Abstract
Eaf1 (for Esa1-associated factor 1) and Eaf2 have been identified as stable subunits of NuA4, a yeast histone H4/H2A acetyltransferase complex implicated in gene regulation and DNA repair. While both SWI3-ADA2-N-CoR-TF IIIB domain-containing proteins are required for normal cell cycle progression, their depletion does not affect the global Esa1-dependent acetylation of histones. In contrast to all other subunits, Eaf1 is found exclusively associated with the NuA4 complex in vivo. It serves as a platform that coordinates the assembly of functional groups of subunits into the native NuA4 complex. Eaf1 shows structural similarities with human p400/Domino, a subunit of the NuA4-related TIP60 complex. On the other hand, p400 also possesses an SWI2/SNF2 family ATPase domain that is absent from the yeast NuA4 complex. This domain is highly related to the yeast Swr1 protein, which is responsible for the incorporation of histone variant H2AZ in chromatin. Since all of the components of the TIP60 complex are homologous to SWR1 or NuA4 subunits, we proposed that the human complex corresponds to a physical merge of two yeast complexes. p400 function in TIP60 then would be accomplished in yeast by cooperation between SWR1 and NuA4. In agreement with such a model, NuA4 and SWR1 mutants show strong genetic interactions, NuA4 affects histone H2AZ incorporation/acetylation in vivo, and both preset the PHO5 promoter for activation. Interestingly, the expression of a chimeric Eaf1-Swr1 protein recreates a single human-like complex in yeast cells. Our results identified the key central subunit for the structure and functions of the NuA4 histone acetyltransferase complex and functionally linked this activity with the histone variant H2AZ from yeast to human cells.
Chromatin is a very dynamic structure, and it plays an intricate regulatory role in DNA replication, transcription, and repair. Two major types of activities regulating chromatin structure and function have been studied extensively over the past few years and have been functionally linked together in diverse nuclear processes (65). ATP-dependent chromatin-remodeling complexes of the SWI2/SNF2 family disrupt DNA-histone contacts within nucleosomes, increasing DNA accessibility and nucleosome mobility (25). Histone-modifying complexes target specific residues on histones for acetylation, methylation, phosphorylation, and ubiquitinylation. These modifications can affect the level of DNA compaction but also serve as markers identifying the chromatin state of specific genomic loci. Different histone modifications influence each other and create a specific local signature that can be recognized by protein domains present in various regulators, e.g., bromodomains for acetylated lysines and chromodomains for methylated lysines. These posttranslational modifications are reversible and highly dynamic during cell growth (36). Diverse ATP-dependent remodelers and histone modifiers have been shown to be recruited to specific loci through direct interactions with DNA-bound factors. For example, histone acetyltransferase (HAT) and histone deacetylase (HDAC) complexes are recruited to specific promoter regions by transcriptional activators or repressors (61). The local incorporation of specific histone variants is an additional mechanism that regulates chromatin function. Activities responsible for these incorporations recently have been identified, and a specific class of ATP-dependent remodelers has been implicated in this process (31, 40).
Nucleosome acetyltransferase of H4 (NuA4) is a multisubunit HAT complex that is highly conserved in eukaryotes and plays important roles in transcription and DNA repair (2, 3, 18, 62). Its catalytic subunit, Esa1, is the only HAT protein essential for viability in Saccharomyces cerevisiae and is responsible for the bulk of histone H4 and H2A acetylation in vivo (18). NuA4 is recruited to the promoter region of many highly transcribed genes (54), regulates the expression of ribosomal protein genes (53), and presets the PHO5 promoter for chromatin remodeling and transcription activation by creating a region of hyperacetylated chromatin (50). Furthermore, a functional subcomplex of NuA4, Piccolo NuA4 (picNuA4), is formed by the Esa1/Epl1/Yng2 trimer, can be independently purified, and is responsible for the global nontargeted acetylation of chromatin by Esa1 (9). Rapid global acetylation-deacetylation of chromatin creates a highly dynamic equilibrium that can be pushed in either direction by the local recruitment of HAT or HDAC activity (32). The NuA4 complex also harbors two of the three chromodomain-containing proteins in budding yeast (21, 60), and it was shown that histone H3 methylation regulates the Esa1-dependent H4 acetylation of the MET16 promoter during transcription activation (47). The ATM family protein Tra1 is shared by the NuA4 and Spt-Ada-Gcn5 acetyltransferase HAT complexes and is implicated as an interaction surface for recruitment by specific transcription activators (11). The NuA4 complex also has been directly implicated in the repair of DNA double-strand breaks (2, 7, 17). It was shown to be recruited to DNA breaks in vivo, to interact with the surrounding phosphorylated H2AX through its Arp4 subunit, and to regulate the subsequent recruitment of ATP-dependent remodelers (17). Finally, NuA4 also plays a role in the establishment of the heterochromatin/euchromatin boundary near telomere regions (9, 69). The NuA4 complex is highly homologous to the TIP60 HAT complex in higher eukaryotes, which has been implicated in transcription regulation by numerous transcription factors (including Myc, E2F, NF-κB, and p53) and in cell transformation, development, apoptosis, and DNA repair (4, 18, 19, 62).
