Abstract
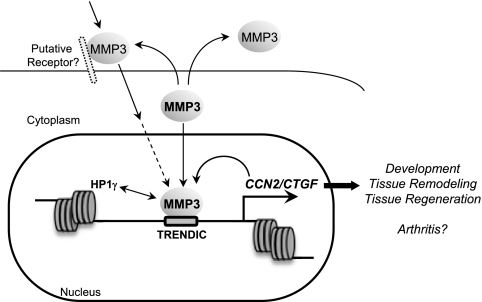
Matrix metalloproteinase 3 (MMP3) is well known as a secretory endopeptidase that degrades extracellular matrices. Recent reports indicated the presence of MMPs in the nucleus (A. J. Kwon et al., FASEB J. 18:690-692, 2004); however, its function has not been well investigated. Here, we report a novel function of human nuclear MMP3 as a trans regulator of connective tissue growth factor (CCN2/CTGF). Initially, we cloned MMP3 cDNA as a DNA-binding factor for the CCN2/CTGF gene. An interaction between MMP3 and transcription enhancer dominant in chondrocytes (TRENDIC) in the CCN2/CTGF promoter was confirmed by a gel shift assay and chromatin immunoprecipitation. The CCN2/CTGF promoter was activated by overexpressed MMP3, whereas a TRENDIC mutant promoter lost the response. Also, the knocking down of MMP3 suppressed CCN2/CTGF expression. By cytochemical and histochemical analyses, MMP3 was detected in the nuclei of chondrocytic cells in culture and also in the nuclei of normal and osteoarthritic chondrocytes in vivo. The nuclear translocation of externally added recombinant MMP3 and six putative nuclear localization signals in MMP3 also were shown. Furthermore, we determined that heterochromatin protein gamma coordinately regulates CCN2/CTGF by interacting with MMP3. The involvement of this novel role of MMP3 in the development, tissue remodeling, and pathology of arthritic diseases through CCN2/CTGF regulation thus is suggested.
Connective tissue growth factor (CTGF/CCN2) is a member of the CCN family of matricellular proteins and also has been designated Hcs24, FISP12, IGFBP8, IGFBP-rP2, βIG-M2, and ecogenin. The other CCN proteins include Cyr61/CCN1, NOV/CCN3, WISP1/CCN4, WISP2/CCN5, and WISP3/CCN6 (5, 26, 38, 39) as well, and they are structurally and functionally related glycoproteins involved in cell differentiation, proliferation, adhesion, migration, and the formation of the extracellular matrix. These matricellular functions of CCNs are involved in physiological processes such as wound healing, angiogenesis, morphogenesis, and embryogenesis as well as in pathological states including fibrotic disorders, cancer, and arthritis.
Earlier we showed that CCN2 promotes endochondral ossification by acting on chondrocytes, osteoblasts, and endothelial cells (35, 37, 46). For example, CCN2 promotes physiological chondrocytic proliferation and extracellular matrix (ECM) formation. We also reported the regeneration of defects in articular cartilage in rat knee joints following treatment with recombinant CCN2 (36). Furthermore, ctgf-null mice were dead on delivery and were characterized by defective angiogenesis, the derangement of endochondral ossification, and dysmorphisms that occurred as a result of impaired chondrocyte proliferation and an abnormal ECM composition within the hypertrophic zone (24).
Matrix metalloproteinases (MMPs) are zinc-dependent endopeptidases that are involved in the remodeling and turnover of the ECM in physiological processes such as angiogenesis, wound healing, embryogenesis, and morphogenesis as well as in pathological states including cancers, myocardial infarction, fibrotic disorders, rheumatism, and osteoarthritis (33, 49). Cartilage is a connective tissue that is constructed by chondrocytes embedded within an ECM predominantly composed of collagens and proteoglycans. ECM remodeling is achieved by regulating the production and degradation of specific ECM components. MMPs, which comprise a large family of enzymes with differential abilities to degrade specific ECM components, play a vital role in this process. MMPs also cleave growth factors and their binding proteins, thereby activating or inhibiting specific signaling events (15). Of note, the expression and role of MMP3 have been investigated in the pathological status of articular cartilage, such as in osteoarthritis and rheumatism (1, 52).
Recent study has demonstrated the existence and functions of intracellular MMPs and tissue inhibitors of metalloproteinases (TIMPs). TIMP-1 accumulates in the cellular nuclei in association with the cell cycle (54). Alternative splicing and promoter usage generate an intracellular MMP11 isoform directly translated as an active MMP (31). MMP2 is found in the nuclei of cardiac myocytes and is capable of cleaving poly(ADP-ribose) polymerase (PARP) in vitro (28). MMP3 also is detected in the nuclei of hepatocytes and is involved in apoptosis (47). MT1-MMP exhibits an intracellular cleavage function and causes chromosome instability, and it cleaves centrosomal pericentrin in human cells but not in murine cells (12).
The mechanisms of CCN2/CTGF induction/production have been well investigated (4, 26, 29); however, there have been few approaches to identify directly binding regulatory proteins of the CCN2/CTGF gene. Recently, we investigated cell type-specific mechanisms of CCN2/CTGF gene regulation and found a cis-acting element, transcription enhancer dominant in chondrocytes (TRENDIC), between positions −202 and −180 from the transcription start site of CCN2/CTGF (7), a region that previously had been predicted to contain an NF-1-like site (7, 13). In our previous study, the strong production of CCN2 from chondrocytic cells was estimated to be mediated by TRENDIC rather than by a juxtaposing Smad-binding element (SBE) (7, 23). In this present study, we subsequently cloned the cDNAs encoding TRENDIC-binding factors and unexpectedly found MMP3/stromelysin-1 to be one of them. We then investigated whether or not MMP3 is localized in the nuclei of chondrocytes in vitro and in vivo. Having found such localization, we also examined if the nuclear MMP3 could bind with enhancer sequences in the CCN2/CTGF promoter and activate CCN2/CTGF transcription, and we showed that it did so. Finally, we evaluated the properties of MMP3 as a transcription factor by analyzing nuclear MMP3-associated proteins (NuMAPs).
MATERIALS AND METHODS
HCS-2/8 cDNA phage library.
The cDNA phage library was constructed by using a ZAP cDNA synthesis kit (Toyobo, Osaka, Japan) and a ZAP express predigested Gigapack cloning kit (Stratagene, La Jolla, CA) according to the manufacturer's protocols. Total RNA was prepared from HCS-2/8 cells by an acid guanidinium-phenol-chloroform (AGPC) method. Polyadenylated RNA was prepared from the total RNA by using an Oligotex-dT30 Super mRNA purification kit (Takara, Otsu, Japan). First-strand cDNA was synthesized by using linker primer 5′-CTCGAGTTTTTTTTTTTT and 5-methyl-dCTP. Second-strand cDNA was synthesized by using RNase H and DNA polymerase I. The double-strand cDNA was blunted by using Pfu DNA polymerase. The cDNA was added to a phosphorylated EcoRI adapter, digested by XhoI, and inserted in a ZAP express vector digested with EcoRI and XhoI. The average length of the insert cDNA of the library was confirmed by PCR to be approximately 2 kb.
Southwestern screening.
We performed Southwestern screening as previously described, with a slight modification (53). The Escherichia coli XL1-Blue MRF′ strain was infected with the lambda phage library and then spread on NZY agar plates containing 0.5 M isopropyl-β-d-thiogalactopyranoside (IPTG). Formed plaques were lifted onto nitrocellulose membranes (GE Healthcare, Uppsala, Sweden). The double-stranded DNA (dsDNA) of four tandem repeats of TRENDIC was prepared by annealing the following oligonucleotides: 4× TRENDIC-s, 5′-ACG CGT [(CTG TGA GCT GGA GTG TGC CAGC) CAG]4 CTC GAG-3′; 4× TRENDIC-as, 5′-CTC GAG [CTG (GCT GGC ACA CTC CAG CTC ACAG)]4 ACG CGT-3′. The annealed probe was end labeled with [γ-32P]ATP and was applied to the membranes in binding buffer (10 mM HEPES, pH 7.9, containing 50 mM KCl, 10 mM MgCl2, 0.1 mM EDTA, 1 mM dithiothreitol [DTT], 0.25% skim milk, and 50 μg/ml of denatured and undenatured salmon sperm DNA). After the membranes had been washed three times with a wash buffer (10 mM HEPES, pH 7.9, containing 250 mM KCl, 10 mM MgCl2, 0.1 mM EDTA, 1 mM DTT, and 0.25% skim milk), they were exposed to X-ray films. The positive plaques were picked up for subsequent secondary and tertiary screenings as well. The final cDNA clones in a pBK-cytomegalovirus vector were obtained by in vivo excision using the E. coli XLOLR strain.
DNA sequencing and computer analysis.
The cDNA cloned into each plasmid was sequenced by the dideoxy chain termination method (42) with a BigDye Terminator cycle sequencing ready reaction kit, version 2.0 (Applied Biosystems, Foster City, CA), and an ABI PRISM 310 genetic analyzer (Applied Biosystems). The sequences were analyzed by using BLASTn, Evidence Viewer, and UniGene online services at the NCBI website (http://www.ncbi.nlm.nih.gov/). These data on the cloned genes are summarized in Table 1.
TABLE 1.
cDNA clones of TRENDIC binding factor candidatesa
| Length of cloned cDNA | Gene name | Expression profile in UniGene | Chromosomal position | GenBank accession no. |
|---|---|---|---|---|
| The last 1,857 bp of a total of 3,733 bp | FLJ14525 | Ubiquitous | 1q42.13-q43 | NM_032800 |
| 1,821 bp (FL) | MMP3 | Connective tissue, cartilage, bone, osteoarthritis cartilage, and nonneoplasia | 11q22.3 | NM_002422 |
| 2,036 bp (FL) | DDOST | Ubiquitous | 1p36.1 | NM_005216 |
The DNA sequences were 100% matched to the known genes in the NCBI GenBank database. Their GenBank accession numbers, expression tendency as determined by the UniGene expressed sequence tag database, and their chromosomal positions are shown. FL, full-length cDNA.
Cell culture.
The following human-derived cells were used: human chondrosarcoma-derived chondrocytic HCS-2/8 cells (40, 51), MDA-MB-231 breast carcinoma cells, HeLa cells derived from human cervical cancer, and SaOS-2 osteosarcoma-derived cells. A COS7 monkey kidney-derived cell line also was used. Cells were cultured in Dulbecco's modified Eagle's minimum essential medium (DMEM) supplemented with 10% fetal bovine serum (FBS) in humidified air containing 5% CO2 at 37°C. For immunocytochemistry and internalization studies, glass chamber slides (4 or 8 well) were coated with 50 μg/ml of collagen (Cellmatrix type I-C; Nitta-Gellatin, Osaka, Japan) for 30 min before cells were seeded.
Antibodies.
We used the following anti-MMP3 antibodies: anti-MMP3 N-terminal region (5052; Sigma, St. Louis, MO), anti-MMP3 hinge region (4802; Sigma), anti-MMP3 C-terminal domain (4927; Sigma), and anti-MMP3 catalytic (CAT) domain antibody (4190; Sigma). We also used anti-CCN2/CTGF (AF660; R&D), anti-histone H3 (06-599; Upstate), anti-Sox6 (S7193; Sigma), anti-α-tubulin immunoglobulin G1 (IgG1) (T9026; Sigma), anti-cathepsin D (C-20; Santa Cruz Biotech, Santa Cruz, CA), anti-lamin A/C monoclonal IgM (sc-7293; Santa Cruz), and anti-β-actin (AC-74; Sigma). We used the following antibodies to detect tags: anti-Flag M2 (Sigma), anti-Myc tag (Abcam, Cambridge, United Kingdom), and anti-glutathione S-transferase (anti-GST) (GE Healthcare) antibodies. An antidigoxigenin alkaline phosphatase-conjugated Fab (Roche, Basel, Switzerland) was used in electrophoresis mobility shift assays (EMSA). For Western blotting, we used horseradish peroxidase-conjugated secondary antibodies against anti-mouse IgG (Amersham) and anti-rabbit IgG (Dako, Copenhagen, Denmark). An anti-rabbit IgG rhodamine conjugate (Sigma) was used for immunostaining. We used these antibodies at the concentrations instructed by the manufacturer. The MMP3 enzyme-linked immunosorbent assay (ELISA) kit was purchased from Daiichi Pure Chemicals (Tokyo, Japan).
Preparation of subcellular fraction proteins.
The nuclear and cytoplasmic proteins were prepared by using a CelLytic NuCLEAR extraction kit (Sigma) according to the manufacturer's protocol. The subcellular fractions were prepared by using a ProteoExtract subcellular proteome extraction kit (Calbiochem, San Diego, CA) according to the manufacturer's protocol. Total cell lysate was prepared by using a CelLytic M reagent (Sigma) according to the manufacturer's protocols. The protease inhibitor cocktail (Sigma) was added at the appropriate steps.
SDS-PAGE and Western blot analysis.
Extracted proteins were heated at 95°C for 5 min in sodium dodecyl sulfate (SDS) sample buffer in the presence of 5% 2-mercaptoethanol and separated by SDS-polyacrylamide gel electrophoresis (PAGE) in 12% polyacrylamide gel. Alternatively, proteins were heated at 70°C for 10 min in lithium dodecyl sulfate sample buffer containing 50 mM DTT and were separated in a 10% Bis-Tris NuPAGE gel (Invitrogen) in morpholinepropanesulfonic acid running buffer containing an antioxidant. Semidry electroblotting was carried out using a polyvinylidene difluoride membrane (Hybond P; GE Healthcare). The membrane was blocked in Tris-buffered saline containing 0.05% Tween 20 (TBST) and 5% skim milk for 30 min at room temperature (RT). After being blocked, the membrane was incubated with the primary antibody overnight at 4°C and subsequently was incubated with the secondary antibody for 1 h at RT in the blocking solution. The blot was visualized by using an enhanced chemiluminescence (ECL) Western blotting analysis system (GE Healthcare) with chemiluminescence detection. The photogram was obtained by autoradiography or by using an ECL minicamera (GE Healthcare). The band signals obtained by Western blotting were quantified by using ImageJ, version 1.37 (Wayne Rasband, NIH).
Recombinant proteins, MMP3 activator, and inhibitors.
A recombinant human proenzyme MMP3 (rhMMP3; R&D) was purchased and used for the molecular weight control and the internalization assay. For other experiments, we used our purified rhMMP3, prepared as described below. Organomercurial (4-aminophenyl)mercuric acetate (APMA) (Sigma) was dissolved in 50 mM NaOH. Active MMP3 was prepared by incubating proenzyme MMP3 (final concentration, 83.3 nM) with 1 mM of APMA in Tris buffer (50 mM Tris-HCl, pH 7.5, containing 5 mM CaCl2 and 0.05% Triton X-100) at 37°C for 4 h. All MMP inhibitors were purchased from Calbiochem and were dissolved and stored in dimethyl sulfoxide (DMSO). Before use, they were diluted 100-fold once in a cell culture medium (1% DMSO) and then added to cultured cells (final concentration, 0.01% DMSO). Transforming growth factor β (TGF-β) (10 ng/ml) was purchased from Peprotech (Rocky Hill, NJ).
Stromelysin endopeptidase activity assay.
The stromelysin endopeptidase activity assay was carried out by using the fluorescence resonance energy transfer peptide substrate Mca-RPKPVE-Nval-WRK(Dnp)-NH2 fluorogenic substrate II (R&D, Minneapolis, MN) (32). The substrate (10 μM) was mixed with enzyme or nuclear extract in a Tris buffer (50 mM Tris-HCl, pH 7.5, containing 5 mM CaCl2 and 0.05% Triton X-100) and incubated at 37°C. The relative fluorescence units (RFUs) of the reactant in a 96-well black plate were measured by using a Fluoroskan AscentFL (Labsystem, Helsinki, Finland).
Immunocytochemistry.
