Abstract
Although chromatin structure is known to affect transcriptional activity, it is not clear how broadly patterns of changes in histone modifications and nucleosome occupancy affect the dynamic regulation of transcription in response to perturbations. The identity and role of chromatin remodelers that mediate some of these changes are also unclear. Here, we performed temporal genome-wide analyses of gene expression, nucleosome occupancy, and histone H4 acetylation during the response of yeast (Saccharomyces cerevisiae) to different stresses and report several findings. First, a large class of predominantly ribosomal protein genes, whose transcription was repressed during both heat shock and stationary phase, showed strikingly contrasting histone acetylation patterns. Second, the SWI/SNF complex was required for normal activation as well as repression of genes during heat shock, and loss of SWI/SNF delayed chromatin remodeling at the promoters of activated genes. Third, Snf2 was recruited to ribosomal protein genes and Hsf1 target genes, and its occupancy of this large set of genes was altered during heat shock. Our results suggest a broad and direct dual role for SWI/SNF in chromatin remodeling, during heat shock activation as well as repression, at promoters and coding regions.
The eukaryotic genome is packaged into a protein-DNA complex known as chromatin, with nucleosomes as its basic structural unit (52, 57). Dynamic changes in the structure of chromatin permit localized decondensation and remodeling that in turn affect the regulation of transcription, DNA replication, recombination, and repair (8, 35).
Chromatin remodelers are large, multisubunit, biochemically diverse protein complexes that play a central role in nucleosome dynamics. These complexes affect the regulation of gene expression either by covalently modifying the N-terminal tails of histones, eviction of histones in trans, and movement of histones in cis or by exchanging one or more core histones with histone variants (33). Posttranslational modifications of histones can act in a sequential or combinatorial manner to bring about unique downstream consequences. One of the best characterized of these modifications that is strongly correlated with transcriptional activity is histone acetylation (22, 46).
Chromatin remodelers are recruited to promoters by gene-specific transcriptional activators or repressors (31, 34, 59). Although specific histone modifications are strongly correlated with steady-state transcriptional status, it is not clear how broadly specific changes in chromatin structure affect dynamic regulation of transcription in response to physiological signals. Simultaneous temporal genome-wide analyses of changes in chromatin as well as transcription status in response to perturbation provides a means of analyzing the relationship between short-term chromatin states and downstream transcriptional effects. Such dynamic analysis can reveal global patterns, and thus the general principles, if any, underlying the relationship between chromatin changes and transcriptional changes and potentially shed light on the causal connections between these two processes.
Yeast (Saccharomyces cerevisiae) cells adapt to environmental stresses such as heat shock, oxidative stress, nutrient starvation, and exposure to chemicals and heavy metals by rapid alterations in global gene expression patterns (12). Transcriptional activation of many stress-inducible genes is mediated in part by heat shock factor 1 (HSF1) (32) and is known to involve chromatin changes. Hsf1 is constitutively bound to some heat shock gene promoters but also exhibits stress-inducible binding to a majority of its targets (16). Studies on the HSP82 promoter have shown that upon heat shock, there is domain-wide disassembly of nucleosomes, and that prior to their eviction, the nucleosomes are transiently hyperacetylated (61). However, a study of HSP12, HSP82, and SSA4 indicates that the extents and timings of histone H3 acetylation and nucleosome displacement differ for these three heat shock gene promoters (10). Given that stresses, including heat shock, transcriptionally activate nearly 5% of the yeast genome (12), it is of interest to understand the relationship between chromatin changes and transcriptional activation after heat shock. Another response to stress is the downregulation of ribosomal protein (RP) genes (42, 54). In rapidly growing cells, RP gene transcription accounts for about 50% of transcription by RNA polymerase II (Pol II). There is a marked reduction of transcription of these genes in response to different stresses (9, 12, 55).
Several genomic maps of nucleosome localization (58), variant histones (15, 38, 60), and covalently modified histones (24-26, 36) in yeast have been published recently. These studies provide a good framework for understanding the dynamics of eukaryotic chromatin. However, several questions remain regarding the role of chromatin remodeling during global transcriptional reprogramming of a cell. First, do nucleosome occupancy and histone acetylation states at large classes of genes correlate exclusively with their transcriptional status or do distinctive chromatin states appear depending on how a gene is activated and repressed? Second, what are the kinetics of chromatin remodeling relative to transcriptional changes? Third, what is the role of known chromatin remodeling complexes at the genes that are transcriptionally affected?
To address these questions, we studied two different types of stress conditions in yeast: heat shock and stationary-phase stress. Heat shock causes immediate, but transient, transcriptional changes within minutes of exposure to stress, whereas stationary-phase stress, in contrast, results in more long-term and steady-state levels of gene expression. There is considerable overlap between genes regulated by these two stresses (12). In order to determine the relationship between genome-wide chromatin states and transcriptional regulation of genes when yeast cells responded to these two different stress conditions, we systematically evaluated promoter nucleosome occupancy and histone H4 acetylation status by chromatin immunoprecipitation (ChIP)-chip using histone-specific and acetylation-specific antibodies and, in parallel, monitored the changes in expression of their downstream genes in response to the two stress conditions. We show the following. First, heat shock and stationary-phase stresses result in contrasting chromatin acetylation states. Second, SWI/SNF regulates the magnitude of gene activation as well as repression following heat shock. Third, chromatin remodeling at heat shock-activated and -repressed genes is accompanied by changes in the occupancy of SWI/SNF at promoters as well as coding regions, indicating a direct mechanistic role for SWI/SNF during transcription initiation as well as elongation. Finally, we propose a model for chromatin remodeling at both activated and repressed genes after heat shock stress.
MATERIALS AND METHODS
Yeast strains, medium, and culture conditions.
The S. cerevisiae strains used in this study are YKY6B (UCC1111 background; MATα ade2::his3-Δ200 elu2-Δ0 hht2::MET15 hhf1-hht1::LEU2 pRS412(ADE2 CEN ARS)-MYC-HHF2-HHT2) (25) and FY2103 (MATα ura3-Δ0 trp1-Δ63 his3-Δ200 lys2-Δ0 SNF2-C18MYC::TRP1) (28). The deletion strains shown in Fig. S3 in the supplemental material are from the yeast knockout collection from Open Biosystems (56). We generated the YKY6B-snf2Δ strain from YKY6B by use of a long flanking homology-PCR-based gene disruption using a kanMX selectable marker (27, 53). The gene disruption was confirmed by genomic PCR. Cells were cultured in synthetic complete medium (yeast nitrogen base, a complete mix of amino acid supplements, and 2% glucose) for all experiments. Cells were grown to log phase (optical density [OD] at 600 nm of 0.6 to 0.8), and typically, one half of the cells was used to isolate mRNA for expression profiling. The other half was used for formaldehyde cross-linking followed by ChIP. For heat shock treatment, cells were grown to an OD of 0.7 at 30°C, centrifuged at 3,000 rpm for 5 min, and resuspended in an equal volume of synthetic complete medium prewarmed to 39°C. Heat shock was carried out either for 15 min or for the times specified in the time course experiments (see Fig. 3 and 4 below; also see Fig. S5 in the supplemental material) in a 39°C water bath. Cells were harvested at specified time points and flash frozen in liquid nitrogen for mRNA extraction or cross-linked for ChIP. Stationary-phase stress was carried out by allowing the cells to grow to an OD of 5.0 in synthetic complete medium.
FIG. 3.