Here, we present the characterization of two NuA4 subunits carrying a SWI3-ADA2-N-CoR-TF IIIB (SANT) domain, a conserved region found in many chromatin regulators and implicated in interactions with histone tails, DNA, or other proteins (10). While both subunits play important roles for NuA4 function in cell cycle progression, gene regulation, and DNA repair, they do not affect the global Esa1-dependent acetylation of chromatin. We show that Eaf1 (for Esa1-associated factor 1) is the only subunit exclusively found in the NuA4 complex and that it is the platform on which the different functional groups of other subunits are assembled into the native complex. This identifies Eaf1 as the only tool to specifically analyze the native NuA4 complex in vivo. Based on homology with the TIP60 complex in higher eukaryotes and the fact that NuA4 shares four subunits with the SWR1 ATP-dependent chromatin remodeler, we also present a functional and structural analysis of interactions between NuA4, SWR1, and the histone variant H2AZ. Our results indicate that the NuA4-dependent acetylation of chromatin cooperates with the incorporation of H2AZ by SWR1. We also demonstrate that the TIP60 complex corresponds to an exact physical merge of two distinct protein complexes in yeast, NuA4 and SWR1.
MATERIALS AND METHODS
Yeast strains and reagents.
All yeast manipulations, cultures, transformations, sporulations, and matings and all bacterial manipulations were performed according to standard protocols. The deletion mutants of Eaf1 (Δ1-538, Δ329-538, and Δ604-815; created by restriction enzyme digestion) were amplified by PCR from the EAF1-containing pSK plasmids and cloned into a BFG1 high-copy-number plasmid under the control of the PGK promoter (24). The full-length EAF2 as well as its promoter were amplified from yeast genomic DNA and cloned sequentially into autonomous replicating sequence/centromere (ARS/CEN) vectors in order to introduce a hemagglutinin (HA) tag at the N terminus of EAF2 (pPAN107) as described previously (49). The truncated version of Eaf2 was created by first amplifying the sequence coding for the amino acids (aa) 1 to 285 of Eaf2. The amplified segment was digested and ligated into an ARS/CEN vector (pPNE2). Strain QY208 was obtained by the transformation of the diploid strain 24632 (Open Biosystems) with pAN107, followed by sporulation and tetrad dissection. To place EAF2 under the control of the inducible promoter GAL1, the GAL1 sequence (linked to three HA tags) was amplified from pFA6a-kanMX6-PGAL-3HA (42). The amplified product was transformed into QY204 (49). The epl1(1-380)Δswr1 and the epl1(1-380)Δhtz1 strains were obtained by the deletion of the SWR1 or HTZ1 open reading frame (ORF) in QY142 (9) with the HIS3 marker. Cells then were grown on medium containing 5-fluoroorotic acid (5-FOA) to chase the URA3 plasmid harboring the wild-type EPL1 gene. The yDomino Eaf1-Swr1 fusion expression vector was created by amplifying the Swr1 ATPase domain and cloning it in the NheI site of EAF1 (aa 538) and cloned as described above.
Polyclonal antibodies against Eaf1(1-538) were made by cloning the cDNA fragment in pET15b followed by the expression/purification and injection of the purified recombinant protein into two rabbits. Collected serum was purified by ammonium sulfate precipitation and carboxymethyl-Affi-Gel blue as described previously (1). Tra1, Esa1, Arp1, and AcK14 Htz1 antibodies have been previously described (1, 24, 29, 45). Anti-methyl-K4 H3, anti-AcK8 H4, anti-AcK12 H4, anti-AcH2A, anti-AcH3, and anti-Htz1 (C terminus) were purchased from Upstate; anti-H3 (C terminus), anti-Htz1 (N terminus), and anti-Yaf9 were purchased from Abcam; and anti-AcK5 H4 and anti-AcK16 H4 were purchased from Serotec. The anti-AcK14 Htz1, anti-Arp4, anti-Tra1, anti-Yng2, and anti-Eaf3 antibodies were kindly provided by the laboratories of M. Grunstein, D. J. Stillman, J. L. Workman, S. Tan, and J. C. Lucchesi, respectively. Anti-Rvb1 and anti-Rvb2 were generous gifts of Y. Makino.