For immunocytochemistry, cells were fixed in 4% (wt/vol) paraformaldehyde in phosphate-buffered saline (PBS) for 15 min, washed in TBS, and permeabilized with 0.2% Triton X-100 for 15 min. Incubation with primary and secondary antibodies was performed in PBS containing 1.5% normal goat serum and 0.05% Triton X-100 at RT for 1 h, respectively. After three washes with TBS, the mounting and DNA staining were performed by using ProLong gold antifade reagents with 4′,6-diamino-2-phenylindole (DAPI) (Molecular Probes, Invitrogen). Cells were observed with an immunofluorescence microscope (Biozero; Keyence, Osaka, Japan) or a Radiance 2100 laser scanning system (Bio-Rad).
Conjugation of Cy3 to rhMMP3.
For the conjugation of Cy3 to rhMMP3, the Tris-based solvent of rhMMP3 was replaced with sodium carbonate-sodium bicarbonate buffer (pH 9.3) by use of a Microcon YM-30 (Millipore, Billerica, MA), and the concentration was adjusted to 100 μg/ml. MMP3 (50 μl) was added to a vial of Cy3 monoreactive dye (GE Healthcare) and was incubated at RT for 30 min with mixing every 10 min. The labeled MMP3 was desalted and separated from the unconjugated Cy3 on a MicroSpin G-25 column (GE Healthcare) preequilibrated with PBS.
Internalization studies.
The cells in chamber slides were washed once with prewarmed serum-free medium and incubated with Cy3-MMP3 (1 μg/ml) in the serum-free medium at 37°C for 5, 10, 15, 30, or 60 min and subsequently were fixed with 4% paraformaldehyde in PBS for 15 min. The cells were stained with 10 nM Sytox green nucleic acid stain (Molecular Probes, Invitrogen) in TBS for 20 min and mounted in ProLong antifade reagents (Molecular Probes, Invitrogen). The cells were observed by confocal laser microscopy as described below, and Cy3-positive cells were classified into three groups: cells with cytoplasm-dominant signals, cells with both cytoplasmic and nuclear signals, and cells with nucleus-dominant signals.
Confocal laser scanning microscopy.
Confocal laser scanning was carried out with a Radiance 2100 laser scanning system equipped with an argon and krypton laser (excitation wavelengths of 488 and 568 nm) (Bio-Rad) through an Eclipse TE2000-U microscope (Nikon) with a ×60 objective lens. The scan was visualized with the software Laser Sharp 2000 (Bio-Rad).
NLS analysis.
Nuclear localization signals (NLSs) in MMP3 were predicted by the PSORTII program (http://psort.nibb.ac.jp/helpwww2.html#src) (34). Several arginine- and lysine-rich sequences in MMP3 also were selected for analysis. pEGFP-NLSs were constructed as described below. COS7 cells were cultured in the 4-well chamber slides and were transfected with 200 ng of a series of pEGFP-NLS constructs by the aid of Fugene 6 transfection reagent (Roche). After being cultured for 24 h, cells were fixed with 4% formaldehyde in PBS and mounted with a fluorescent mounting medium (Dako). The subcellular localization of the enhanced green fluorescent protein (EGFP) was visualized with confocal laser scanning microscopy.
EMSA.
EMSA was carried out as described previously (7) by using a digoxigenin system (Roche). For preparing the probes, sense and antisense oligonucleotides were annealed in a TEN buffer (10 mM Tris, 1 mM EDTA, 100 mM NaCl, pH 8.0) and labeled with digoxigenin-11-UTP by using terminal transferase (Roche). The following pairs of oligonucleotides were annealed: TRENDIC-s (5′-CTG TGA GCT GGA GTG TGC CAG C-3′) and TRENDIC-as (5′-GCT GGC ACA CTC CAG CTC ACA G-3′); BCE1/TbRE-s (5′-CTG AGT GTC AAG GGG TCA GGA-3′) and BCE1/TbRE-as (5′-TCC TGA CCC CTT GAC ACT CAG-3′); and Seq2-s (5′-GAA TCA GGA GTG GTG CGA AG-3′) and Seq2-as (5′-CTT CGC ACC ACT CCT GAT TC-3′). The cellular or nuclear extract (5 μg) was preincubated with antibodies for 15 min at RT. Subsequently, 60 fmol of probes was added to the 0.5× binding buffer [10 mM HEPES, pH 7.6, containing 0.5 mM EDTA, 5 mM (NH4)2SO4, 0.5 mM DTT, 1% Tween 20, and 15 mM KCl] with 5 ng/ml of poly(dI-dC) for 15 min at RT. rhMMP3 (300 ng) was incubated with 45 fmol of probe, 50 ng/ml of poly-l-lysine, and 5 ng/ml of poly(dI-dC) in the binding buffers for 15 min. Electrophoresis was performed in a 6% polyacrylamide Tris-borate-EDTA (TBE) gel in a 0.5× TBE buffer for 60 to 100 min. Electroblotting was carried out on a positively charged nylon membrane (GE Healthcare). The UV cross-linking and detection of digoxigenin were carried out according to the manufacturer's protocols (Roche).
Probe CCN2p160, which contains a CCN2 promoter DNA fragment between positions −292 and −137, was prepared from a plasmid template by PCR and labeled with digoxigenin. For EMSA performed with rhMMP3, MMP3 inhibitor (0.5 mM) was added to each reaction. For supershift assays, rhMMP3 and an antibody (2 μg) were preincubated at 4°C for 40 min before the addition of the probes. For competitive EMSA, the recombinant protein and competitor were preincubated at 4°C for 5 min before the addition of the probe. Poly-l-lysine (0.5 or 1.0 μg/reaction) was used.
ChIP assay.
The chromatin immunoprecipitation (ChIP) assay was carried out according to the manufacturer's protocol (Upstate) with a slight modification. A half million HCS-2/8 cells were seeded in each well of a 6-well plate and were cultured for 48 h with a medium change at 24 h. Formaldehyde (final concentration, 1%) was added to the medium, and the cells were incubated for 10 min at 37°C. The cells were washed twice and scraped in ice-cold PBS containing a protease inhibitor cocktail (Sigma) and then centrifuged for 4 min at 200 × g at 4°C. Subsequently, the cells were lysed in 200 μl of SDS lysis buffer (50 mM Tris-HCl, pH 8.1, containing 10 mM EDTA and 1% SDS) for 10 min on ice. DNA was sheared by three 10-s rounds of sonication on ice by using a Handy Sonic model UR-20P (Tomy Seiko, Tokyo, Japan) at 30% of maximum power. After centrifugation at 10,000 × g at 4°C for 10 min, the supernatant was diluted 10-fold in ChIP dilution buffer (16.7 mM Tris-HCl, pH 8, containing 167 mM NaCl, 1.2 mM EDTA, 0.01% SDS, 1.1% Triton X-100, and protease inhibitor cocktail), and 1% of it was retained as the input. The sample was precleared by rotating it with 80 μl (4%) of a 50% salmon sperm DNA-50% protein A-agarose slurry (Upstate) for 30 min at 4°C. After a brief centrifugation, the supernatant was rotated with or without the anti-MMP3 CAT domain antibody (1:500; Sigma) overnight at 4°C and was further incubated after adding 60 μl (3%) of the 50% salmon sperm DNA-50% protein A-agarose slurry at 4°C for 1 h. The protein A-agarose-antibody-antigen complex was centrifuged at 300 × g at 4°C for 1 min, and the pellet was washed once with low-salt wash buffer (20 mM Tris-HCl, pH 8.1, containing 150 mM NaCl, 2 mM EDTA, 0.1% SDS, and 1% Triton X-100), once with high-salt wash buffer (20 mM Tris-HCl, pH 8.1, containing 500 mM NaCl, 2 mM EDTA, 0.1% SDS, and 1% Triton X-100), once with LiCl wash buffer (10 mM Tris-HCl, pH 8.1, containing 0.25 M LiCl, 1% NP-40, 1% deoxycholate, and 1 mM EDTA), and twice with TE buffer (10 mM Tris-HCl, pH 8.0, 1 mM EDTA). The antigen complex was eluted by being mixed and rotated in 250 μl of freshly prepared elution buffer (1% SDS, 0.1 M NaHCO3). The elution step was repeated, and the eluates were combined. A 4% volume of 5 M NaCl was added to the eluate and to the input sample. The cross-linked chromatin complex was reversed by being heated at 65°C for 4 h, and DNA was purified using QIAquick spin columns (Qiagen, Hilden, Germany). PCR was carried out using Prime STAR HS DNA polymerase (Takara) according to the manufacturer's protocol. Ten percent of the total purified DNA was used for the PCR in 50 μl of reaction mixture. The 204 bp of CCN2 enhancer fragment between −292 and −88 was amplified by using the primer pair Seq2-s (5′-GAA TCA GGA GTG GTG CGA AG-3′) and 110bp-as (5′-ATT CCT CGC ATT CCT CCC CAC CT-3′) in 30 cycles of PCR under the following conditions: 98°C for 10 s, 62°C for 30 s, and 72°C for 30 s. The 215-bp cDNA of GAPDH was amplified by using the primer pair GAPDH NEO LCL (5′-GCC AAA AGG GTC ATC ATC TC-3′) and GAPDH NEO LCR (5′-GTC TTC TGG GTG GCA GTG AT-3′) in 30 cycles of PCR under the following conditions: 98°C for 10 s, 65°C for 20 s, and 72°C for 20 s. The PCR products were analyzed by 2% agarose gel electrophoresis.
Plasmid constructs.
The cDNA of MMP3 cloned in pBK-CMV by Southwestern screening was subcloned into the p3xFlag-myc vector (Sigma) via a PCR-based method, and it was designated p3Flag-MMP3-myc. A series of CTGF/CCN2 promoter-luciferase reporter constructs were described previously (7-9). The cDNA of MMP3 was subcloned from the p3Flag-MMP3-myc vector to the pCold-TFII vector for expression in E. coli. Expression plasmids for MMP3 domains (full length [FL] and active forms, the prodomain, the combination of the CAT and Hinge domains [CAT+Hinge], and hemopexin-like repeat [PEX]) and MMP3 point mutants (H218R, H228R, and H218/228R) were constructed via a PCR-mediated mutagenesis method. The cDNA of GST was recombined from the pGEX-6P-3 vector (Amersham) to the pCold-TFII vector (Takara) using HindIII and SalI sites by a PCR-mediated method to constitute pCold-GST. The cDNAs of heterochromatin protein 1α (HP1α), HP1γ, NF45, nuclear receptor corepressor 1 (NCoR1), and chromatin assembly factor p48/retinoblastoma binding protein 4/7 (CAFp48/RBBP4) were prepared from mRNA of HCS-2/8 cells via reverse transcription-PCR and were subcloned into the pCold-GST vector. For the preparation of pEGFP-NLS constructs, sense and antisense oligonucleotides for NLSs were annealed and subcloned into the pEGFP-C1 vector (Clontech, Takara) between XhoI and EcoRI sites. These constructs are designated pEGFP-NLS0 to pEGFP-NLS5. All constructs were confirmed by DNA sequencing.
pcDNA3.1(−)-3HA (p3xHA) was constructed by inserting a three-hemagglutinin (HA) tag in the pcDNA3.1(−) vector (Invitrogen). pcDNA3.1(−)HP1γ-3HA, pcDNA3.1(−)NF45-3HA, pcDNA3.1(−)RBBP4-3HA, and pcDNA3.1(−)DDOST-3HA were constructed via PCR-mediated subcloning. All constructs were confirmed by DNA sequencing and Western blotting.
DNA transfection and luciferase assay.
Cationic liposome-mediated DNA transfection was carried out with a Fugene 6 transfection reagent according to the manufacturer's optimized methodology (Roche). For reporter gene assays, cells were seeded in 12-well plates and cultured for 12 to 24 h. Subsequently, cells were transfected with a total of 1 μg of plasmid DNA in the reporter/effecter/control ratio of 2:1:1 or 10:10:1. Medium was changed 20 h after the transfection. After being cultured for a further 24 h, cells were lysed in 200 μl of 1× passive lysis buffer (Promega, Madison, WI) with gentle rocking for 20 min and were collected. Luciferase assays were carried out by using a Dual Glo luciferase assay system (Promega), as described previously, on a smaller scale (17). Relative luciferase activities were calculated as the ratios of firefly luciferase activity to Renilla luciferase activity.
siRNA and gene knockdown studies.
The specific short interfering RNAs (siRNAs) were predicted and synthesized by iGENE (Sapporo, Japan). The siRNA targeting MMP3 (siM3-1369) was an oligoduplex of 5′-GAG UUU GAC CCA AAU GCA AAG AAA G-AG-3′ (sense) and 5′-CUU UCU UUG CAU UUG GGU CAA ACU C-AU-3′ (antisense). The control was an oligoduplex of 5′-UUA GUG GUG AAU AUA ACA AGC UCU C-AG-3′ (sense) and 5′-GAG AGC UUG UUA UAU UCA CCA CUA A-AU-3′ (antisense). This control double-strand RNA (dsRNA) was predicted not to be homologous to known mRNA sequences of vertebrates. For transient gene knockdown studies, cells were seeded in 12-well plates and cultured for 24 h. Cells were transfected with siRNA (25 nM) with the aid of 1 μl of Lipofectamine 2000 (Invitrogen, Carlsbad, CA) and were further cultured for 24 h. After being trypsinized, the cells were collected by centrifugation at 300 × g for 5 min. Cytoplasmic RNA was isolated by using RNeasy minicolumns (Qiagen) according to the manufacturer's protocol.
For reporter gene assays, 500 ng of the reporter construct and 50 ng of the phRL-TK(int−) control vectors (Promega) were transfected at 8 h after the siRNA transfection. The medium was changed 24 h later, and the cells were lysed at 48 h after the reporter transfection. The luciferase assay was performed as described above.
A retroviral vector (pSINsi-hU6; Takara, Otsu, Japan) that expresses the short hairpin RNA (shRNA) targeting MMP3 (shMMP3) or scrambled shRNA was constructed by inserting the following sense-loop-antisense DNA sequence: shMMP3, 5′-G AGT TTG ACC CAA ATG CAA AGA AAG GTG TGC TGT CCC TTT CTT TGC ATT TGG GTC AAA CTC-3′, or scrambled shRNA, 5′-T TAG GGG ATA AGT ACG GTT GAA TCT GTG TGC TGT CCA GAT TCA ACC GTA CTT ATC CCC TAA-3′. 293T cells were cotransfected with an established combination of viral protein expression vectors, and then the conditioned media were prepared. HCS-2/8 cells (2 × 105/well of 6-well plates) were cultured overnight, and then the medium was changed to 1 ml of DMEM containing 10% FBS, 8 μg Polybrene, and 40 μl of the virus-containing conditioned medium. One milliliter of DMEM containing 10% FBS was added 6 h after the infection. The medium was changed every 3 days. Total RNA was prepared 2 weeks after the infection by using Isogen (Nippongene, Tokyo, Japan). cDNA synthesis and real-time PCR were performed as described below.
Reverse transcription and real-time PCR.
Reverse transcription was carried out with 500 ng of the cytoplasmic RNA by using Omniscript reverse transcriptase (Qiagen) and oligo(dT) according to the manufacturer's protocol. The real-time PCR was carried out as described previously (7) using a LightCycler system (Roche) with Sybr green (Toyobo) according to the manufacturer's directions. The PCR conditions were as follows: primary denaturation at 95°C for 30 s, followed by 45 cycles of PCR at 95°C for 5 s, 65°C (for MMP3 and CCN2) or at 60°C (for GAPDH) for 10 s, and 72°C for 15 s. The signals were collected at 72°C in every cycle. The specific primers for real-time PCR were designed and synthesized by NGRL (Sendai, Japan) as MMP3LCL (5′-CAG GCT TTC CCA AGC AAA TA-3′) and MMP3LCR (5′-GTG CCC ATA TTG TGC CTT CT-3′) for human MMP3. The nucleotide sequences of the primer pairs for CCN2 and GAPDH were previously described (7).
Coimmunoprecipitation and LC-MS.
