Abstract
Ewing's family tumor (EFT) is a rare pediatric tumor of unclear origin that occurs in bone and soft tissue. Specific chromosomal translocations found in EFT cause EWS to fuse to a subset of ets transcription factor genes (ETS), generating chimeric EWS/ETS proteins. These proteins are believed to play a crucial role in the onset and progression of EFT. However, the mechanisms responsible for the EWS/ETS-mediated onset remain unclear. Here we report the establishment of a tetracycline-controlled EWS/ETS-inducible system in human bone marrow-derived mesenchymal progenitor cells (MPCs). Ectopic expression of both EWS/FLI1 and EWS/ERG proteins resulted in a dramatic change of morphology, i.e., from a mesenchymal spindle shape to a small round-to-polygonal cell, one of the characteristics of EFT. EWS/ETS also induced immunophenotypic changes in MPCs, including the disappearance of the mesenchyme-positive markers CD10 and CD13 and the up-regulation of the EFT-positive markers CD54, CD99, CD117, and CD271. Furthermore, a prominent shift from the gene expression profile of MPCs to that of EFT was observed in the presence of EWS/ETS. Together with the observation that EWS/ETS enhances the ability of cells to invade Matrigel, these results suggest that EWS/ETS proteins contribute to alterations of cellular features and confer an EFT-like phenotype to human MPCs.
Ewing's family tumor (EFT) is a rare childhood cancer arising mainly in bone and soft tissue. Since EFT has a poor prognosis, it is important to elucidate the underlying pathogenic mechanisms for establishing a more effective therapeutic strategy. EFT is characterized by the presence of chimeric genes composed of EWS and ets transcription factor genes (ETS) formed by specific chromosomal translocations, i.e., EWS/FLI1, t(11;22)(q24;q12); EWS/ERG, t(21;22)(q12;q12); EWS/ETV1, t(7;22)(p22;q12); EWS/E1AF, t(17;22)(q12;q12); and EWS/FEV, t(2;22)(q33;q12) (26). The products of these chimeric genes behave as aberrant transcriptional regulators and are believed to play a crucial role in the onset and progression of EFT (3, 36). Indeed, recent studies have revealed that the induction of EWS/FLI1 proteins can trigger transformation in certain cell types, including NIH 3T3 cells (36), C2C12 myoblasts (12), and murine primary bone marrow-derived mesenchymal progenitor cells (MPCs) (6, 45, 52). However, studies have also indicated that overexpression of EWS/FLI1 provokes apoptosis and growth arrest in mouse normal embryonic fibroblasts and primary human fibroblasts (10, 31), hence hampering understanding of the precise role of EWS/ETS proteins in the development of EFT. The function of EWS/ETS proteins would be greatly influenced by cell type, and thus the cells that can originate EFTs might be more susceptible to the tumorigenic effects of EWS/ETS.
Although the cell origin of EFT is still unknown, the expression of neuronal markers in spite of the occurrence in bone and soft tissues has kept open the debate as to a potential mesenchymal or neuroectodermal origin. As described above, ectopic expression of EWS/FLI1 results in dramatic changes in morphology and the formation of EFT-like tumors in murine primary bone marrow-derived MPCs but not in murine embryonic stem cells (6, 45, 52), supporting the notion that MPCs are a plausible cell origin of EFT (45). However, others argue that MPCs cannot be considered progenitors of EFT without further evidence of similarity between human EFT and MPC-EWS/FLI1-induced tumors in mice (29, 46).
The development of experimental systems using murine species is useful for elucidating the mechanisms behind the pathogenesis of EFT. However, several differences between human and murine systems cannot be ignored; these differences include the expression patterns of surface antigens in MPCs, for instance (7, 44, 51, 53). Moreover, human cells are difficult to transform in vitro, and the transformed cells of mice seem to produce a more aggressive tumor than those of humans (1). The findings suggest the existence of undefined cell-autonomous mechanisms that render human cells resistant to malignant transformation. Therefore, the use of human cell models is ideal for clarifying how EFT develops. Models of the onset of EFT have been generated using primary fibroblasts (31) and rhabdomyosarcoma cells (23). However, these cell types are not appropriate for studying the origins of EFT, and a model that precisely recapitulates EWS/ETS-mediated EFT formation is required.
UET-13 cells are obtained by prolonging the life span of human bone marrow stromal cells by use of the retroviral transgenes hTERT and E7 (38, 50), retain the ability to differentiate into not only mesodermal derivatives but also neuronal progenitor-like cells, and are considered a good model for studying the cellular events in human MPCs. Therefore, we have examined the biological effect of EWS/ETS in human MPCs by use of UET-13 cells by exploiting tetracycline-inducible systems for expressing EWS/ETS (EWS/FLI1 and EWS/ERG). Here we report that overexpression of EWS/ETS mediates an EFT-like phenotype, including morphology, immunophenotype, and gene expression profile, with enhancement of the Matrigel invasion ability of UET-13 cells.
MATERIALS AND METHODS
Cell cultures and establishment of UET-13TR-EWS/ETS cell lines.
UET-13 cells were cultured in Dulbecco's modified Eagle's medium (DMEM) with 10% Tet system approved fetal bovine serum (T-FBS) (Takara) at 37°C under a humidified 5% CO2 atmosphere. EFT cell lines (EES-1 [20], SCCH196 [21], RD-ES and SK-ES1 [5], NCR-EW2 and NCR-EW3 [19], and W-ES [13]) and neuroblastoma (NB) cell lines (NB69 and NB9 [15] and GOTO [47]) were cultured in RPMI 1640 with 10% FBS. A rhabdomyosarcoma cell line, NRS-1 (40), was cultured in Eagle's minimal essential medium with 10% FBS. The cell lines used in this study are listed in Table 1.
TABLE 1.
Cell lines used in this study and fusion transcript types
| Cell line | Diagnosis | Fusion transcript type | Reference |
|---|---|---|---|
| EES-1 | EFT | EWS/FLI1 type I | 20 |
| SCCH196 | EFT | EWS/FLI1 type I | 21 |
| RD-ES | EFT | EWS/FLI1 type II | 5 |
| SK-ES1 | EFT | EWS/FLI1 type II | 5 |
| NCR-EW2 | EFT | EWS/FLI1 type II | 19 |
| NCR-EW3 | EFT | EWS/E1AF | 19 |
| W-ES | EFT | EWS/ERG | 13 |
| NB69 | NB | 15 | |
| NB9 | NB | 15 | |
| GOTO | NB | 47 | |
| NRS-1 | RMS | PAX3/FKHR | 40 |
UET-13 cells were seeded at a density of 5 × 104 cells per well in 24-well tissue culture plates 1 day prior to transfection. For introducing the tetracycline-inducible system, UET-13 cells were transfected with pcDNA6-TR (Invitrogen) by use of Lipofectamine 2000 (Invitrogen). After 72 h, the medium was replaced with fresh medium containing 200 μg/ml of blasticidin S (Invitrogen). Individual resistant clones were selected for a month and designated UET-13TR cells. UET-13TR cells were further transfected with pcDNA4-EWS/ETSs constructed as described below, and individual resistant clones were selected in DMEM containing 10% T-FBS and 200 to 300 μg/ml of Zeocin (Invitrogen). The Zeocin-resistant clones were expanded and tested for the induction of EWS/ETS expression upon the addition of tetracycline by use of reverse transcription-PCR (RT-PCR) as described below.
Plasmid construction.
A gateway cassette (bases 1 to 1705) was amplified from pBLOCK-iT3-DEST (Invitrogen) by PCR, and the PCR product was inserted into the EcoRV site of pcDNA4-TO (Invitrogen) (termed pcDNA4-DEST). Since the type II EWS/FLI1 is a stronger transactivator than the type I product (32), we used the type II variant in the present study. EWS/ERG was isolated from W-ES, an EFT cell line, joining EWS exon 7 and ERG exon 9. Full-length EWS/FLI1 type II and EWS/ERG cDNAs were amplified from cDNAs prepared from NCR-EW2 and W-ES cells, respectively, by PCR as described below and cloned into the XmnI-EcoRV sites of pENTR11 (Invitrogen). The resulting pENTR11-EWS/ETSs were recombined with pcDNA4-DEST by use of LR recombination reaction as instructed by the manufacturer (Invitrogen) to construct the tetracycline-inducible EWS/ETS expression vector pcDNA4-EWS/ETSs.
Western blot analysis.
