of mitochondria and the expression of genes involved in fat oxidation. Consistent with this idea, the expression of a collection of genes involved in mitochondrial biogenesis, including those of both the nuclear and mitochondrial genomes, were greater in adipose tissue from LXRα−/− mice than in that from WT animals (Fig. 2A and C). In addition to Ucp1, which was increased up to 10-fold in WAT of LXRα−/− mice, several other genes of particular interest that were significantly increased in both BAT and WAT of LXRα−/− mice include those for the brown adipocyte marker CIDE-A, mitochondrial transcription factor A, and subunits of nuclear respiratory factor 2. These data suggest increased brown adipocyte numbers as well as mitochondrial density within these adipose depots. Immunostained sections from BAT or gonadal WAT for UCP1 readily show that the cells in BAT from LXRα−/− mice tend to be smaller and more intensely stained for UCP1, as indicated by the red-brown color. They also contain smaller lipid droplets than WT mice (Fig. 2B). Similarly, in white adipocytes from LXRα-null animals one can see significantly greater punctuate red-brown UCP1 staining, as shown in Fig. 2D.
FIG. 2.
Increased expression of mitochondrial and brown fat markers in BAT and WAT of LXRα−/− mice. RT-PCR analysis of genes in BAT (A) and WAT (C) of WT and LXRα−/− mice at 25°C. Data are expressed relative to those for the WT mice. Symbols: *, P < 0.05; **, P < 0.001; ***, P < 0.0001 versus corresponding gene in WT (two-tailed t test). Immunohistochemical staining for UCP1 in BAT (B) and gonadal WAT (D) from WT and LXRα−/− mice. All results are means ± SEM for five mice for each genotype.
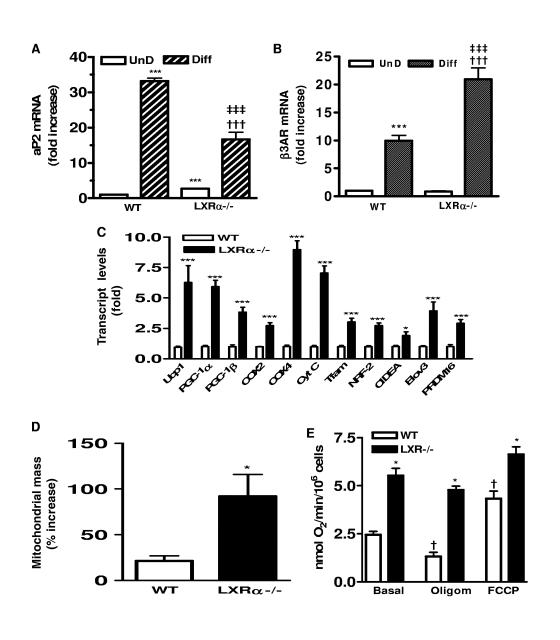
Since adipose tissue contains other cell types in addition to adipocytes, MEFs derived from the WT or LXRα−/− mice were subject to the standard adipocyte differentiation protocol, and the expression levels of the same panel of genes were examined. Cells from both genotypes could differentiate into adipocytes. Morphologically, both WT and LXRα-null cells accumulated lipid droplets, and the expression of adipocyte marker genes such as aP2 and β3AR were increased compared with what was seen for untreated fibroblasts (Fig. 3A and B). While the expression of aP2 is lower in the LXRα−/− adipocytes, the expression of β3AR is higher. Figure 3C shows that the pattern of mitochondrial and brown fat marker gene expression observed in adipose tissues from the WT and LXRα−/− mice was preserved in the cultured cells. Transcripts were elevated for each of the brown adipocyte marker genes in the adipocytes derived from the LXRα−/− MEFs. Some were increased 2- to 3-fold while others, such as those for Elovl3, Ucp1, PGC-1α, Cox4, and cytochrome c, were increased between 5- and 10-fold. Again, it is important to note that this is under basal conditions without added cAMP or other stimulators. Next, mitochondrial content was measured before and after differentiation of MEFs into adipocytes by use of MitoTracker green coupled with flow cytometry. Figure 3D shows that upon differentiation from the fibroblast stage, mitochondrial mass increased by approximately 20% in WT cells. However, in the LXRα-null cells the increase in mitochondrial mass following differentiation was more than four times greater than that seen for the WT. Thus, even in the unstimulated, basal state, LXRα−/− adipocytes are endowed with significantly more mitochondria. These adipocytes from LXRα−/− mice also exhibit increased oxygen consumption and uncoupled respiration. As shown in Fig. 3E, the basal oxygen consumption of LXR−/− cells is twofold higher than the level observed for WT cells. In the presence of FCCP, which raises the respiratory activity to maximal levels, oxygen consumption in the LXRα−/− cells was still significantly higher than that of WT cells. This indicates that LXRα−/− cells have a greater mitochondrial electron transport capacity, consistent with the increased mitochondrial mass. The fact that FCCP induced a twofold increase in the respiratory rate of WT cells but no significant increase in LXRα−/− cells implies that the basal electron transport activity of the LXRα−/− cells is nearly maximal. Oligomycin inhibits ATP synthetase to suppress only oxidative phosphorylation-associated respiration. As a result, all residual respiration is due to uncoupling. With oligomycin treatment, LXRα−/− cells still maintained a threefold increase in oxygen consumption over that of the WT cells. In addition, in WT cells there was a twofold-decreased respiratory rate in the presence of oligomycin, whereas in LXRα−/− cells most respiration was oligomycin insensitive. These results reflect a net increase in the uncoupling capacity of LXRα−/− cells. Since LXRα−/− adipocytes have increases not only in UCP1 expression but also in mitochondrial mass, respiration, and mitochondrial uncoupling, this finding is consistent with the data in Fig. 2 showing increased UCP1 and mitochondrial gene expression in adipose tissue from LXRα−/− mice. Altogether, this supports the idea that increased respiration and uncoupling reduces fat storage in LXRα−/− mice. In terms of events at the cellular level, the data also indicate that in the absence of LXRα there is a greater predilection for the adipocytes to differentiate into what resembles a brown adipocyte phenotype.
FIG. 3.
Increased mitochondrial mass, respiration, and brown adipocyte markers in LXRα−/− adipocytes. Expression of adipogenic marker genes aP2 (A) and β3AR (B) in undifferentiated (UnD) and differentiated (Diff) WT and LXRα−/− MEFs. Symbols: ***, P < 0.001 versus WT (UnD); †††, P < 0.001 versus LXRα−/− (UnD); ‡‡‡, P < 0.001 versus WT (Diff) (one-way ANOVA; n = 6). (C) Expression levels of genes related to mitochondrial biogenesis and brown fat in adipocytes derived from WT and LXRα−/− MEFs. ***, P < 0.001 versus the corresponding gene in the WT (two-tailed t test; n = 6). (D) Percent increase in mitochondrial mass upon differentiation of WT and LXRα−/− MEFs measured by MitoTracker green and flow cytometry. *, P < 0.05 versus WT (two-tailed t test; n = 2). (E) Oxygen consumption in adipocytes derived from WT and LXRα−/− MEFs by using a Clark-type oxygen electrode under basal conditions or following ATP synthetase inhibitor oligomycin (2 μg/ml; uncoupled respiration) or FCCP (8 μM; maximum respiration). Symbols: *, P < 0.05; **, P < 0.01; ***, P < 0.001 versus corresponding treated WT; †, P < 0.01; †††, P < 0.001 versus basal untreated WT (one-way ANOVA; n = 3). All data are means ± SEM.