MMS, rapamycin, and hydroxyurea sensitivity and telomere-silencing assays.
Yeast strains were grown overnight to stationary phase in yeast extract-peptone-dextrose (YPD) medium, diluted to an optical density at 600 nm (OD600) of 0.25, and incubated for a further 3 h. From the diluted culture, 7 μl of serial 10-fold dilutions were spotted onto YPD solid medium containing either 0.03% methyl methanesulfonate (MMS) (Sigma), 25 mM rapamycin (Aldrich), or 130 mM hydroxyurea (Sigma), and the samples were incubated at 30°C. For telomere-silencing assays, EAF1 and YAF9 ORFs were deleted from UCC1001 (a gift of D. Gottschling) and tested as described before (9) on Hartwell complete medium and Hartwell complete medium-5-FOA.
NuA4/TAP purifications and peptide sequencing.
The classical purification of the NuA4 complex was performed as described previously (1). Peptide sequences of Eaf1 and Eaf2 isolated from immunopurified NuA4 were obtained by ion trap mass spectrometry (MS) and protein sequencing as described previously (21). The purifications of the NuA4 and SWR1 complexes using the tandem affinity purification (TAP) system were performed as described previously (51). Up to 6 liters of yeast cultures expressing the desired TAP-tagged protein was grown to an OD600 of 2.5. Cell extracts were precleared against Sepharose and fractionated sequentially on 100 μl of immunoglobulin G and calmodulin resins for every liter of culture, and the final elution of purified TAP-protein complex was done with 10 mM EGTA-containing buffer. Protein complexes were visualized on sodium dodecyl sulfate (SDS)-polyacrylamide gels and stained with silver. The different TAP tag PCR amplifications for each target gene were done using the plasmids pBS1479 and pBS1539 (51) in an S288c background. The endogenous EAF2 locus was directly modified in order to express either full-length Eaf2 or Eaf2(1-285) tagged with TAP at the C terminus. For peptide sequencing, the gel was stained using colloidal blue (Bio-Rad). The appropriate bands were destained, excised, and submitted to mass spectrometry (MS/MS) at the Centre de Protéomique de l'Est du Québec.
HAT assays and Western/Northern blot analyses.
Liquid HAT assays were performed, according to a method described previously(28), using 0.5 μg of free core histones or oligonucleosomes, protein fractions, and 0.125 μCi of [3H]acetyl coenzyme A (4.7 Ci/mmol; Amersham Biosciences). The acetylation specificity was determined by loading the reaction onto an SDS-18% polyacrylamide gel electrophoresis (PAGE) followed by fluorography with Enhance (DuPont NRN). For Western blot analysis, protein factions were separated on an SDS-10% PAGE, transferred to a nitrocellulose membrane, and screened with antibodies in 1% nonfat dry milk in phosphate-buffered saline (PBS)-0.1% Tween 20 for 3 h at room temperature. RNA from yeast cells was isolated using the hot phenol method, and 15 to 20 μg was analyzed by Northern blotting as described previously (24). The probes used were ORFs from HIS4, PHO5, TRP4, RPS11b, GAL1, and ACT1, which were obtained by PCR and radiolabeled with random primers (Amersham Biosciences).
Flow cytometry analysis.
One million freshly grown cells were harvested and fixed in 50 mM Tris (pH 5.0), 70% ethanol and incubated at 4°C for 15 h. Cells were washed and resuspended in 500 μl of 50 mM Tris (pH 7.5), incubated with 0.5 mg of RNase A at 37°C for 2 h, and then incubated with 50 μg of proteinase K at 50°C for 1 h. The cells subsequently were stained with 1 ml of 2.5-mg/ml propidium iodide in 50 mM Tris (pH 7.5), incubated in the dark for 15 h at 4°C, and then analyzed with a Becton Dickinson FACScan. The G1 arrest was achieved using the α-mating factor at 1.66 μg/ml. The block was maintained for a period of time equivalent to two generations. Subsequently, cells were released by being washed and were incubated in fresh medium.
ChIP.