UET-13 transfectants were cultivated with or without 3 μg/ml of tetracycline for 72 h. Western blot analysis was performed as previously described (37). Briefly, the cell lysates were prepared and separated on a 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis gel and transferred onto a polyvinylidene difluoride membrane. The membranes were blocked with 5% skimmed milk in phosphate-buffered saline (PBS) containing 0.01% Tween 20 (Sigma) and incubated with primary antibodies. As the primary antibodies, anti-Fli-1, anti-Erg-1/2/3 (Santa Cruz Biotechnology), and anti-actin (Sigma) were used. Horseradish peroxidase-conjugated anti-rabbit or anti-mouse immunoglobulin G (IgG) antibodies (DakoCytomation) were used as secondary antibodies. Blots were detected by chemiluminescence using an ECL Plus Western blotting detection system (GE Healthcare Bio-Science Corp.) and exposed to X-ray film (Kodak) for 5 to 30 min.
MTT assay and detection of apoptosis.
Growth curves of UET-13 transfectants were determined using the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay as described previously (18). The apoptosis was detected using an annexin V-fluorescein isothiocyanate (FITC) apoptosis detection kit (Biovision) according to the manufacturer's instructions and analyzed by flow cytometry (Cytomics FC500; Beckman Coulter).
Immunofluorescence analysis.
After 1 week of culture in the absence or presence of tetracycline, UET-13 cells and the transfectants were harvested with 0.25% trypsin plus EDTA (IBL). The cells (2 × 105) were incubated with mouse monoclonal antibodies for 20 min. In the case of fluorescence-labeled antibodies, the cells were washed with PBS and then analyzed. In the case of primary unconjugated mouse antibodies, the cells were washed and then incubated with FITC-conjugated goat anti-mouse IgG antibody (Jackson ImmunoResearch Laboratories) for 20 min. Cell fluorescence was detected using a Cytomics FC500 instrument as described previously (27).
Antibodies against the following human antigens were used: CD10, CD13, CD14, CD29, CD34, CD40, CD44, CD45, CD49e, CD54, CD56, CD61, CD90, CD105, CD117, and CD166 from Beckman Coulter; CD73 from BD Biosciences-Pharmingen; CD55 from Abcam; CD59 from Cedarlane Laboratories; and CD133 and CD271 from Miltenyi Biotec GmbH.
Immunocytochemistry.
Cells were grown on collagen type I-coated cover glasses (Iwaki). After 72 h with or without tetracycline, cells were fixed for 30 min in 4% paraformaldehyde and permeabilized in PBS containing 0.2% Triton X-100 (Sigma) for 30 min. Subsequently, they were washed with PBS and blocked in PBS containing 0.1% Triton X-100 and 1% bovine serum albumin (Sigma) for 30 min before being incubated with a monoclonal anti-CD99 antibody, i.e., 12E7 (1:100) (DakoCytomation) or O13 (1:200) (Thermo), and polyclonal anti-Fli-1 antibody (1:100) (Santa Cruz) for 1 h. Bound antibodies were visualized with appropriate secondary antibodies, i.e., Alexa Fluor 488 goat anti-mouse IgG (heavy plus light chains) highly cross-adsorbed and Alexa Fluor 546 goat anti-rabbit IgG (heavy plus light chains) highly cross-adsorbed (Invitrogen) for 1 h at 1:300. Nuclei were counterstained with 4′,6′-diamidino-2-phenylindole (DAPI) or propidium iodide (PI) (Sigma). For the visualization of whole cells, cells were treated with Celltracker Blue (Invitrogen) for 30 min and then fixed. Fluorescence was observed and analyzed using a confocal laser scanning microscope and image software (either FV500 from Olympus or LSM510 from Carl Zeiss). Precise measurements of cell size, nuclear size, and the nucleus-to-cytoplasm (N/C) ratio were performed using Image J (16).
RT-PCR analysis.
Total RNA was extracted from cells by use of an RNeasy kit (Qiagen) and reverse transcribed using a first-strand cDNA synthesis kit (GE Healthcare Bio-Science Corp). RT-PCR was performed with a HotstarTaq master mix kit (Qiagen). As an internal control, human GAPDH cDNA was also amplified. The sequences of gene-specific primers for RT-PCR were as follows: for EWS/FLI1 (forward), 5′-ATGGCGTCCACGGATTACAGTACCT-3′; for EWS/FLI1 (reverse), 5′-GGGTCTTCTTTGACACTCAATCG-3′; for EWS/ERG (forward), 5′-ATGGCGTCCACGGATTACAGTACCT-3′; for EWS/ERG (reverse), 5′-TTAGTAGTAAGTGCCCAGATGAGAA-3′; for GAPDH (forward), 5′-CCACCCATGGCAAATTCCATGGCA-3′; and for GAPDH (reverse), 5′-TCTAGACGGCAGGTCAGGT/CCACC-3′. PCR products were electrophoresed with a 1% agarose gel and stained with ethidium bromide.
Real-time RT-PCR.
Real-time RT-PCR was performed using TaqMan universal PCR master mix and TaqMan gene expression assays and an inventoried assay on an ABI Prism 7900HT sequence detection system (Applied Biosystems) according to the manufacturer's instructions. The human GAPDH gene was used as an internal control for normalization.
DNA microarray analysis.
Total RNA isolated from cells was reverse transcribed and labeled using one-cycle target labeling and control reagents as instructed by the manufacturer (Affymetrix). The labeled probes were hybridized to the human genome U133 Plus 2.0 array (Affymetrix). The arrays were performed in a single experiment and analyzed using GeneChip operating software, version 1.2 (Affymetrix). Background subtraction, normalization, and principal component analysis (PCA) were performed by GeneSpring GX 7.3 software (Agilent Technologies). Signal intensities were prenormalized based on the median of all measurements on that chip. To account for the difference in detection efficiencies between the spots, prenormalized signal intensities on each gene were normalized to the median of prenormalized measurements for that gene. The data were filtered using the following steps. (i) Genes that were scored as absent in all samples were eliminated. (ii) Genes for which the signal intensities were lower than 100 were eliminated. (iii) Performing cluster analysis using filtering genes, we selected the genes that exhibited increased expression or decreased expression in tetracycline-treated cells. Accession numbers for the microarray data are given below.
Invasion assay.
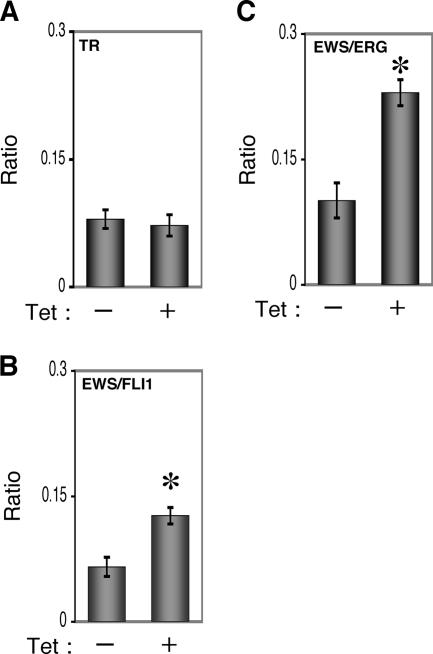
The invasion assay was performed using Matrigel (BD Bioscience) according to the previous description (34) with some modification. Polycarbonate filter inserts containing 8-μm pores (BD Falcon) were coated with 50 μl of a 6:1 mixture of culture medium and Matrigel and placed into 24-well culture plates containing DMEM supplemented with 10% T-FBS as chemoattractants. Cells (2.5 × 104) treated with or without tetracycline for 72 h were suspended in DMEM containing 0.01% T-FBS and plated on top of each filter insert. After 20 h in culture in the presence or absence of tetracycline, noninvading cells were removed from upper surface of the filter with a cotton swab. The invading cells on the lower surface of the filter were fixed with formalin, stained with hematoxylin-eosin, and counted in five fields per membrane with light microscopy. As a control, cells were also cultured on uncoated filter inserts. The invasion efficiency was presented as the ratio of the number of invading cells on Matrigel-coated inserts to that on uncoated inserts. Experiments were performed in triplicate, and the means with standard deviations of the values are shown in the graphs in Fig. 8.
FIG. 8.
Effects of EWS/ETS expression on the Matrigel invasion ability of UET-13 cells. UET-13TR (A), UET-13TR-EWS/FLI1 (B), and UET-13TR-EWS/ERG (C) cells were cultured in the absence or presence of tetracycline (Tet) for 72 h and then plated (2.5 × 104) on Matrigel-coated or uncoated filter inserts. After 20 h of culture, invading cells were stained with hematoxylin-eosin and counted in five fields per membrane as described in Materials and Methods. *, P < 0.05.
Microarray data accession numbers.
Microarray data have been deposited in the Gene Expression Omnibus database GEO (www.ncbi.nlm.nih.gov/geo) (accession numbers GSE8665 and GSE8596).