LXRα suppresses βAR agonist- and FSK-induced Ucp1 gene expression.
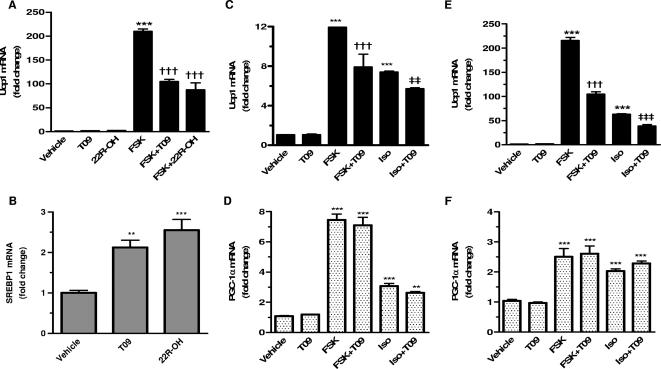
In order to explore the mechanism by which LXRα affects Ucp1 gene expression, we treated differentiated HIB-1B brown fat cells with two different LXR ligands: the oxysterol 22R-OH and the synthetic ligand T09. As shown in Fig. 4A, there was no effect of the LXR ligands on basal levels of Ucp1 gene transcripts, which were very low. Following provision of the adenylyl cyclase activator FSK, there was a very robust stimulation of Ucp1 gene expression, which was suppressed by both LXR ligands. In the same cells, however, the expression of SREBP1, a well-known LXR target gene, was increased by both 22R-OH and T09 (Fig. 4B). The effects of T09 and 22R-OH on SREBP1 were not significantly different. These results indicate that LXR is capable of both stimulating and repressing different target genes within the same cell. While the ability of LXRs to directly bind and activate the transcription of various target genes is well known, this unique suppression of Ucp1 gene expression by LXR activation could be either direct or indirect.
FIG. 4.
LXR agonists selectively suppress cAMP-stimulated Ucp1 gene expression in brown adipocytes. Expression levels of Ucp1(A) and SREBP1 (B) in differentiated HIB-1B cells treated for 6 h with vehicle (DMSO), 10 μM FSK, 1 μM 22R-OH, and 5 μM T09 as indicated. Transcripts were measured by RT-PCR using TaqMan probes. Results are means ± SEM (n = 4) relative to values for vehicle-treated cells. (C to F) Ucp1 and PGC-1α transcripts were measured by RT-PCR by TaqMan probes in differentiated primary brown adipocytes (C and D) and HIB-1B cells (E and F). Cells were treated for 6 h with vehicle (DMSO), 10 μM FSK, 10 μM Iso, and 5 μM T09 as indicated. Results from three experiments were performed in duplicate and are presented as means ± SEM relative to values for vehicle-treated cells. Symbols: **, P < 0.01; ***, P < 0.001 versus vehicle; †††, P < 0.001 versus FSK; ‡‡, P < 0.01, ‡‡‡; P < 0.001 versus Iso (one-way ANOVA).
To examine this inhibitory activity of LXR on UCP1 in more detail, we extended these initial studies into primary brown adipocytes as well as WT and LXRα−/− MEFs. In the first experiment, fully differentiated primary cultures of brown fat cells and HIB-1B immortalized brown adipocytes were treated with the β-agonist Iso or with FSK to induce Ucp1 gene expression. As shown to the left of Fig. 4C and E, in both cell models T09 alone had no effect on Ucp1 gene expression but, as for results depicted in Fig. 4A, it eliminated at least half of the increase due to either Iso or FSK. Interestingly, although we and others have consistently seen a close temporal and mechanistic parallel in the responses of the Ucp1 and PGC-1α genes to β-adrenergic stimulation (6, 41, 57), in this case the addition of T09 did not affect basal or cAMP-induced PGC-1α gene expression (Fig. 4D and F, right).
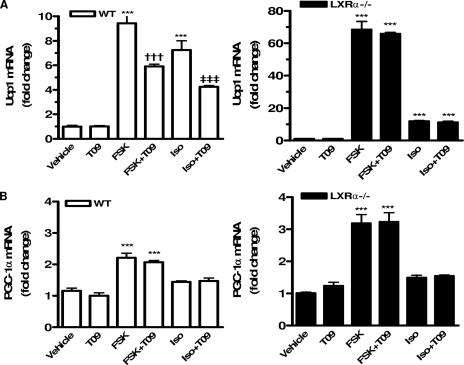
The next experiments using MEFs derived from the WT and LXRα−/− mice provided a genetic approach to the role of LXRα on cAMP-induced Ucp1 gene expression. As depicted in Fig. 5A, TaqMan RT-PCR results comparing WT and LXRα-null cells showed that Ucp1 gene expression was strongly induced in response to FSK in cells from both genotypes. Note that in LXRα−/− cells this induction was almost 10-fold higher than that in the WT cells. The same pattern was observed when cells were treated with Iso. The ability of T09 to suppress cAMP-dependent Ucp1 gene expression was observed only in WT cells, not in LXRα-null cells. Since even under nonstimulated conditions the expression of Ucp1 is already 8- to 10-fold higher in LXRα−/− cells, this indicates that under stimulated conditions the absolute differences in expression of Ucp1 between WT and LXRα−/− cells are several hundredfold. By contrast, the absence of LXRα had a small effect on cAMP-dependent increases in the expression of the PGC-1α gene (Fig. 5B). These results are similar to the data from primary brown adipocytes and HIB-1B cells (Fig. 4). Altogether, the results support a model in which activated LXRα antagonizes the cAMP-dependent transcription of the Ucp1 gene. Since LXRβ is still present in these LXRα−/− cells, and LXRα−/− LXRβ−/− cells behaved in a fashion essentially identical to that of the LXRα−/− cells (not shown), the results also indicate that, at least for regulating Ucp1 gene expression, LXRβ does not compensate for the absence of LXRα. Thus, it appears that the effect of LXR agonists to blunt Ucp1 gene expression is a property of LXRα alone.
FIG. 5.
Suppression of cAMP-stimulated Ucp1 expression is absent from LXRα−/− adipocytes. Differentiated MEFs were treated for 6 h with vehicle (DMSO), 5 μM T09, 10 μM FSK, and FSK plus T09 as indicated. RT-PCR analysis of Ucp1(A) and PGC-1α (B) gene expression in WT and LXRα−/− adipocytes. Results are means ± SEM relative to vehicle-treated cells for each genotype. Symbols: ***, P < 0.001 versus vehicle-treated control; †††, P < 0.001 versus FSK; ‡‡‡, P < 0.001 versus Iso (one-way ANOVA; n = 4 to 6).