The anti-MMP3 antibody affinity columns were prepared by using antibodies described above and a ProFound mammalian coimmunoprecipitation kit (Pierce, Rockford, IL) according to the manufacturer's protocol. Eluate (20% of the total) was used for the subsequent tryptic digestion using an in-solution tryptic digestion and guanidination kit (Pierce). The eluted proteins were reduced in ultrapure water containing 5 mM DTT and 25 mM ammonium bicarbonate at 95°C for 5 min. To minimize disulfate bond formation and side chain modification, the alkylation of the samples was carried out by adding iodoacetamide (IAA; 10 mM at final concentration) at RT for 20 min in the dark. The protein samples were digested by adding activated trypsin (final concentration, 3 ng/μl) and incubating them at 30°C overnight. Formic acid was added (final concentration, 0.1%). One microliter of the peptide samples was applied to nanospray high-performance liquid chromatography chip-ion trap-mass spectrometry/mass spectrometry (MS/MS) with an Agilent 1100 liquid chromatography (LC)/MSD Trap XCT Ultra series system (Agilent Technologies, Santa Clara, CA). The MS and MS/MS data were analyzed by the data analysis software Spectrum Mill, version 3.3 (Agilent). The cutoff score for proteins was 8.0. The score was defined based on the covering rate for amino acid sequences and the frequency of detected fragments. To remove the background signals, we excluded the data obtained with a control IgG column from those obtained with the anti-MMP3 columns.
Recombinant protein preparation.
Recombinant proMMP3, GST, GST-fused HP1α, HP1γ, NF45, NCoR1(1-152), and CAFp48/RBBP4 were prepared by a cold shock system (Takara) according to the manufacturer's protocol. These recombinant proteins were designed to be fused with a trigger factor (Takara), which improves protein solubility. E. coli Rosetta2(DE3)pLysS-competent cells (Invitrogen) were transformed by the pCold-derived vectors described above. Transformed clones were cultured in 2 ml of Luria-Bertani broth containing 500 μg/ml of carbenicillin at 37°C for 8 h. Cells were further cultured in 10 to 500 ml of the broth at 37°C until an optical density at 600 nm of between 0.4 and 0.5 was reached. The induction of the protein synthesis was carried out by cooling the E. coli at 15°C for 30 min and further shaking the sample for 24 h at 15°C after the addition of 0.5 mM IPTG. Cellular pellets were obtained by centrifugation, frozen at −80°C, melted, suspended in appropriate volumes of lysis buffer (50 mM Tris, pH 8.0, containing 500 mM NaCl, 1% Triton X-100, 1 μM pepstatin A, and 1 mM phenylmethylsulfonyl fluoride [PMSF]), disrupted by four 30-s sets of sonication, and centrifuged at 17,000 × g for 15 min at 4°C. To confirm the recombinant protein production in the supernatants, we carried out SDS-10% PAGE and Coomassie brilliant blue (CBB) staining using CBB R-250 (Sigma). Bovine serum albumin (BSA) was used as a concentration standard.
GST pull-down assay.
The GST pull-down assay was carried out by using the soluble fractions including recombinant proMMP3, GST, and GST-fused proteins as described above. Twenty microliters of glutathione-Sepharose 4B beads (GE Healthcare) was washed once with 500 μl of the Tris buffer (50 mM Tris-HCl, pH 7.5, containing 150 mM NaCl, 1 mM EDTA, and 1% Triton X-100) with protease inhibitors (0.5 mM PMSF and 1 μM pepstatin A). The beads were mixed with the soluble fractions containing 3, 10, or 30 μg of GST-fused recombinant proteins in 500 μl of the Tris buffer, and the soluble fraction containing 10 μg of recombinant proMMP3 tagged with Flag3 was added. The mixture was rotated for 2 h at 4°C, and the precipitant was washed four times with 500 μl of the Tris buffer. The precipitant was dissolved in 20 μl of 2× SDS sample buffer containing β-mercaptoethanol, was boiled at 95°C for 5 min, and was separated by SDS-10% PAGE. Western blot analysis was performed using an anti-Flag M2 antibody (Sigma) or anti-GST antibody (GE Healthcare).
Animals, tissue preparation, and immunohistochemistry.
The hind limbs of 8-week-old female BALB/c nu/nu mice (Clea, Tokyo, Japan) were fixed with 10% neutral phosphate-buffered formalin. The experimental osteoarthritis was induced by injecting monoiodoacetic acid (MIA; Sigma) into the intraarticular spaces of 6-month-old Wister rats, as described previously (36). These procedures were approved by the Animal Committee of the Okayama University Dental School. The animals were processed for histological analysis at 6 weeks after the injections. The specimens were decalcified in a 14% EDTA solution for 2 to 3 weeks and then embedded in paraffin, and the sections were prepared. After deparaffinization, the antigen in the section was activated by immersion in 10 mM target retrieval solution (Dako) in a microwave oven operating for 2 min. The sections were blocked in TBS with 10% FBS for 10 min and incubated with the anti-MMP3 C-terminal antibody (1:100) in TBS containing 3% BSA at 4°C overnight. Afterwards, the sections were incubated with anti-rabbit IgG tetramethyl rhodamine isothiocyanate conjugate (1:100) for 1 h in TBS containing 3% BSA and subsequently were incubated with 10 nM Sytox green nucleic acid stain (Molecular Probes, Invitrogen) in TBS for 1 h. Fluorescent medium (Dako) was used for mounting. The sections were observed by confocal laser scanning microscopy.
Statistics.
Data were expressed as means ± standard deviations, and the statistical significance of differences in mean values was assessed by Student's unpaired t tests. Differences among the mean values were considered significant at a P of <0.05.
RESULTS
MMP3 in the cell nucleus.
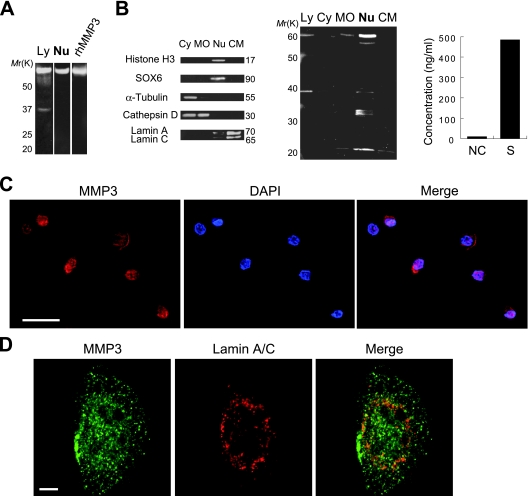
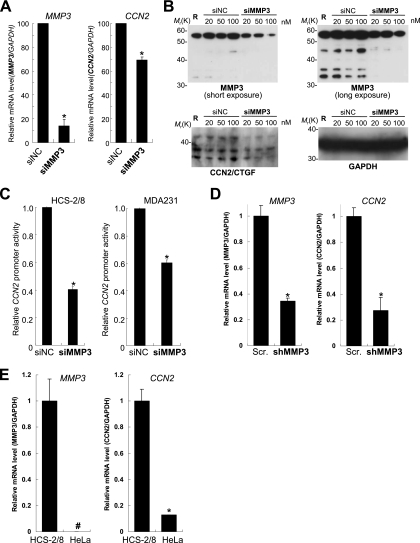
To identify TRENDIC-binding nuclear factors, we performed Southwestern screening by using a cDNA library of HCS-2/8 cells, since TRENDIC was first identified as an enhancer element dominant in these cells (7). Among the three genes cloned (Table 1), we further analyzed MMP3, because MMP3/stromelysin-1 is known to have an important role in cartilaginous tissues, although it had been recognized to act in the extracellular environment. In order to investigate if MMP3 was present in the cell nucleus, we prepared subcellular fractions of HCS-2/8 cells and analyzed the distribution of MMP3 by Western blotting. Surprisingly, MMP3 was detected in the nuclear extract of HCS-2/8 cells (Fig. 1A). The molecular weight of the major nuclear MMP3 was confirmed to be identical to that of rhMMP3 by Western blotting with antibody against the MMP3 CAT domain (Fig. 1A). Next, we further analyzed MMP3 in a variety of subcellular fractions by using an anti-MMP3 C-terminal antibody. Initially, the quality of subcellular fractions was verified. We detected histone H3 as well as the transcription factor Sox6 specifically in the nuclear fraction, α-tubulin in the cytosolic fraction, cathepsin D in cytosolic and membrane/organelle fractions, and lamin A/C in the cytoskeletal matrix fraction (Fig. 1B, left). The distinct localization of these marker proteins verified the purity of the fractions. Among these fractions, FL and putative fragments of MMP3 were distinctly detected in the nuclear fraction (Fig. 1B, center). MMP3 also was detected immunocytochemically in the nuclei of HCS-2/8 cells. The signal of MMP3 overlapped with that of the nuclear stain DAPI (Fig. 1C). Finally, the existence of MMP3 inside the circle of lamin A/C, which is a component of the nuclear lamina, was confirmed by confocal laser scanning microscopy (Fig. 1D). We also detected a significant amount of secreted MMP3 in the cell culture supernatant of HCS-2/8 as evaluated by an ELISA (480 ng/ml) (Fig. 1B, right). Taking these findings together, we confirmed that MMP3 was actively synthesized in HCS-2/8 cells and was secreted or/and translocated into the nuclei.
FIG. 1.
Detection of nuclear MMP3 by using its specific antibodies. (A) Immunoblotting of MMP3 in the lysate (Ly) or in the nuclear extract (Nu) of HCS-2/8 cells. The lysate, nuclear extract, or rhMMP3 (control) was analyzed by immunoblotting using an anti-MMP3 antibody (4190; Sigma). (B) Immunoblotting analysis of cytosolic (Cy), membrane and cellular organelle (MO), nuclear (Nu), and cytoskeletal matrix (CM) subcellular fractions prepared from HCS-2/8 cells. Successful fractionation was confirmed by the immunoblotting of fraction marker molecules (left). MMP3 in the subcellular fractions was detected by immunoblotting using the anti-MMP3 PEX antibody (center). FL and shorter forms of MMP3 were detected. The total lysate from HCS-2/8 cells also was tested. The level of MMP3 in cell culture supernatant (S) and DMEM containing 10% FBS (NC) was quantified by using ELISA (right). (C) Immunocytochemistry of MMP3 in HCS-2/8 cells. Cells were stained with the anti-MMP3 C-terminal antibody (red), and DNA was stained with DAPI (blue). MMP3 colocalized with DNA (purple in the merged view). Bar, 50 μm. (D) Immunocytochemistry of MMP3 (green) and lamin A/C (red) observed by confocal laser scanning microscopy. MMP3 detected by a specific antibody (M4190; Sigma) was observed in the circle of lamin A/C, indicating its localization in the nucleus. Bar, 5 μm.
DNA-binding ability of MMP3.
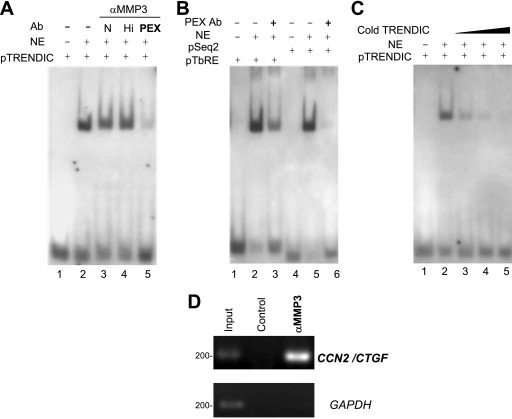
We originally cloned MMP3 as a DNA-binding factor (Table 1). To examine whether MMP3 could bind to TRENDIC or not, we first performed EMSAs. The TRENDIC probe (pTRENDIC) was shifted in the gel with nuclear proteins of HCS-2/8 cells (Fig. 2A, lanes 1 and 2), as observed previously (7). The band shift was attenuated by the anti-MMP3 PEX repeat antibody (Fig. 2A, lane 5) but not by the addition of other anti-MMP3 antibodies (Fig. 2A, lanes 3 and 4). The control anti-rabbit IgG also did not interrupt the protein-TRENDIC interaction (data not shown). We also examined the interruptive effect of the PEX antibody on the interaction of nuclear proteins with BCE1/TbRE and Seq2 probes, which are other enhancers in the CCN2/CTGF promoter (7, 13). The band shift of pBCE1/TbRE or pSeq2 also was attenuated by the PEX antibody (Fig. 2B, lanes 3 and 6), indicating that MMP3 also is involved in the gene regulation mediated by these elements.
FIG. 2.
DNA-binding ability of MMP3. (A) The binding of the nuclear protein to pTRENDIC was interrupted by anti-MMP3 PEX antibody (lane 5). Nuclear extracts from HCS-2/8 cells were preincubated with anti-MMP3 antibodies, and then the pTRENDIC probe was added to the reaction mixture. Anti-MMP3 antibody abbreviations are the following: N, anti-N-terminal; Hi, anti-Hinge region; PEX, anti-PEX. The blot shown is representative of three different experiments with similar results. Ab, antibody. NE, nuclear extract. (B) The MMP3 PEX antibody also blocked the binding of pBCE1/TbRE or pSeq2 by nuclear proteins (lanes 3 and 6). The blot shown is representative of three different experiments with similar results. (C) The TRENDIC binding of a protein in the nuclear extracts was blocked by cold TRENDIC in a concentration-dependent manner. (D) The genomic CCN2 enhancer region was coimmunoprecipitated by the anti-MMP3 antibody (4190; Sigma). After the chromatin from HCS-2/8 cells was cross-linked, immunoprecipitation was carried out with or without the anti-MMP3 CAT domain antibody, and subsequently PCR was performed for the detection of the genomic CCN2 enhancer region between −292 and −88. The genomic CCN2 enhancer region was specifically amplified by the ChIP assay with the anti-MMP3 antibody but was not detected without the antibody (control). The ORF of GAPDH was not amplified by the ChIP with or without the anti-MMP3 antibody (αMMP3). Both the CCN2 enhancer region and GAPDH were detected in the chromatin before immunoprecipitation (positive control).
To confirm the specificity of the TRENDIC-MMP3 binding, a competitive EMSA was performed. The probe-protein interaction was inhibited by the addition of cold TRENDIC in a dose-dependent manner (10-, 100-, or 1,000-fold excess of cold TRENDIC) (Fig. 2C).
To further examine the interaction between the genomic CCN2 enhancer and the nuclear MMP3, we carried out a ChIP assay. In the chromatin immunoprecipitate obtained from HCS-2/8 cells by using anti-MMP3 antibody, the genomic CCN2 enhancer region between positions −292 and −88, which contains enhancers such as TRENDIC, BCE1/TbRE, and Seq2, was detected by the ChIP assay (Fig. 2E). No such signal was detected without the anti-MMP3 antibody (Fig. 2E). The open reading frame (ORF) fragment of GAPDH was not detected by the ChIP, either with or without the anti-MMP3 antibody (Fig. 2E). Both the genomic CCN2/CTGF enhancer region and the ORF fragment of GAPDH were detected in the cross-linked chromatin sample before immunoprecipitation (Fig. 2E). These results indicate that MMP3 exists in the protein complex, binding to the CCN2 enhancer region. Taken together, these results indicate that MMP3 not only interacted in vitro with dsDNA having the enhancer sequences but also interacted in situ with the genomic CCN2/CTGF enhancer region in the HCS-2/8 cells.
Effects of intracellular MMP3 on CCN2 promoter activity.