RESULTS
EWS/ETS expression results in morphological changes in UET-13 cells.
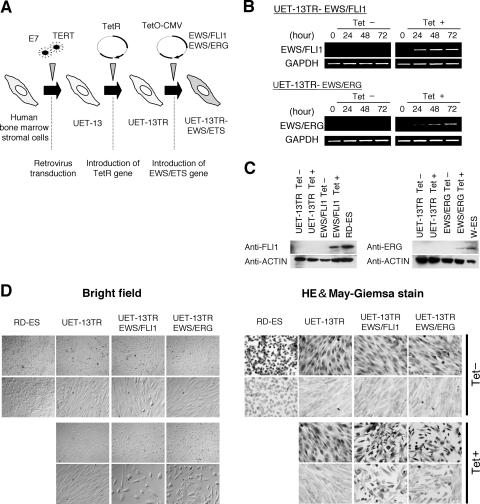
To investigate how the expression of EWS/ETS affects human MPCs, we used UET-13 cells as a model of human MPCs and expressed EWS/FLI1 (UET-13TR-EWS/FLI1) and EWS/ERG (UET-13TR-EWS/ERG) in a tetracycline-inducible manner (Fig. 1A). As shown in Fig. 1B and C, we confirmed that the tetracycline treatment could induce EWS/ETS expression by RT-PCR analysis and Western blotting. The inducibility upon the addition of doxycycline was comparable to that upon the addition of tetracycline.
FIG. 1.
The effect of EWS/ETS on the morphology of UET-13 cells. (A) The establishment of a tetracycline-inducible EWS/ETS expression system in UET-13 cells. CMV, cytomegalovirus. (B) Analyses for confirming the inducible expression of EWS/ETS genes. EWS/ETS mRNAs were detected in UET-13 transfectants UET-13TR-EWS/FLI1 and UET-13TR-EWS/ERG by RT-PCR. These cells were treated with or without 3 μg/ml of tetracycline (Tet) for the indicated periods. As an internal control, a human GAPDH gene was used. (C) Analyses for confirming the inducible expression of EWS/ETS proteins. The cells were treated as described for panel B and subjected to Western blotting for the detection of EWS/ETS proteins. The extracts of RD-ES and W-ES cells were also examined as positive controls. Membranes were reprobed with anti-actin antibody as a loading control. (D) Morphological change after tetracycline treatment of UET-13 transfectants. UET-13 cells and the transfectants were cultured in the absence or presence of tetracycline for 72 h and observed by light microscopy. Magnification, ×40 (top); ×200 (bottom). Cells were also examined using hematoxylin-eosin (HE) (top) and May-Giemsa (bottom) staining (magnification, ×200).
Using these cell systems, first we examined the effect of EWS/ETS expression on morphology in UET-13 transfectants. When tetracycline was added to the culture, the morphologies of both UET-13TR-EWS/FLI1 and UET-13TR-EWS/ERG cells were dramatically changed (Fig. 1D). Tetracycline-treated UET-13TR-EWS/ETS cells consisted of a mixture of small round-to-polygonal cells and short spindle cells. The cell morphology resembled that of EFT cell lines. To assess the reproducibility of this phenotypic change, other UET-13TR-EWS/ETS clones were examined, and similar morphological changes were observed. Since tetracycline treatment did not affect the morphology of UET-13TR cells (Fig. 1D), it was suggested that the morphological alteration in UET-13 cells from a mesenchymal cell shape to small round cells, one of the characteristics of EFT, can be attributed to EWS/ETS expression.
EWS/ETS expression inhibits cell growth in UET-13 cells.
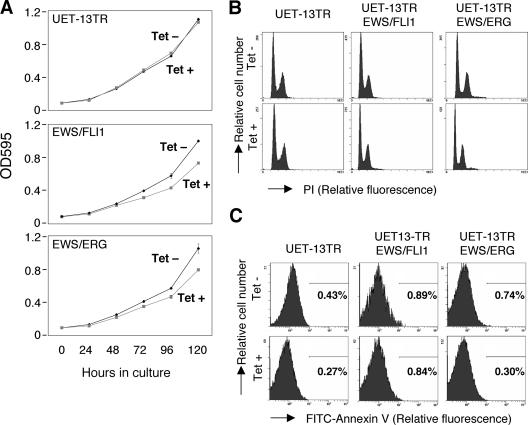
Next, the effect of EWS/ETS expression on the growth of UET-13 cells was analyzed. As shown in Fig. 2A, an MTT assay revealed that the addition of tetracycline had no effect on the growth of UET-13TR cells but slightly inhibited that of UET-13TR-EWS/ETS cells. We also assessed the cell growth of UET-13 transfectants after tetracycline addition by cell counting and obtained results well in accord with those from the MTT assay (data not shown). To determine the mechanism of this inhibition, DNA content and the binding of annexin V to UET-13 transfectants were examined. No significant increase in either sub-G1-phase cells (Fig. 2B) or annexin V binding cells (Fig. 2C) was detected, suggesting that EWS/ETS-mediated growth inhibition in UET-13 cells was not due to the activation of an apoptotic pathway. Moreover, no significant decrease in S-G2-phase cells was observed (Fig. 2B).
FIG. 2.
Effects of EWS/ETS on cell growth in UET-13 cells. (A) Growth curve for UET-13 transfectants. Cells were seeded at 103/well and cultured as described for Fig. 1. The increase in cell number was analyzed by MTT assay. Values are means with the standard errors (SE) from three independent experiments. Diamond symbols indicate UET-13 transfectants in the absence of tetracycline (Tet); box symbols indicate UET-13 transfectants in the presence of tetracycline. (B) Cells were cultured as described for panel A in the absence or presence of tetracycline for 3 days and then stained with PI, and DNA contents were analyzed by flow cytometry (x axis, relative intensity of fluorescence; y axis, relative cell number). (C) Cells treated as described for panel B were stained with FITC-annexin V and analyzed.
Effect of EWS/ETS on CD99 expression in UET-13 cells.
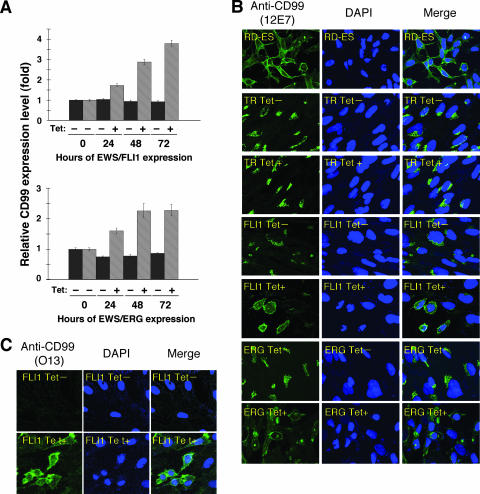
The p30/32MIC-2 gene product, CD99, is a cell surface glycoprotein expressed in EFT with a strong membranous staining pattern and thus constitutes a useful marker for EFT (2, 30). Knowing the dramatic change of morphology in UET-13 cells, we next investigated the mRNA level of CD99 in tetracycline-treated and untreated UET-13 transfectants by quantitative real-time RT-PCR. CD99 levels were clearly elevated by tetracycline treatment in both UET-13TR-EWS/FLI1 and UET-13TR-EWS/ERG cells in a time-dependent manner (Fig. 3A).
FIG. 3.
Effects of tetracycline-mediated EWS/ETS expression on the expression and distribution of CD99 in UET-13 cells. (A) Relative CD99 levels in UET-13 transfectants in the absence or presence of tetracycline (Tet). UET-13 transfectants were treated with or without 3 μg/ml of tetracycline for the indicated periods. Real-time RT-PCR was performed to investigate the expression pattern of CD99. Signal intensities of CD99 were normalized using those of a control housekeeping gene (human GAPDH gene). Data are relative values with standard deviations from triplicate wells and are normalized to the mRNA level at 0 h, which is arbitrarily set to 1 in the graphical presentation. (B and C) Immunocytostaining of CD99 in UET-13 transfectants. Cells were cultured on coverslips in the absence or presence of tetracycline for 72 h and then stained with anti-CD99 antibody 12E7 (B) or O13 (C) as described in Materials and Methods. RD-ES cells were also examined as a positive control. For the staining of nuclei, DAPI was used.
We also examined the protein expression of CD99 by immunostaining using 12E7 antibody, which is most widely used as an anti-CD99 antibody. An EFT cell line, RD-ES, showed strong membranous staining of CD99 (Fig. 3B), while neither UET-13TR cells nor UET-13 cells had such a staining. Of note is the fact that although 12E7 reactivity was observed only in the cytoplasm in perinuclear regions in both UET-13TR (Fig. 3B) and UET-13 (data not shown) cells, this antibody is well known to cross-react with a cytoplasmic protein not yet characterized. Since another anti-CD99 antibody, O13, did not react with either UET-13TR (Fig. 3C) or UET-13 (data not shown) cells, we concluded that the perinuclear staining of 12E7 mentioned above was a cross-reaction with unrelated proteins.