Overnight cultures were diluted and grown to an OD600 of 0.5 to 1.0. Cells were fixed and chromatin was isolated, immunoprecipitated, and analyzed by real-time PCR essentially as described previously (17, 50). Immunoprecipitations were carried out using 100 μg of cross-linked chromatin (sonicated to 200 to 600 bp) and 3 μl of anti-Htz1 (N terminus) (Abcam) (see Fig. 4), 4 μl of anti-Htz1 (C terminus) (Upstate) (see Fig. 5), 1 μl anti-AcK14 Htz1 (45), 3 μl of anti-Eaf1 (17 and this study), 0.5 μl anti-AcK8 H4 (Upstate), or 1 μl anti-H3 (C terminus) (Abcam). PCR primers for the UAS2 region of PHO5 were analyzed for specificity, efficiency, and linearity range by real-time PCR with a LightCycler (Roche), and their sequences/features are available upon request. All of the chromatin immunoprecipitation (ChIP) experiments were repeated from different cultures with similar results. The results shown are based on two independent experiments with standard errors. Data are corrected for the nucleosome occupancy using the total H3 signal (ratio of immunoprecipitate [IP]/input) and are presented relative to the values for the wild-type strain (set to 1).
FIG. 4.
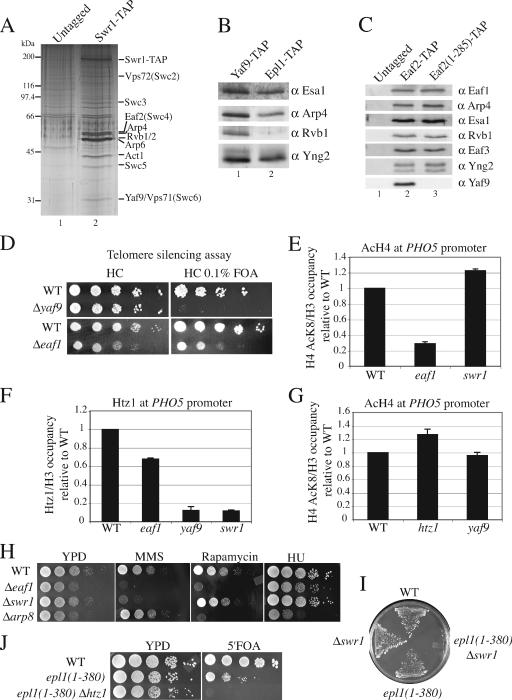
Structural and functional relationships between NuA4 and SWR1 complexes. (A) The SWR1 complex shares subunits with NuA4. Purified material from untagged strains and Swr1-TAP strains was loaded onto gradient gels and visualized by silver staining. Bands corresponding to the SWR1 subunits are indicated on the right. (B) Yaf9 is a subunit common to the NuA4 and Swr1 complexes. Shown is a Western blot analysis using the final elution of YAF9-TAP and EPL1-TAP purifications using the indicated antibodies (α). (C) Yaf9 requires the C-terminal portion of Eaf2 for its association with NuA4 or SWR1. Purified material from untagged strains and EAF2-TAP and Eaf2(1-285)-TAP strains was loaded onto gradient gels and analyzed by Western blotting using the indicated antibodies. (D) The Δyaf9 and Δeaf1 mutants show a defect in telomere silencing. Wild-type (WT), Δyaf9, and Δeaf1 strains were plated on 5-FOA to verify the expression of the URA3 marker positioned in a silent telomeric region. The absence of growth on 5-FOA indicates URA3 marker expression. (E) Deletion of EAF1 cripples H4 acetylation (AcH4) at the PHO5 promoter, while SWR1 deletion has no effect. Shown is a ChIP analysis with anti-H3 (C terminus) and anti-H4AcK8 in wild-type, Δeaf1, and Δswr1 strains. Precipitated DNA was analyzed by PCR with primers corresponding to chromosomal regions at the PHO5 promoter (UAS2 region). Data are presented as an IP ratio of the amount of AcK8 H4 on total H3 (to correct for a change in nucleosome occupancy) relative to that of the wild-type strain. The values are based on two independent experiments. (F) Incorporation of Htz1 at the PHO5 promoter depends on SWR1 but also is affected by Eaf1. ChIP analysis was performed as described for panel E with anti-H3 and anti-Htz1 in wild-type, Δeaf1, Δyaf9, and Δswr1 strains. (G) SWR1/Htz1 do not influence H4 acetylation at the PHO5 promoter. ChIP analysis was done as described for panel E with wild-type, Δhtz1, and Δyaf9 strains. (H) NuA4 and SWR1 mutants show common and distinct phenotypes. Serial 10-fold dilutions of Δeaf1, Δswr1, Δarp8 (specific to the INO80 complex), and wild-type mid-log-phase cultures were grown in the presence or absence of 0.03% MMS, 25 nM rapamycin, or 130 mM hydroxyurea (HU). (I) swr1/htz1 mutants show synthetic lethality with mutant cells lacking normal NuA4 complex but containing picNuA4 global HAT activity. Wild-type, Δswr1, epl1(1-380), and epl1(1-380)Δswr1 cells were plated onto solid medium containing 5-FOA (to lose an episomal copy of wild-type EPL1 on a URA3 vector). (J) Serial 10-fold dilutions of wild-type, epl1(1-380), and epl1(1-380)Δhtz1 strains carrying an episomal copy of wild-type EPL1 on a URA3 vector were plated onto YPD with or without 5-FOA.