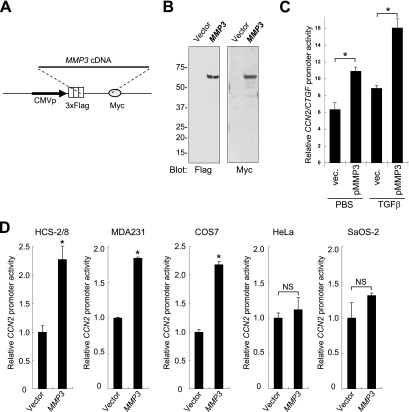
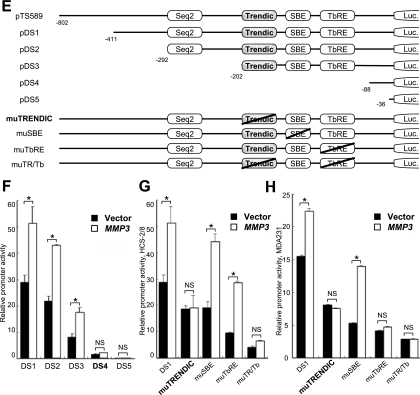
To clarify if MMP3 could modulate the CCN2 promoter activity, we cotransfected several cell lines with an MMP3 expression vector (p3xFlagMMP3-myc; schemed in Fig. 3A) and CCN2 promoter reporter constructs and then quantified the promoter activity by conducting luciferase assays. MMP3 expression by transient transfection was confirmed by immunoblotting (Fig. 3B). The intracellular overexpression of MMP3 activated the CCN2/CTGF promoter (Fig. 3C, D). TGF-β and intracellular MMP3 synergistically activated the CCN2/CTGF promoter (Fig. 3C). The CCN2 promoter activity with MMP3 overexpression was 2.2-fold higher in HCS-2/8 cells, 1.8-fold higher in MDA231 cells, and 2.2-fold higher in COS7 cells than in their controls (Fig. 3D). In contrast, the effect of overexpressed MMP3 was not significant in HeLa or SaOS-2 cells (Fig. 3D). It should be noted that the relative CCN2 promoter activity in HCS-2/8 and MDA231 cells was over 30-fold higher than that in SaOS-2 and HeLa cells (data not shown). To further investigate this mechanism, we utilized mutants of the CCN2 promoter (Fig. 3E). Mutants pDS4 and pDS5, lacking the element between −202 and −88, lost the response to overexpressed MMP3 in HCS-2/8 cells (Fig. 3F). The response of the CCN2 promoter to overexpressed MMP3 was diminished by the mutagenesis of TRENDIC but not by that of SBE or BCE1/TbRE in HCS-2/8 cells (Fig. 3G). Also, in MDA231 cells the TRENDIC mutant of the CCN2 promoter lost the response to overexpressed MMP3 (Fig. 3H). However, in contrast to the result obtained with HCS-2/8 cells, mutations in BCE1/TbRE deprived the CCN2 promoter of the response to overexpressed MMP3 in MDA231 cells (Fig. 3H). Based on these results, we concluded that TRENDIC mediated the effect of MMP3 on the activation of the CCN2 promoter in chondrocytic cells. Interestingly, our data also indicated that MMP3 could enhance the CCN2 promoter activity in collaboration with another enhancer element, BCE1/TbRE, in MDA231 cells but not in HCS-2/8 cells.
FIG. 3.
MMP3 overexpression activated the CCN2 promoter via TRENDIC. (A) Schematic representation of the MMP3 expression construct p3xFlag-MMP3-myc. This construct did not include the coding sequence for the secretion peptide signal. (B) Production of MMP3 from HCS-2/8 cells transfected with p3xFlag-MMP3-myc. HCS-2/8 cells were transfected with the MMP3 expresser or an empty vector. The overexpressed MMP3 in the cell lysate was immunoblotted by using anti-Flag and anti-Myc tag antibodies. (C) Synergy of MMP3 and TGF-β on CCN2/CTGF promoter activation. Intracellular MMP3 overexpression activated the CCN2/CTGF promoter. The MMP3 activation of the CCN2/CTGF promoter was enhanced by the addition of TGF-β. *, P < 0.05 (n = 4). vec., vector. (D) Effect of MMP3 overexpression on the CCN2 promoter. Different types of cell lines were cotransfected with the p3xFlag-MMP3-myc or empty vector and the CCN2 promoter-luciferase construct (pTS589). The CCN2 promoter was activated by MMP3 overexpression in HCS-2/8, MDA231, and COS7 cells. No significant response was observed in HeLa or SaOS-2 cells. (E) Schematic representations of the mutants of the CCN2 promoter reporter constructs. The enhancer elements involved are indicated. Mutations are represented by slashes. Luc., luciferase. (F) The response of the CCN2 promoter to MMP3 overexpression was diminished by deleting the elements located between −202 and −88 (compare DS3 and DS4). (G) Mutagenesis of TRENDIC diminished the response of the CCN2 promoter to MMP3 overexpression in HCS-2/8 cells. The SBE mutant (muSBE) or the BCE1/TbRE mutant (muTbRE) still was responsive to MMP3 overexpression in HCS-2/8 cells. These results indicate that MMP3 trans activated the CCN2 gene via TRENDIC in HCS-2/8 cells. (H) Mutagenesis of either TRENDIC or BCE1/TbRE diminished the response of the CCN2 promoter to the overexpressed MMP3 in MDA231 cells. *, P < 0.05 (n = 3 or 4). NS, not significant.
Effects of MMP3 knockdown on CCN2 gene expression.
To investigate if the endogenous MMP3 was involved in the abundant production of CCN2 from HCS-2/8 cells (7, 9), we knocked down MMP3 mRNA by RNA interference technology and evaluated the effect of the siRNA on CCN2 mRNA and protein expression and promoter activity. Transient transfection of siMMP3-1369 (20 nM) decreased MMP3 mRNA expression to 14% of the control dsRNA (siNC) level (Fig. 4A, left), indicating that siMMP3-1369 successfully degraded MMP3 mRNA. Under the condition of MMP3 being knocked down, the CCN2 mRNA level also was decreased to 70% of the control level (Fig. 4A, right).
FIG. 4.
Effects of MMP3 siRNA on CCN2 gene expression. (A) Knockdown of MMP3 also downregulated CCN2 gene expression. mRNA expression of MMP3 or CCN2 was quantified after the transfection of HCS-2/8 cells with siMMP3 or negative control dsRNA (siNC). siMMP3 knocked down the MMP3 mRNA to 15% of the level of the control while decreasing the amount of CCN2 mRNA to 70% of the control. (B) The knockdown of MMP3 also decreased the amount of cell-associated CCN2/CTGF protein. Negative control RNA (siNC) or siMMP3 was transfected at a final concentration of 20, 50, or 100 nM into HCS-2/8 cells. MMP3, CCN2, or GAPDH protein in the cell lysate at 48 h after the transfection was analyzed by Western blotting. R indicates the transfection-reagent-only control. Levels of FL (left upper panel, short exposure) or fragments (right upper panel, long exposure) of MMP3 were decreased by the knockdown. Under the MMP3 knocked-down condition, CCN2/CTGF protein also was decreased (left lower panel), indicating that MMP3 regulates CCN2. (C) CCN2 promoter activities under the condition of the MMP3 knockdown, as quantified by luciferase assays. siMMP3 also downregulated the CCN2 promoter activities in HCS-2/8 cells (to 40% of the control level) and also in MDA231 cells (to 60% of the control). (D) Retrovirus-mediated MMP3 knockdown decreased CCN2 mRNA expression. HCS-2/8 cells were infected by a retroviral vector that expressed shMMP3 or scrambled shRNA (Scr.), and then the mRNA expression levels of MMP3 and CCN2 at 2 weeks after the infection were analyzed by real-time PCR. MMP3 mRNA expression was successfully knocked down to 35% of the control level (Scr.). The CCN2 mRNA level under the MMP3 knockdown condition was 28% of the control level (Scr.). (E) Relative MMP3 and CCN2 expression levels in HCS-2/8 and HeLa cells. MMP3 mRNA expression was deficient in HeLa cells (left), while CCN2 mRNA expression in HeLa cells was 13% of that in the HCS-2/8 cells. These results indicate that MMP3 is a major regulator for CCN2 gene expression. *, P < 0.05 (n = 3 or 4); #, below the detection limit.
Next, to evaluate the effect of the MMP3 siRNA (siMMP3) on MMP3 and CCN2 protein production, 0, 20, 50, or 100 nM of siMMP3-1302 or control dsRNA (siNC) was transfected into HCS-2/8 cells, and MMP3, CCN2, and GAPDH protein production in the lysate was evaluated by Western blotting. As a result, the levels of FL MMP3 (Fig. 4B, upper left) and MMP3 fragments (Fig. 4B, upper right) decreased in a dose-dependent manner, while the levels of these MMP3 molecules were not decreased by control dsRNA transfection. Under the MMP3 knockdown conditions, the CCN2 protein level also was decreased (Fig. 4B, lower left) compared to that with control dsRNA. The GAPDH protein production level was not affected by the knockdown of MMP3 (Fig. 4B, lower right). These results indicate that MMP3 regulates CCN2 gene expression in HCS-2/8 cells. In agreement with this finding, the CCN2 promoter activity was attenuated down to 40% of the control level in HCS-2/8 cells (Fig. 4C, left) and 60% of the control level in MDA231 cells (Fig. 4C, right) by the siMMP3-1369 transfection.
To further confirm the regulation of CCN2 gene expression by MMP3 in HCS-2/8 cells, a retroviral vector that expressed shMMP3 or scrambled shRNA was constructed and infected into HCS-2/8 cells, and MMP3 and CCN2 expression was evaluated 2 weeks after the infection. The shMMP3 retroviral vector successfully knocked down the level of expression of MMP3 mRNA to 35% of that of the control (Fig. 4D, left). In the retrovirus-mediated long-term MMP3 knockdown condition, CCN2 mRNA expression was as low as 28% of the control level (Fig. 4D, right).
If MMP3 is a critical and general regulator of CCN2, the expression of CCN2 is anticipated to be significantly lower in other cells without MMP3 expression. To confirm this, we comparatively analyzed the expression of CCN2 and MMP3 in HCS-2/8 and HeLa cells. In contrast to its expression in HCS-2/8 cells, MMP3 mRNA expression was deficient in HeLa cells (Fig. 4E). As we expected, the CCN2 expression level in HeLa cells was strikingly low (12%) compared to that in HCS-2/8 cells. These results indicate that abundant MMP3 regulates CCN2 gene expression in HCS-2/8 cells, while CCN2 expression in HeLa was low because of the deficiency of MMP3, a crucial factor in CCN2 induction. Additionally, both MMP3 and CCN2 expression levels in SaOS2 osteoblastic cells were quite low (data not shown). Taken together, these data demonstrated that cellular MMP3 regulates CCN2 gene expression at the transcription level.
MMP3 domains required for CCN2/CTGF promoter activation.
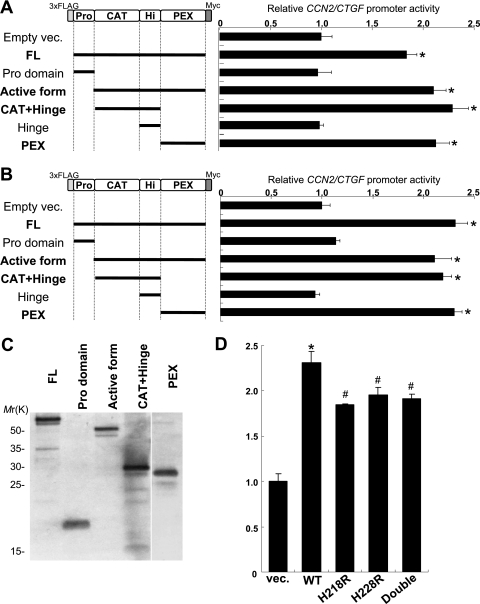
In order to determine the involvement of individual domains of MMP3 in the CCN2/CTGF trans-activation function, several assemblages of MMP3 domains were expressed in COS7 and HCS-2/8 cells, and the CCN2/CTGF promoter reporter assay was carried out. The expression of the MMP3 domain deletion mutants in COS7 cells was confirmed by Western blotting (Fig. 5C). The FL and active forms of MMP3 trans activated the CCN2/CTGF promoter, while the prodomain alone did not (Fig. 5A). Interestingly, the PEX domain of MMP3 lacking a CAT domain trans activated the CCN2/CTGF promoter in the cells, and the trans activation ability of the MMP3 PEX domain was comparable to that of the FL and active form (Fig. 5A). Moreover, CAT+Hinge also trans activated the CCN2/CTGF promoter, while Hinge alone had no effect for trans activation (Fig. 5A). Similar results of MMP3 domain functions were obtained in HCS-2/8 cells (Fig. 5A and B). These results clarified that not only the FL and active forms of MMP3 but also a few MMP3 fragments, such as PEX and CAT+Hinge domains, could trans activate the CCN2/CTGF promoter.
FIG. 5.
Effects of MMP3 domains for CCN2/CTGF promoter regulation. (A) Schemes of MMP3 domains expressed from p3Flag-myc vector (vec.) in COS7 cells (left). Effects of the MMP3 domains on the CCN2/CTGF promoter in COS7 cells were quantified (graph on the right). The FL and active forms of MMP3 trans activated the CCN2/CTGF promoter, while the prodomain alone did not (compare empty vec., FL, prodomain, and active-form results). Interestingly, the PEX domain of MMP3 lacking the CAT domain trans activated the CCN2/CTGF promoter in the cells. Moreover, the CAT+Hinge domain also trans activated the CCN2/CTGF promoter, while Hinge alone had no effect on trans activation (compare CAT+Hinge and Hinge results). *, P < 0.05 (n = 3 to 4). (B) Schemes of MMP3 domains expressed in HCS-2/8 (left). The PEX domain and the CAT+Hinge domain also activated the CCN2/CTGF promoter in HCS-2/8 cells (right). *, P < 0.05 (n = 3 or 4). (C) The expression of the MMP3 domains in COS7 cells was confirmed by Western blotting. (D) Catalytically dead mutants of MMP3 expression resulted in the decreasing of the trans-activation ability for the CCN2 promoter. To remove the catalytic activity, H218R, H228R, and H218R/H228R (Double) mutations in the HEXXHXXGXXH zinc-binding motif were introduced in the pMMP3 construct. These results clarified that not only the FL and active forms of MMP3 but also individual MMP3 domains, such as PEX and CAT+Hinge, can trans activate the CCN2/CTGF promoter. *, P < 0.05 (n = 3 or 4); #, P < 0.05 (n = 3 or 4). WT, wild type.
A proteolytic activity of MMPs has been known to be zinc dependent. In the part of the CAT domain forming the active site, a Zn2+ ion is coordinated by a chelate bond with three histidine residues found in the conserved sequence HEXXHXXGXXH and is called a zinc-binding motif. The change of any one of the histidines to arginine was reported to cause the loss of the proteolytic activity of MMPs (27). To clarify if the trans-activation ability of MMP3 for CCN2 expression is dependent on proteolytic activity or not, we constructed plasmids expressing catalytically dead mutants of MMP3. As a result, the alteration of a histidine residue at position 218 (H218) to arginine (H218R mutant) resulted in a decrease of the trans-activation ability of MMP3 for the CCN2 promoter (Fig. 5D). The H228R mutant and the double mutant (H218R/H228R) yielded similar results (Fig. 5D). Both the CAT domain and the PEX domain lacking the CAT domain hold the trans-activation ability (Fig. 5A, B). Taking these findings together, the trans-activation ability of MMP3 for the CCN2 promoter is partly dependent on the catalytic activity, and the PEX domain greatly contributes to the activation of the CCN2 promoter.
Uptake and nuclear translocation of extracellular MMP3.