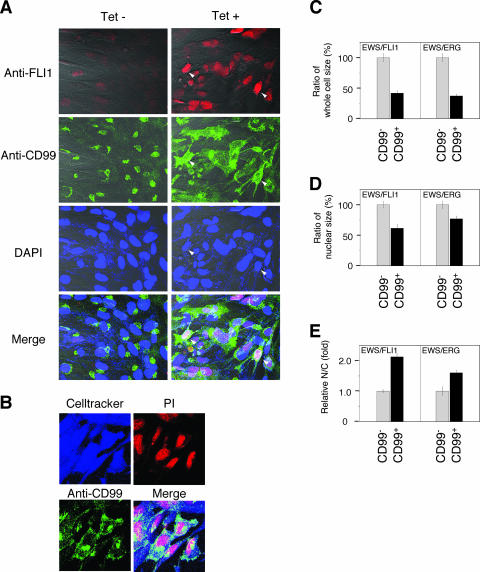
In the absence of tetracycline, both UET-13TR-EWS/FLI1 and UET-13TR-EWS/ERG cells were also negative with anti-CD99 antibodies (a pattern designated CD99−), similar to UET-13 cells. Surprisingly, however, tetracycline induced a membranous staining pattern (designated CD99+) in UET-13TR-EWS/FLI1 and UET-13TR-EWS/ERG cells, and some CD99+ cells had irregularly contoured nuclei (Fig. 3B). The same results were observed with another anti-CD99 antibody, O13 (Fig. 3C), indicating that the membranous staining observed for UET-13 transfectants with the anti-CD99 antibodies was really CD99 derived. Despite the fact that cells were single colony derived, there was a heterogeneous response to tetracycline treatment in UET-13TR-EWS/FLI1 and UET-13TR-EWS/ERG cells, but most of the CD99+ cells had a small round morphology, one of the characteristics of EFT. To assess the correlation between EWS/FLI1 expression and the change of the CD99 expression pattern, we performed immunofluorescence studies using anti-Fli1 and anti-CD99 antibodies. As shown in Fig. 4A, tetracycline treatment induced a marked enhancement of nuclear staining with anti-Fli1 antibodies in a large number of UET-13TR-EWS/FLI1 cells, indicating the induction of EWS/FLI1 proteins. Furthermore, we observed that the cells with a strong signal for Fli1 tended to reveal a membranous staining pattern with anti-CD99 antibodies and a small round morphology (Fig. 4A). To further verify the correlation between CD99 expression pattern and cell morphology, we estimated the size of cells by triple staining using Celltracker Blue, PI, and anti-CD99 antibody (Fig. 4B). As presented in Fig. 4C and D, the results clearly showed that the majority of CD99+ cells were significantly smaller in both whole-cell size and nuclear size than the CD99− cells. Moreover, CD99+ cells also had a substantially increased N/C ratio (Fig. 4E). These results indicated that EWS/ETS expression promoted CD99 expression in UET-13 cells, and CD99 expression status is correlated with the degree of morphological change.
FIG. 4.
EWS/ETS expression, alteration of CD99 distribution, and cell morphological changes in UET-13 cells. (A) Immunofluorescence studies using anti-Fli1 (red), anti-CD99 (green), and DAPI (blue). UET-13TR-EWS/FLI1 cells were cultured on coverslips in the absence or presence of tetracycline (Tet) for 72 h and then stained as described in Materials and Methods. White arrowheads indicate CD99+ cells that have a strong staining pattern with anti-Fli1 antibodies and also have remarkable CD99 expression and morphological features. (B) Immunofluorescence analysis by triple staining with whole cells (Celltracker; blue), CD99 (anti-CD99; green), and nuclei (PI; red). UET-13TR-EWS/FLI1 cells were cultured as described for panel A and then stained as described in Materials and Methods. (C to E) Measurements of whole-cell size (C), nuclear size (D), and N/C ratio (E) in tetracycline-treated UET-13 transfectants. UET-13TR-EWS/FLI1 and UET-13TR-EWS/ERG cells were cultured on coverslips in the presence of tetracycline for 72 h and then stained as described in Materials and Methods. These samples were analyzed by the image analysis software Image J (n = 50). (C and D) Data are relative values with the SE and are normalized to the size of CD99− cells, which is arbitrarily set to 100. (E) Data are relative values with the SE and are normalized to the size of CD99− cells, which is arbitrarily set to 1.
EWS/ETS expression altered the immunophenotype of UET-13 cells.
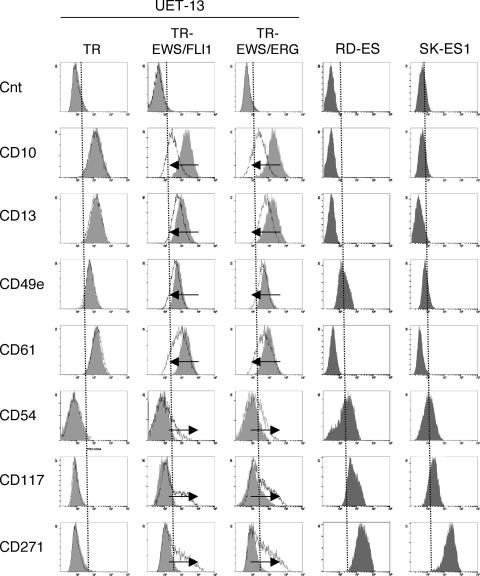
Human MPCs reveal a characteristic expression of several surface antigens and can be identified on the basis of the reactivity with a set of monoclonal antibodies against CD antigens (25, 42). On the other hand, some CD antigens are characteristically expressed on EFT cells (17, 28, 33). Using the combinations of these antibodies listed in Table 2, which are useful for the immunodetection of either MPCs or EFT cells, we further examined whether EWS/ETS expression affects the immunophenotype of UET-13 cells and compared its effect with that on the immunophenotype of EFT cell lines (Table 2 and Fig. 5). As shown in Table 2, UET-13 cells express most of the human primary MPCs markers. Some of the antigens expressed in MPCs, namely, CD29, CD59, CD90, CD105, and CD166, were also found to be expressed in EFT cell lines, but others, namely, CD10, CD13, CD44, CD61, and CD73, were not. In contrast, antigens recognized to be present in EFT cells, including CD40, CD56, and CD133, were absent from UET-13 cells. Interestingly, when the effect of tetracycline-mediated EWS/ETS expression on the immunophenotype of UET-13 cells was tested, levels of some of the antigens present in UET-13 cells, such as CD10, CD13, and CD61, were found to be decreased (Fig. 5). In contrast, some of the markers found in EFT cells, i.e., CD54, CD117, and CD271, became positive in UET-13TR-EWS/ETS cells after tetracycline treatment. Because UET-13TR cells did not show such immunophenotypic change upon treatment with tetracycline, these results indicated that, at least in part, the immunophenotype of UET-13 cells was changed in favor of EFT in the presence of EWS/ETS.
TABLE 2.
Immunophenotypic characterization of UET-13 transfectants and EFT cells
| MPC statusa | CD marker | Result forb:
|
EFT statusc | SK-ES1 | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| UET-13
|
UET-13TR
|
UET-13TR- EWS/FLI1
|
UET-13TR- EWS/ERG
|
RD-ES | |||||||
| Tet− | Tet+ | Tet− | Tet+ | Tet− | Tet+ | ||||||
| M+ | CD29 | + | + | + | + | + | + | + | + | + | |
| M+ | CD59 | + | + | + | + | + | + | + | + | + | |
| M+ | CD90 | + | + | + | + | + | + | + | + | + | E+ |
| M+ | CD105 | + | + | + | + | + | + | + | + | + | |
| M+ | CD166 | + | + | + | + | + | + | + | + | + | |
| M+ | CD44 | + | + | + | + | + | + | + | − | − | |
| M+ | CD73 | + | + | + | + | + | + | + | − | − | |
| M+ | CD10 | + | + | + | + | Down | + | Down | − | − | |
| M+ | CD13 | + | + | + | + | Down | + | Down | − | − | |
| M+ | CD49e | + | + | + | + | Down | + | Down | + | − | |
| M+ | CD61 | + | + | + | + | Down | + | Down | − | − | |
| M+ | CD55 | + | + | + | + | Down | + | + | + | − | |
| M+ | CD54 | − | − | − | − | Up | − | Up | + | + | E+ |
| M(−) | CD117 | − | − | − | − | Up | − | Up | + | + | E+ |
| M+/− | CD271 | − | − | − | − | Up | − | Up | + | + | E+ |
| CD40 | − | − | − | − | − | − | − | + | + | E+ | |
| CD56 | − | − | − | − | − | − | − | + | + | E+ | |
| M(−) | CD133 | − | − | − | − | − | − | − | + | + | |
| M(−) | CD14 | − | − | − | − | − | − | − | − | − | |
| M(−) | CD34 | − | − | − | − | − | − | − | − | − | |
| M(−) | CD45 | − | − | − | − | − | − | − | − | − | |
M(−), negative for MPCs; M+/−, positive for BM-derived MPCs but negative after in vitro culture; M+, positive for MPCs.