FIG. 5.
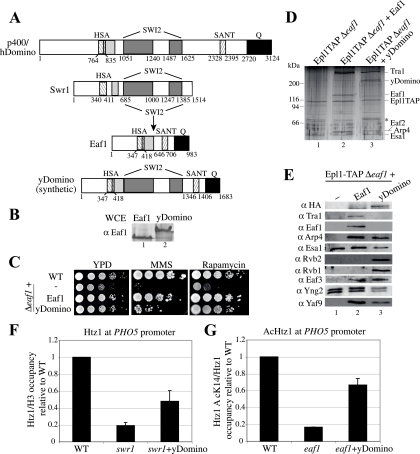
Fusion of Eaf1 and Swr1 proteins reconstitutes a human TIP60-like complex in yeast cells. (A) Construction scheme of yDomino. The SWI2 domain of Swr1 was introduced by restriction endonucleases into Eaf1 to produce a yeast version of human p400/Domino (p400/hDomino). (B) Synthetic construct of a yeast Domino-like protein is expressed at levels similar to that of Eaf1 in vivo. The Western blot analysis of whole-cell extracts (WCE) from strains carrying the indicated expression vectors using anti-Eaf1 is shown. (C) The Eaf1-Swr1 fusion protein suppresses the MMS and rapamycin sensitivity of eaf1 mutant cells. Serial 10-fold dilutions of mid-log-phase Δeaf1, Δeaf1+Eaf1, and Δeaf1+yDomino cultures were spotted on medium containing 0.03% MMS or 25 nM rapamycin. (D) Reconstitution of the human NuA4/TIP60-like complex in yeast. TAP-purified material from strains EPL1-TAPΔeaf1 and EPL1-TAPΔeaf1 expressing Eaf1 or yDomino from a vector was visualized on a gel after silver staining. The asterisk corresponds to nonspecific bands known to purify with TAP-tagged protein. (E) Rvb1/2 helicases associate with NuA4 harboring the yDomino fusion protein. Shown is a Western blot analysis of purified material described for panel C using the indicated antibodies (α), which reveals normal NuA4 assembly in the presence of yDomino and the association of helicases Rvb1/2 with the complex. (F) yDomino-containing complex increases the amount of Htz1 at the PHO5 promoter in the absence of Swr1. ChIP analysis was performed as described in the legend to Fig. 4E with anti-H3 and anti-Htz1 in wild-type (WT), Δswr1, and Δswr1+yDomino strains. Data are presented as IP ratios of the amount of Htz1 on total H3 relative to that of the wild-type strain. (G) yDomino-containing complex allows the acetylation of Htz1 (AcHtz1) at the PHO5 promoter in the absence of Eaf1. ChIP analysis was performed as described in the legend to Fig. 4E with anti-AcK14 Htz1 and anti-Htz1 (C terminus) in wild-type, Δeaf1, and Δeaf1+yDomino strains. Data are presented as IP ratios of the amount of AcK14 Htz1 on total Htz1 (to correct for the effect of Eaf1 on Htz1 incorporation [Fig. 4F]) relative to that of the wild-type strain.
RESULTS
Eaf1 is a stable and specific subunit of the NuA4 HAT complex and is critical for its targeted functions.
In our initial purification of the yeast NuA4 complex, we identified two components carrying a SANT domain, named Eaf1 and Eaf2. Cellular Eaf1 (also known as Vid21) copurifies tightly with the NuA4 complex throughout many chromatographic steps, including gel filtration (Fig. 1A). On the basis of stoichiometry and MS analysis, TAP of Eaf1 itself indicates that it is exclusively associated with NuA4 in vivo (Fig. 1B). This is a very important feature of Eaf1, since all of the other 12 subunits of NuA4 also are present in distinct protein complexes (1, 9, 13, 17, 33, 35, 37; N. Lacoste and J. Côté, unpublished data). In addition to its SANT domain, Eaf1 carries an HSA (for helicase/SANT-associated) domain (16), a highly charged region, and a C-terminal glutamine-rich region (Fig. 1C).