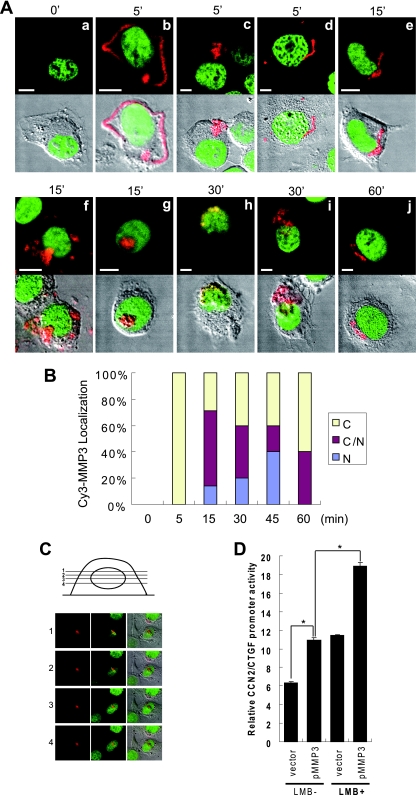
Next, to clarify the subcellular dynamics of MMP3, we prepared Cy3-labeled rhMMP3 (Cy3-MMP3), added it to the medium of HCS-2/8 cells in culture, and observed its behavior with a confocal laser scanning microscope. The Cy3-MMP3 signals emanated from the cellular membrane or cytoplasm between 5 and 60 min after the addition of Cy3-MMP3 to the cell culture (Fig. 6A, images b to f, i, and j). Of note, Cy3-MMP3 signals also were observed in the cell nuclei between 15 and 30 min after the addition (Fig. 6, images f to i). The Cy3-MMP3 signal was barely detected at 60 min after the addition, indicating the possibility of degradation or resecretion. The Cy3-MMP3 localization in the Cy3-MMP3-positive cell population was quantitatively analyzed (Fig. 6B). The Cy3-MMP3 was taken up into the cells within 5 min after the addition, showing dominant localization in the cytoplasm, which was followed by the translocation into the nucleus between 15 and 60 min after the addition. Additionally, to demonstrate that MMP3-Cy3 was certainly in the nucleus, a sequential horizontal view of the MMP3-Cy3 signal and DNA in a cell was presented (Fig. 6C). These observations clarified that extracellular MMP3 is taken up into the HCS-2/8 cells and subsequently translocated into the nucleus.
FIG. 6.
Extracellular MMP3 was internalized into the cells and translocated into the nucleus. (A) Cy3-MMP3 was added to the culture medium of HCS-2/8 cells. The cells were fixed at 0 (a), 5 (b to d), 15 (e to g), 30 (h, i), or 60 min (j) after the addition of MMP3 and then were observed under a confocal laser scanning microscope. Red signals indicate Cy3-MMP3, and green signals indicate DNA. Cy3-MMP3 was observed on the cell surface or in the cytoplasm between 5 and 60 min after the addition (b to f, i, and j). Cy3-MMP3 was observed in the nuclei at 15 and 30 min (f to i). Bars, 5 μm. (B) Quantitative analysis of the Cy3-MMP3-positive cells in relation to subcellular localization. N, nucleus; C, cytoplasm; N/C, both. n = 4 to 7. (C) Sequential view of the cells by using confocal laser scanning microscopy revealed that MMP3-Cy3 (red) is inside the nucleus filled with DNA (green). (D) Effect of LMB on the activation of the CCN2/CTGF promoter by intracellular MMP3 expression. The addition of LMB (20 ng/ml) enhanced both the basal level and the trans-activation effect of MMP3 for the CCN2/CTGF promoter. *, P < 0.05 as indicated by brackets (n = 4). vec., vector.
Effect of LMB on the induction of CCN2/CTGF by MMP3.
In order to demonstrate that nuclear translocated MMP3 trans activates the CCN2/CTGF promoter, leptomycin B (LMB) was employed. LMB is an antibiotic with membrane permeability, and it has been known to inhibit the nuclear export function of CRM1/exportin 1 by directly binding to its cysteine residue. We hypothesized that the nuclear export of MMP3 could be CRM1 dependent, and thus LMB could cause the nuclear accumulation of MMP3 and the enhancement of the activation of the CCN2/CTGF promoter by nuclear MMP3. Exactly as we predicted, LMB enhanced the trans-activation effect of MMP3 on the CCN2/CTGF promoter (Fig. 6D). The increase in the CCN2 promoter activity without MMP3 overexpression may represent the effect of LMB on endogenous MMP3. These results represent that nuclear-cytoplasmic trafficking is crucial in the trans activation of the CCN2/CTGF promoter by MMP3.
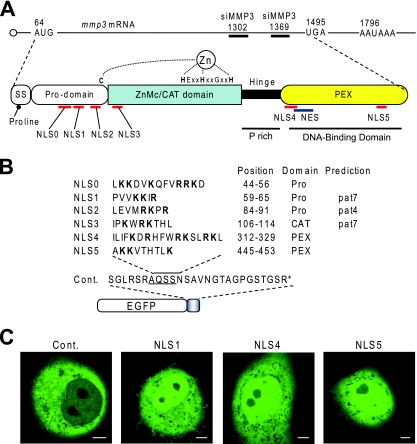
Domain structure and nuclear localization signals of MMP3.
In addition to the classical domain structure of MMP3, we found and predicted several novel domains and signal sequences in MMP3 (Fig. 7A). As an anti-MMP3 PEX antibody blocked protein-DNA interactions, a DNA-binding domain can be included in the PEX domain. The Hinge region has been known to be just a linker; however, it contains proline-rich sequences found in some transcription factors. The signal sequence at the amino terminus of MMP3 contains proline at the fifth position. It can work as a helix breaker, causing the partition sorting of MMP3 to the endoplasmic reticulum (ER)-Golgi secretion pathway as well as to the cytoplasm (44). A leucine-rich sequence in the PEX domain can work as a nuclear export signal (NES).
FIG. 7.
Domain structure of MMP3 and putative nuclear trafficking signals. (A) MMP3 mRNA is schematized at the top with the first AUG codon, the UAG stop codon, and the positions of the siRNA targets (siMMP3-1369 and siMMP3-1302). The domain structure of MMP3 is illustrated in the middle. SS indicates the secretion signal sequence containing the proline helix breaker at the fifth amino acid. This may result in sorting MMP3 to the ER-Golgi secretion pathway as well as the cytoplasm after the signal sequence emerges from the ribosome. The prodomain (Pro) also is designated a putative peptidoglycan-binding domain. The CAT domain also is designated the ZnMc domain, which is conserved among zinc-dependent metalloproteinases. The hinge region is a linker domain characterized by a proline-rich (P rich) structure. The PEX domain was revealed to be a DNA-binding domain that forms a propeller-like structure. Putative NLSs in MMP3 are mapped beneath the structure. A putative NES containing four leucines also was found. (B) List of NLSs in MMP3 and schematically depicted EGFP-NLS fusion proteins. The arginine (R)- and lysine (K)-rich sequences were picked up from the FL amino acid sequence of MMP3 and were designated NLS0 to NLS5. The cDNAs encoding these NLSs were cloned into the pEGFP-C1 vector. Arginine and lysine residues are indicated by boldface. The positions of the NLSs in proMMP3 and the domain in which the NLSs were found are shown. PEX, hemopexin-like repeat. NLS1, NLS2, and NLS3 in MMP3 also were predicted by using PSORTII pattern 4 (pat4) or pat7. (C) Subcellular localization of the GFP-NLSs. The subcellular localization of EGFP-NLSs and the control (Cont.) was examined in COS7 cells by confocal laser microscopy. EGFP-NLS1, EGFP-NLS4, and EGFP-NLS5 were observed in the nuclei as well as in the cytoplasm. Similar results were obtained for EGFP-NLS0, EGFP-NLS2, and EGFP-NLS3 (data not shown). EGFP-MCS (control) was observed only in the cytoplasm. Scale bars, 5 μm. The photogram is representative of 10 independent cells with similar results.
Classically, the nuclear import of proteins is mediated by importins α and β, which recognize basic amino acid clusters, designated NLSs, on the protein to be imported (3, 34). In order to assess if MMP3 entered the nucleus via this classical pathway, we sought putative NLSs in MMP3. Six lysine- and arginine-rich sequences were found in the amino acid sequence of MMP3, and we designated them NLS0 to NLS5 (Fig. 7A, B). The putative NLSs were scattered among all of the domains of MMP3. Since MMP3 can be (auto)cleaved, the scattered NLSs can function individually in the resultant fragments. In order to investigate the function of individual NLSs, we constructed the EGFP-NLS expression plasmids for all six putative NLSs in MMP3, overexpressed them in COS7 cells, and observed their subcellular localizations. The native EGFP (control) was observed only in the cytoplasm and not in the nucleus (Fig. 7C). Surprisingly, all EGFP-NLSs were observed in the nuclei as well as in the cytoplasm, as shown in Fig. 7C (data not shown for NLS0, NLS2, and NLS3). These results indicate the possible involvement of the NLS-mediated classical pathway in the nuclear translocation of MMP3 even for the FL and the fragments of MMP3.
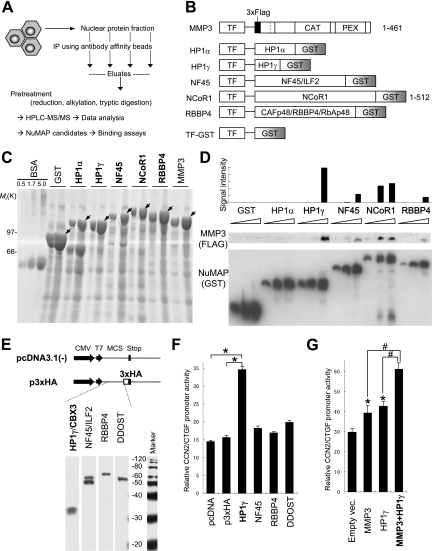
Identification of NuMAPs.
If MMP3 plays a certain role in the nuclei, it should be associated with other proteins to exhibit the downstream phenomenon, whether it cleaves them or not. To clarify this point, we applied the nuclear extract of HCS-2/8 cells to anti-MMP3 antibody columns and then eluted the bound proteins and analyzed them by using an LC-MS/MS system (Fig. 8A). Identified proteins included heterochromatin proteins, transcription activators/repressors, RNA polymerase II, nucleosome/chromatin assembly protein, and others (Table 2 and data not shown). Among them, we confirmed the mRNA expression of HP1α, HP1γ, NF45/interleukin enhancer binding factor 2 (ILF2), NCoR1, and CAFp48/RBBP4/RbAp48 in the cells; thereafter, we prepared their GST-fused recombinant proteins and recombinant MMP3 (Fig. 8B, C). The bindings of HP1γ, NF45, NCoR1, and CAFp48 to MMP3 were confirmed from the results of a GST pull-down assay (Fig. 8D); thus, these proteins were designated NuMAPs. These data suggest that MMP3 associated with these NuMAPs in the nucleus in exerting its nuclear function.
FIG. 8.
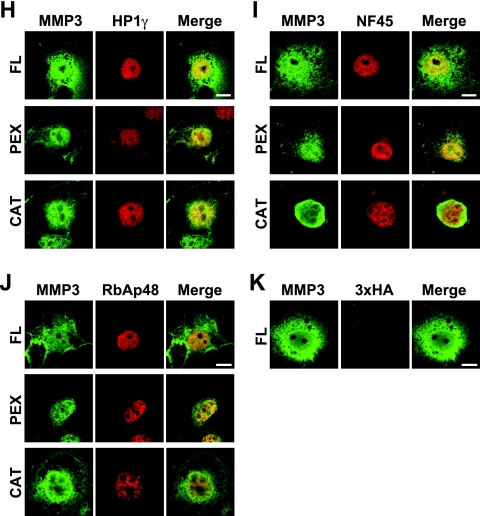
Identification of NuMAPs. (A) The methodology to identify the NuMAPs. The nuclear proteins extracted from HCS-2/8 cells were immunoprecipitated by using the anti-MMP3 affinity beads for the MMP3 CAT domain (Cat.), Hinge region, or PEX antibody or IgG (control). The eluted proteins were digested with trypsin at the C-terminal peptide bonds of lysine and arginine. Samples were applied to the nanoflow high-performance liquid chromatography (HPLC) chip MS/MS, and the data were analyzed to identify the proteins. The NuMAP candidates are shown in Table 2. (B) Structures of the recombinant MMP3 and several GST-fused NuMAP candidates. Trigger factor (TF) was added to all of the proteins to increase their solubility. (C) CBB staining of the recombinant proteins. The soluble fractions (1 or 5 μl) or the BSA control was electrophoresed in 10% polyacrylamide gels and stained. Arrows indicate the recombinant proteins. (D) MMP3-NuMAP interactions. Flag-tagged MMP3 (10 μg) was mixed with the GST-fused NuMAP candidates (3, 10, or 30 μg) and then pulled down with glutathione-Sepharose beads. The pulled down proteins were analyzed by immunoblotting using an anti-Flag or anti-GST antibody. The direct interaction of MMP3 with HP1γ, NF45, NCoR1, and RBBP4 was confirmed. (E) Expression of HA3-tagged HP1γ, NF45, RBBP4, and DDOST in COS7 cells. Schemes of the expression constructs are illustrated on the top. Their expressions were confirmed by examining the lysates by Western blotting (lower panels). (F) Trans activation of the CCN2/CTGF promoter by HP1γ among NuMAPs, revealed by using reporter assays. *, P < 0.05 as indicated by brackets (n = 4). (G) Cooperative trans activation of the CCN2/CTGF promoter by MMP3 and HP1γ. HCS-2/8 cells were cotransfected with p802CCN2promoter-luc (400 ng), phRL-TK(int−) (100 ng), p3Flag-myc or pMMP3 (1 μg), and p3xHA or pHP1γ (1 μg) in each well of a 12-well plate for the luciferase assay. *, P < 0.05 (n = 4); #, P < 0.05 as indicated by brackets (n = 4). (H to K) Colocalization of HA3-tagged HP1γ (H), NF45 (I), or RBBP4 (J) with the FL, PEX domain, or CAT domain of MMP3. Nuclear localization of HP1γ, NF45, and RBBP4 was observed. The FL, PEX domain, or CAT domain of MMP3 was observed to be colocalized with these NuMAPs, and these MMP3 also were observed in the cytoplasm. (K) The HA3 tag was faintly observed to be diffused. Scale bars, 5 μm. IP, immunoprecipitation; vec., vector.
TABLE 2.
List of NuMAPsa
| NuMAP(s) | Putative function(s) | Antibody type | Score | GenBank accession no. |
|---|---|---|---|---|
| HP1α/CBX5 | Heterochromatin, etc. | CAT | 17.81 | 6912292 |
| HP1γ/CBX3 | Heterochromatin, etc. | PEX | 9.82 | 15680098 |
| NCoR1 | trans repression | CAT | 10.64 | 35193218 |
| CAFp48/RBBP4/RbAp48 | Chromatin assembly, etc. | Hinge | 9.3 | 62897117 |
| ILF2/NF45 | trans activation | Hinge | 10.9 | 55962125 |
The partial list of NuMAPs was identified by coimmunoprecipitation and LC-MS/MS. The binding abilities of these NuMAPs with MMP3 were confirmed and are shown in Fig. 8. Anti-MMP3 antibodies used for coimmunoprecipitation were against CAT, Hinge, and C-terminal PEX domains, and subsequent LC-MS/MS analysis was performed with the data-analyzing software Spectrum Mill. The cutoff score for proteins and peptides was 8.0. We subtracted the background data obtained from a control experiment with IgG from those obtained with anti-MMP3 antibody.
To clarify the function of these NuMAPs and the collaborative function with intracellular MMP3 for CCN2/CTGF transcription, mammalian NuMAP expression plasmids were constructed (Fig. 8E) and coexpressed in HCS-2/8 cells for a reporter gene assay. HP1γ recently was found to exhibit multiple functions beyond heterochromatin formation (19). In our study, the overexpression of HP1γ activated the CCN2/CTGF promoter (to a level 2.5-fold above that of the control), while the expression of other NuMAPs (NF45, RBBP4, and DDOST) caused no substantial change compared to that of the controls [pcDNA3.1(−) and pcDNA3.1(−)3HA] (Fig. 8F). In addition, the coexpression of HP1γ and MMP3 synergistically activated the CCN2/CTGF promoter (Fig. 8G). These results suggested that HP1γ, a member of the NuMAPs, regulates CCN2/CTGF transcription in cooperation with nuclear MMP3.