+, most cells positive; −, negative; Up, up-regulated by tetracycline treatment; Down, down-regulated by tetracycline treatment. Boldface indicates the antigens the immunophenotypes of which were changed in favor of EFT. Tet−, tetracycline negative; Tet+, tetracycline positive.
E+, positive for EFTs.
FIG. 5.
Immunophenotypic change on induction of EWS/ETS expression in UET-13 cells. UET-13 transfectants were cultured with or without 3 μg/ml of tetracycline for 1 week and flow cytometric analyses were performed by using a set of antibodies as indicated. The histograms of UET-13 transfectants with (empty) and without (gray) tetracycline treatment were overlaid. Dotted lines indicate fluorescence intensities in negative control panels (Cnt). Arrows indicate the immunophenotypic change caused by tetracycline. The immunophenotypes of the EFT cell lines RD-ES and SK-ES1 were also examined.
EWS/ETS in UET-13 cells modulates EFT-like gene expression.
To further examine the molecular mechanism of EWS/ETS-dependent cellular modulation in human mesenchymal progenitor background, we performed DNA microarray-based expression profiling using the Affymetrix human genome U133 Plus 2.0 array. As a first step to this approach, we validated our experimental systems by analyzing the sequential changes of known EWS/ETS target genes, i.e., inhibitor of differentiation 2 (ID2) (14, 39), NK2 transcription factor related, locus 2 (NKX2.2) (9, 48), and insulin-like growth factor binding protein 3 (IGFBP3) (41). Consistent with previous reports, levels of ID2 and NKX2.2 increased with the expression of EWS/ETS in a time-dependent manner, whereas the expression level of IGFBP3 decreased (Fig. 6A). Employing the same procedure, we also examined whether the change of surface antigen expression was regulated at the transcriptional level and determined the mRNA expression levels of some surface antigens in UET-13 transfectants with or without tetracycline treatment. In accordance with the results of immunocytometric and immunohistological experiments, the mRNA expression levels of CD10, CD13, CD49e, and CD61 were decreased, while those of CD54, CD99, CD117, and CD271 were markedly increased in tetracycline-treated UET-13TR-EWS/ETS cells (Fig. 6B and C), indicating that the expression of these antigens is controlled at the transcriptional level in the presence of EWS/ETS.
FIG. 6.
The change of expression profile on induction of EWS/ETS in UET-13 cells. UET-13TR-EWS/FLI1 and UET-13TR-EWS/ERG cells were cultured in the absence or presence of tetracycline (Tet) for the indicated periods and analyzed using the Affymetrix human genome U133 Plus 2.0 array as described in Materials and Methods. (A) The sequential changes of ID2, NKX2.2, and IGFBP3 mRNA levels in UET-13 transfectants upon treatment with or without tetracycline. Diamond symbols indicate UET-13 transfectants in the absence of tetracycline; box symbols indicate UET-13 transfectants in the presence of tetracycline. (B and C) Microarray studies for the determination of expression profiles of surface antigens in UET-13 transfectants. UET-13 transfectants were treated with or without 3 μg/ml of tetracycline for 72 h. mRNA levels were determined with the Affymetrix human genome U133 Plus 2.0 array.
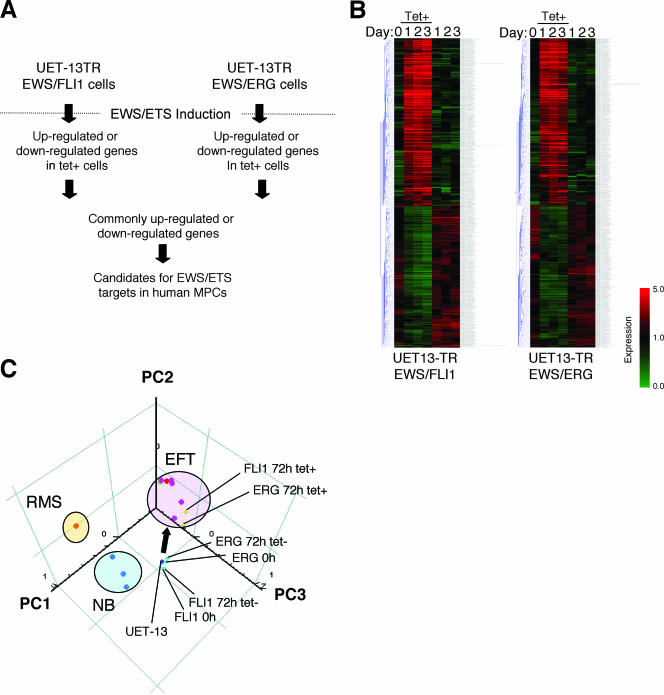
We next investigated the candidate genes whose expression is regulated by EWS/ETS in human MPCs. First, we selected the genes with up-regulated or down-regulated expression by EWS/ETS induction using gene cluster analysis (Fig. 7A; UET-13TR-EWS/FLI1 up, 4,294 probes; down, 4,103 probes; UET-13TR-EWS/ERG up, 3,358 probes; down, 3,705 probes). To reduce the number of the candidate genes, we selected up-regulated genes that are expressed in tetracycline-treated cells at least 1.5-fold higher than in untreated cells (UET-13TR-EWS/FLI1, 1,137 probes; UET-13TR-EWS/ERG, 835 probes). Similarly, the down-regulated genes that are expressed in tetracycline-treated cells at least 0.75-fold lower than in untreated cells (UET-13TR-EWS/FLI1, 1,803 probes; UET-13TR-EWS/ERG, 773 probes). By selecting common probes in both cells, we finally identified a group of candidate genes significantly controlled by EWS/ETS induction in the human mesenchymal progenitor background. Since microarray analysis was performed as a global screening in a single experiment, it is likely that there is a fair bit of noise in the derived gene profiles due to the lack of replicate data. This may account in part for the limited overlap between the profiles induced by EWS-FLI1 and EWS-ERG, whereas we still identified 349 probes of common up-regulated genes and 293 probes of common down-regulated genes (see the supplemental material). In addition to the EFT-specific genes mentioned above, these contained those previously described as EFT-specific genes, such as those for OB-cadherin/cadherin-11 (31), Janus kinase 1 (JAK1) (49), keratin 18, and six-transmembrane epithelial antigen of the prostate (STEAP) (22). The expression pattern of these genes (642 probes) in UET-13 transfectants in the absence or presence of tetracycline is shown in the gene cluster in Fig. 7B. The expression of these genes was indeed changed significantly after EWS/ETS expression in both cells. They included genes associated with signal transduction (such as those for epidermal growth factor receptor, FAS [CD95], and fibroblast growth factor receptor 1) and development (such as jagged-1 and frizzled-4, -7, and -8). Interestingly, in addition to the surface antigens presented in Fig. 6B and C, the expression profiling of EWS/ETS-expressing UET-13 cells displayed the modulation of several genes associated with cell adhesion, cytoskeletal structure, and membrane trafficking, such as those for collagen-11 and -21, ephrin receptor-A2, -B2, and -B3, ephrin-B1, claudin-1, integrin-α11, -αM, and -β2, CD66 (carcinoembryonic antigen-related cell adhesion molecule-1), and CD102 (intercellular cell adhesion molecule-2). They also included genes of chemokines CCL-2 and -3. These data raise the possibility that EWS/ETS can contribute to the membrane condition in human MPCs via the regulation of these cell surface molecules and chemokines.
FIG. 7.