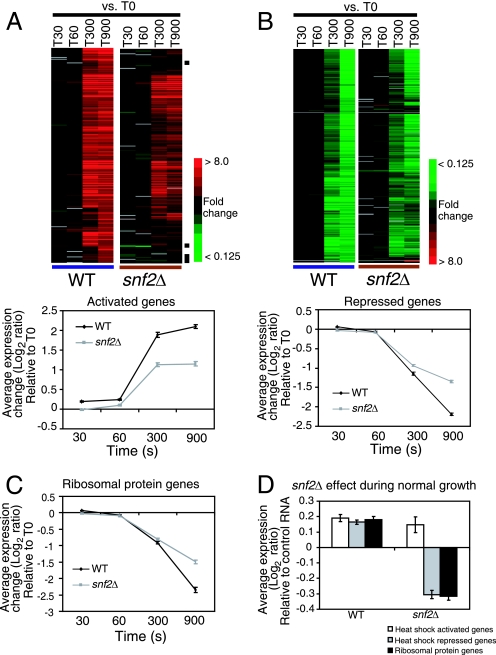
Time course gene expression profiles of heat shock-activated and -repressed genes in WT and snf2Δ cells. mRNAs were harvested before heat shock (T0) and at T30, T60, T300, and T900 after heat shock. The expression levels of all the genes from the different time points in WT and snf2Δ cells were normalized to the expression levels in the respective T0 samples. (A) The expression profiles of heat shock-activated genes were clustered hierarchically and are displayed using a red-green heat map. There were 22 genes that were not upregulated in snf2Δ cells even after 15 min of heat shock and are indicated with black bars on the right. The graph below shows the average expression levels of all the activated genes at each of the time points. (B) The expression profiles of heat shock-repressed genes are displayed on top, and a graph showing the average expression levels at each of the time points for these genes is plotted below. Error bars represent the standard error of the mean. (C) Average expression levels of the RP genes at different time points after heat shock in WT and snf2Δ cells. Error bars represent the standard errors of the means. (D) Deletion of SNF2 does not affect the basal level expression of heat shock-activated genes during normal growth conditions. However, there was a 1.4-fold decrease in the expression of heat shock-repressed genes and RP genes for snf2Δ cells compared to what was seen for WT cells during normal growth at 30°C.
FIG. 4.
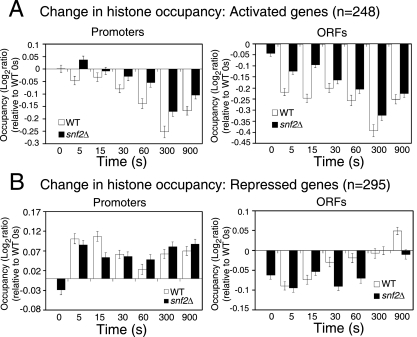
Kinetics of the change in histone H4 occupancy at the promoters and coding regions of heat shock-activated and -repressed genes. WT and snf2Δ cells were subjected to heat shock and cells were cross-linked at T5, T15, T30, T60, T300, and T900 after temperature shift. Histone occupancies at all time points in both strains were normalized to the occupancy in WT cells at time point T0. Data represent the average of three biological replicates and the error bars represent the standard errors of the means. (A) Kinetics of histone depletion from the promoters and coding regions (ORFs) of activated genes. (B) Kinetics of increase in histone occupancy at the promoters and coding regions of heat shock-repressed genes.
RNA isolation and cDNA labeling.
Cells from a 50-ml culture were resuspended in 8 ml of AE buffer (50 mM sodium acetate, pH 5.2, 10m M EDTA, 1.7% sodium dodecyl sulfate), and total RNA was extracted using the acid phenol lysis protocol described previously (45). Poly(A)+ RNA was purified by allowing the mRNA to bind to oligo(dT) cellulose for 90 min in the presence of 500 mM NaCl. The oligo(dT) cellulose was washed three times with 10 mM Tris-Cl, pH 7.5, 500 mM NaCl and two times with 10 mM Tris-Cl, pH 7.5, 250 mM NaCl and then eluted in 10 mM Tris-Cl, pH 7.5, prewarmed to 65°C. Reverse transcription using oligo(dT) primer and cDNA labeling were carried out as described previously (16).
ChIP and labeling.
Proteins were cross-linked to DNA by adding formaldehyde to the culture to a final concentration of 1% before heat shock, at the indicated time points after heat shock at 39°C (see Fig. 4 below; also see Fig. S5 in the supplemental material), and during stationary phase. The cross-linked DNA-protein complex was isolated, sheared by sonication, and immunoprecipitated (21) using the following antibodies: anti-Myc (9E10), anti-acetyl-histone H4 antibody (detects acetylation on K5, 8, 12, and 16), anti-acetyl-histone H4 K16, anti-acetyl-histone H3 K9/18 from Upstate Biotechnology, and anti-histone H3 from Abcam. The cross-links were reversed and the immunoprecipitated and reference input or genomic DNA was amplified by PCR and coupled to Cy3 and Cy5 fluorophores according to published protocols (20).
DNA microarrays.
Microarrays containing nearly every open reading frame (ORF) and intergenic region were manufactured as described previously (19). The amplified input and immunoprecipitated DNA, labeled with Cy3 and Cy5, respectively, were hybridized onto DNA microarrays for 12 to 16 h, washed, scanned with an Axon 4000B scanner (Molecular Devices), and quantitated using GenePix Pro software. The data were uploaded into Acuity microarray informatics software (Molecular Devices) and filtered on basic quality control measures, such as low signal intensity before analysis.
Microarray data analysis.
Intergenic spots on the microarray were assigned as a promoter for a downstream gene based on current annotations from SGD. For expression profiling, the median of ratios (Cy5/Cy3) was calculated for every ORF and the log2-transformed ratio was clustered and plotted as red-green heat maps. The activated and repressed genes shown in Fig. 1 to 5 were those showing ≥2.5-fold expression change compared to unstressed wild-type (WT) cells. For ChIP-chip experiments, data from at least three and up to five independent replicate experiments were used for analysis and to filter for consistency. All data shown for acetylated histone H4 were normalized for underlying histone H4 occupancy.
FIG. 1.
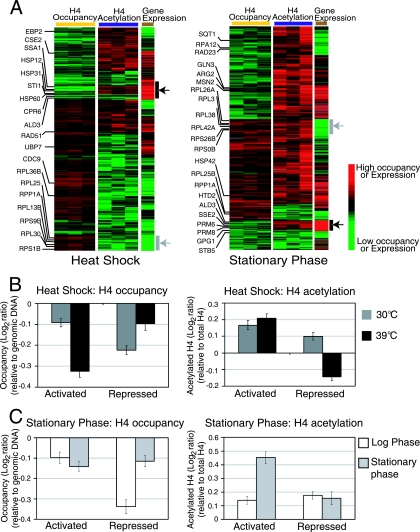
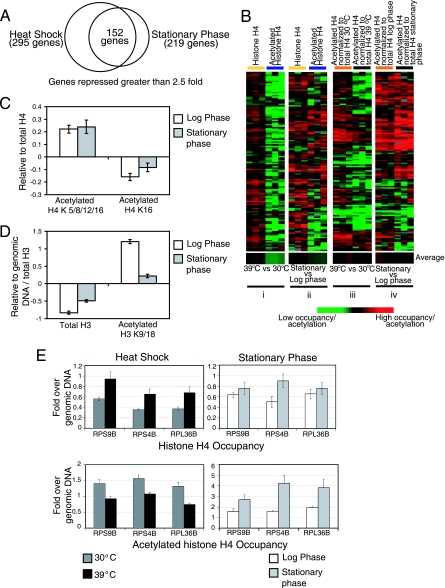
Gene expression levels correlate positively with histone H4 acetylation and negatively with histone H4 occupancy. (A) Comparison of gene expression changes with change in histone H4 occupancy and acetylation levels at promoters after heat shock and stationary-phase stress. In the two left panels of the heat map (H4 occupancy and acetylation), each column represents an immunoprecipitation performed from an independent cross-linking experiment. The gene expression column represents the median expression change calculated from three independent biological replicates. ChIP-chip data were filtered for consistent changes (positive or negative log2 ratio) in occupancy and acetylation across the three biological replicates and clustered hierarchically along with the gene expression data. The black and gray arrows indicate the most strongly upregulated and downregulated genes, respectively. (B and C) Average histone H4 occupancy and acetylation levels at the promoters of genes activated and repressed more than 2.5-fold after heat shock and stationary-phase stress, respectively. (B) The bars represent the average H4 occupancy and acetylation at the promoters of 248 activated and 295 repressed heat shock genes at the indicated temperatures. (C) The white bars represent the average H4 occupancy and acetylation at the promoters of 280 activated and 219 repressed stationary-phase genes at the indicated phases of growth. The y axes show the average change in the occupancy or acetylation measured as the log2 ratio. Error bars represent the standard error of the mean.
FIG. 5.