MMP3 can be (auto)cleaved into the CAT domain and the PEX domain in or out of the cells, and the PEX domain and CAT+Hinge domain possess a trans-activation ability for the CCN2/CTGF promoter at a level comparable to that of FL MMP3. In addition, both contain putative NLSs. To clarify the localization of NuMAPs (HP1γ, NF45, and RBBP4) with MMP3s (FL, PEX, and CAT domains), these proteins or domains were coexpressed in COS7 cells, and then the colocalization was investigated by using confocal microscopy. Firstly, HP1γ, NF45, and RBBP4 were observed mainly to be localized only in the nucleus (Fig. 8H to J), while the HA3 tag was observed to be faintly diffused (Fig. 8K). The FL, PEX, or CAT domain of MMP3 was observed to be localized in the nucleus as well as in the cytoplasm. Neither NuMAPs (HP1γ, NF45, and RBBP4) nor MMP3s (FL, PEX, and CAT domains) were observed in the nucleolus (Fig. 8H to K). These findings indicated that HP1γ, NF45, and RBBP4 can interact with MMP3 in the nucleus in vivo, as demonstrated by in vitro GST pull-down assays (Fig. 8D). Taking these results together, among all NuMAPs tested, HP1γ can trans activate the CCN2/CTGF promoter by interacting with MMP3 in the nucleus.
Subcellular localization of MMP3 in normal and osteoarthritic cartilaginous tissues.
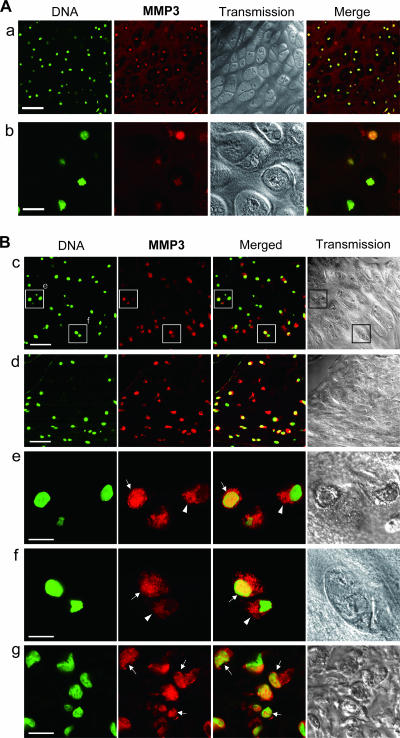
To investigate the relationship between subcellular MMP3 localization and the pathophysiology of articular cartilage in animals, we immunohistochemically examined MMP3 in normal and osteoarthritic articular cartilages with a confocal laser microscope. MMP3 was immunopositive in the normal articular cartilage of 2-month-old mice (Fig. 9A, row a), while control IgG brought no significant signal. The colocalization of the MMP3 signals with DNA was very evident at high-power magnification (Fig. 9A, row b). In a rat osteoarthritic cartilage, the articular and semilunar chondrocytes were positively stained by the MMP3 antibody (Fig. 9B). We also observed the colocalization of the MMP3 staining with DNA in articular chondrocytes at high-power magnification (Fig. 9B, rows e and f). MMP3 in the cytoplasm also was observed in some osteoarthritic articular chondrocytes (Fig. 9B, rows e and f; yellow in merged view). It should be noted that the fibrochondrogenic cells, in which CCN2 would be expressed and involved in the tissue remodeling, showed MMP3 existed alongside DNA (Fig. 9B, row g; yellow in merged view). The data that MMP3 is localized in the nuclei of normal developing and osteoarthritic articular chondrocytes in animals suggest a role of the nuclear MMP3 in the development and regeneration of articular cartilage.
FIG. 9.
Immunohistochemical analysis of MMP3 in articular cartilage of animals. Sections were stained by using the anti-MMP3 C-terminal antibody (red) with a fluorescent DNA dye (green) and were observed by confocal microscopy. (A) Articular cartilage of a 2-month-old mouse (a) and a high-power magnification view (b). Scale bars, 50 (a) and 10 μm (b). The nuclear accumulation of MMP3 in the articular chondrocytes was clearly evident (yellow in merged views). (B) MMP3 in osteoarthritic articular (c) and semilunar (d) cartilage in the knee joint of a 6-month-old rat. Two subregions of row c (enclosures) also were viewed under high-power magnification (e and f). (g) Fibrochondrogenic cells also were observed under high-power magnification. Scale bars, 50 (c and d) and 10 μm (e to g). MMP3 (red) was colocalized with DNA (green) in the nuclei (arrows) or was distributed in the cytoplasm (arrowheads).
Effects of MMP inhibitors on CCN2/CTGF promoter activity and on nuclear stromelysin-like endopeptidase activity.
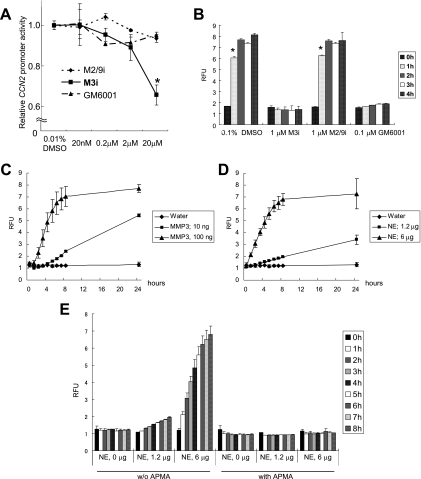
To investigate if the proteinase activity of MMP3 was associated with the observed transcriptional activation of CCN2, we added MMP inhibitors to the culture medium of HCS-2/8 cells and subsequently quantified the CCN2 promoter activities. A specific inhibitor of human MMP3 (MMP3 inhibitor II) suppressed the CCN2 promoter activity in a dose-dependent manner (Fig. 10A). In contrast, a specific inhibitor of MMP2 and MMP9 did not suppress the CCN2 promoter activity (Fig. 10A), nor did GM6001, a well-known broad-spectrum hydroxamic acid inhibitor of MMPs (Fig. 10A). These results indicate that the structural hindrance caused by the specific inhibitor blocked the interaction between MMP3 and its binding target, thus strongly suggesting the involvement of the CAT domain in the transcriptional activation events.
FIG. 10.
Characterization of stromelysin-like activity in HCS-2/8 nuclei. (A) Effects of MMP inhibitors on CCN2 promoter activity in HCS-2/8 cells. MMP2/MMP9 inhibitor I (M2/9i), MMP3 inhibitor II (M3i), or GM6001 was added to the culture medium of HCS-2/8 cells at a final concentration of 0, 0.02, 0.2, 2, or 20 μM. The cells were transfected with the CCN2/CTGF promoter reporter 8 h after the addition, and then reporter gene expression was evaluated 48 h after the transfection. Only MMP3 inhibitor II at a concentration of 20 μM suppressed the CCN2/CTGF promoter activity (to 66% of the control level); however, neither GM6001 nor the gelatinase inhibitor affected the promoter activity. These results indicate that endogenous MMP3 regulated CCN2/CTGF gene expression via the CCN2/CTGF promoter. *, P < 0.05 (n = 4). (B) Establishment of the stromelysin activity assay in vitro. A stromelysin-specific peptide substrate, Mca-RPKPVE-Nval-WRK(Dnp)-NH2 fluorogenic substrate, was tested for cleavage by rhMMP3 as well as by a nuclear extract of HCS-2/8 cells, and the endopeptidase activity was quantified by measuring the RFUs. Active MMP3 (8.3 nM) fully cleaved the peptide in 2 h, and the endopeptidase activity was perfectly blocked by MMP3 inhibitor II (M3i) or GM6001 but not by the MMP2/MMP9 inhibitor (M2/9i). *, P < 0.05 (n = 4). (C) Proenzyme MMP3 (1.7 nM; 100 ng) cleaved the substrate as well, suggesting autoactivation along the assay. (D) Nuclear extract (NE) of HCS-2/8 cells (100 ng) also cleaved the substrate. (E) The endopeptidase activity of the nuclear extract was perfectly blocked by 1 mM APMA. w/o, without.
To determine further if the nuclei of HCS-2/8 cells exerted stromelysin endopeptidase activity, we tested the ability of a nuclear extract of HCS-2/8 cells to cleave a fluorogenic substrate of stromelysin (32). Initially, the stromelysin activity of MMP3 (8.3 nM) activated by 1 mM APMA was successfully detected by measuring the number of RFUs emitted by the cleavage of the peptide substrate (Fig. 10B). This stromelysin endopeptidase activity of active MMP3 was blocked by MMP3 inhibitor II or GM6001 but not by the MMP2/MMP9 inhibitor (Fig. 10B). The endopeptidase activity of proenzyme MMP3 also was tested. Proenzyme MMP3 (1.7 nM) cleaved the fluorogenic substrate relatively slowly (Fig. 10C), suggesting that proenzyme MMP3 was becoming activated in the reaction buffer at 37°C. Instead of the recombinant MMP3, we next tested the stromelysin endopeptidase activity of the nuclear extract prepared from HCS-2/8 cells. This extract also cleaved the peptide substrate in a dose-dependent manner; 6 μg of the nuclear extract exerted enough activity to cleave the peptide to the same extent as 100 ng (1.7 nM) of proenzyme MMP3 (Fig. 10D). These findings support our idea that active forms of FL or/and smaller forms of MMP3 exist in the nuclei of chondrocytes. Unexpectedly, this stromelysin activity of HCS-2/8 nuclear extracts was perfectly blocked by 1 mM APMA (Fig. 10E) but not by any inhibitor tested, i.e., GM6001, MMP3 inhibitor II, and the MMP2/MMP9 inhibitor (data not shown). Previously, PEX domain-deleted mini-stromelysin mutants were revealed to show an altered response to APMA, in that precursor processing was inhibited rather than accelerated (11). In our data, FL and 35-kDa MMP3 were detected by Western blotting using an anti-MMP3 CAT domain antibody (Fig. 1A). FL and several shorter MMP3s also were detected by using anti-MMP3 C-terminal domain antibody (Fig. 1B). Thus, we speculate that the FL or shorter MMP3s in the nucleus possess stromelysin activity, which was blocked by APMA. In contrast to the results of the endopeptidase activity test using nuclear extracts in vitro, in a reporter gene assay, MMP3 inhibitor II inhibited the CCN2 promoter activity, whereas GM6001 did not (Fig. 10E). Therefore, GM6001 may not be able to efficiently enter the cells, and MMP3 inhibitor II may have blocked the MMP3 action in any of the ensuing steps, e.g., the release from ECM, endocytosis, or nuclear import, rather than in the nucleus. These findings suggest that the possible modification of MMP3 or some interaction partner of MMP3 in the nucleus protects MMP3 from MMP inhibitors to sustain stromelysin activity.
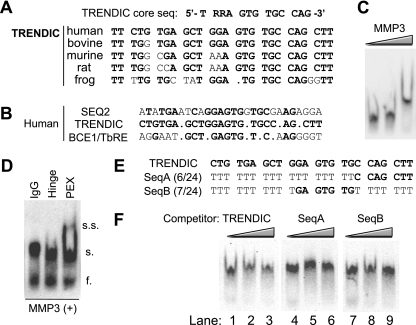
Specificity of MMP3-TRENDIC interaction and its consensus sequence.
The DNA-binding ability of MMP3 was already demonstrated in Fig. 2, in which we showed that TRENDIC and the other cis elements were able to bind MMP3 in the nuclear extract. To further investigate the sequence specificity of MMP3-targeted DNA, we initially performed in silico analyses and, subsequently, a competitive EMSA (Fig. 11). TRENDIC sequences from different species were aligned and compared (GenBank accession numbers were shown in a previous study) (Fig. 11A) (7). Obviously, the TRENDIC sequence is highly conserved among vertebrates (Fig. 11A). The TRENDICs were 92% conserved between human and bovine species. The TRENDICs also were highly conserved in rodents, particularly mice and rats (96%). The amphibian TRENDIC sequence was relatively different from those of other vertebrates (77% conserved between humans and frogs). Nevertheless, a sequence (5′-TRRAGTGTGCCAG-3′) can be found that is mostly conserved among all of the species. Next, we aligned TRENDIC, BCE1/TbRE, and Seq2 sequences shown to interact with nuclear MMP3 (Fig. 2A and B and 11B). By the alignment, the core sequence 5′-GAGTG-3′ and several flanking nucleotides were found to be conserved. Importantly, this core sequence also is involved in the TRENDIC consensus motif described above. In order to further evaluate the DNA-binding property of MMP3, we established an EMSA system by using rhMMP3 and a p160 probe containing TRENDIC, BCE1/TbRE, and Seq2. The p160 probe was shifted by adding rhMMP3 (Fig. 11C) and supershifted by further adding the anti-PEX antibody for MMP3 (Fig. 11D), demonstrating that rhMMP3 bound to p160. This MMP3-p160 probe complex was stably present even in the presence of a vast amount of poly(dI-dC), ruling out the possibility that the MMP3-p160 interaction is nonspecific. Moreover, based on the result of the sequence alignment, we designed mutant TRENDIC sequences (Fig. 11F) and performed competitive EMSAs to examine the specificity of MMP3-TRENDIC interactions. The SeqA sequence contains 5′-CCAGCTT at the downstream end, whereas SeqB contains 5′-GAGTGTG-3′ (the core sequence) in the center. The dsDNA of wild-type TRENDIC competed with the MMP3-p160 binding in a concentration-dependent manner (Fig. 11G, lanes 1 to 3). Perfect competition was not achieved, probably due to the involvement of SEQ2 and BCE1/TbRE in p160 or a possible DNA interaction with the zinc-dependent metazincine (ZnMc)/CAT domain. In contrast, SeqA dsDNA completely failed to compete with the p160-MMP3 binding (Fig. 11G, lanes 4 to 6). SeqB was not very competitive with the probe (lanes 7 to 12), indicating that the core sequence is necessary but not sufficient to warrant the specific interaction with MMP3. All of these results demonstrated that the flanking nucleotide sequences in TRENDIC and the other elements, as well as the core sequence, determine the specific binding of certain DNAs to MMP3.
FIG. 11.
Sequence specificity of MMP3-TRENDIC binding. (A) Sequence conservation of TRENDIC among species. Human, bovine, murine, rat, and frog TRENDIC sequences are aligned. The nucleotides conserved between human TRENDIC and those of other species are shown in boldface. The TRENDIC core sequence predicted from the TRENDIC sequences of the species is shown at the top. (B) Alignment of the cis elements in the human CCN2 promoter. The nucleotides identical to those in TRENDIC are shown in boldface. (C) Specific binding of MMP3 to the CCN2 enhancer sequence in the presence of an excess of nonspecific competitor. Probe CCN2p160, which contains the three enhancers shown in panel B, was shifted with recombinant MMP3 in a dose-dependent manner in the presence of 0.5 μg of poly(dA-dT). (D) The MMP3-enhancer complex was supershifted with anti-PEX antibody but not with anti-Hinge or control IgG. Poly(dA-dT) (1 μg/reaction) was used. f., free probe; s., shifted probe; s.s., supershifted probe. (E) Sense sequences of modified TRENDIC competitors. The corresponding dsDNAs were used for the competitive EMSA shown in panel F. (F) The MMP3-p160 interaction was interfered with by TRENDIC but not by its modified sequences. The competitors (TRENDIC, SeqA, and SeqB; 1-, 10-, 100-fold excesses of probe) were preincubated with rhMMP3 before the addition of the probe p160 with poly(dI-dC) (0.1 μg/reaction). TRENDIC competed with p160 for binding with rhMMP3 (lanes 1 to 3), whereas the SeqA dsDNA failed to compete with the binding reaction (lanes 4 to 6). SeqB was modestly competitive with the probe (lanes 7 to 9).
DISCUSSION
In this report, we demonstrated a novel aspect of MMP3; i.e., that it acted as a transcription factor in cell nuclei. First, we cloned MMP3 as a gene encoding a TRENDIC-binding factor (Table 1). Next, the existence of MMP3 in the nuclei (Fig. 1 and 2) was indicated in vitro and also in vivo (Fig. 9). Interestingly, both the nuclear uptake of extracellular MMP3 (Fig. 6) and the nuclear targeting ability of putative NLSs in MMP3 (Fig. 7) were shown. The binding of MMP3 to the DNA cis elements, especially to TRENDIC, was confirmed by EMSA (Fig. 2 and 11). Moreover, the interaction between MMP3 and the genomic CCN2 enhancer region including TRENDIC was confirmed by the ChIP assay (Fig. 2E). The functional requirement for the TRENDIC element in the CCN2 promoter for MMP3-mediated activation was demonstrated by the mutagenesis experiments (Fig. 3). Finally, the regulation of CCN2 gene expression by MMP3 and by its domains was demonstrated by overexpression and gene knockdown studies (Fig. 3, 4, and 5). In addition, HP1γ was revealed to be a coregulator with MMP3 for CCN2/CTGF trans activation (Fig. 8 and Table 2). An MMP3 inhibitor specifically suppressed the CCN2/CTGF promoter activity (Fig. 10). All of these data together support our contention that MMP3 is translocated to the cell nucleus and, once there, trans activates the CCN2 gene.