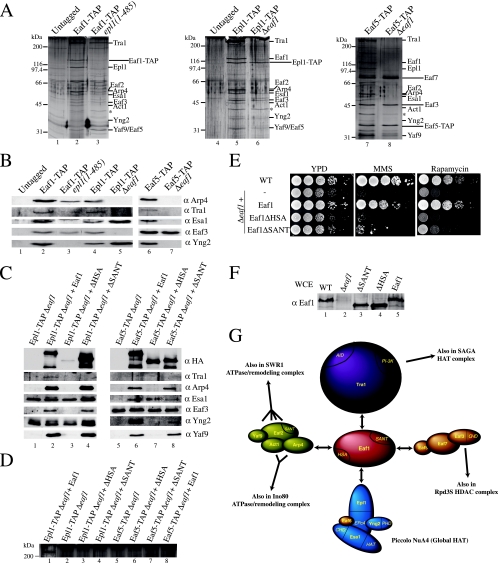
FIG. 1.
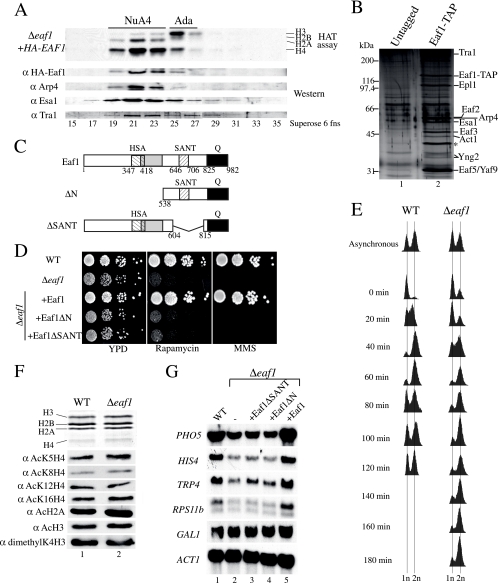
Eaf1 is found exclusively in the NuA4 HAT complex and is required for its functions but not for global histone H4/H2A acetylation. (A) Eaf1 coelutes with the HAT activity of NuA4 as well as with subunits of the NuA4 HAT complex. An extract from Δeaf1 cells harboring an episomal HA-EAF1 gene was fractionated on nickel and Mono Q columns. The desired fractions (fns) were pooled and loaded on a Superose-6 gel filtration column. Western blots show Arp4, Tra1, and Esa1 coelution with NuA4 HAT activity (fractions 19 to 23). α, anti. (B) Eaf1 is specific to the NuA4 complex. The TAP of Eaf1 shows the association with NuA4 subunits. Purified material from untagged and EAF1-TAP strains was loaded onto gradient gels and visualized by silver staining. Bands corresponding to the NuA4 subunits are indicated on the right. The asterisk corresponds to a nonspecific band known to purify with TAP-tagged protein. (C) Schematic representation of Eaf1. Eaf1 contains an HSA domain,a charged domain (gray) partially overlapping the HSA domain, and a SANT domain at the positions indicated. The C terminus of Eaf1 is enriched in glutamine (Q) (black). (D) Full-length Eaf1 is required for growth in the presence of DNA damage or crippled ribosome biogenesis. Serial 10-fold dilutions of Δeaf1, eaf1ΔN, eaf1ΔSANT, and isogenic wild-type (WT) strains were grown in the presence or absence of 0.03% MMS or 25 nM rapamycin. (E) Deletion of Eaf1 results in a slow G2/M passage. Liquid cultures of wild-type and Δeaf1 cells were blocked in G1 by the addition of α-factor. DNA content was quantified by fluorescent-activated cell sorter analysis. The 1n peak represents cells in the G1/S stage, whereas the 2n peak represents cells in the G2/M stage. (F) Deletion of Eaf1 does not affect global acetylation by NuA4. Nucleosomal histones from isogenic wild-type and Δeaf1 strains were purified and analyzed by Western blotting using antibodies indicated on the left (the top panel shows Coomassie-stained histones). (G) Eaf1 is implicated in gene-specific regulation. RNAs from isogenic wild-type, Δeaf1, eaf1ΔSANT, eaf1ΔN, and EAF1 cells were isolated, and Northern blot analyses were performed using the probes indicated on the left.