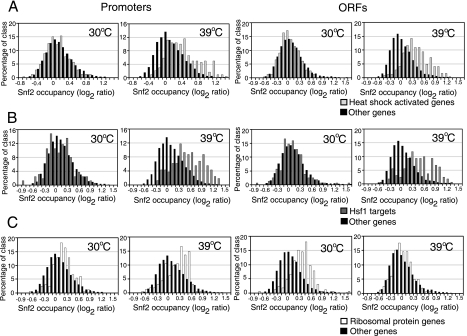
Snf2 is recruited to RP genes under normal (unstressed) growth conditions and to Hsf1 target genes upon heat shock. Cy5-labeled Snf2 ChIP DNA was hybridized onto microarrays along with Cy3-labeled input DNA. The distributions of log2 ratios for heat shock-activated gene promoters and coding regions (A), Hsf1 target gene promoters (16) and coding regions (B), and RP gene promoters and coding regions (C) are shown. The distributions of the log2 ratios for the remainder of the genes on the array (other genes) are also plotted in each of the graphs.
qPCR verification.
Primer pairs used in Fig. 2 were designed to amplify approximately 60-bp regions in the promoters of the RPS9B, RPS4B, and RPL36B genes. Control primers used for normalization were designed for the promoter of YSN1. Five heat shock-responsive genes that showed an increase in Snf2 occupancy at their promoters were selected for verification (see Fig. 6 below). Three genes that did not show an increase in Snf2 occupancy were selected as negative controls. The normalization primer pair amplified the GET2 promoter where there is no Snf2 binding (28). Table S3 in the supplemental material lists the sequences of all the quantitative real-time PCR (qPCR) primers used in this study. qPCR was performed using SYBR green chemistry on an ABI 7900 instrument. Enrichment of target loci in the ChIP sample relative to either genomic (Fig. 2) or input (see Fig. 6) DNA is shown, after normalization to the reference primers.
FIG. 2.
Genes repressed after both heat shock and stationary-phase stress show inverse histone H4 acetylation levels. (A) Venn diagram indicating the total number of genes repressed more than 2.5-fold after heat shock and stationary-phase stress and the number of genes common to both sets. (B) Change in histone H4 occupancy and acetylation at the promoters of the 152 genes repressed after both heat shock and stationary-phase stress. Panels i and ii show the change in H4 occupancy and acetylation after stress, whereas panels iii and iv show the change in acetylated histone H4 occupancy before and after stress, normalized to the underlying nucleosome occupancy. The average H4 occupancy and acetylation levels from the heat map are shown at the bottom. Heat shock is accompanied by a decrease in histone H4 acetylation at the promoters of the 152 genes, whereas stationary-phase stress is accompanied by an increase in H4 acetylation at the same promoters. The heat map was gen-erated after hierarchical clustering. (C) Average H4 acetylation levels at K5, 8, 12, and 16 and acetylation levels specifically at H4 K16 during log and stationary phases at the promoters of genes repressed after both heat shock and stationary-phase stress. The high levels of acetylation seen at the H4 N-terminal tail are not due to H4 K16 acetylation. (D) H3 acetylation at the promoters of the 152 repressed genes during stationary-phase stress. There was an increase in total H3 occupancy at these promoters but a strong reduction in H3 K9/18 acetylation upon stationary-phase stress. Total H3 occupancy was measured relative to genomic DNA and the acetyl-H3 K9/18 occupancy was normalized to total H3 occupancy. (E) q-PCR verification of the change in histone H4 occupancy and acetylation at the promoters of 3 of the 152 repressed genes chosen at random. Error bars represent the standard deviation across three replicates.
FIG. 6.
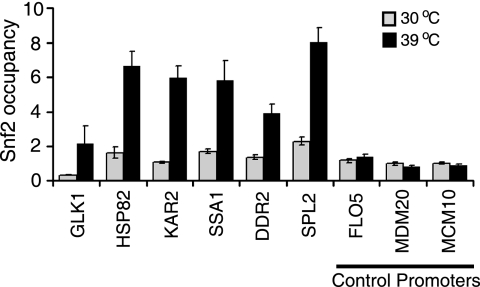
Quantitative real-time verification of Snf2 ChIP-chip data. Five genes (GLK1, HSP82, KAR2, SSA1, DDR2, and SPL2) that showed an increase in Snf2 occupancy after heat shock and three control genes (FLO5, MDM20, and MCM10) that did not show an increase after heat shock in the microarray experiments were selected. Primers were designed to amplify 60 to 80 bp of their promoters, and qRT-PCR was carried out. Error bars show the standard deviations from three replicate reactions.
Microarray data accession number.
Microarray data are available from the Gene Expression Omnibus (GEO) under accession number GSE7665 at http://www.ncbi.nlm.nih.gov/geo/.
RESULTS
Relationship between transcription, nucleosome occupancy, and H4 acetylation during the stress response.
We subjected yeast cells to stress and compared changes in whole-genome expression profiles with changes in nucleosome occupancy and in histone H4 acetylation levels (at K5, 8, 12, and 16). We selected heat shock stress (39°C, 15 min) and stationary-phase stress (OD at 600 nm of 5.0), both of which are known to perturb the expression of hundreds of genes, some common and some distinct to each of the stress conditions. We determined the change in nucleosome occupancy by labeling with Cy5 the histone H4-associated DNA immunoprecipitated by an anti-Myc tag antibody from cells subjected to stress, labeling with Cy3 the parallel ChIP DNA from unstressed cells, and hybridizing them simultaneously to microarrays representing almost all genes and promoters in yeast. Because nucleosomes are not uniformly distributed across chromosomes and in promoter regions, we measured changes in histone H4 acetylation levels after normalizing them to underlying nucleosome occupancy. We accomplished this by hybridizing differentially labeled ChIP DNA by use of an anti-acetylated histone H4 antibody and the ChIP DNA by use of the anti-Myc tag antibody (which immunoprecipitates total histone H4-DNA complex) from each yeast culture to the same microarray. We calculated the change in histone acetylation levels after transcriptional perturbation by dividing the acetylated histone H4/total histone H4 ratio in stressed cells by the corresponding ratio in unstressed cells. We considered only promoters showing consistent Cy5/Cy3 ChIP ratios across three independent biological replicates. The Pearson correlation coefficients for the biological replicates averaged 0.91 for gene expression arrays, 0.895 for H4 occupancy arrays, and 0.73 for acetylated H4 occupancy arrays.
Our results confirmed previous studies (2, 25) showing that nucleosome occupancy at promoters is inversely correlated with gene expression (see Fig. S1 in the supplemental material). Each of the stresses affected the expression of more than 400 genes. Heat shock resulted in 248 genes upregulated and 295 genes downregulated more than 2.5-fold relative to what was seen for unstressed cells (see Table S1 in the supplemental material). Stationary-phase stress resulted in 280 genes upregulated and 219 genes downregulated more than 2.5-fold relative to what was seen for log-phase cells (see Table S2 in the supplemental material). There were 75 genes activated more than 2.5-fold and 152 genes that were repressed more than 2.5-fold common to both stress conditions, consistent with previous observations and the regulation of genes during both stresses by common transcription factors such as Msn2/Msn4 (3, 12). We visualized relationships between nucleosome occupancy, H4 acetylation, and gene expression using heat maps where ChIP data for promoters were aligned with the expression data for their downstream genes, followed by clustering to organize the data. Genes that were strongly upregulated after either stress showed a reduction in nucleosome occupancy and higher histone H4 acetylation at their promoters (Fig. 1A). Conversely, genes that were strongly repressed showed an increase in nucleosome occupancy at their promoters (Fig. 1A). A striking difference between the cellular responses to the two stress conditions was that in response to stationary-phase stress, most promoters throughout the genome showed an increase in histone H4 acetylation, regardless of the expression status of the downstream gene (Fig. 1A; also see Fig. S1B in the supplemental material).
The relationship between nucleosome occupancy, histone acetylation, and gene expression depended on the perturbation and differed for activation versus repression. During heat shock, there was a strong decrease in nucleosome occupancy and a moderate increase in histone H4 acetylation at the promoters of activated genes. The promoters of repressed genes showed the inverse effect in that the decrease in histone H4 acetylation was more pronounced than the increase in nucleosome occupancy (Fig. 1B). During stationary phase, the decrease in nucleosome occupancy was less pronounced than the increase in histone H4 acetylation at activated promoters (Fig. 1C). However, histone H4 acetylation levels at promoters repressed after stationary-phase stress showed an increase relative to basal levels rather than a decrease as observed for heat shock.
Previous studies have shown that the HSP82 promoter is open and sensitive to DNase I even in the absence of heat shock as a result of Hsf1 binding to the high-affinity heat shock element upstream of the HSP82 gene (11, 14). Heat shock-induced histone loss at the HSP82 promoter and coding region is dependent on Hsf1 and occurs within seconds of temperature shift (61). It has also been shown that Hsf1 is preloaded at the HSP82 and SSA4 promoters in an inactive form and is activated upon heat shock, thereby mediating chromatin remodeling and RNA Pol II recruitment (10).