Absence of LXRα does not alter the activity of βAR-regulated kinases.
To address the possibility that the LXRα−/− cell phenotype is due to changes in the amount or activity of more-proximal elements in the βAR signaling cascade that lead ultimately to Ucp1 gene transcription, we examined the βAR-dependent activation of PKA and p38 MAPK, two key factors involved in the transcription of the Ucp1 gene (6, 44, 56). A comparison of PKA and p38 MAPK activities under both basal and stimulated (10 μM Iso) conditions in WT and LXRα−/− adipocytes differentiated from MEFs is given elsewhere (see http://www.thehamner.org/docs/collins-mcb08.pdf). Relative to what was seen for the untreated WT cells, it is clear that there are no differences in basal PKA and p38 MAPK activities in the LXRα−/− cells and minimal differences between genotypes in response to Iso.
LXRα represses cAMP-dependent Ucp1 gene transcription through a DR-4 element in its enhancer region.
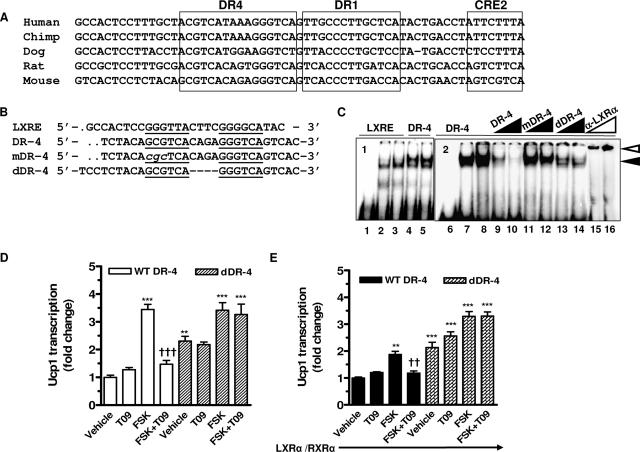
Within the powerful enhancer region of the Ucp1 promoter that was originally defined by Kozak et al. (28) and Cassard-Doulcier et al. (7), we noticed a previously unidentified conserved DR-4 sequence element (Fig. 6A). To test the idea that this DR-4 in the Ucp1 enhancer could be responsible for the effects of ligand-activated LXRα to antagonize cAMP-dependent transactivation, we first determined whether LXRα is capable of binding to this DR-4 sequence. Shown in Fig. 6B are the sequences used in gel shift experiments. The LXRE used as a positive control is from the mouse GLUT4 gene (13). Two mutations in the Ucp1 DR-4 were made: an internal deletion that removed the four-base spacer between the repeats (“dDR-4”), and a substitution mutation within the upstream repeat (“mDR-4”). Nuclear extract from HIB-1B cells (treated with T09 or not as indicated) produced DNA-protein complexes with the radiolabeled LXRE and the DR-4 of the Ucp1 enhancer (Fig. 6C, lanes 1 to 5). When the Ucp1 DR-4 sequence was used as a probe (Fig. 6C, lanes 6 to 16), this binding could be eliminated by increasing concentrations of unlabeled WT sequence, while the mutants were substantially impaired in their ability to compete for binding. The identity of LXRα as the binding species was confirmed by the inclusion of anti-LXRα antiserum, resulting in the supershifted species indicated. These results demonstrate that LXRα can bind the Ucp1 DR-4 sequence in vitro. It also implies that LXRβ does not bind, since the antiserum is specific for LXRα (58).
FIG. 6.
Activated LXRα suppresses cAMP-stimulated Ucp1 enhancer activity through a DR-4 element. (A) Alignment of DR-4, DR-1, and CRE2 in the enhancer regions of the Ucp1 genes from five species. (B) Design of synthetic oligonucleotides containing LXRE from mGlut4 (13) and DR-4 from the mouse Ucp1 enhancer. (C) Gel shift assays using nuclear extracts (5 μg) from HIB-1B cells incubated with radiolabeled LXRE or DR-4 in the absence or presence of increasing molar amounts (10×, 100×) of the unlabeled WT and mutant competitor oligonucleotides shown in panel B. For supershift analysis, anti-LXRα antiserum (1 μl, 2 μl) was added to the binding reaction mixture. Lanes 1 and 6 contain no nuclear protein. The black arrowhead indicates the protein-DNA complex. The white arrowhead indicates the supershifted complex. (D and E) Transient transfection of HIB-1B cells with tk-CAT reporter genes containing the 220-bp enhancer element from the mouse Ucp1 gene with WT or mutant (dDR-4) DR-4 sequences shown in panel B plus β-actin-luc as an internal control, without (D) or with (E) cotransfected LXRα/RXRα. Results presented are means ± SEM. Symbols: **, P < 0.01; ***, P < 0.001 versus basal untreated WT-DR-4; ††, P < 0.01; †††, P < 0.001 versus FSK-treated WT-DR-4 (one-way ANOVA; n = 3).
The next experiments were designed to determine whether the DR-4 sequence is directly involved in the inhibition of Ucp1 gene expression by activated LXRα. Deletion of the 4-base spacer region of the Ucp1 DR-4 element was constructed in the 220-bp enhancer sequence of the mouse Ucp1 gene and used in promoter-reporter assays as performed previously (5, 6). The WT and dDR-4 constructs were transfected into differentiated HIB-1B cells, and their activity in response to FSK and T09 was assessed. As shown in Fig. 6D, T09 blocked FSK-induced transactivation of the WT Ucp1 enhancer. There was no effect of T09 under basal conditions, similar to findings for cultured adipocytes (Fig. 4, 5, and 6). However, the basal activity of the dDR-4 construct was elevated more than twofold, and T09 was unable to blunt the increase in response to FSK. The same results were obtained for a construct containing the substitution mutation in DR-4 (not shown). In a separate approach, LXRα and RXRα were overexpressed together with the cotransfected Ucp1 promoter constructs (Fig. 6E). In this case, a similar inhibitory effect of LXRα on FSK-stimulated Ucp1 enhancer activity was observed, but compared with the results in Fig. 6D, the ability of FSK to transactivate the WT Ucp1 enhancer was greatly diminished. Moreover, overexpression of LXRα and RXRα had no effect on the activity of the dDR-4 mutant. Altogether, these findings are consistent with the view that activated LXRα suppresses Ucp1 gene transcription through DR-4 in the enhancer of the Ucp1 gene.
The corepressor RIP140 is required for the suppression of Ucp1 gene transcription by LXRα.