Identification of candidates for the target of EWS/ETS in human MPCs by use of a microarray. UET-13TR-EWS/FLI1 and UET-13TR-EWS/ERG cells were cultured as described for Fig. 6 and analyzed using the Affymetrix human genome U133 Plus 2.0 array as described in Materials and Methods. (A) Scheme for the analysis of microarray data. (B) Gene cluster analysis of UET-13 transfectants in the absence or presence of tetracycline by use of 642 candidate genes for targets of EWS/ETS in human MPCs. (C) Visualization of sequential change by the gene expression profile in UET-13 transfectants following tetracycline-mediated EWS/ETS expression based on a PCA of 642 candidate genes. Deep blue plots indicate UET-13 cells. Light blue plots indicate UET-13 transfectants in the absence of tetracycline for 72 h. Yellow plots indicate UET-13 transfectants in the presence of tetracycline for 72 h. The pink circle indicates EFT cell lines expressing EWS/FLI1 (purple plots), EWS/ERG (red plot), and EWS/E1AF (light green plot). The light blue circle with blue plots indicates NB cell lines. The yellow circle with an orange plot indicates a rhabdomyosarcoma (RMS) cell line. Cutoff induction and repression levels are 1.5-fold and 0.75-fold, respectively. Tet, tetracycline.
Using these genes, we performed a PCA to visualize the shift in the gene expression pattern among the 642 probes. As shown in Fig. 7C, the plots of UET-13 transfectants treated with tetracycline became closer to those of EFT cells than to those of UET-13 transfectants without tetracycline treatment. These results indicated that the expression pattern of these genes was altered from that of UET-13 cells to that of EFT cells in an EWS/ETS-dependent manner. Since the gene expression profile of UET-13 cells is similar to those of other cell types of mesenchymal origin (data not shown), our results highlighted that the phenotypic alteration from mesenchyme to EFT-like cells in UET-13 cells induced by tetracycline treatment was accompanied by a change in the global gene expression profile.
EWS/ETS expression enhances the Matrigel invasion of UET-13 cells.
To assess the role of EWS/ETS in malignant transformation in human MPCs, UET-13 transfectants were examined by invasion assay. As shown in Fig. 8A, tetracycline treatment did not affect the Matrigel invasion ability of UET-13TR cells. When examined similarly, however, tetracycline treatment resulted in an apparently increased invasion (P < 0.05) for both UET-13TR-EWS/FLI1 (Fig. 8B) and UET-13TR-EWS/ERG (Fig. 8C) cells. The results indicated that EWS/ETS expression can induce Matrigel invasion properties in human MPCs.
DISCUSSION
In the present study, using UET-13 cells as a model of human MPCs, we demonstrated that ectopic expression of EWS/ETS promoted the acquisition of an EFT-like phenotype, including cellular morphology, immunophenotype, and gene expression profile. Moreover, EWS/ETS expression enhances the ability of UET-13 cells to invade Matrigel. This assay is thought to mimic the early steps of tumor invasion in vivo (34), and the ability to penetrate the Matrigel has been positively correlated with invasion potential in several studies. Therefore, we concluded that EWS/ETS expression could mediate a part of the feature of tumor transformation in human MPCs. Thus, our culture system would provide a good model for testing the effects of EWS/ETS in human MPCs.
Several lines of evidence have indicated the transforming ability of EWS/FLI1, whereas that of EWS/ERG is not yet to be clarified. Therefore, it is noteworthy that our data demonstrated that EWS/ERG could promote an EFT-like phenotype in UET-13 cells similarly to EWS/FLI1. Thus, EWS/ERG also has the ability to induce an EFT-like phenotype in the human system. The major steps in the development of EFT should be commonly regulated by distinct chimeric EWS/ETS proteins. Indeed, several genes are common transcriptional targets of different chimeric EWS/ETS proteins in the murine system (11, 24, 35). Our data also showed that the 642 probes are coregulated in both EWS/FLI1-expressing cells and EWS/ERG-expressing cells. Further comparative studies of both the EWS/FLI1- and the EWS/ERG-mediated onset of EFT could allow us to understand the common functions of EWS/FLI1 and EWS/ERG in EFT. In addition, our systems are also useful for precisely distinguishing between the functions of these chimeric molecules in the development of EFT.
As mentioned above, the immunophenotypic analysis also revealed that the expression profiles of surface antigens in UET-13 cells were changed in favor of EFT cells in the presence of EWS/ETS (Fig. 4). Notably, the expression of CD54 (intercellular cell adhesion molecule-1 [ICAM1]), CD117 (c-kit), and CD271 (low-affinity nerve growth factor receptor [LNGFR]) increased in EWS/ETS-expressing UET-13 cells. These markers are positive in EFT cell lines (17, 28, 33), and in addition, CD117 is detected in about 40% of patient samples (17) and is negative in human primary MPCs (4, 43). Thus, it is reasonable to consider that a phenotypic marker of EFT was induced in UET-13 cells by EWS/ETS expression. On the other hand, CD54 and CD271 are positive in human primary MPCs (8, 25, 42), whereas these markers are negative in UET-13 cells. However, a previous report showed the disappearance of some positive markers, including CD271, from primary human MPCs during the process of ex vivo expansion (25), and it has been speculated that the expression of these molecules in MPCs is induced in vivo via interaction with the bone marrow microenvironment and that the necessary stimuli are absent from ex vivo culture conditions. Therefore, the immunophenotype of UET-13 cells rather might be related to that of ex vivo-expanded primary human MPCs. In addition, it may be possible that EWS/ETS expression led to the reexpression of these disappeared markers in UET-13 cells without the necessary stimuli. In this case, the maintenance of CD271 expression outside of the bone marrow microenvironment might be a characteristic of EFT. Thus, our results proved that both EWS/FLI1 and EWS/ERG can be major causes of the expression of these markers and that human MPCs that precisely recapitulate the expression are strong candidates for the cell origins of EFT cells. The findings also imply that these antigens are suitable targets for diagnostic tools and new therapeutic agents. In fact, imatinib mesylate, which demonstrates anticancer activity against malignant cells expressing BCR-ABL as well as CD117 and platelet-derived growth factor receptor, inhibits proliferation and increases sensitivity to vincristine and doxorubicin in EFT cells (17).
Notably, our results also indicate that UET-13 cells, which have the MPC phenotype, possess the potential to acquire an EFT-like phenotype upon the expression of EWS/ETS. Unlike what is seen for human primary fibroblasts (31), ectopic EWS/ETS expression induces an EFT-like morphological change in human MPCs, suggesting that the cell type affects susceptibility to the events following EWS/ETS expression. In murine MPCs, retrovirally transduced EWS/FLI1 has been reported to induce the expression of CD99, a most useful marker for EFT, though the results are controversial (6, 45). However, our direct evidence obtained with UET-13 cells clearly demonstrated that CD99 expression is induced by EWS/ETS proteins in human MPCs. Moreover, we showed that the expression of CD99 might correlate with EWS/ETS-mediated morphological change, whereas the functional role of CD99 and the correlation between CD99 expression status and EWS/ETS-mediated morphological change in the development of EFT remain unclarified.
Consistent with the morphological and immunophenotypic changes, the expression pattern of a set of genes in EWS/ETS-expressing UET-13 cells shifted to that in EFT cells (Fig. 7C). Although EWS/ETS expression enhanced the ability of UET-13 cells to invade Matrigel, it did not promote migratory ability and surface-independent growth, as assessed by migration assay and soft agar colony formation assay (data not shown). We also failed to develop EFT-like tumors by injecting EWS/ETS-inducing UET-13 cells into irradiated nude mice treated with tetracycline (data not shown). These results imply that EWS/ETS expression is not sufficient to induce the full transformation in UET-13 cells, and other genetic abnormalities not regulated by EWS/ETS could still be required for the full transformation of human MPCs into EFT cells. An identification of these genes will greatly improve our understanding of the additional genetic lesions that occur after EWS/ETS expression. The genes expressed in EFT cell lines but not in EWS/ETS-expressing UET-13 cells would be candidates for such genes.
In summary, we reported the development of an inducible EWS/ETS expression system in UET-13 cells as a model for the development of EFT in MPCs. In our system, the chimeric genes alone are sufficient to confer EFT-like phenotypes, EFT-specific gene expression pattern, and partial but not full features of malignant transformation. Further analysis using our system should elucidate the pathogenic mechanism by which EFTs develop from MPCs, especially the initiating events mediated by EWS/ETS expression. Our system should also aid in the identification of novel targets of the EWS/ETS-mediated pathway as potential anticancer targets.
Supplementary Material
Acknowledgments
This work was supported in part by health and labor sciences research grants (the 3rd-Term Comprehensive 10-Year Strategy for Cancer Control [H19-010], Research on Children and Families [H18-005 and H19-003], Research on Human Genome Tailor Made, and Research on Publicly Essential Drugs and Medical Devices [H18-005[) and a grant for child health and development from the Ministry of Health, Labor and Welfare of Japan, JSPS (Kakenhi 18790263). This work was also supported by a CREST, JST grant from the Japan Health Sciences Foundation for Research on Publicly Essential Drugs and Medical Devices and the Budget for Nuclear Research of the Ministry of Education, Culture, Sports, Science and Technology, based on screening and counseling by the Atomic Energy Commission. Y. Miyagawa is an awardee of a research resident fellowship from the Foundation for Promotion of Cancer Research (Japan) for the 3rd-Term Comprehensive 10-Year Strategy for Cancer Control.