Cells carrying a deletion of the EAF1 gene are viable but have a slow-growth phenotype (Fig. 1D, left). In addition, eaf1 mutant cells are nonviable in the presence of DNA break-inducing agents like MMS and phleomycin but not hydroxyurea (Fig. 1D, right; also see Fig. 4H) (data not shown), indicating that Eaf1 is critical for NuA4 function in the repair of DNA double-strand breaks. EAF1-deleted cells also have been identified in genome-wide screens for yeast mutants that are the most highly sensitive to ionizing radiation (6). EAF1 deletion also provokes high sensitivity to the TOR pathway-targeting drug rapamycin (Fig. 1D, middle), which likely reflects the role of NuA4 in the transcription of ribosomal protein genes (53, 55). The deletion of large regions encompassing the HSA or SANT domain creates growth phenotypes identical to that of the full deletion, suggesting important roles for these regions (Fig. 1D). To characterize the slow-growth phenotype of eaf1 cells, we synchronized cultures by α-factor blockage and release. A cell cycle analysis of the cells at different time points after release clearly indicates that Eaf1 is important for passage through G2/M (Fig. 1E). These growth phenotypes are very similar to what has been reported for other important NuA4 subunits, like Esa1 and Epl1 (9, 15). These proteins also have been shown to be part of picNuA4 and to be required for the global nontargeted acetylation of H4 and H2A in vivo (9). In contrast, Eaf1 has no major effect on the global acetylation of chromatin in vivo (Fig. 1F). These data indicate that roles in cell cycle progression through G2/M, DNA repair, and transcription of ribosomal genes are attributed to the NuA4 complex independently of global acetylation by picNuA4. This is again reflected in the Northern blot analysis of eaf1 mutant cells, which shows gene-specific transcription defects previously reported for NuA4 mutants, including PHO5 and RPS11b (Fig. 1G) (50, 53).
SANT domain protein Eaf2 is essential for cell viability but is required for a subset of NuA4 functions.
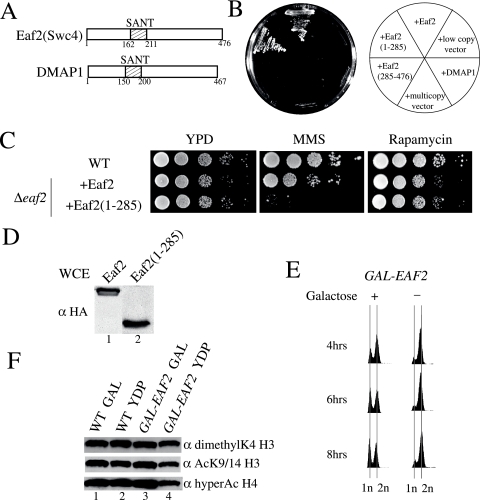
The other SANT domain present in the NuA4 complex is located within the Eaf2 protein. Eaf2 (also known as Swc4) is shared between NuA4 and the SWR1 ATP-dependent chromatin-remodeling complex, an activity responsible for the incorporation of the histone variant H2AZ in chromatin (17, 35, 38, 46, and data not shown) (see Fig. 4). Eaf2 is highly related to human DMAP1 (Fig. 2A), a protein implicated in DNA replication and a stable subunit of the TIP60 HAT complex (12, 19, 56). In parallel with our analysis of Eaf1, we studied the role of Eaf2 in NuA4 function. Eaf2 is essential for yeast cell viability, but the SANT domain-containing half of the protein is sufficient to support growth (Fig. 2B). On the other hand, the expression of homologous human DMAP1 in yeast cells cannot complement the loss of Eaf2. This also is the case for other human orthologs of essential NuA4 subunits, like Tip60 (Esa1) and EPC1 (Epl1) (Y. Doyon and J. Côté, unpublished data). The deletion of the Eaf2 C-terminal region creates a sensitivity to MMS, as such deletions do for other NuA4 mutants, but it also creates a sensitivity to hydroxyurea (Fig. 2C and data not shown). No sensitivity to rapamycin was detected, which is the opposite of findings for eaf1 mutant cells. The downregulation of Eaf2 in vivo by placing it under the control of the GAL1 promoter indicates that, as is the case for other NuA4 subunits, it is required for cell cycle progression through G2/M (Fig. 2E). Similarly to what is seen with Eaf1, Eaf2 is not required for the global Esa1-dependent acetylation of histone H4 in vivo (Fig. 2F) (the small decrease detected is nonspecific due to the accumulation of cells in G2/M; see reference 63). Thus, it appears that Eaf2 is involved in specific functions of NuA4, like the DNA damage response and passage through G2/M, but not in the transcription of ribosomal protein genes. This was expected, since Eaf2 is located in a specific functional subcomplex within NuA4 (see below). It will be interesting to define the importance of the SANT domains present in Eaf1 and Eaf2 proteins. SANT domains have been implicated in histone binding within HAT and HDAC complexes as well as in other protein-protein interactions (10). A first attempt at mutating conserved residues in Eaf1 and Eaf2 SANT domains did not show obvious growth phenotypes (data not shown). These results could be explained by the redundancy between Eaf1 and Eaf2 SANT domains or if we mutated only noncritical residues. Additional work will be needed to characterize the role of the two SANT domains present in the NuA4 complex.