The results from the gel shift experiments and the effects of the DR-4 mutations on Ucp1 expression suggested that LXRα is interfering with the transactivation of the Ucp1 enhancer. In earlier studies, we and others discussed the importance of the DR-1 PPARγ binding site (6), which is a target of the PKA/p38 MAPK-dependent transactivation of the Ucp1 gene (6). Since this DR-1 is immediately adjacent to DR-4, with only one base between them (Fig. 6A), this suggested that occupancy of DR-4 by LXRα might somehow interfere with the cAMP-stimulated binding and function of PPARγ at the DR-1 for the transactivation of the Ucp1 gene. To explore this possibility, ChIP assays were performed with HIB-1B cells that had been treated with FSK or T09 as indicated. Figure 7A shows representative gels and Fig. 1B presents the PCR quantitation. T09 alone did not cause recruitment of LXRα to the Ucp1 enhancer. However, binding was recovered from cells treated with FSK. PPARγ was also recruited to the Ucp1 enhancer in response to FSK, as we have previously observed (6, 44), but, importantly, with the addition of T09 this interaction was lost. These data also illustrate that LXRα is recruited to the Ucp1 enhancer by cAMP, but it is the presence of the LXR agonist that is necessary for interference with the cAMP-dependent recruitment of PPARγ. The results indicate that when LXRα is in its ligand-activated conformation, a competition or exclusion exists between these two nuclear factors for occupancy of this region of the Ucp1 enhancer. As shown in Fig. 7C, results from ChIP assays performed with LXR-null cells show that the dismissal of PPARγ from the Ucp1 enhancer in response to the LXR agonist is abolished.
The action of transcription factors depends on the recruitment of additional elements to the target genes where they play distinct roles. Along with the activity of various chromatin remodeling enzymes and the polymerase II transcriptional machinery, gene activation is orchestrated by recruitment of coactivators, and the process of transcriptional activation can be reversed by recruitment of corepressors. PGC-1α is a coactivator for Ucp1, and we and others have shown previously that it is recruited to the Ucp1 enhancer. Interestingly, several aspects of the phenotype of mice lacking the corepressor RIP140 mirror the LXRα−/− phenotype (8, 30). Therefore, since RIP140 has been found on the Ucp1 gene promoter (8, 30), we set out to determine whether LXRα recruits RIP140 to function as the corepressor on the Ucp1 enhancer upon activation by T09. ChIP assay results are shown in Fig. 7D, and the quantification is shown in Fig. 7E. In the presence of T09 alone, immunoprecipitation with the RIP140 antisera did not recover the Ucp1 enhancer. In the presence of FSK, in which LXRα is present at the enhancer, there was a small increase in RIP140, as detected in the quantitation (Fig. 7E). However, upon the addition of T09 in the presence of cAMP, the recruitment of RIP140 on the Ucp1 enhancer was dramatically increased. Also as shown in Fig. 7E, the LXR ligand did not cause the recruitment of PGC-1α binding, nor did it affect PGC-1α recruitment induced by FSK. Obviously, this does not eliminate the possibility that upon the addition of LXR ligand there is some modification of PGC-1α that affects its coactivator function, but in any event these data show that the loss of PGC-1α is not a necessary requirement for the repressive activity. Finally, as shown in Fig. 7F, data from ChIP assays performed with adipocytes differentiated from RIP140-null cells show that, as observed for LXR-null cells (Fig. 7C), the loss of PPARγ from the Ucp1 enhancer with the LXR ligand was abolished. These results further suggest that the transrepression of the Ucp1 gene mediated by activated LXR requires corepressor RIP140 and its regulated clearance of PPARγ.
In order to further investigate whether RIP140 can functionally modulate LXRα-mediated repression of Ucp1 gene transcription, gene silencing experiments were performed using siRNA. Pilot RNA interference (RNAi) experiments (http://www.thehamner.org/docs/figures.pdf) showed complete knockdown of RIP140. Under these conditions, in HIB-1B cells transiently transfected with the Ucp1 enhancer/reporter plasmid, the scrambled sequence had no effect on the ability of ligand-activated LXRα to block FSK-stimulated transactivation of the Ucp1 enhancer (Fig. 7G). In the absence of RIP140, the LXR ligand failed to suppress FSK-stimulated activity. We performed the same RNAi knockdown experiment and examined the expression of the endogenous Ucp1 gene. As shown in Fig. 7H, the results were essentially the same as those observed for the reporter constructs with the HIB-1B cells, as shown in Fig. 7G. In the absence of RIP140, the basal expression of Ucp1 was ∼5-fold greater than in cells provided the scrambled sequence, and FSK-stimulated expression was not blocked by T09. All the data together lead to the conclusion that RIP140 is required for the suppression of Ucp1 gene transcription by activated LXRα.
DISCUSSION
The ability to express UCP1 in WAT depots is associated with resistance to obesity, as shown in genetic studies with various mouse strains as well as overexpression studies (1, 12, 18, 27, 49). In the latter case, simply overexpressing UCP1 in WAT appears sufficient to promote mitochondrial biogenesis (49). Recently, several reports have indicated that mitochondrially derived signals such as those that stem from changes in mitochondrial membrane potential or ATP production can promote mitochondrial biogenesis by a process known as “retrograde signaling,” but the molecular basis of this is not clear (31). Therefore, because of the evidence for decreased mitochondrial function in metabolic disease (34) and the search for mechanisms to combat obesity, understanding the molecular basis of brown versus white adipocyte differentiation continues to be of interest.
What factors and pathways control the differentiation of brown versus white adipocytes has been a long-standing question (22). More recently, there has been a debate in the field, based on several compelling observations, as to whether brown and white adipocytes have the capacity to interchangeably “transdifferentiate” between these two cytotypes (10, 35). The SNS is the main driver for the development and maintenance of the brown adipocyte phenotype. It is well established that under conditions in which SNS activity is minimized, such as when animals are maintained at thermoneutrality, brown adipocytes lose their characteristic features of UCP1 expression and their high density of mitochondria (9). If maintained in these conditions long enough, these cells can begin to produce typical white adipocyte markers, such as leptin (3). When SNS function is reestablished, expression of UCP1 and mitochondrial biogenesis is restored. Therefore, brown adipocytes might be able to morphologically resemble white adipocytes but retain a genetic “memory” that allows them to reacquire the brown adipocyte phenotype when stimulated.
In the pursuit of understanding the molecular basis for the brown versus white fat fate, investigators have searched for the pathways and factors that activate or promote brown adipocyte differentiation. For example, PGC-1α was discovered as a factor that was highly enriched in brown but not white adipocytes (41) and now appears to be important for mitochondrial biogenesis per se in many cell types (16). Another molecule called prdm16 has recently been proposed as a determining factor for brown adipocyte differentiation (51), but its mechanism is unclear. In addition to these approaches, it is equally useful to identify those elements that actively inhibit or repress brown adipocyte differentiation or its thermogenic activity. In that regard, targeted disruption in mice of several genes directly involved in energy storage and fat accumulation have been shown to lead to a lean phenotype and dramatic increases in UCP1 expression in adipocytes, particularly in white fat depots (see reference 20 for a review). For example, the apoptotic family member CIDE-A is highly expressed in brown fat, where it appears to interact with UCP1 to suppress its uncoupling function. In the absence of CIDE-A, there is unrestrained uncoupling of respiration, and the animals exhibit a lean phenotype (61). There are also several nuclear transcription factors whose absence also leads to a similar phenotype: these include the LXRs, ERRα, retinoblastoma protein, and RIP140 (20).