In order to examine the relationship between Hsf1 binding and nucleosome occupancy more globally, we compared histone H4 occupancies at all promoters occupied by Hsf1 at 30°C (16) (see Fig. S2A in the supplemental material). This analysis showed that Hsf1 target promoters that had higher occupancy of Hsf1 at 30°C had comparatively lower nucleosome occupancy and that promoters with lower occupancy of Hsf1 had higher nucleosome occupancy (see Fig. S2B in the supplemental material). Overall, nucleosome occupancy of promoters occupied by Hsf1 was low. We also examined the correlation between RNA Pol II occupancy (37) and Hsf1 occupancy. Promoters with higher Hsf1 occupancy had a greater tendency to have prebound RNA Pol II, while promoters with lower Hsf1 occupancy had lower RNA Pol II occupancy (see Fig. S2B in the supplemental material). This depletion of nucleosomes at promoters bound by Hsf1 suggests that binding of Hsf1 is followed by chromatin remodeling at the promoter regions of heat shock-responsive genes even in the absence of heat shock, resulting in RNA Pol II recruitment and thus enabling those genes to be poised for rapid transcriptional activation within minutes after heat shock (see below).
The H4 acetylation pattern of genes repressed after stationary-phase stress is the opposite of that after heat shock stress.
We examined the genes repressed after heat shock and stationary-phase stress in greater detail. Out of the 295 heat shock-repressed genes and 219 stationary-phase-repressed genes, there were 152 genes in common (Fig. 2A). A majority of these commonly repressed genes (112 genes out of 152, or 74%) were RP genes. After heat shock, there was a decrease in histone H4 acetylation at the promoters of these repressed genes but in contrast, after stationary-phase stress, there was an increase in H4 acetylation at most of the same promoters (Fig. 2Bi and ii). When histone H4 acetylation levels at each of the 152 promoters were normalized to the underlying nucleosome occupancy, it was apparent that histone H4 acetylation decreased after heat shock stress but increased after stationary-phase stress (Fig. 2Biii and iv and C). This difference in acetylation status is striking given that these 152 genes, largely comprising RP genes, were transcriptionally repressed by both stresses.
The anti-acetylated histone H4 antibody used in the preceding experiments did not distinguish between acetylation at K5, K8, K12, or K16 in the N-terminal tail. To further probe the nature of the H4 acetylation increases we observed in response to stationary-phase stress, we used an antibody specific for H4 K16 acetylation for ChIP-chip in normally growing and stationary-phase cells. H4 K16 acetylation is well characterized in yeast and has a role in transcriptional activation and the maintenance of euchromatin (43). H4 K16 is thought to be a unique acetylation site of histone tails, functioning as a dual switch for higher-order chromatin structure and protein-histone interactions. Acetylation on K5, K8, and K12 is dispensable for the folding of nucleosomal arrays. ChIP-chip with the anti-K16 acetyl antibody showed that indeed, during stationary phase, the increase in acetylation at repressed promoters was not at H4 K16 and could therefore be attributed to acetylation of K5, K8, or K12 (Fig. 2C). We also performed ChIPs against acetylated histone H3 to examine whether the increase in histone acetylation at repressed promoters during stationary phase reflected a general increase in acetylation or was specific to the three residues in histone H4. Although there were high levels of acetylated histone H3 at these promoters during log phase, there was a dramatic reduction in acetylated H3 occupancy at these promoters during stationary phase (Fig. 2D). We detected a modest increase in histone H3 occupancy at these promoters, similar to what was seen for histone H4.
In order to quantitatively and independently verify the results obtained using whole-genome microarrays, we carried out qRT-PCR using promoter-specific primers for three randomly selected RP genes, RPS9B, RPS4B, and RPL36B. This experiment confirmed that repression of many RP genes during stationary-phase stress is accompanied by an increase in histone H4 acetylation at their promoters, but heat shock, which also represses these same genes, results in a decrease in histone H4 acetylation (Fig. 2E).
A role for SWI/SNF in mediating the global transcriptional response to heat shock.
The reduction in nucleosome occupancy relative to histone acetylation at activated genes during the rapid heat shock response, which was stronger than that seen for the slower stationary-phase stress, suggested that rapid clearance of nucleosomes from the promoter is important for transcriptional activation following heat shock. To identify the factor that might mediate chromatin remodeling after heat shock, we examined global gene expression changes after heat shock in strains lacking one of four different ATP-dependent chromatin remodeler genes known to be involved in transcriptional regulation: snf2Δ, chd1Δ, isw1Δ, and isw2Δ. Snf2 is the catalytic subunit of the SWI/SNF chromatin remodeling complex (29), while Chd1 is a member of the CHD family of proteins (50). Isw1 is a member of the ISWI (imitation-switch) class of proteins and forms a complex with Ioc2 and Ioc4 to regulate transcription elongation and forms a complex with Ioc3 to repress transcription initiation. Isw2 is also a member of the ISWI class and forms a complex with Itc1 and is required for the repression of MATa-specific genes, INO1, and early meiotic genes during mitotic growth (30). We compared global gene expression in the WT and the four knockout strains before and after heat shock in three ways. First, we compared the genome-wide changes in gene expression in the WT strain and deletion mutants after heat shock. Second, we compared the expression levels of all genes between different pairs of deletion mutants at 30°C. Third, we compared the expression levels of all genes at 39°C between different pairs of deletion mutants. We then calculated the pair-wise Pearson correlation coefficients for all the experiments. This analysis revealed that gene expression changes in the snf2Δ cells showed the greatest differences from WT cells (see Fig. S3A in the supplemental material). In addition, at both 30°C and 39°C, gene expression in the snf2Δ strain was clearly different from that of all the other deletion strains (see Fig. S3B in the supplemental material). These results indicate that in comparison to the other key chromatin remodelers, Snf2 plays a more important role in mediating the transcriptional response to heat shock.
Next, we compared the kinetics of the transcriptional response to heat shock in WT and snf2Δ cells, focusing on the genes that were most strongly activated or repressed in WT cells. We analyzed RNA samples at 30, 60, 300, and 900 s (T30, T60, T300, and T900, respectively) after temperature shift of WT and snf2Δ cells and compared them to RNA from unstressed WT cells (see Fig. S4 in the supplemental material). In order to visualize the snf2Δ effect on the activation and repression of genes after heat shock, we also compared the RNA samples at T30, T60, T300, and T900 after temperature shift in both WT and snf2Δ cells and normalized them to RNA from the respective unstressed cells. Although most genes activated by heat shock in WT cells were also activated in snf2Δ cells, the extent of activation was distinctly lower, in that the average transcript levels at all time points after heat shock were lower for the snf2Δ cells than for WT cells (Fig. 3A). At T900, transcript levels of activated genes were twofold lower on average in snf2Δ cells than in WT cells. Out of the 243 heat shock-activated genes at T900, 22 did not show an increase in expression even after 15 min of heat shock in snf2Δ cells (Fig. 3A).
Repression of genes by heat shock was similarly affected in the snf2Δ strain, though not as markedly. mRNA levels in the snf2Δ strain declined between T60 and T300 after heat shock, but this rate slowed down by T900. In contrast, mRNA levels continued to decline rapidly in WT cells until T900, such that the average repression in WT cells was about 1.8-fold more than in snf2Δ cells (Fig. 3B). When we considered the expression decrease of the RP genes as a group upon heat shock, we saw that the average repression of the RP genes in WT cells was about 1.75-fold more than in snf2Δ cells (Fig. 3C). Interestingly, Snf2 was also required for the normal transcription of genes that were repressed by heat shock (Fig. 3D). Thus, Snf2 is required not only for high-level expression of RP genes during normal growth but also for the activation and repression of distinct sets of genes after heat shock.
Snf2 affects the kinetics of nucleosome remodeling in response to stress.