We are grateful to T. Motoyama for the NRS-1 cell line. We respectfully thank S. Yamauchi for her secretarial work and M. Itagaki for many helpful discussions and support.
Footnotes
Published ahead of print on 22 January 2008.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Akagi, T. 2004. Oncogenic transformation of human cells: shortcomings of rodent model systems. Trends Mol. Med. 10542-548. [DOI] [PubMed] [Google Scholar]
- 2.Ambros, I. M., P. F. Ambros, S. Strehl, H. Kovar, H. Gadner, and M. Salzer-Kuntschik. 1991. MIC2 is a specific marker for Ewing's sarcoma and peripheral primitive neuroectodermal tumors. Evidence for a common histogenesis of Ewing's sarcoma and peripheral primitive neuroectodermal tumors from MIC2 expression and specific chromosome aberration. Cancer 671886-1893. [DOI] [PubMed] [Google Scholar]
- 3.Arvand, A., and C. T. Denny. 2001. Biology of EWS/ETS fusions in Ewing's family tumors. Oncogene 205747-5754. [DOI] [PubMed] [Google Scholar]
- 4.Bertani, N., P. Malatesta, G. Volpi, P. Sonego, and R. Perris. 2005. Neurogenic potential of human mesenchymal stem cells revisited: analysis by immunostaining, time-lapse video and microarray. J. Cell Sci. 1183925-3936. [DOI] [PubMed] [Google Scholar]
- 5.Bloom, E. T. 1972. Further definition by cytotoxicity tests of cell surface antigens of human sarcomas in culture. Cancer Res. 32960-967. [PubMed] [Google Scholar]
- 6.Castillero-Trejo, Y., S. Eliazer, L. Xiang, J. A. Richardson, and R. L. Ilaria, Jr. 2005. Expression of the EWS/FLI-1 oncogene in murine primary bone-derived cells results in EWS/FLI-1-dependent, Ewing sarcoma-like tumors. Cancer Res. 658698-8705. [DOI] [PubMed] [Google Scholar]
- 7.Colter, D. C., I. Sekiya, and D. J. Prockop. 2001. Identification of a subpopulation of rapidly self-renewing and multipotential adult stem cells in colonies of human marrow stromal cells. Proc. Natl. Acad. Sci. USA 987841-7845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Conget, P. A., and J. J. Minguell. 1999. Phenotypical and functional properties of human bone marrow mesenchymal progenitor cells. J. Cell. Physiol. 18167-73. [DOI] [PubMed] [Google Scholar]
- 9.Davis, S., and P. S. Meltzer. 2006. Ewing's sarcoma: general insights from a rare model. Cancer Cell 9331-332. [DOI] [PubMed] [Google Scholar]
- 10.Deneen, B., and C. T. Denny. 2001. Loss of p16 pathways stabilizes EWS/FLI1 expression and complements EWS/FLI1 mediated transformation. Oncogene 206731-6741. [DOI] [PubMed] [Google Scholar]
- 11.Deneen, B., S. M. Welford, T. Ho, F. Hernandez, I. Kurland, and C. T. Denny. 2003. PIM3 proto-oncogene kinase is a common transcriptional target of divergent EWS/ETS oncoproteins. Mol. Cell. Biol. 233897-3908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eliazer, S., J. Spencer, D. Ye, E. Olson, and R. L. Ilaria, Jr. 2003. Alteration of mesodermal cell differentiation by EWS/FLI-1, the oncogene implicated in Ewing's sarcoma. Mol. Cell. Biol. 23482-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fujii, Y., Y. Nakagawa, T. Hongo, Y. Igarashi, Y. Naito, and M. Maeda. 1989. Cell line of small round cell tumor originating in the chest wall: W-ES. Hum. Cell 2190-191. (In Japanese.) [PubMed] [Google Scholar]
- 14.Fukuma, M., H. Okita, J. Hata, and A. Umezawa. 2003. Upregulation of Id2, an oncogenic helix-loop-helix protein, is mediated by the chimeric EWS/ets protein in Ewing sarcoma. Oncogene 221-9. [DOI] [PubMed] [Google Scholar]
- 15.Gilbert, F., G. Balaban, P. Moorhead, D. Bianchi, and H. Schlesinger. 1982. Abnormalities of chromosome 1p in human neuroblastoma tumors and cell lines. Cancer Genet. Cytogenet. 733-42. [DOI] [PubMed] [Google Scholar]
- 16.Girish, V., and A. Vijayalakshmi. 2004. Affordable image analysis using NIH Image/ImageJ. Indian J. Cancer 4147. [PubMed] [Google Scholar]
- 17.Gonzalez, I., E. J. Andreu, A. Panizo, S. Inoges, A. Fontalba, J. L. Fernandez-Luna, M. Gaboli, L. Sierrasesumaga, S. Martin-Algarra, J. Pardo, F. Prosper, and E. de Alava. 2004. Imatinib inhibits proliferation of Ewing tumor cells mediated by the stem cell factor/KIT receptor pathway, and sensitizes cells to vincristine and doxorubicin-induced apoptosis. Clin. Cancer Res. 10751-761. [DOI] [PubMed] [Google Scholar]
- 18.Hansen, M. B., S. E. Nielsen, and K. Berg. 1989. Re-examination and further development of a precise and rapid dye method for measuring cell growth/cell kill. J. Immunol. Methods 119203-210. [DOI] [PubMed] [Google Scholar]
- 19.Hara, S., E. Ishii, S. Tanaka, J. Yokoyama, K. Katsumata, J. Fujimoto, and J. Hata. 1989. A monoclonal antibody specifically reactive with Ewing's sarcoma. Br J. Cancer 60875-879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hatori, M., H. Doi, M. Watanabe, H. Sasano, M. Hosaka, S. Kotajima, F. Urano, J. Hata, and S. Kokubun. 2006. Establishment and characterization of a clonal human extraskeletal Ewing's sarcoma cell line, EES1. Tohoku J. Exp. Med. 210221-230. [DOI] [PubMed] [Google Scholar]
- 21.Homma, C., Y. Kaneko, K. Sekine, S. Hara, J. Hata, and M. Sakurai. 1989. Establishment and characterization of a small round cell sarcoma cell line, SCCH-196, with t(11;22)(q24;q12). Jpn. J. Cancer Res. 80861-865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hubert, R. S., I. Vivanco, E. Chen, S. Rastegar, K. Leong, S. C. Mitchell, R. Madraswala, Y. Zhou, J. Kuo, A. B. Raitano, A. Jakobovits, D. C. Saffran, and D. E. Afar. 1999. STEAP: a prostate-specific cell-surface antigen highly expressed in human prostate tumors. Proc. Natl. Acad. Sci. USA 9614523-14528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu-Lieskovan, S., J. Zhang, L. Wu, H. Shimada, D. E. Schofield, and T. J. Triche. 2005. EWS-FLI1 fusion protein up-regulates critical genes in neural crest development and is responsible for the observed phenotype of Ewing's family of tumors. Cancer Res. 654633-4644. [DOI] [PubMed] [Google Scholar]
- 24.Im, Y. H., H. T. Kim, C. Lee, D. Poulin, S. Welford, P. H. Sorensen, C. T. Denny, and S. J. Kim. 2000. EWS-FLI1, EWS-ERG, and EWS-ETV1 oncoproteins of Ewing tumor family all suppress transcription of transforming growth factor beta type II receptor gene. Cancer Res. 601536-1540. [PubMed] [Google Scholar]
- 25.Jones, E. A., S. E. Kinsey, A. English, R. A. Jones, L. Straszynski, D. M. Meredith, A. F. Markham, A. Jack, P. Emery, and D. McGonagle. 2002. Isolation and characterization of bone marrow multipotential mesenchymal progenitor cells. Arthritis Rheum. 463349-3360. [DOI] [PubMed] [Google Scholar]
- 26.Khoury, J. D. 2005. Ewing sarcoma family of tumors. Adv. Anat. Pathol. 12212-220. [DOI] [PubMed] [Google Scholar]