In the case of the LXRs, they normally promote energy storage through the expression of the lipogenic transcription factor SREBP1c, while the genetic absence of both subtypes of LXR leads to ectopic expression of UCP1 in muscle and WAT, which is more pronounced following consumption of a fat- and cholesterol-containing “Western” diet (26). Here in this study we show that, unlike what was found in the study of Kalaany et al., mice lacking only LXRα have a significantly higher basal body temperature that is consistent with the greater levels of expression of UCP1 in both BAT and WAT. LXRα−/− mice also possess more mitochondria and express a broad range of genes that encode mitochondrial proteins and specific markers of brown adipocytes. Thus, it would seem that in adipose tissue LXRα functions as an inhibitor of the common SNS and βAR signaling pathways that promote both diet-induced and cold-induced thermogenesis. Since the results for mice and cultured fat cells lacking LXRα alone were indistinguishable from results obtained with LXR double knock-out samples, this strongly suggests that LXRα is the operationally important isoform for this phenotype of increased basal UCP1 expression and mitochondrial biogenesis.
In the current work, we demonstrate by a number of different approaches that agonist-activated LXRα directly antagonizes transcription of the Ucp1 gene through a DR-4 element in the Ucp1 gene promoter to block the induction that results from SNS activation. First, using several different adipocyte models, we observed that the suppression of Ucp1 gene expression was evident only when both LXRα and its selective agonist were present. Adipocytes differentiated from WT and LXRα−/− MEFs showed that cAMP-stimulated UCP1 expression could be blocked only in WT cells and not in LXRα-null cells. An important point is that LXRα−/− adipocytes expressed significantly more UCP1 even under unstimulated basal conditions, and there was an enormous increase in expression in response to FSK or the βAR agonist Iso. Importantly, we determined that cultured adipocytes from the LXRα−/− mice had higher mitochondrial density, oxygen consumption, and uncoupled respiration, which was also reflected in the increased expression of a panel of mitochondrial and brown adipocyte marker genes in these cells.
An important factor shown to promote Ucp1 expression in brown adipocytes, and mitochondriogenesis in general, is the coactivator PGC-1α. PGC-1α participates together with PPARγ in Ucp1 gene transcription in response to β-adrenergic stimulation, through a cAMP/p38 MAPK pathway (15). Transcription of both the Ucp1 and PGC-1α genes in response to β-adrenergic stimulation often shows close temporal and mechanistic parallels. However, the inhibitory effect of LXRα is specifically on the Ucp1 gene and not a primary consequence of changes in PGC-1α itself. Instead, we identified a DR-4 element in the Ucp1 gene enhancer to which LXRα binds. As shown in ChIP assays, this site is immediately adjacent to the DR-1 site that becomes occupied by PPARγ upon elevations in cAMP (6). Curiously, we observed that conditions that elevate cAMP also result in LXRα being recruited to the enhancer, even in the absence of its ligand. It is unclear whether LXRα is chemically modified in some way and what the function of this cAMP-dependent binding of LXRα is for the enhancer, since the genetic absence of LXR obviously does not hamper the ability to transactivate the Ucp1 gene; instead, the expression of Ucp1 is enormously increased. In any event, the significant point is that upon agonist activation of LXRα, the cAMP-induced binding of PPARγ is lost. This suggests that the dismissal of PPARγ is involved in the suppression of Ucp1 transcription. Other interpretations were considered. These included a possible direct interaction between LXRα and PPARγ, but this is unlikely for two reasons. First, in response to cAMP, which is the stimulus for Ucp1 transcriptional activation, LXRα and PPARγ are nevertheless both found on the Ucp1 enhancer. Second, reports of cross talk between PPAR and LXR have appeared and, at least with cell culture models of forced overexpression, show that these two nuclear receptors can interfere with each other's ability to activate target genes by competing for limiting amounts of RXR (23, 60). However, for our studies, this explanation is unlikely, since the provision of excess receptors did not eliminate the ability of LXR to repress Ucp1 gene expression. Finally, it is known that thyroid hormone augments the process of nonshivering thermogenesis and UCP1 expression in BAT (55), but the exact mechanism is still not clear. Although thyroid hormone receptors can bind DR-4 elements (21), we can find no support for an involvement of a thyroid hormone receptor from either gene transfection or in vitro DNA-protein binding experiments that would explain the DR4-dependent regulation of Ucp1 gene expression (data not shown).
Since the control of gene expression by nuclear receptors requires the recruitment of coregulator complexes, RIP140 was a good candidate to investigate for several reasons. The phenotype of RIP140-null mice is remarkably similar to that of LXRα−/− mice (8, 30). These animals are reported to be lean and resistant to obesity induced by a high-fat diet, due to a higher mitochondrial uncoupling-derived energy expenditure. RIP140 is also highly induced during white adipocyte differentiation (39). Interestingly, this is driven by the nuclear receptor ERRα, the absence of which in mice has also been reported to produce a hypermetabolic, lean mouse (32). Finally, RIP140 was previously found on the Ucp1 enhancer (8), although there was no particular association with any nuclear receptor. Recently, Debevec et al. proposed that RIP140 regulates Ucp1 gene transcription with the involvement of ERRα and PPARα (14), but no specific binding site was proposed. We clearly show by ChIP assays that RIP140 appears on the Ucp1 promoter only in response to the agonist activation of LXR, when PPARγ is eliminated from the promoter. In addition, RNAi silencing of RIP140 expression eliminated the ability of the LXR agonist to suppress Ucp1 expression; results for RIP140 RNAi in LXRα−/− cells were similar. Other corepressors, such as nuclear receptor corepressor and the silencing mediator of retinoic acid and thyroid hormone receptors, might be considered to play a role in the repression mediated by LXRα, since they have been shown in another system to be involved in LXR ligand-independent transcriptional repression at the SREBP1 promoter (59). However, repression of the Ucp1 promoter by LXRα occurs only in response to cAMP and in the presence of the LXRα ligand. Thus, with the discovery of the DR-4 in the Ucp1 enhancer that mediates this LXRα repression, there is clearly a need for further work to gain a better mechanistic understanding of the complex molecular interactions that are taking place on this functionally “crowded” enhancer region of the Ucp1 promoter.
In summary, our results show a novel role for LXRα to function as a direct transcriptional repressor. It also suggests that LXRα is functioning as a “brake” on the expression of genes that promote the differentiation process toward the brown adipocyte phenotype. It is unclear at this point whether the molecular basis of this phenomenon is due to (i) the ability of LXRα to repress a network of genes that contribute to the brown adipocyte phenotype or (ii) the robust expression of UCP1, now unrestrained by that lack of LXRα, creating a mitochondrially derived signal that promotes mitochondrial biogenesis by retrograde signaling. Both mechanisms are plausible, and this question will need to be addressed in future studies. On a physiological level, LXRα is equally abundant in both BAT and WAT, but UCP1 and a rich density of mitochondria are found mainly in BAT. This raises the question as to whether the pivotal regulatory factor by which LXRα regulates Ucp1 and mitochondrial biogenesis involves the availability of an LXR ligand. Of the enzymes that are presently known to be involved in the synthesis of oxysterols, cholesterol 24-hydroxylase is confined to the central nervous system, leaving cholesterol 25-hydroxylase (CH25H) as a relevant enzyme expressed in adipose tissue, although the exact nature of the true endogenous ligands are still a matter of debate (D. Russell, personal communication). Interestingly, when expression levels of CH25H were measured, there was 30-fold more CH25H in WAT than in BAT (http://www.thehamner.org/docs/collins-mcb08.pdf). These observations would appear to suggest that under physiological conditions when conditions favor the storage of lipid-rich energy, it would be preferable for a system such as that we have shown for LXRα to suppress energy-consuming events such as UCP1-dependent uncoupling and wasting of energy through thermogenesis. Altogether, this study provides an interesting new avenue of investigation into understanding the signals and mechanisms that control the differentially expressed genes in white and brown adipocytes.