In this study, most of the data were obtained by using HCS-2/8 chondrocytic cells, a cell line that has been a useful model in chondrocyte biology. The cells have been used as a model of normal and osteoarthritis chondrocytes in a number of studies (30, 40, 51). Of note, CCN2 originally was cloned as a hypertrophic chondrocyte-specific gene product 24 (Hcs24) from HCS-2/8 cells and was demonstrated to function as an endochondral ossification-promoting factor (50). Indeed, HCS-2/8 cells produced abundant amounts of CCN2 compared to the amount produced by other cell types (7, 9). The HCS-2/8 cell line was established from a chondrosarcoma, but the expression of CCN2 in cartilaginous tumors is negatively related with the level of their malignancy (45), indicating that HCS-2/8 cells can be a model of a normal phenotype of chondrocytes. This cell line also has been used for arthritis studies, because HCS-2/8 produces MMP3 and matrix degradation-related molecules in response to tumor necrosis factor alpha (16, 17, 41). In addition, MMP3 immunopositive cells were specifically expressed in normal articular cartilage of a 14-year-old human, while MMP9, MMP13, and ADAMTS4 were not detected (H. I. Roach, personal communication). We observed nuclear MMP3 in both normal and osteoarthritic cartilages (Fig. 8). In addition, the strong expression of CCN2/CTGF in chondrocytes in osteoarthritic tissues already has been reported (36). Thus, nuclear MMP3 may function when chondrocytes are in a physiological or pathological status.
The trans activation of the CCN2/CTGF promoter by the nuclearly translocated MMP3 is a major discovery in this study. One piece of evidence for this finding is that the knockdown of MMP3 resulted in a decrease in CCN2/CTGF expression (Fig. 4). Moreover, a major CCN2/CTGF-producing cell line, HCS-2/8, was found to produce MMP3, while MMP3 expression was undetectable in HeLa cells that produce small amounts of CCN2/CTGF (Fig. 4) (7-9). MMP3 is known as a secretory proteinase that degrades ECM proteins such as proteoglycans and collagens, while MMP3 also cleaves and activates other MMPs, such as gelatinases, and growth factors, such as TGF-β. In an analysis of the signal sequence at the amino terminus of MMP3, proline was found at the fifth position of the sequence. The proline in the signal sequence can work as a helix breaker and may result in bisorting between the ER-Golgi secretion pathway and cytoplasm after the signal peptide emerges from the ribosome (44). If this partition trafficking occurred, some MMP3 would be located in the cytosol for further transport to the nucleus, while other MMP3 would be secreted through the ER-Golgi complex. This may result in both direct nuclear transportation from the cytoplasm and the internalization-nuclear transportation of MMP3.
The TGF-β/Smad signal regulates CCN2/CTGF gene expression at the level of transcription, and the activation of TGF-β by MMP3 in the extracellular microenvironment can indirectly activate CCN2/CTGF through TGF-βR/Smad signaling (7); however, the intracellular overexpression of MMP3 from the pMMP3 vector trans activated the CCN2/CTGF promoter (Fig. 3). Therefore, we consider that nuclearly translocated MMP3 directly trans activates the CCN2/CTGF gene. Interestingly, the overexpression of intracellular MMP3 and TGF-β synergistically activated the CCN2/CTGF promoter (Fig. 3C). If the Smads and intracellular MMP3 signals are activated simultaneously, some signal cross talk may occur, because the MMP3-binding site as well as TRENDIC is located immediately upstream of the Smad binding site (SBE) in the CCN2/CTGF promoter (7). Thus, MMP3 can regulate CCN2/CTGF transcription directly by itself and through activating TGF-β/Smad signaling. Further investigation about this signaling cross talk is under way.
MMP3-null mice have been reported to show delayed wound healing, joint inflammation/osteoarthritis, abnormal CD4+ physiology, increased susceptibility to bacterial infection, abnormal neuromuscular synapse morphology, and abnormal miniature end plate potential (48). CCN2/CTGF has been known to be a key player in the wound-healing process. Together with our discovery that nuclear MMP3 regulates CCN2/CTGF, the insufficient supply of CCN/CTGF in MMP3-null mice should result in a delayed wound-healing phenotype. Further investigation by using MMP3-null mice is under way.
We should be careful to explain the mechanism of trans activation by MMP3 and the novel domain structure of MMP3, because MMP3 can cleave and activate itself. The overexpression of the MMP3 deletion mutants revealed that the PEX domain and CAT+Hinge domain can trans activate the CCN2/CTGF promoter per se as well as the FL or active form of MMP3 (Fig. 5A). Catalytically dead mutants of MMP3 decreased the trans activation ability for the CCN2 promoter compared to that of the wild-type MMP3 (Fig. 5D). These results indicate that both the CAT domain and PEX domain of MMP3 can activate the CCN2 promoter independently. The catalytic activity partially contributed to the trans activation of the CCN2 promoter; however, it is not essential for the trans activation. For the mechanisms of activation, the involvement of NuMAPs was strongly suggested (Fig. 8D to G). Indeed, HP1γ synergistically trans activated the CCN2/CTGF promoter with FL MMP3 (Fig. 8E-G). The cell type-specific trans-activation mode of MMP3 for CCN2/CTGF may be ascribed to the cell type-specific involvement of cofactors such as HP1γ. Locating the HP1γ-binding domain in MMP3 also is important and is under investigation (Fig. 7A).
The DNA-binding domain of MMP3 was revealed to be in the PEX domain, because the TRENDIC-nuclear protein interaction was blocked by the anti-PEX antibody (Fig. 2A, B). In sharp contrast, the anti-N-terminal, anti-Hinge antibodies caused no significant change in the protein-DNA interactions (Fig. 2A). Moreover, no upper-shift band was observed with the other anti-MMP3 antibodies by the supershift assay (Fig. 2A). Of note, in another experiment with purified rhMMP3, the anti-PEX antibody made an upper shift with the MMP3-p160 complex (Fig. 11D). Several mechanisms can be considered in accounting for the differential effects of the anti-PEX antibody in the EMSA. For example, (i) the ZnMc/CAT domain can also bind to p160, and such a secondary DNA-binding domain may be hidden by other factors, such as NuMAPs, in the nuclear extracts; (ii) the DNA-binding ZnMc/CAT domain of nuclear MMP3 may have been cleaved away from the PEX domain; or (iii) enhanced MMP3 dimerization under the purified condition may yield a new MMP3 complex that binds to both DNA and the anti-PEX antibody.
In our study, recombinant MMP3 was translocated from outside of the cells into the nuclei (Fig. 6). Also, endogenous MMP3 was in the cells and on the cell surface (Fig. 1C and D and 9). Recent studies indicated that proMMP2 made a complex with TIMP1 and low-density lipoprotein-related protein 1 (LRP1) and then was internalized into the cells (6, 14). The LRP1/proMMP2/TIMP2 complex also was reported to be internalized (10). LRP1 is abundantly expressed in HCS-2/8 cells (25). We also observed that LRP1 was colocalized with MMP3 in HCS-2/8 cells by using laser confocal microscopy (data not shown). Thus, MMP3 or the MMP3/TIMP complex may bind to the trans-membrane type of receptor molecules, such as LRPs, for internalization into cells. The delivery of certain molecules into the nucleus from the plasma membrane by endocytosis already has been reported (3). Therefore, MMP3 may be transported to the nucleus through such a pathway. The regulation of MMP3 internalization is under investigation by using LRP1 and its inhibitor receptor-associated protein.
We also found six putative NLSs in MMP3 for nuclear entry through the nucleopores (Fig. 7). In addition, we found one putative NES in MMP3 (Fig. 7). Indeed, externally added rhMMP3 translocated to the nucleus in 30 min, whereas the amount was decreased at 60 min after the addition (Fig. 6). This loss of nuclear MMP3 may have been caused by degradation or nuclear export. LMB, which suppresses CRM1-dependent nuclear export, caused the activation of the CCN2/CTGF promoter (Fig. 6E). This result may be due to the nuclear accumulation of MMP3 caused by the blockade of CRM1-mediated nuclear export. Therefore, MMP3 can shuttle back to the cytoplasm after being translocated to the nucleus.
Of note, Si-Tayeb et al. (47) also reported an NLS in MMP3 that is the same as one of the NLSs identified in our study (Fig. 7B). It is possible that multiple NLSs are recognized synergistically for efficient nuclear translocation. Also, an NLS must be exposed on the surface of the molecule to be transported for recognition by importins for the nuclear translocation from the cytoplasm. The posttranslational modification of MMP3 can remove or hide the primary NLS and expose another NLS on the molecular surface, suggesting the differential use of multiple NLSs. We also identified a RAN-binding protein as an MMP3-associated protein. This protein is involved in the nuclear import event. As such, two independent pathways to the nucleus are indicated at present. Further investigation will specify the precise mechanism of nuclear translocation of MMP3.
We detected FL MMP3 dominantly in the nuclear fraction of HCS-2/8 cells (Fig. 1) with minor signals for its fragments, which can exert stromelysin proteolytic activity (Fig. 5). In addition to stromelysin activity in the nuclear extract of HCS-2/8 cells, there have been two reports of stromelysin activity in the nucleus (47, 54). Si-Tayeb et al. (47) reported a 35-kDa nuclear MMP3 in HepG2 cells that displayed stromelysin activity. Of note, a specific MMP3 inhibitor repressed the promoter activity of CCN2 (Fig. 4). Thus, the proteinase activity may be involved in the trans activation of CCN2 by MMP3. We speculate that an appropriate amount of nuclear MMP3 is under the control of TIMPs, so that it can work properly as a transcription factor, since we also detected TIMP-1 in the nuclear fraction of HCS-2/8 cells (data not shown). It was previously reported that TIMP-1 accumulates in the cell nuclei in association with the cell cycle (54), and multiple roles of TIMPs other than their roles as proteinase inhibitors have been indicated, such as stimulating cell growth and acting as an antiapoptotic molecule (18). Thus, it is anticipated that MMP3 interacts with TIMPs in the cells as well as in the extracellular environment. It was reported that the intracellular overexpression of MMP3 induced apoptosis (47). Here, we have speculated about the mechanism of the apoptosis elicited by cellular MMP3. One possibility is that a vast amount of cellular/nuclear MMP3 induces apoptosis by digesting cytoplasmic and intranuclear protein components. A death-associated protein that was identified as one of the NuMAPs may be involved in the apoptosis process (data not shown). Another possibility is that the nuclear MMP3 induces the overexpression of CCN2/CTGF, which subsequently induces cellular apoptosis (22).
We identified several NuMAPs by performing the coimmunoprecipitation study (Fig. 8; Table 2). These putative cofactors included heterochromatin proteins, transcription coactivators, corepressors, RNA polymerase II, chromatin assembly factors, and others. One may expect to find MMP3 in the proteins pulled down by the beads with anti-MMP3, but we found no MMP3. We believe that the elution condition we employed dissociated MMP3-NuMAP complexes but did not interfere with stronger antigen-antibody interactions. It also is possible that TIMPs have not been detected for a similar reason. One of the NuMAPs, HP1γ, was demonstrated to interact with MMP3 and to coactivate the CCN2/CTGF promoter with MMP3 (Fig. 8). Although multiple functions of heterochromatin proteins as transcription factors and as binding targets of RNA interference machinery (19, 21) have been reported, heterochromatin proteins are known to recognize the trimethylated ninth lysine of histone H3 (H3K9), resulting in chromatin condensation (2). Moreover, abundant trimethylated H3K9, representing the active form of chromatin, was detected in the promoter regions but not in the enhancer regions (20, 43). Hence, it is possible that nuclear MMP3 binds to HP1/Swi6, one of the NuMAPs, on the histone of the CCN2 enhancer region to trans activate the CCN2 gene. Indeed, we detected an interaction between MMP3 and the genomic CCN2 proximal promoter region by ChIP assay (Fig. 4E). Therefore, native HP1γ may collaborate with MMP3 on the genomic DNA locus in the cells.
As a NuMAP, NCoR1 also was identified, which is a transcription repressor that acts by promoting chromatin condensation and by preventing access to the gene by the transcription machinery. Another possible mechanism of CCN2 gene trans activation is that MMP3 degrades NCoR1 to prevent transcription repression. Recently, MMP2 was found in the nuclei of cardiac myocytes and is capable of cleaving PARP in vitro (28). Thus, PARP can be inactivated or activated by MMP3 itself, or MMP2 can be activated by MMP3. PARP has been known to perform poly(ADP-ribosyl)ation of the chromatin DNA for chromatin DNA maintenance. The cleavage and inactivation of PARP result in the inhibition of poly(ADP-ribosyl)ation and expose the DNase hypersensitive region. The exposure of such naked DNA can supply an MMP3-accessible region on DNA. Further genetic and epigenetic approaches will clarify the precise mechanisms of CCN2 induction by MMP3 acting with such cofactors.
In this report, we showed that MMP3 from outside of the cells and in the cytoplasm was translocated into the nucleus, where MMP3 bound to DNA/chromatin and trans activated the CCN2 gene (Fig. 12). During this process, some of the NuMAPs may have contributed to this transcription factor-like function of MMP3. Based on our present findings and numerous past reports, we propose a dual role for MMP3, i.e., its functioning both extra- and intracellularly. In this model, MMP3 acts to degrade ECM outside, whereas it induces CCN2 and other matrix-related proteins in the nucleus. This extra- and intracellular, dual-functioning MMP3 may play an important role in the development and matrix remodeling of cartilage and bone. Abnormalities in the MMP3 dynamics may be involved in the MMP/CCN2-related matrix diseases, e.g., osteoarthritis and rheumatism, and in fibrotic diseases such as systemic sclerosis and atherosclerosis. Clarifying and controlling the behavior of extracellular/intracellular MMP3 may assist in the establishment of a new therapeutic approach to combat such matrix-related diseases.
FIG. 12.
Novel transcription factor-like function of MMP3. The internalization of MMP3 from the cell surface into the cells via a putative receptor and the successive transportation into the nucleus were indicated. Another possible pathway is that MMP3 directly translocates to the nucleus after translation. MMP3 binds to the TRENDIC motif for trans regulation of the CCN2/CTGF gene. This trans regulation can modify the signal in cartilage development, tissue remodeling, and tissue regeneration through CCN2 molecules and may be involved in some arthritic diseases. HP1γ and certain other NuMAPs contribute to the transcription factor-like function of MMP3.
Acknowledgments
We thank Takako Hattori, Takashi Nishida, Eriko Aoyama, Takeshi Yanagita, Harumi Kawaki, and Mamoru Ohuchida for their useful discussions and technical assistance. We acknowledge Yuki Nonami for secretarial assistance and thank Seiji Tamaru for technical assistance with the LC-MS/MS analysis. We appreciate Derek C. Radisky, Motomi Enomoto-Iwamoto, Kinichi Nakashima, and Koichi Ikuta for useful discussions and helpful suggestions. We thank H. I. Roach for informing us of unpublished data from a human case and Cindy Farach-Carson for analyzing signal sequences and for useful discussions. We thank the members of ODR, NILS, and NCGG for thoughtful actions in completing this work.
This work was supported by Research Fellowships for Young Scientists of the Japan Society for the Promotion of Science to T.E., Grants-in-Aid for Scientific Research (S) to M.T. and (C) to S.K. and a Grant-in-Aid for Exploratory Research to M.T. from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (MEXT), and by grants from the Sumitomo Foundation to M.T. and the Foundation of Sanyo Broadcasting to S.K.
Footnotes
Published ahead of print on 2 January 2008.