- 27.Kiyokawa, N., Y. Kokai, K. Ishimoto, H. Fujita, J. Fujimoto, and J. I. Hata. 1990. Characterization of the common acute lymphoblastic leukaemia antigen (CD10) as an activation molecule on mature human B cells. Clin. Exp. Immunol. 79322-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Konemann, S., T. Bolling, A. Schuck, J. Malath, A. Kolkmeyer, K. Horn, D. Riesenbeck, S. Hesselmann, R. Diallo, J. Vormoor, and N. A. Willich. 2003. Effect of radiation on Ewing tumour subpopulations characterized on a single-cell level: intracellular cytokine, immunophenotypic, DNA and apoptotic profile. Int. J. Radiat. Biol. 79181-192. [DOI] [PubMed] [Google Scholar]
- 29.Kovar, H., and A. Bernard. 2006. CD99-positive “Ewing's sarcoma” from mouse bone marrow-derived mesenchymal progenitor cells? Cancer Res. 669786. [DOI] [PubMed] [Google Scholar]
- 30.Kovar, H., M. Dworzak, S. Strehl, E. Schnell, I. M. Ambros, P. F. Ambros, and H. Gadner. 1990. Overexpression of the pseudoautosomal gene MIC2 in Ewing's sarcoma and peripheral primitive neuroectodermal tumor. Oncogene 51067-1070. [PubMed] [Google Scholar]
- 31.Lessnick, S. L., C. S. Dacwag, and T. R. Golub. 2002. The Ewing's sarcoma oncoprotein EWS/FLI induces a p53-dependent growth arrest in primary human fibroblasts. Cancer Cell 1393-401. [DOI] [PubMed] [Google Scholar]
- 32.Lin, P. P., R. I. Brody, A. C. Hamelin, J. E. Bradner, J. H. Healey, and M. Ladanyi. 1999. Differential transactivation by alternative EWS-FLI1 fusion proteins correlates with clinical heterogeneity in Ewing's sarcoma. Cancer Res. 591428-1432. [PubMed] [Google Scholar]
- 33.Lipinski, M., K. Braham, I. Philip, J. Wiels, T. Philip, C. Goridis, G. M. Lenoir, and T. Tursz. 1987. Neuroectoderm-associated antigens on Ewing's sarcoma cell lines. Cancer Res. 47183-187. [PubMed] [Google Scholar]
- 34.Lochter, A., A. Srebrow, C. J. Sympson, N. Terracio, Z. Werb, and M. J. Bissell. 1997. Misregulation of stromelysin-1 expression in mouse mammary tumor cells accompanies acquisition of stromelysin-1-dependent invasive properties. J. Biol. Chem. 2725007-5015. [DOI] [PubMed] [Google Scholar]
- 35.May, W. A., A. Arvand, A. D. Thompson, B. S. Braun, M. Wright, and C. T. Denny. 1997. EWS/FLI1-induced manic fringe renders NIH 3T3 cells tumorigenic. Nat. Genet. 17495-497. [DOI] [PubMed] [Google Scholar]
- 36.May, W. A., S. L. Lessnick, B. S. Braun, M. Klemsz, B. C. Lewis, L. B. Lunsford, R. Hromas, and C. T. Denny. 1993. The Ewing's sarcoma EWS/FLI-1 fusion gene encodes a more potent transcriptional activator and is a more powerful transforming gene than FLI-1. Mol. Cell. Biol. 137393-7398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miyagawa, Y., J. M. Lee, T. Maeda, K. Koga, Y. Kawaguchi, and T. Kusakabe. 2005. Differential expression of a Bombyx mori AHA1 homologue during spermatogenesis. Insect Mol. Biol. 14245-253. [DOI] [PubMed] [Google Scholar]
- 38.Mori, T., T. Kiyono, H. Imabayashi, Y. Takeda, K. Tsuchiya, S. Miyoshi, H. Makino, K. Matsumoto, H. Saito, S. Ogawa, M. Sakamoto, J. Hata, and A. Umezawa. 2005. Combination of hTERT and bmi-1, E6, or E7 induces prolongation of the life span of bone marrow stromal cells from an elderly donor without affecting their neurogenic potential. Mol. Cell. Biol. 255183-5195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nishimori, H., Y. Sasaki, K. Yoshida, H. Irifune, H. Zembutsu, T. Tanaka, T. Aoyama, T. Hosaka, S. Kawaguchi, T. Wada, J. Hata, J. Toguchida, Y. Nakamura, and T. Tokino. 2002. The Id2 gene is a novel target of transcriptional activation by EWS-ETS fusion proteins in Ewing family tumors. Oncogene 218302-8309. [DOI] [PubMed] [Google Scholar]
- 40.Ogose, A., T. Motoyama, T. Hotta, and H. Watanabe. 1995. In vitro differentiation and proliferation in a newly established human rhabdomyosarcoma cell line. Virchows Arch. 426385-391. [DOI] [PubMed] [Google Scholar]
- 41.Prieur, A., F. Tirode, P. Cohen, and O. Delattre. 2004. EWS/FLI-1 silencing and gene profiling of Ewing cells reveal downstream oncogenic pathways and a crucial role for repression of insulin-like growth factor binding protein 3. Mol. Cell. Biol. 247275-7283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Quirici, N., D. Soligo, P. Bossolasco, F. Servida, C. Lumini, and G. L. Deliliers. 2002. Isolation of bone marrow mesenchymal stem cells by anti-nerve growth factor receptor antibodies. Exp. Hematol. 30783-791. [DOI] [PubMed] [Google Scholar]
- 43.Reyes, M., T. Lund, T. Lenvik, D. Aguiar, L. Koodie, and C. M. Verfaillie. 2001. Purification and ex vivo expansion of postnatal human marrow mesodermal progenitor cells. Blood 982615-2625. [DOI] [PubMed] [Google Scholar]
- 44.Reyes, M., and C. M. Verfaillie. 2001. Characterization of multipotent adult progenitor cells, a subpopulation of mesenchymal stem cells. Ann. N. Y. Acad. Sci. 938231-235. [DOI] [PubMed] [Google Scholar]
- 45.Riggi, N., L. Cironi, P. Provero, M. L. Suva, K. Kaloulis, C. Garcia-Echeverria, F. Hoffmann, A. Trumpp, and I. Stamenkovic. 2005. Development of Ewing's sarcoma from primary bone marrow-derived mesenchymal progenitor cells. Cancer Res. 6511459-11468. [DOI] [PubMed] [Google Scholar]
- 46.Riggi, N., M. L. Suva, and I. Stamenkovic. 2006. Ewing's sarcoma-like tumors originate from EWS-FLI-1-expressing mesenchymal progenitor cells. Cancer Res. 669786.17018640 [Google Scholar]
- 47.Sekiguchi, M., T. Oota, K. Sakakibara, N. Inui, and G. Fujii. 1979. Establishment and characterization of a human neuroblastoma cell line in tissue culture. Jpn. J. Exp. Med. 4967-83. [PubMed] [Google Scholar]
- 48.Smith, R., L. A. Owen, D. J. Trem, J. S. Wong, J. S. Whangbo, T. R. Golub, and S. L. Lessnick. 2006. Expression profiling of EWS/FLI identifies NKX2.2 as a critical target gene in Ewing's sarcoma. Cancer Cell 9405-416. [DOI] [PubMed] [Google Scholar]
- 49.Staege, M. S., C. Hutter, I. Neumann, S. Foja, U. E. Hattenhorst, G. Hansen, D. Afar, and S. E. Burdach. 2004. DNA microarrays reveal relationship of Ewing family tumors to both endothelial and fetal neural crest-derived cells and define novel targets. Cancer Res. 648213-8221. [DOI] [PubMed] [Google Scholar]
- 50.Takeda, Y., T. Mori, H. Imabayashi, T. Kiyono, S. Gojo, S. Miyoshi, N. Hida, M. Ita, K. Segawa, S. Ogawa, M. Sakamoto, S. Nakamura, and A. Umezawa. 2004. Can the life span of human marrow stromal cells be prolonged by bmi-1, E6, E7, and/or telomerase without affecting cardiomyogenic differentiation? J. Gene Med. 6833-845. [DOI] [PubMed] [Google Scholar]
- 51.Tondreau, T., N. Meuleman, A. Delforge, M. Dejeneffe, R. Leroy, M. Massy, C. Mortier, D. Bron, and L. Lagneaux. 2005. Mesenchymal stem cells derived from CD133-positive cells in mobilized peripheral blood and cord blood: proliferation, Oct4 expression, and plasticity. Stem Cells 231105-1112. [DOI] [PubMed] [Google Scholar]
- 52.Torchia, E. C., S. Jaishankar, and S. J. Baker. 2003. Ewing tumor fusion proteins block the differentiation of pluripotent marrow stromal cells. Cancer Res. 633464-3468. [PubMed] [Google Scholar]
- 53.Woodbury, D., E. J. Schwarz, D. J. Prockop, and I. B. Black. 2000. Adult rat and human bone marrow stromal cells differentiate into neurons. J. Neurosci. Res. 61364-370. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.