Acknowledgments
We sincerely thank Jean-Marc A. Lobaccaro for providing anti-LXRα antisera, Leslie Kozak and Thomas Gettys for anti-UCP1 antisera, David Russell for discussions about cholesterol hydroxylases, and Barbara Cannon and Martin Brand for helpful advice about respiration experiments. We also thank Wenhong Cao for his advice and comments on the manuscript and other members of the Collins lab for their support.
This work was supported by grants from the NIH to S.C. (R01-DK53092) and D.J.M. (U19DK62434), the Robert A. Welch Foundation (grant I-1275 to D.J.M.), the Howard Hughes Medical Institute (D.J.M.), and a research fund of The Hamner Institutes (S.C.).
Footnotes
Published ahead of print on 14 January 2008.
REFERENCES
- 1.Almind, K., M. Manieri, W. I. Sivitz, S. Cinti, and C. R. Kahn. 2007. Ectopic brown adipose tissue in muscle provides a mechanism for differences in risk of metabolic syndrome in mice. Proc. Natl. Acad. Sci. USA 1042366-2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barbera, M. J., A. Schluter, N. Pedraza, R. Iglesias, F. Villarroya, and M. Giralt. 2001. Peroxisome proliferator-activated receptor alpha activates transcription of the brown fat uncoupling protein-1 gene. A link between regulation of the thermogenic and lipid oxidation pathways in the brown fat cell. J. Biol. Chem. 2761486-1493. [DOI] [PubMed] [Google Scholar]
- 3.Cancello, R., M. C. Zingaretti, R. Sarzani, D. Ricquier, and S. Cinti. 1998. Leptin and UCP1 genes are reciprocally regulated in brown adipose tissue. Endocrinology 1394747-4750. [DOI] [PubMed] [Google Scholar]
- 4.Cannon, B., and J. Nedergaard. 2004. Brown adipose tissue: function and physiological significance. Physiol. Rev. 84277-359. [DOI] [PubMed] [Google Scholar]
- 5.Cao, W., A. V. Medvedev, K. W. Daniel, and S. Collins. 2001. β-Adrenergic activation of p38 MAP kinase in adipocytes: cAMP induction of the uncoupling protein-1 (UCP1) gene requires p38 MAP kinase. J. Biol. Chem. 27627077-27082. [DOI] [PubMed] [Google Scholar]
- 6.Cao, W., J. Robidoux, P. Puigserver, K. W. Daniel, A. V. Medvedev, X. Bai, L. M. Floering, B. M. Spiegelman, and S. Collins. 2004. p38 MAP kinase is the central regulator of cyclic AMP-dependent transcription of the brown fat uncoupling protein 1 gene. Mol. Cell. Biol. 243057-3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cassard-Doulcier, A.-M., C. Gelly, N. Fox, J. Schrementi, S. Raimbault, S. Klaus, C. Forest, F. Bouillard, and D. Ricquier. 1993. Tissue-specific and β-adrenergic regulation of the mitochondrial uncoupling protein gene: control by cis-acting elements in the 5′-flanking region. Mol. Endocrinol. 7497-506. [DOI] [PubMed] [Google Scholar]
- 8.Christian, M., E. Kiskinis, D. Debevec, G. Leonardsson, R. White, and M. G. Parker. 2005. RIP140-targeted repression of gene expression in adipocytes. Mol. Cell. Biol. 259383-9391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cinti, S. 1999. The adipose organ. Editrice Kurtis, Milan, Italy.
- 10.Cinti, S. 2005. The adipose organ. Prostaglandins Leukot. Essent. Fatty Acids 739-15. [DOI] [PubMed] [Google Scholar]
- 11.Collins, S., W. Cao, and J. Robidoux. 2004. Learning new tricks from old dogs: beta-adrenergic receptors teach new lessons on firing up adipose tissue metabolism. Mol. Endocrinol. 182123-2131. [DOI] [PubMed] [Google Scholar]
- 12.Collins, S., K. W. Daniel, A. E. Petro, and R. S. Surwit. 1997. Strain-specific response to β3-adrenergic receptor agonist treatment of diet-induced obesity in mice. Endocrinology 138405-413. [DOI] [PubMed] [Google Scholar]
- 13.Dalen, K. T., S. M. Ulven, K. Bamberg, J.-A. Gustafsson, and H. I. Nebb. 2003. Expression of the insulin-responsive glucose transporter GLUT4 in adipocytes is dependent on liver X receptor {alpha}. J. Biol. Chem. 27848283-48291. [DOI] [PubMed] [Google Scholar]
- 14.Debevec, D., M. Christian, D. Morganstein, A. Seth, M. Parker, and R. White. 2007. Receptor interacting protein 140 regulates expression of uncoupling protein 1 in adipocytes through specific peroxisome proliferator activated receptor isoforms and estrogen related receptor alpha. Mol. Endocrinol. 211581-1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.del Mar Gonzalez-Barroso, M., C. Pecqueur, C. Gelly, D. Sanchis, M. C. Alves-Guerra, F. Bouillaud, D. Ricquier, and A. M. Cassard-Doulcier. 2000. Transcriptional activation of the human ucp1 gene in a rodent cell line. Synergism of retinoids, isoproterenol, and thiazolidinediones is mediated by a multipartite response element. J. Biol. Chem. 27531722-31732. [DOI] [PubMed] [Google Scholar]
- 16.Goffart, S., and R. Wiesner. 2003. Regulation and co-ordination of nuclear gene expression during mitochondrial biogenesis. Exp. Physiol. 8833-40. [DOI] [PubMed] [Google Scholar]
- 17.Gonzalez-Barroso, M. M., C. Fleury, I. Arechaga, P. Zaragoza, C. Levi-Meyrueis, S. Raimbault, D. Ricquier, F. Bouillaud, and E. Rial. 1996. Activation of the uncoupling protein by fatty acids is modulated by mutations in the C-terminal region of the protein. Eur. J. Biochem. 239445-450. [DOI] [PubMed] [Google Scholar]
- 18.Guerra, C., R. A. Koza, H. Yamashita, K. Walsh, and L. P. Kozak. 1998. Emergence of brown adipocytes in white fat in mice is under genetic control: effects on body weight and adiposity. J. Clin. Investig. 102412-420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Handschin, C., and B. M. Spiegelman. 2006. PGC-1 coactivators, energy homeostasis, and metabolism. Endocr. Rev. 27728-735. [DOI] [PubMed] [Google Scholar]
- 20.Hansen, J. B., and K. Kristiansen. 2006. Regulatory circuits controlling white versus brown adipocyte differentiation. Biochem. J. 398153-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harbers, M., G. Wahlstrom, and B. Vennstrom. 1996. Transactivation by the thyroid hormone receptor is dependent on the spacer sequence in hormone response elements containing directly repeated half-sites. Nucleic Acids Res. 242252-2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hull, D., and M. M. Segall. 1966. Distinction of brown from white adipose tissue. Nature 212469-472. [DOI] [PubMed] [Google Scholar]