REFERENCES
- 1.Aigner, T., B. Kurz, N. Fukui, and L. Sandell. 2002. Roles of chondrocytes in the pathogenesis of osteoarthritis. Curr. Opin. Rheumatol. 14578-584. [DOI] [PubMed] [Google Scholar]
- 2.Bannister, A. J., P. Zegerman, J. F. Partridge, E. A. Miska, J. O. Thomas, R. C. Allshire, and T. Kouzarides. 2001. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410120-124. [DOI] [PubMed] [Google Scholar]
- 3.Benmerah, A., M. Scott, V. Poupon, and S. Marullo. 2003. Nuclear functions for plasma membrane-associated proteins? Traffic 4503-511. [DOI] [PubMed] [Google Scholar]
- 4.Blom, I. E., R. Goldschmeding, and A. Leask. 2002. Gene regulation of connective tissue growth factor: new targets for antifibrotic therapy? Matrix Biol. 21473-482. [DOI] [PubMed] [Google Scholar]
- 5.Brigstock, D. R. 2003. The CCN family: a new stimulus package. J. Endocrinol. 178169-175. [DOI] [PubMed] [Google Scholar]
- 6.Chirco, R., X. W. Liu, K. K. Jung, and H. R. Kim. 2006. Novel functions of TIMPs in cell signaling. Cancer Metastasis Rev. 2599-113. [DOI] [PubMed] [Google Scholar]
- 7.Eguchi, T., S. Kubota, K. Kawata, Y. Mukudai, T. Ohgawara, K. Miyazono, K. Nakao, S. Kondo, and M. Takigawa. 2007. Different transcriptional strategies for ccn2/ctgf gene induction between human chondrocytic and breast cancer cell lines. Biochimie 89278-288. [DOI] [PubMed] [Google Scholar]
- 8.Eguchi, T., S. Kubota, S. Kondo, T. Kuboki, H. Yatani, and M. Takigawa. 2002. A novel cis-element that enhances connective tissue growth factor gene expression in chondrocytic cells. Biochem. Biophys. Res. Commun. 295445-451. [DOI] [PubMed] [Google Scholar]
- 9.Eguchi, T., S. Kubota, S. Kondo, T. Shimo, T. Hattori, T. Nakanishi, T. Kuboki, H. Yatani, and M. Takigawa. 2001. Regulatory mechanism of human connective tissue growth factor (CTGF/Hcs24) gene expression in a human chondrocytic cell line, HCS-2/8. J. Biochem. (Tokyo) 13079-87. [DOI] [PubMed] [Google Scholar]
- 10.Emonard, H., G. Bellon, L. Troeberg, A. Berton, A. Robinet, P. Henriet, E. Marbaix, K. Kirkegaard, L. Patthy, Y. Eeckhout, H. Nagase, W. Hornebeck, and P. J. Courtoy. 2004. Low density lipoprotein receptor-related protein mediates endocytic clearance of pro-MMP-2.TIMP-2 complex through a thrombospondin-independent mechanism. J. Biol. Chem. 27954944-54951. [DOI] [PubMed] [Google Scholar]
- 11.Galazka, G., L. J. Windsor, H. Birkedal-Hansen, and J. A. Engler. 1999. Spontaneous propeptide processing of mini-stromelysin-1 mutants blocked by APMA [(4-aminophenyl)mercuric acetate]). Biochemistry 381316-1322. [DOI] [PubMed] [Google Scholar]
- 12.Golubkov, V. S., S. Boyd, A. Y. Savinov, A. V. Chekanov, A. L. Osterman, A. Remacle, D. V. Rozanov, S. J. Doxsey, and A. Y. Strongin. 2005. Membrane type-1 matrix metalloproteinase (MT1-MMP) exhibits an important intracellular cleavage function and causes chromosome instability. J. Biol. Chem. 28025079-25086. [DOI] [PubMed] [Google Scholar]
- 13.Grotendorst, G. R., H. Okochi, and N. Hayashi. 1996. A novel transforming growth factor beta response element controls the expression of the connective tissue growth factor gene. Cell Growth Differ. 7469-480. [PubMed] [Google Scholar]
- 14.Hahn-Dantona, E., J. F. Ruiz, P. Bornstein, and D. K. Strickland. 2001. The low density lipoprotein receptor-related protein modulates levels of matrix metalloproteinase 9 (MMP-9) by mediating its cellular catabolism. J. Biol. Chem. 27615498-15503. [DOI] [PubMed] [Google Scholar]
- 15.Hashimoto, G., I. Inoki, Y. Fujii, T. Aoki, E. Ikeda, and Y. Okada. 2002. Matrix metalloproteinases cleave connective tissue growth factor and reactivate angiogenic activity of vascular endothelial growth factor 165. J. Biol. Chem. 27736288-36295. [DOI] [PubMed] [Google Scholar]
- 16.Hattori, T., H. Kawaki, S. Kubota, Y. Yutani, B. de Crombrugghe, K. von der Mark, and M. Takigawa. 2003. Downregulation of a rheumatoid arthritis-related antigen (RA-A47) by ra-a47 antisense oligonucleotides induces inflammatory factors in chondrocytes. J. Cell. Physiol. 19794-102. [DOI] [PubMed] [Google Scholar]
- 17.Hattori, T., S. Kubota, Y. Yutani, T. Fujisawa, T. Nakanishi, K. Takahashi, and M. Takigawa. 2001. Change in cellular localization of a rheumatoid arthritis-related antigen (RA-A47) with downregulation upon stimulation by inflammatory cytokines in chondrocytes. J. Cell. Physiol. 186268-281. [DOI] [PubMed] [Google Scholar]
- 18.Hayakawa, T. 2002. Multiple functions of tissue inhibitors of metalloproteinases (TIMPs): a new aspect involving osteoclastic bone resorption. J. Bone Miner. Metab. 201-13. [DOI] [PubMed] [Google Scholar]
- 19.Hediger, F., and S. M. Gasser. 2006. Heterochromatin protein 1: don't judge the book by its cover! Curr. Opin. Genet. Dev. 16143-150. [DOI] [PubMed] [Google Scholar]
- 20.Heintzman, N. D., R. K. Stuart, G. Hon, Y. Fu, C. W. Ching, R. D. Hawkins, L. O. Barrera, S. Van Calcar, C. Qu, K. A. Ching, W. Wang, Z. Weng, R. D. Green, G. E. Crawford, and B. Ren. 2007. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 39311-318. [DOI] [PubMed] [Google Scholar]
- 21.Hiragami, K., and R. Festenstein. 2005. Heterochromatin protein 1: a pervasive controlling influence. Cell. Mol. Life Sci. 622711-2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hishikawa, K., T. Nakaki, and T. Fujii. 2000. Connective tissue growth factor induces apoptosis via caspase 3 in cultured human aortic smooth muscle cells. Eur. J. Pharmacol. 39219-22. [DOI] [PubMed] [Google Scholar]
- 23.Holmes, A., D. J. Abraham, S. Sa, X. Shiwen, C. M. Black, and A. Leask. 2001. CTGF and SMADs, maintenance of scleroderma phenotype is independent of SMAD signaling. J. Biol. Chem. 27610594-10601. [DOI] [PubMed] [Google Scholar]
- 24.Ivkovic, S., B. S. Yoon, S. N. Popoff, F. F. Safadi, D. E. Libuda, R. C. Stephenson, A. Daluiski, and K. M. Lyons. 2003. Connective tissue growth factor coordinates chondrogenesis and angiogenesis during skeletal development. Development 1302779-2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kawata, K., T. Eguchi, S. Kubota, H. Kawaki, M. Oka, S. Minagi, and M. Takigawa. 2006. Possible role of LRP1, a CCN2 receptor, in chondrocytes. Biochem. Biophys. Res. Commun. 345552-559. [DOI] [PubMed] [Google Scholar]
- 26.Kubota, S., and M. Takigawa. 2007. Role of CCN2/CTGF/Hcs24 in bone growth. Int. Rev. Cytol. 2571-41. [DOI] [PubMed] [Google Scholar]
- 27.Kurisu, G., T. Kinoshita, A. Sugimoto, A. Nagara, Y. Kai, N. Kasai, and S. Harada. 1997. Structure of the zinc endoprotease from Streptomyces caespitosus. J. Biochem. (Tokyo) 121304-308. [DOI] [PubMed] [Google Scholar]
- 28.Kwan, J. A., C. J. Schulze, W. Wang, H. Leon, M. Sariahmetoglu, M. Sung, J. Sawicka, D. E. Sims, G. Sawicki, and R. Schulz. 2004. Matrix metalloproteinase-2 (MMP-2) is present in the nucleus of cardiac myocytes and is capable of cleaving poly (ADP-ribose) polymerase (PARP) in vitro. FASEB J. 18690-692. [DOI] [PubMed] [Google Scholar]
- 29.Leask, A., and D. J. Abraham. 2006. All in the CCN family: essential matricellular signaling modulators emerge from the bunker. J. Cell Sci. 1194803-4810. [DOI] [PubMed] [Google Scholar]
- 30.Lin, Z., C. Willers, J. Xu, and M. H. Zheng. 2006. The chondrocyte: biology and clinical application. Tissue Eng. 121971-1984. [DOI] [PubMed] [Google Scholar]
- 31.Luo, D., B. Mari, I. Stoll, and P. Anglard. 2002. Alternative splicing and promoter usage generates an intracellular stromelysin 3 isoform directly translated as an active matrix metalloproteinase. J. Biol. Chem. 27725527-25536. [DOI] [PubMed] [Google Scholar]
- 32.Nagase, H., C. G. Fields, and G. B. Fields. 1994. Design and characterization of a fluorogenic substrate selectively hydrolyzed by stromelysin 1 (matrix metalloproteinase-3). J. Biol. Chem. 26920952-20957. [PubMed] [Google Scholar]
- 33.Nagase, H., and J. F. Woessner, Jr. 1999. Matrix metalloproteinases. J. Biol. Chem. 27421491-21494. [DOI] [PubMed] [Google Scholar]
- 34.Nakai, K., and P. Horton. 1999. PSORT: a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem. Sci. 2434-36. [DOI] [PubMed] [Google Scholar]
- 35.Nishida, T., S. Kubota, T. Fukunaga, S. Kondo, G. Yosimichi, T. Nakanishi, T. Takano-Yamamoto, and M. Takigawa. 2003. CTGF/Hcs24, hypertrophic chondrocyte-specific gene product, interacts with perlecan in regulating the proliferation and differentiation of chondrocytes. J. Cell. Physiol. 196265-275. [DOI] [PubMed] [Google Scholar]
- 36.Nishida, T., S. Kubota, S. Kojima, T. Kuboki, K. Nakao, T. Kushibiki, Y. Tabata, and M. Takigawa. 2004. Regeneration of defects in articular cartilage in rat knee joints by CCN2 (connective tissue growth factor). J. Bone Miner. Res. 191308-1319. [DOI] [PubMed] [Google Scholar]
- 37.Nishida, T., S. Kubota, T. Nakanishi, T. Kuboki, G. Yosimichi, S. Kondo, and M. Takigawa. 2002. CTGF/Hcs24, a hypertrophic chondrocyte-specific gene product, stimulates proliferation and differentiation, but not hypertrophy of cultured articular chondrocytes. J. Cell. Physiol. 19255-63. [DOI] [PubMed] [Google Scholar]
- 38.Perbal, B. 2004. CCN proteins: multifunctional signalling regulators. Lancet 36362-64. [DOI] [PubMed] [Google Scholar]
- 39.Perbal, B., and M. Takigawa. 2005. CCN proteins: a new family of cell growth and differentiation regulators, 1st ed. Imperial College Press, London, United Kingdom.
- 40.Saas, J., K. Lindauer, B. Bau, M. Takigawa, and T. Aigner. 2004. Molecular phenotyping of HCS-2/8 cells as an in vitro model of human chondrocytes. Osteoarthritis Cartilage 12924-934. [DOI] [PubMed] [Google Scholar]
- 41.Sakai, T., F. Kambe, H. Mitsuyama, N. Ishiguro, K. Kurokouchi, M. Takigawa, H. Iwata, and H. Seo. 2001. Tumor necrosis factor alpha induces expression of genes for matrix degradation in human chondrocyte-like HCS-2/8 cells through activation of NF-kappaB: abrogation of the tumor necrosis factor alpha effect by proteasome inhibitors. J. Bone Miner. Res. 161272-1280. [DOI] [PubMed] [Google Scholar]
- 42.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 745463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schübeler, D. 2007. Enhancing genome annotation with chromatin. Nat. Genet. 39284-285. [DOI] [PubMed] [Google Scholar]
- 44.Shaffer, K. L., A. Sharma, E. L. Snapp, and R. S. Hegde. 2005. Regulation of protein compartmentalization expands the diversity of protein function. Dev. Cell 9545-554. [DOI] [PubMed] [Google Scholar]
- 45.Shakunaga, T., T. Ozaki, N. Ohara, K. Asaumi, T. Doi, K. Nishida, A. Kawai, T. Nakanishi, M. Takigawa, and H. Inoue. 2000. Expression of connective tissue growth factor in cartilaginous tumors. Cancer 891466-1473. [PubMed] [Google Scholar]
- 46.Shimo, T., T. Nakanishi, T. Nishida, M. Asano, M. Kanyama, T. Kuboki, T. Tamatani, K. Tezuka, M. Takemura, T. Matsumura, and M. Takigawa. 1999. Connective tissue growth factor induces the proliferation, migration, and tube formation of vascular endothelial cells in vitro, and angiogenesis in vivo. J. Biochem. 126137-145. [DOI] [PubMed] [Google Scholar]
- 47.Si-Tayeb, K., A. Monvoisin, C. Mazzocco, S. Lepreux, M. Decossas, G. Cubel, D. Taras, J. F. Blanc, D. R. Robinson, and J. Rosenbaum. 2006. Matrix metalloproteinase 3 is present in the cell nucleus and is involved in apoptosis. Am. J. Pathol. 1691390-1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Somerville, R. P., S. A. Oblander, and S. S. Apte. 2003. Matrix metalloproteinases: old dogs with new tricks. Genome Biol. 4216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sternlicht, M. D., and Z. Werb. 2001. How matrix metalloproteinases regulate cell behavior. Annu. Rev. Cell Dev. Biol. 17463-516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Takigawa, M., T. Nakanishi, S. Kubota, and T. Nishida. 2003. Role of CTGF/HCS24/ecogenin in skeletal growth control. J. Cell. Physiol. 194256-266. [DOI] [PubMed] [Google Scholar]
- 51.Takigawa, M., T. Okawa, H. Pan, C. Aoki, K. Takahashi, J. Zue, F. Suzuki, and A. Kinoshita. 1997. Insulin-like growth factors I and II are autocrine factors in stimulating proteoglycan synthesis, a marker of differentiated chondrocytes, acting through their respective receptors on a clonal human chondrosarcoma-derived chondrocyte cell line, HCS-2/8. Endocrinology 1384390-4400. [DOI] [PubMed] [Google Scholar]
- 52.van Bilsen, J. H., J. P. Wagenaar-Hilbers, M. C. Grosfeld-Stulemeijer, M. J. van der Cammen, M. E. van Dijk, W. van Eden, and M. H. Wauben. 2004. Matrix metalloproteinases as targets for the immune system during experimental arthritis. J. Immunol. 1725063-5068. [DOI] [PubMed] [Google Scholar]
- 53.Yoshimura, T., J. Fujisawa, and M. Yoshida. 1990. Multiple cDNA clones encoding nuclear proteins that bind to the tax-dependent enhancer of HTLV-1: all contain a leucine zipper structure and basic amino acid domain. EMBO J. 92537-2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhao, W. Q., H. Li, K. Yamashita, X. K. Guo, T. Hoshino, S. Yoshida, T. Shinya, and T. Hayakawa. 1998. Cell cycle-associated accumulation of tissue inhibitor of metalloproteinases-1 (TIMP-1) in the nuclei of human gingival fibroblasts. J. Cell Sci. 1111147-1153. [DOI] [PubMed] [Google Scholar]