The fact that the deletion of SNF2 caused transcriptional defects in genes activated as well as repressed by heat shock raised the question of whether the effect was directly due to defects in chromatin remodeling at the promoters and/or the coding regions of these genes. To investigate this, we measured genome-wide changes in nucleosome occupancy in WT and snf2Δ cells to determine how rapidly nucleosomes were displaced after heat shock. We carried out a high-resolution time course study on the order of seconds to identify chromosomal changes immediately after heat shock. We cross-linked cells at T5, T15, T30, T60, T300, and T900 following a shift to higher temperature. In WT cells, nucleosome depletion was apparent within 5 s of temperature shift, at the promoters as well as the coding regions of activated genes (Fig. 4A). Histone depletion increased consistently until T300, after which there was a small relative increase in histone occupancy. In the snf2Δ strain, there was a delay in loss of histone H4 at promoters, with depletion beginning only 15 s after heat shock. Interestingly, there was an initial increase in histone H4 occupancy 5 s after heat shock in the snf2Δ strain that was not seen for WT cells. These data, obtained from three independent biological replicate experiments, establish that although the histone H4 occupancy changes were small, they were consistent across hundreds of genes and multiple time points and further corroborate the results from our time course gene expression analysis.
At the promoters of genes that were repressed by heat shock, there was an increase in histone H4 occupancy within 5 s of heat shock for both WT and snf2Δ cells (Fig. 4B). For WT cells, over the coding regions of repressed genes, there was a gradual increase in nucleosome occupancy following an initial decrease, until 15 min after heat shock. However, for snf2Δ cells, nucleosome occupancy over the coding regions of repressed genes tended to be lower than for WT cells (Fig. 4B). This is consistent with the expression data, where in comparison to WT cells, snf2Δ cells showed higher average mRNA levels (Fig. 3B).
It is possible that nucleosome displacement from DNA is preceded by covalent histone modification. At the promoters of the heat shock genes HSP82, HSP12, and SSA4, a transient increase in histone acetylation precedes nucleosome displacement upon heat shock (10, 61). In order to ascertain whether there was any general relationship between histone H4 acetylation and nucleosome clearance in the absence of Snf2, we performed ChIP using the anti-acetyl-H4 antibody on the same cell lysates used for the analysis of histone H4 occupancy in WT and snf2Δ strains. We hybridized ChIP DNA from all time points for WT and snf2Δ cells to arrays along with a genomic DNA reference and calculated the change in acetylated histone H4 occupancy by dividing the Cy5/Cy3 ratios for all the time points for both WT and snf2Δ cells by the corresponding ratio in the WT at zero time point. For WT cells, the kinetics of acetyl-H4 depletion at promoters followed a pattern similar to that of the depletion of total histone H4 (see Fig. S5 in the supplemental material), with a transient increase in acetylation at some of the promoters, including HSP82. However, for snf2Δ cells, where nucleosome depletion was delayed relative to what was seen for WT cells and the histone H4 occupancy at T900 was higher than in WT cells (Fig. 4A), there was a clear enrichment of acetylated histone H4 (hyperacetylation) at many of the promoters across all time points, and this was consistent both before and after normalization to total H4 at each time point (see Fig. S5 in the supplemental material). These results suggest that when yeast cells are subjected to heat shock stress, histone H4 at the promoters of activated genes is transiently hyperacetylated prior to Snf2-dependent chromatin remodeling (see Discussion).
The genomic targets occupied by Snf2 vary before and after heat shock stress.
To investigate whether the SWI/SNF complex is directly involved in chromatin remodeling during heat shock, we examined whether Snf2 was recruited to the genomic loci undergoing remodeling and whether its occupancy changed during heat shock and chromatin remodeling. We subjected yeast cells harboring a C18-Myc-tagged Snf2 to heat shock for 15 min, followed by formaldehyde cross-linking, immunoprecipitation with an anti-Myc antibody, and microarray hybridization. We carried out five independent replicate cross-linking experiments under unstressed and heat shock conditions and combined the results from the five experiments in our analysis. We calculated the median log ratio indicating Snf2 occupancy from the five replicates and compared the occupancies of Snf2 at the promoters and coding regions (ORFs) of (i) genes activated by heat shock in WT cells, (ii) Hsf1 target genes (16), and (iii) RP genes, with the occupancy of Snf2 at the background set of promoters and ORFs. At 30°C, Snf2 occupancy at both promoters and coding regions of heat shock-activated as well as Hsf1 target genes was no different from what was seen for the background (Fig. 5A and B, 30°C). Upon heat shock, the occupancy of Snf2 increased robustly at both promoters and, strikingly, at the coding regions of these genes, indicating that Snf2 was recruited to these regions within 15 min of heat shock (Fig. 5A and B, 39°C). In contrast, Snf2 occupancy was high over the coding regions of RP genes and moderately high at their promoters during unstressed conditions relative to the background (Fig. 5C, 30°C). This is consistent with our previous finding that there was a decrease in the average levels of transcription of the RP genes in an snf2Δ background at 30°C (Fig. 3D). Therefore, Snf2 occupancy of the RP genes is likely to be involved in maintaining the high levels of transcription of the RP genes at 30°C. Upon heat shock (Fig. 5C, 39°C), the occupancy of Snf2 at the coding region of the RP genes was reduced to background levels, while its occupancy increased at their promoters, suggesting that Snf2 directly mediates the remodeling of chromatin at the promoters of RP genes after heat shock to bring about a repressive chromatin state.
We independently verified the heat shock-dependent changes in Snf2 occupancy at the promoters of six activated genes (GLK1, HSP82, KAR2, SSA1, DDR2, and SPL2) by q-PCR (Fig. 6). We selected three promoters, FLO5, MDM20, and MCM10, as negative controls, as they did not show any change in Snf2 occupancy upon heat shock in our microarray experiments. The qPCR data showed that there was a 3- to 6.5-fold increase in the occupancy of Snf2 at the six heat shock-responsive promoters that we tested and no increase at the promoters of the three negative-control genes, corroborating the ChIP-chip results.
Taken together, these results indicate that the SWI/SNF complex has a direct role in (i) the transcription of RP genes during unstressed growth conditions, (ii) the repression of RP gene expression after heat shock stress, and (iii) the transcriptional activation of heat shock-responsive and Hsf1 target genes upon heat shock.
We further carried out an Snf2 ChIP-chip experiment with stationary-phase cells to investigate whether Snf2 is recruited to genes activated by stationary-phase stress. Our results showed that unlike what is seen for heat shock, Snf2 was not recruited to these promoters during stationary-phase stress (not shown). Moreover, Snf2 did not show increased occupancy at the promoters of RP genes upon stationary-phase stress, indicating that the dual role of Snf2 in activating heat shock-responsive genes and repressing RP gene expression is specific to heat shock stress and cannot be generalized as a response to all other stress conditions.
DISCUSSION
Yeast cells respond differently to heat shock and stationary-phase stress.
Our analysis shows that the global relationship between transcriptional changes and changes in chromatin states can differ depending on the physiological trigger or mechanism of transcriptional activation. Although more than 150 genes were repressed after both heat shock and stationary-phase stress alike in yeast, the histone H4 acetylation levels at their promoters showed contrasting patterns (Fig. 2). In response to heat shock, transcriptional repression of these genes was accompanied by an increase in nucleosome occupancy at their promoters and a decrease in H4 acetylation. In contrast, during the response to stationary-phase stress, although there was a similar increase in histone occupancy at the promoters of these same transcriptionally repressed genes, there was an increase in histone H4 acetylation.
There could be three explanations for this difference. First, it is possible that different transcriptional repressors mediate the repression of these genes during the two stress conditions. Histone acetylation at activated promoters is differentially affected by specific activators. For example, CUP1 is induced by Hsf1 upon heat shock and by Ace1 during copper induction. Hsf1-dependent induction of CUP1 is accompanied by increased H4 acetylation, while Ace1-dependent induction of the same gene is accompanied by decreased H4 acetylation (5). For human HSP70, heat shock activation causes acetylation of histone H4, whereas activation by sodium arsenite produces both H4 acetylation and H3 phosphorylation (49). These examples illustrate that the differential response of acetylation at regulated promoters may be due to two different regulatory pathways. Of the 152 genes that were repressed by both stresses, 112 were RP genes. A majority of RP genes are bound by the transcription factor Rap1 and also by cofactors Fhl1 and Ifh1. When transcription of the RP genes is repressed by nutrient starvation, heat shock, osmotic stress, or the diauxic shift, Ifh1 leaves the promoter but Rap 1 and Fhl1 remain bound (41, 42, 54). However, it is not known if there are additional mechanisms of transcriptional repression of RP genes during environmental stress. It is possible that multiple pathways lead to the repression of RP genes after heat shock and stationary-phase stress, especially given that the former response, like all the other conditions where RP gene repression by transcription factors has been examined, is a rapid or transient response, whereas repression during stationary-phase stress is more long term.