- 23.Ide, T., H. Shimano, T. Yoshikawa, N. Yahagi, M. Amemiya-Kudo, T. Matsuzaka, M. Nakakuki, S. Yatoh, Y. Iizuka, S. Tomita, K. Ohashi, A. Takahashi, H. Sone, T. Gotoda, J. Osuga, S. Ishibashi, and N. Yamada. 2003. Cross-talk between peroxisome proliferator-activated receptor (PPAR) alpha and liver X receptor (LXR) in nutritional regulation of fatty acid metabolism. II. LXRs suppress lipid degradation gene promoters through inhibition of PPAR signaling. Mol. Endocrinol. 171255-1267. [DOI] [PubMed] [Google Scholar]
- 24.Janowski, B. A., M. J. Grogan, S. A. Jones, G. B. Wisely, S. A. Kliewer, E. J. Corey, and D. J. Mangelsdorf. 1999. Structural requirements of ligands for the oxysterol liver X receptors LXRalpha and LXRbeta. Proc. Natl. Acad. Sci. USA 96266-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Juvet, L. K., S. M. Andresen, G. U. Schuster, K. T. Dalen, K. A. R. Tobin, K. Hollung, F. Haugen, S. Jacinto, S. M. Ulven, K. Bamberg, J.-A. Gustafsson, and H. I. Nebb. 2003. On the role of liver X receptors in lipid accumulation in adipocytes. Mol. Endocrinol. 17172-182. [DOI] [PubMed] [Google Scholar]
- 26.Kalaany, N. Y., K. C. Gauthier, A. M. Zavacki, P. P. Mammen, T. Kitazume, J. A. Peterson, J. D. Horton, D. J. Garry, A. C. Bianco, and D. J. Mangelsdorf. 2005. LXRs regulate the balance between fat storage and oxidation. Cell Metab. 1231-244. [DOI] [PubMed] [Google Scholar]
- 27.Kopecky, J., M. Rossmeisl, Z. Hodny, I. Syrovy, M. Horakova, and P. Kolarova. 1996. Reduction of dietary obesity in aP2-Ucp transgenic mice: mechanism and adipose tissue morphology. Am. J. Physiol. 270E776-E786. [DOI] [PubMed] [Google Scholar]
- 28.Kozak, U. C., J. Kopecky, J. Teisinger, S. Enerback, B. Boyer, and L. P. Kozak. 1994. An upstream enhancer regulating brown-fat-specific expression of the mitochondrial uncoupling protein gene. Mol. Cell. Biol. 1459-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Laffitte, B. A., L. C. Chao, J. Li, R. Walczak, S. Hummasti, S. B. Joseph, A. Castrillo, D. C. Wilpitz, D. J. Mangelsdorf, J. L. Collins, E. Saez, and P. Tontonoz. 2003. Activation of liver X receptor improves glucose tolerance through coordinate regulation of glucose metabolism in liver and adipose tissue. Proc. Natl. Acad. Sci. USA 1005419-5424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leonardsson, G., J. H. Steel, M. Christian, V. Pocock, S. Milligan, J. Bell, P. W. So, G. Medina-Gomez, A. Vidal-Puig, R. White, and M. G. Parker. 2004. Nuclear receptor corepressor RIP140 regulates fat accumulation. Proc. Natl. Acad. Sci. USA 1018437-8442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu, Z., and R. A. Butow. 2006. Mitochondrial retrograde signaling. Annu. Rev. Genet. 40159-185. [DOI] [PubMed] [Google Scholar]
- 32.Luo, J., R. Sladek, J. Carrier, J. A. Bader, D. Richard, and V. Giguere. 2003. Reduced fat mass in mice lacking orphan nuclear receptor estrogen-related receptor alpha. Mol. Cell. Biol. 237947-7956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Medvedev, A. V., S. K. Snedden, S. Raimbault, D. Ricquier, and S. Collins. 2001. Transcriptional regulation of the mouse uncoupling protein-2 gene. Double E-box motif is required for peroxisome proliferator-activated receptor-gamma-dependent activation. J. Biol. Chem. 27610817-10823. [DOI] [PubMed] [Google Scholar]
- 34.Morino, K., K. F. Petersen, and G. I. Shulman. 2006. Molecular mechanisms of insulin resistance in humans and their potential links with mitochondrial dysfunction. Diabetes 55S9-S15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morroni, M., A. Giordano, M. C. Zingaretti, R. Boiani, R. De Matteis, B. B. Kahn, E. Nisoli, C. Tonello, C. Pisoschi, M. M. Luchetti, M. Marelli, and S. Cinti. 2004. Reversible transdifferentiation of secretory epithelial cells into adipocytes in the mammary gland. Proc. Natl. Acad. Sci. USA 10116801-16806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nechad, M., P. Kuusela, C. Carneheim, P. Bjorntorp, J. Nedergaard, and B. Cannon. 1983. Development of brown fat cells in monolayer culture. I. Morphological and biochemical distinction from white fat cells in culture. Exp. Cell Res. 149105-118. [DOI] [PubMed] [Google Scholar]
- 37.Nechad, M., P. Kuusela, C. Carneheim, P. Bjorntorp, J. Nedergaard, and B. Cannon. 1983. Development of brown fat cells in monolayer culture. II. Ultrastructural characterization of precursors, differentiating adipocytes and their mitochondria. Exp. Cell Res. 149119-127. [DOI] [PubMed] [Google Scholar]
- 38.Nedergaard, J., T. Bengtsson, and B. Cannon. 2007. Unexpected evidence for active brown adipose tissue in adult humans. Am. J. Physiol. Endocrinol. Metab. 293E444-E452. [DOI] [PubMed] [Google Scholar]
- 39.Nichol, D., M. Christian, J. H. Steel, R. White, and M. G. Parker. 2006. RIP140 expression is stimulated by estrogen-related receptor {alpha} during adipogenesis. J. Biol. Chem. 28132140-32147. [DOI] [PubMed] [Google Scholar]
- 40.Powelka, A. M., A. Seth, J. V. Virbasius, E. Kiskinis, S. M. Nicoloro, A. Guilherme, X. Tang, J. Straubhaar, A. D. Cherniack, M. G. Parker, and M. P. Czech. 2006. Suppression of oxidative metabolism and mitochondrial biogenesis by the transcriptional corepressor RIP140 in mouse adipocytes. J. Clin. Investig. 116125-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Puigserver, P., Z. Wu, C. Park, R. Graves, M. Wright, and B. Spiegelman. 1998. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell 92829-839. [DOI] [PubMed] [Google Scholar]