A second possibility is that acetylations of different lysine residues in histone H4 at the promoters of RP genes have contrasting transcriptional consequences. Histone H4 K16 acetylation serves as a switch for changing chromatin from a repressive to a transcriptionally active state by affecting higher-order chromatin compaction (43). However, the roles of K5, K8, and K12 acetylation are less clear. It is known that H4-dependent histone deposition requires at least one of K5, K8, or K12 to be acetylated (44). Mutational analysis of these latter lysine residues has shown that their effect on gene expression is nonspecific and cumulative (6). Our experiments showed that the increase in histone H4 acetylation at repressed genes during stationary-phase stress was not because of H4 K16 acetylation. Moreover, acetylation at histone H3 K9/18 decreased strongly during stationary-phase stress. Therefore, it is likely that acetylation at H4 K5, K8, and/or K12 is specifically important for the repression of genes during stationary-phase stress but not upon heat shock stress. This highlights an interesting difference in the mechanisms of chromatin repression of the same set of genes by different pathways.
A third possibility is that H4 hyperacetylation is unrelated to the immediate transcriptional status of the repressed genes during stationary phase and instead serves a chromatin mark for other reasons. RNA Pol II is known to occupy even silent promoters during stationary phase in readiness for reactivation of transcription upon release from stationary phase (37). It is possible that histone H4 K5, K8, and/or K12 hyperacetylation during stationary phase, which is a long-term stable physiological state as opposed to the transient heat shock, facilitates this dormant occupancy of promoters by the transcriptional machinery.
A novel role for Snf2 in transcriptional regulation during heat shock.
The SWI/SNF complex is involved in activation and repression of transcription by RNA Pol II (29) and is also known to have a role in repressing ribosomal DNA transcription (7). Based on our whole-genome expression profiling of snf2Δ cells and the in vivo binding distribution of Snf2 in WT cells before and after heat shock, it is clear that the SWI/SNF complex also plays a pivotal role specifically in the transcription of both RP genes and Hsf1 target genes that are activated by heat shock. Interestingly, we observed changes in Snf2 occupancy at both the promoters and the coding regions of genes transcriptionally regulated by stress. Transcription of RP genes is dependent on the activators Rap1, Fhl1, and Ifh1. In addition, the Esa1 subunit of the NuA4 histone acetyltransferase (HAT) complex (an H4-specific HAT) binds to RP gene promoters in a Rap1-dependent manner and is required for RP gene transcription (39). Esa1 occupancy at RP promoters decreases during amino acid starvation and after heat shock. Our data integrate well into these previous studies, given that histone acetylation by either the SAGA (an H3-specific HAT) or NuA4 HAT complex increases the occupancy of SWI/SNF on promoters (17). Snf2 is the bromodomain-containing component of the SWI/SNF complex that recognizes and binds to acetylated histones (18). Therefore, it is possible that SWI/SNF is recruited to RP genes via transcriptional activators but may be stabilized by NuA4 acetylation of histone H4. Upon heat shock, SWI/SNF may direct nucleosome reassembly at the promoters of the RP genes, perhaps in conjunction with presently unknown transcriptional repressors, thereby shutting down transcription of these genes.
Data presented here and in a previous study of histone H3 occupancy after heat shock (25) show that the activation of genes upon heat shock results in nucleosome displacement at promoters. Our data also show that Snf2 occupies the promoters and the coding regions of all of the activated genes within 15 min of heat shock. This is consistent with previously published data on a model heat shock gene showing an increase in the abundance of Snf2 at the promoter, the coding region, and the 3′ untranscribed region following heat shock (61). However, ChIP-chip analysis of stationary-phase cells showed that Snf2 occupancy does not increase at the promoters or coding regions of genes that were activated by stationary phase, suggesting a different mechanism of transcriptional activation of these genes during stationary-phase stress response. Therefore, we propose that the SWI/SNF complex, in addition to having specific roles in the regulation of MATα-specific genes (47), PHO8 (13), and SUC2 (1), also has a broad genomic role in the transcriptional activation of heat shock and Hsf1 target genes.
Human HSF1 (hHSF1) has been shown to interact with the BRG1 subunit of SWI/SNF and recruit this complex to chromatin templates in vitro (48). In vivo, the recruitment of SWI/SNF by hHSF1 remodels nucleosomes in front of the paused polymerase to facilitate the production of full-length hsp70 mRNA, indicating a role for SWI/SNF in both initiation and elongation of this gene (4). In addition, Hsf4b bound to heat shock elements in human cells recruits Brg1 complexes to the promoters of heat shock proteins under physiological growth conditions and in a cell cycle stage-dependent manner (51). These observations with human cells and the increase in Snf2 occupancy at Hsf1 target genes observed in this study suggest that Hsf1 interacts directly or indirectly with Snf2 to recruit the yeast SWI/SNF complex to the promoters of heat shock protein genes. The fact that Snf2 is not essential for the heat shock stress response and that transcription of these genes occurs, albeit at lower levels, even in the absence of Snf2, is discussed below.
Time line of events leading to transcriptional response to heat shock stress.
Unlike nucleosome displacement at the PHO5 promoter, which takes at least an hour (40), our results show that chromatin remodeling in heat shock-responsive genes begins within seconds and is complete within 15 min after temperature upshift. This nucleosome displacement at the heat shock-activated gene promoters is one of the most rapid chromatin changes yet observed. On average, the transcript levels of the heat shock-activated genes for an snf2Δ strain were approximately twofold lower than those for WT cells. Our measurements of histone H4 occupancy revealed that nucleosome displacement at promoters does occur in the absence of Snf2 but is impaired relative to WT cells, indicating that an alternate mechanism is used for histone displacement in snf2Δ cells. In addition, hyperacetylation was detectable at many of the promoters when chromatin remodeling was artificially delayed in an snf2Δ strain (see Fig. S5 in the supplemental material). A previous report showed that domain-wide nucleosome displacement at the HSP82 promoter was independent of prominent remodelers such as Gcn5 (an H3-specific HAT), Set1, the Paf1 elongation complex, and the chromatin assembly protein Asf1 (61). This report also showed that although inactivation of Snf2 had a sixfold effect of the transcription of HSP82, Snf2 was dispensable for domain-wide chromatin remodeling. It remains to be determined whether hyperacetylation, perhaps due to NuA4, is sufficient for the displacement of promoter nucleosomes at heat shock genes. Taken together, our results show that deletion of SNF2 has a marked effect on transcription and a subtle effect on chromatin remodeling at the promoters of a broad set of heat shock-activated genes. Given that the SWI/SNF complex has partially redundant roles with not only Gcn5 but also Chd1 (50) and that a chd1Δ snf2Δ strain is inviable, it is possible that Chd1 contributes to nucleosome remodeling at the heat shock-responsive genes in the absence of Snf2. Thus, Chd1 or another remodeler may partially compensate for the remodeling defect in the absence of Snf2, but it is possible that this is not sufficient to restore transcriptional regulation to normal levels.
Models for gene activation and repression after heat shock stress.
Based on the above-described features of chromatin remodeling and transcription of heat shock-responsive genes and the data described in this study, we propose the following models for activation and repression of these genes (Fig. 7).
FIG. 7.
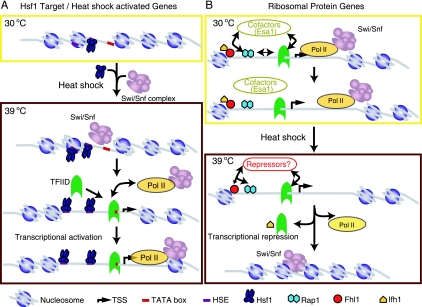
Model for chromatin changes during transcriptional activation and repression after heat shock stress. (A) An activation model for a prototypical heat shock gene. Upon heat shock, Hsf1 bound to the promoter-proximal heat shock element recruits the SWI/SNF complex to the promoter. SWI/SNF displaces nucleosomes in the promoter, exposing the TATA box and making the promoter more accessible for TFIID binding (23). Once TFIID binds, it recruits the holo-RNA Pol II complex and facilitates active preinitiation complex assembly. This leads to activated transcription of the heat shock genes at 39°C. Although not shown in the model, it is possible that Hsf1 recruits a HAT (39) and its action precedes the recruitment of SWI/SNF. (B) Repression model for RP genes. RP genes are actively transcribed at 30°C, and Snf2 recruitment to their coding regions facilitates transcription elongation. Rap1 binds to the promoters of RP genes and directly or indirectly (via cofactors) recruits Esa1 (39). Rap1 also recruits Fhl1 and Ifh1 to the RP genes, and their binding leads to preinitiation complex assembly (41, 42, 54, 62). Upon heat shock, Rap1 recruits presently unknown repressors to the promoter of the RP genes. These repressors downregulate transcription in a SWI/SNF-dependent manner, probably by bringing about nucleosome reassembly at the promoter and the coding region.
The model for activation (Fig. 7A) suggests that the repressive effect of chromatin at Hsf1 target genes is alleviated by the recruitment of the SWI/SNF complex either by Hsf1 or by other coactivators upon heat shock. The SWI/SNF complex binds to NuA4 acetylated histone H4 or SAGA acetylated histone H3 (or both) at the promoter, displaces nucleosomes, and makes the promoter more accessible to transcription factor binding and preinitiation complex assembly. At promoters that are bound by Hsf1 during unstressed conditions, this step occurs prior to heat shock, and the promoters are constitutively poised for rapid transcriptional activation. This is evident by the increased RNA Pol II occupancy at the promoters of heat shock genes prebound by Hsf1 (see Fig. S2 in the supplemental material). We propose that the yeast SWI/SNF complex, similar to its human counterpart, disrupts nucleosomes in the coding region, thus facilitating transcriptional elongation by Pol II in addition to its role at the promoter. This model is supported by our data showing that at 39°C, both promoters and coding regions of heat shock-activated Hsf1 target genes show increased occupancy by Snf2 (Fig. 5).
The model for repression (Fig. 7B) suggests that under normal growth conditions, Snf2, perhaps recruited by a component of the RNA Pol II holoenzyme, facilitates the high-level transcription of the RP genes by nucleosome remodeling during elongation. This is reflected in the higher occupancy of RP gene coding sequences by Snf2. In response to heat shock, either Rap1 or Fhl1, which continue to occupy RP gene promoters even after heat shock (41, 42, 54), recruits SWI/SNF either directly or indirectly via other repressors. SWI/SNF then mediates nucleosome remodeling at the promoter to stabilize the repressed state. Taken together, our models suggest an important dual role for Snf2 during the heat shock stress response as both a coactivator and a corepressor at distinct sets of promoters as well as coding regions in the yeast genome.
Supplementary Material
Acknowledgments
We thank J. D. Lieb and F. Winston for providing the yeast strains YKY6B and FY2103, respectively, and members of the Iyer lab for help with the production of microarrays.
This work was supported in part by a grant from the NIH (CA095548) to V.R.I.
Footnotes
Published ahead of print on 22 January 2008.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Abrams, E., L. Neigeborn, and M. Carlson. 1986. Molecular analysis of SNF2 and SNF5, genes required for expression of glucose-repressible genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 63643-3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bernstein, B. E., C. L. Liu, E. L. Humphrey, E. O. Perlstein, and S. L. Schreiber. 2004. Global nucleosome occupancy in yeast. Genome Biol. 5R62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boy-Marcotte, E., G. Lagniel, M. Perrot, F. Bussereau, A. Boudsocq, M. Jacquet, and J. Labarre. 1999. The heat shock response in yeast: differential regulations and contributions of the Msn2p/Msn4p and Hsf1p regulons. Mol. Microbiol. 33274-283. [DOI] [PubMed] [Google Scholar]
- 4.Corey, L. L., C. S. Weirich, I. J. Benjamin, and R. E. Kingston. 2003. Localized recruitment of a chromatin-remodeling activity by an activator in vivo drives transcriptional elongation. Genes Dev. 171392-1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deckert, J., and K. Struhl. 2001. Histone acetylation at promoters is differentially affected by specific activators and repressors. Mol. Cell. Biol. 212726-2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dion, M. F., S. J. Altschuler, L. F. Wu, and O. J. Rando. 2005. Genomic characterization reveals a simple histone H4 acetylation code. Proc. Natl. Acad. Sci. USA 1025501-5506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dror, V., and F. Winston. 2004. The Swi/Snf chromatin remodeling complex is required for ribosomal DNA and telomeric silencing in Saccharomyces cerevisiae. Mol. Cell. Biol. 248227-8235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ehrenhofer-Murray, A. E. 2004. Chromatin dynamics at DNA replication, transcription and repair. Eur. J. Biochem. 2712335-2349. [DOI] [PubMed] [Google Scholar]
- 9.Eisen, M. B., P. T. Spellman, P. O. Brown, and D. Botstein. 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 9514863-14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Erkina, T. Y., and A. M. Erkine. 2006. Displacement of histones at promoters of Saccharomyces cerevisiae heat shock genes is differentially associated with histone H3 acetylation. Mol. Cell. Biol. 267587-7600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Erkine, A. M., C. C. Adams, T. Diken, and D. S. Gross. 1996. Heat shock factor gains access to the yeast HSC82 promoter independently of other sequence-specific factors and antagonizes nucleosomal repression of basal and induced transcription. Mol. Cell. Biol. 167004-7017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gasch, A. P., P. T. Spellman, C. M. Kao, O. Carmel-Harel, M. B. Eisen, G. Storz, D. Botstein, and P. O. Brown. 2000. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell 114241-4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gregory, P. D., A. Schmid, M. Zavari, M. Munsterkotter, and W. Horz. 1999. Chromatin remodelling at the PHO8 promoter requires SWI-SNF and SAGA at a step subsequent to activator binding. EMBO J. 186407-6414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gross, D. S., C. C. Adams, S. Lee, and B. Stentz. 1993. A critical role for heat shock transcription factor in establishing a nucleosome-free region over the TATA-initiation site of the yeast HSP82 heat shock gene. EMBO J. 123931-3945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guillemette, B., A. R. Bataille, N. Gevry, M. Adam, M. Blanchette, F. Robert, and L. Gaudreau. 2005. Variant histone H2A.Z is globally localized to the promoters of inactive yeast genes and regulates nucleosome positioning. PLoS Biol. 3e384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hahn, J. S., Z. Hu, D. J. Thiele, and V. R. Iyer. 2004. Genome-wide analysis of the biology of stress responses through heat shock transcription factor. Mol. Cell. Biol. 245249-5256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hassan, A. H., K. E. Neely, and J. L. Workman. 2001. Histone acetyltransferase complexes stabilize swi/snf binding to promoter nucleosomes. Cell 104817-827. [DOI] [PubMed] [Google Scholar]
- 18.Horn, P. J., and C. L. Peterson. 2001. The bromodomain: a regulator of ATP-dependent chromatin remodeling? Front. Biosci. 6D1019-D1023. [DOI] [PubMed] [Google Scholar]
- 19.Iyer, V. R. 2003. Isolation and amplification of array material from yeast, p. 30-34. In D. Bowtell and J. Sambrook (ed.), DNA microarrays: a molecular cloning manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 20.Iyer, V. R. 2003. Microarray-based detection of DNA-protein interactions: chromatin immunoprecipitation on microarrays, p. 453-463. In D. Bowtell and J. Sambrook (ed.), DNA microarrays: a molecular cloning manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 21.Iyer, V. R., C. E. Horak, C. S. Scafe, D. Botstein, M. Snyder, and P. O. Brown. 2001. Genomic binding sites of the yeast cell-cycle transcription factors SBF and MBF. Nature 409533-538. [DOI] [PubMed] [Google Scholar]
- 22.Jenuwein, T., and C. D. Allis. 2001. Translating the histone code. Science 2931074-1080. [DOI] [PubMed] [Google Scholar]
- 23.Kim, J., and V. R. Iyer. 2004. Global role of TATA box-binding protein recruitment to promoters in mediating gene expression profiles. Mol. Cell. Biol. 248104-8112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kurdistani, S. K., S. Tavazoie, and M. Grunstein. 2004. Mapping global histone acetylation patterns to gene expression. Cell 117721-733. [DOI] [PubMed] [Google Scholar]
- 25.Lee, C. K., Y. Shibata, B. Rao, B. D. Strahl, and J. D. Lieb. 2004. Evidence for nucleosome depletion at active regulatory regions genome-wide. Nat. Genet. 36900-905. [DOI] [PubMed] [Google Scholar]