- 42.Repa, J. J., G. Liang, J. Ou, Y. Bashmakov, J. M. Lobaccaro, I. Shimomura, B. Shan, M. S. Brown, J. L. Goldstein, and D. J. Mangelsdorf. 2000. Regulation of mouse sterol regulatory element-binding protein-1c gene (SREBP-1c) by oxysterol receptors, LXRalpha and LXRbeta. Genes Dev. 142819-2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rim, J. S., and L. P. Kozak. 2002. Regulatory motifs for CREB-binding protein and Nfe2l2 transcription factors in the upstream enhancer of the mitochondrial uncoupling protein 1 gene. J. Biol. Chem. 27734589-34600. [DOI] [PubMed] [Google Scholar]
- 44.Robidoux, J., W. Cao, H. Quan, K. W. Daniel, F. Moukdar, X. Bai, L. M. Floering, and S. Collins. 2005. Selective activation of mitogen-activated protein (MAP) kinase kinase 3 and p38α MAP kinase is essential for cyclic AMP-dependent UCP1 expression in adipocytes. Mol. Cell. Biol. 255466-5479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Robidoux, J., T. L. Martin, and S. Collins. 2004. Beta-adrenergic receptors and regulation of energy expenditure: a family affair. Annu. Rev. Pharmacol. Toxicol. 44297-323. [DOI] [PubMed] [Google Scholar]
- 46.Rosen, E. D., and B. M. Spiegelman. 2006. Adipocytes as regulators of energy balance and glucose homeostasis. Nature 444847-853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rosenfeld, M. G., V. V. Lunyak, and C. K. Glass. 2006. Sensors and signals: a coactivator/corepressor/epigenetic code for integrating signal-dependent programs of transcriptional response. Genes Dev. 201405-1428. [DOI] [PubMed] [Google Scholar]
- 48.Ross, S. R., L. Choy, R. A. Graves, N. Fox, V. Solevjeva, S. Klaus, D. Ricquier, and B. M. Spiegelman. 1992. Hibernoma formation in transgenic mice and isolation of a brown adipocyte cell line expressing the uncoupling protein gene. Proc. Natl. Acad. Sci. USA 897561-7565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rossmeisl, M., G. Barbatelli, P. Flachs, P. Brauner, M. C. Zingaretti, M. Marelli, P. Janovska, M. Horakova, I. Syrovy, S. Cinti, and J. Kopecky. 2002. Expression of the uncoupling protein 1 from the aP2 gene promoter stimulates mitochondrial biogenesis in unilocular adipocytes in vivo. Eur. J. Biochem. 26919-28. [DOI] [PubMed] [Google Scholar]
- 50.Rothwell, N. J., and M. J. Stock. 1979. A role for brown adipose tissue in diet-induced thermogenesis. Nature 28131-35. [DOI] [PubMed] [Google Scholar]
- 51.Seale, P., S. Kajimura, W. Yang, S. Chin, L. Rohas, M. Uldry, G. Tavernier, D. Langin, and B. M. Spiegelman. 2007. Transcriptional control of brown fat determination by PRDM16. Cell Metab. 638-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sears, I. B., M. A. MacGinnitie, L. G. Kovacs, and R. A. Graves. 1996. Differentiation-dependent expression of the brown adipocyte uncoupling protein gene: regulation by peroxisome proliferator-activated receptor gamma. Mol. Cell. Biol. 163410-3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Seo, J. B., H. M. Moon, W. S. Kim, Y. S. Lee, H. W. Jeong, E. J. Yoo, J. Ham, H. Kang, M. G. Park, K. R. Steffensen, T. M. Stulnig, J. A. Gustafsson, S. D. Park, and J. B. Kim. 2004. Activated liver X receptors stimulate adipocyte differentiation through induction of peroxisome proliferator-activated receptor gamma expression. Mol. Cell. Biol. 243430-3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shabalina, I. G., A. Jacobsson, B. Cannon, and J. Nedergaard. 2004. Native UCP1 displays simple competitive kinetics between the regulators purine nucleotides and fatty acids. J. Biol. Chem. 27938236-38248. [DOI] [PubMed] [Google Scholar]
- 55.Silva, J. E. 2006. Thermogenic mechanisms and their hormonal regulation. Physiol. Rev. 86435-464. [DOI] [PubMed] [Google Scholar]
- 56.Skala, J. P., and B. L. Knight. 1977. Protein kinases in brown adipose tissue of developing rats. State of activation of protein kinase during development and cold exposure and its relationship to adenosine 3′:5′-monophosphate, lipolysis, and heat production. J. Biol. Chem. 2521064-1070. [PubMed] [Google Scholar]
- 57.Tsukiyama-Kohara, K., F. Poulin, M. Kohara, C. T. DeMaria, A. Cheng, Z. Wu, A.-C. Gingras, A. Katsume, M. Elchebly, B. M. Spiegelman, M.-E. Harper, M. L. Tremblay, and N. Sonenberg. 2001. Adipose tissue reduction in mice lacking the translational inhibitor 4E-BP1. Nat. Med. 71128-1132. [DOI] [PubMed] [Google Scholar]
- 58.Volle, D. H., J. J. Repa, A. Mazur, C. L. Cummins, P. Val, J. Henry-Berger, F. Caira, G. Veyssiere, D. J. Mangelsdorf, and J. M. Lobaccaro. 2004. Regulation of the aldo-keto reductase gene akr1b7 by the nuclear oxysterol receptor LXRalpha (liver X receptor-alpha) in the mouse intestine: putative role of LXRs in lipid detoxification processes. Mol. Endocrinol. 18888-898. [DOI] [PubMed] [Google Scholar]
- 59.Wagner, B. L., A. F. Valledor, G. Shao, C. L. Daige, E. D. Bischoff, M. Petrowski, K. Jepsen, S. H. Baek, R. A. Heyman, M. G. Rosenfeld, I. G. Schulman, and C. K. Glass. 2003. Promoter-specific roles for liver X receptor/corepressor complexes in the regulation of ABCA1 and SREBP1 gene expression. Mol. Cell. Biol. 235780-5789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yoshikawa, T., T. Ide, H. Shimano, N. Yahagi, M. Amemiya-Kudo, T. Matsuzaka, S. Yatoh, T. Kitamine, H. Okazaki, Y. Tamura, M. Sekiya, A. Takahashi, A. H. Hasty, R. Sato, H. Sone, J. Osuga, S. Ishibashi, and N. Yamada. 2003. Cross-talk between peroxisome proliferator-activated receptor (PPAR) alpha and liver X receptor (LXR) in nutritional regulation of fatty acid metabolism. I. PPARs suppress sterol regulatory element binding protein-1c promoter through inhibition of LXR signaling. Mol. Endocrinol. 171240-1254. [DOI] [PubMed] [Google Scholar]
- 61.Zhou, Z., S. Yon Toh, Z. Chen, K. Guo, C. P. Ng, S. Ponniah, S. C. Lin, W. Hong, and P. Li. 2003. Cidea-deficient mice have lean phenotype and are resistant to obesity. Nat. Genet. 3549-56. [DOI] [PubMed] [Google Scholar]