- 26.Liu, C. L., T. Kaplan, M. Kim, S. Buratowski, S. L. Schreiber, N. Friedman, and O. J. Rando. 2005. Single-nucleosome mapping of histone modifications in S. cerevisiae. PLoS Biol. 3e328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Longtine, M. S., A. McKenzie III, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14953-961. [DOI] [PubMed] [Google Scholar]
- 28.Martens, J. A., and F. Winston. 2002. Evidence that Swi/Snf directly represses transcription in S. cerevisiae. Genes Dev. 162231-2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martens, J. A., and F. Winston. 2003. Recent advances in understanding chromatin remodeling by Swi/Snf complexes. Curr. Opin. Genet. Dev. 13136-142. [DOI] [PubMed] [Google Scholar]
- 30.Mellor, J., and A. Morillon. 2004. ISWI complexes in Saccharomyces cerevisiae. Biochim. Biophys. Acta 1677100-112. [DOI] [PubMed] [Google Scholar]
- 31.Miller, J. A., and J. Widom. 2003. Collaborative competition mechanism for gene activation in vivo. Mol. Cell. Biol. 231623-1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morano, K. A., and D. J. Thiele. 1999. Heat shock factor function and regulation in response to cellular stress, growth, and differentiation signals. Gene Expr. 7271-282. [PMC free article] [PubMed] [Google Scholar]
- 33.Narlikar, G. J., H. Y. Fan, and R. E. Kingston. 2002. Cooperation between complexes that regulate chromatin structure and transcription. Cell 108475-487. [DOI] [PubMed] [Google Scholar]
- 34.Neely, K. E., A. H. Hassan, A. E. Wallberg, D. J. Steger, B. R. Cairns, A. P. Wright, and J. L. Workman. 1999. Activation domain-mediated targeting of the SWI/SNF complex to promoters stimulates transcription from nucleosome arrays. Mol. Cell 4649-655. [DOI] [PubMed] [Google Scholar]
- 35.Osley, M. A., and X. Shen. 2006. Altering nucleosomes during DNA double-strand break repair in yeast. Trends Genet. 22671-677. [DOI] [PubMed] [Google Scholar]
- 36.Pokholok, D. K., C. T. Harbison, S. Levine, M. Cole, N. M. Hannett, T. I. Lee, G. W. Bell, K. Walker, P. A. Rolfe, E. Herbolsheimer, et al. 2005. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell 122517-527. [DOI] [PubMed] [Google Scholar]
- 37.Radonjic, M., J. C. Andrau, P. Lijnzaad, P. Kemmeren, T. T. Kockelkorn, D. van Leenen, N. L. van Berkum, and F. C. Holstege. 2005. Genome-wide analyses reveal RNA polymerase II located upstream of genes poised for rapid response upon S. cerevisiae stationary phase exit. Mol. Cell 18171-183. [DOI] [PubMed] [Google Scholar]
- 38.Raisner, R. M., P. D. Hartley, M. D. Meneghini, M. Z. Bao, C. L. Liu, S. L. Schreiber, O. J. Rando, and H. D. Madhani. 2005. Histone variant H2A.Z marks the 5′ ends of both active and inactive genes in euchromatin. Cell 123233-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reid, J. L., V. R. Iyer, P. O. Brown, and K. Struhl. 2000. Coordinate regulation of yeast ribosomal protein genes is associated with targeted recruitment of Esa1 histone acetylase. Mol. Cell 61297-1307. [DOI] [PubMed] [Google Scholar]
- 40.Reinke, H., and W. Horz. 2003. Histones are first hyperacetylated and then lose contact with the activated PHO5 promoter. Mol. Cell 111599-1607. [DOI] [PubMed] [Google Scholar]
- 41.Rudra, D., Y. Zhao, and J. R. Warner. 2005. Central role of Ifh1p-Fhl1p interaction in the synthesis of yeast ribosomal proteins. EMBO J. 24533-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schawalder, S. B., M. Kabani, I. Howald, U. Choudhury, M. Werner, and D. Shore. 2004. Growth-regulated recruitment of the essential yeast ribosomal protein gene activator Ifh1. Nature 4321058-1061. [DOI] [PubMed] [Google Scholar]
- 43.Shogren-Knaak, M., H. Ishii, J. M. Sun, M. J. Pazin, J. R. Davie, and C. L. Peterson. 2006. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science 311844-847. [DOI] [PubMed] [Google Scholar]
- 44.Sobel, R. E., R. G. Cook, C. A. Perry, A. T. Annunziato, and C. D. Allis. 1995. Conservation of deposition-related acetylation sites in newly synthesized histones H3 and H4. Proc. Natl. Acad. Sci. USA 921237-1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Spellman, P. T., G. Sherlock, M. Q. Zhang, V. R. Iyer, K. Anders, M. B. Eisen, P. O. Brown, D. Botstein, and B. Futcher. 1998. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol. Biol. Cell 93273-3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Strahl, B. D., and C. D. Allis. 2000. The language of covalent histone modifications. Nature 40341-45. [DOI] [PubMed] [Google Scholar]
- 47.Sudarsanam, P., V. R. Iyer, P. O. Brown, and F. Winston. 2000. Whole-genome expression analysis of snf/swi mutants of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 973364-3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sullivan, E. K., C. S. Weirich, J. R. Guyon, S. Sif, and R. E. Kingston. 2001. Transcriptional activation domains of human heat shock factor 1 recruit human SWI/SNF. Mol. Cell. Biol. 215826-5837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thomson, S., A. Hollis, C. A. Hazzalin, and L. C. Mahadevan. 2004. Distinct stimulus-specific histone modifications at hsp70 chromatin targeted by the transcription factor heat shock factor-1. Mol. Cell 15585-594. [DOI] [PubMed] [Google Scholar]
- 50.Tran, H. G., D. J. Steger, V. R. Iyer, and A. D. Johnson. 2000. The chromo domain protein chd1p from budding yeast is an ATP-dependent chromatin-modifying factor. EMBO J. 192323-2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tu, N., Y. Hu, and N. F. Mivechi. 2006. Heat shock transcription factor (Hsf)-4b recruits Brg1 during the G1 phase of the cell cycle and regulates the expression of heat shock proteins. J. Cell. Biochem. 981528-1542. [DOI] [PubMed] [Google Scholar]
- 52.Van Holde, K. E., C. G. Sahasrabuddhe, and B. R. Shaw. 1974. A model for particulate structure in chromatin. Nucleic Acids Res. 11579-1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wach, A. 1996. PCR-synthesis of marker cassettes with long flanking homology regions for gene disruptions in S. cerevisiae. Yeast 12259-265. [DOI] [PubMed] [Google Scholar]
- 54.Wade, J. T., D. B. Hall, and K. Struhl. 2004. The transcription factor Ifh1 is a key regulator of yeast ribosomal protein genes. Nature 4321054-1058. [DOI] [PubMed] [Google Scholar]
- 55.Warner, J. R. 1999. The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci. 24437-440. [DOI] [PubMed] [Google Scholar]
- 56.Winzeler, E. A., D. D. Shoemaker, A. Astromoff, H. Liang, K. Anderson, B. Andre, R. Bangham, R. Benito, J. D. Boeke, H. Bussey, et al. 1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285901-906. [DOI] [PubMed] [Google Scholar]
- 57.Woodcock, C. L. 2006. Chromatin architecture. Curr. Opin. Struct. Biol. 16213-220. [DOI] [PubMed] [Google Scholar]
- 58.Yuan, G. C., Y. J. Liu, M. F. Dion, M. D. Slack, L. F. Wu, S. J. Altschuler, and O. J. Rando. 2005. Genome-scale identification of nucleosome positions in S. cerevisiae. Science 309626-630. [DOI] [PubMed] [Google Scholar]
- 59.Yudkovsky, N., C. Logie, S. Hahn, and C. L. Peterson. 1999. Recruitment of the SWI/SNF chromatin remodeling complex by transcriptional activators. Genes Dev. 132369-2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang, H., D. N. Roberts, and B. R. Cairns. 2005. Genome-wide dynamics of Htz1, a histone H2A variant that poises repressed/basal promoters for activation through histone loss. Cell 123219-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhao, J., J. Herrera-Diaz, and D. S. Gross. 2005. Domain-wide displacement of histones by activated heat shock factor occurs independently of Swi/Snf and is not correlated with RNA polymerase II density. Mol. Cell. Biol. 258985-8999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhao, Y., K. B. McIntosh, D. Rudra, S. Schawalder, D. Shore, and J. R. Warner. 2006. Fine-structure analysis of ribosomal protein gene transcription. Mol. Cell. Biol. 264853-4862. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.