FIG. 2.
Eaf2, another SANT-containing subunit, is essential for cell viability and affects a subset of NuA4 functions. (A) Schematic representation of Eaf2 (Swc4) and human DMAP1. (B) The SANT-containing N-terminal portion of Eaf2 is essential for viability, and human DMAP1 does not complement eaf2 mutants in yeast. Strains in which the EAF2 gene has been deleted but that express an episomal version of EAF2, the truncated EAF2(1-285) or EAF2(285-476), DMAP1, or an empty vector were streaked on solid YPD medium. (C) Deletion of the C-terminal portion of Eaf2 causes sensitivity to MMS but not to rapamycin. Serial 10-fold dilutions of the indicated strains were incubated on solid YPD medium containing either 0.03% MMS or 25 nM rapamycin. (D) Episomal full-length and truncated Eaf2 are expressed at similar levels in vivo. Whole-cell extracts (WCE) from the indicated strains were analyzed by Western blotting with anti-HA (α HA). (E) Eaf2 is essential for cell cycle progression. The depletion of Eaf2 leads to cells being blocked at G2/M. Cells in which the endogenous EAF2 promoter has been replaced by the inducible GAL1 promoter were incubated in liquid medium in the presence or absence of galactose (GAL), and the DNA content was analyzed by flow cytometry. (F) Eaf2 is not essential for the global acetylation of chromatin by NuA4. Cells harboring EAF2 under the control of the GAL1 promoter as well as wild-type (WT) cells were incubated in medium containing galactose or glucose for 12 h at 30°C. Western blot analyses were performed with histones purified from those strains using the indicated antibodies.
Eaf1 is a platform that coordinates NuA4 molecular assembly.
To gain a better understanding of NuA4 architecture and to determine Eaf1 molecular interactions, we affinity purified the complex from cells harboring different subunits that were tagged or mutated. We have previously shown that the truncation of the Epl1 C-terminal region creates cells that contain only the picNuA4 HAT module (Esa1/Epl1/Yng2), suggesting that this region is responsible for the association with the rest of the NuA4 complex in vivo (9). The purification of Eaf1 in this background confirms the model, since a complex is obtained with all NuA4 subunits except the ones present in picNuA4 (Fig. 3A and B, lanes 1 to 3). We then verified the impact of Eaf1 loss on the NuA4 structure by purifying Epl1 from EAF1-deleted cells. In the absence of Eaf1, Epl1 is purified only with Esa1 and Yng2, i.e., as a picNuA4 complex (Fig. 3A, lanes 4 to 6, and B, lanes 4 and 5). These data indicate that the molecular interaction responsible for anchoring the Esa1/Epl1/Yng2 catalytic core to the rest of NuA4 is between the Epl1 C-terminal region and Eaf1. We then affinity purified NuA4 from wild-type and EAF1-deleted cells using a tag on the Eaf5 subunit. Eaf5 allows the association of Eaf7 and Eaf3 subunits to NuA4, and this trimer also exists in a native independent form in vivo (Lacoste and Côté, unpublished). The Eaf3 chromodomain-containing protein also is shared by the Rpd3S HDAC complex and is linked to histone H3 methylation during transcription (13, 33; Lacoste Côté, unpublished). As shown in Fig. 3A, lanes 7 and 8, and B, lanes 6 and 7, Eaf1 is essential for Eaf5 association to NuA4, since in its absence only the Eaf5/7/3 trimer is recovered.
FIG. 3.
Eaf1 serves as a platform for the assembly of four different functional modules into the NuA4 complex. (A) Epl1 interacts with Eaf1 and bridges picNuA4 with the remaining complex, while the association with an Eaf5/7/3 trimer also is dependent on Eaf1. Purified material from untagged strains and the indicated TAP-tagged strains was loaded onto gradient gels and visualized by silver staining. The asterisk corresponds to a nonspecific band known to purify with TAP-tagged proteins. (B) Western blots using the final elution of the TAP purifications shown in panel A were probed with antibodies α against the indicated proteins. (C) The HSA domain region of Eaf1 interacts with the subunits shared with the SWR1/INO80 ATP-dependent chromatin remodeling complexes. Purified material from the indicated TAP-tagged strains supplemented with an episomal version of either EAF1, Eaf1ΔHAS, or Eaf1ΔSANT was loaded onto a gradient gel and analyzed by Western blotting using the antibodies indicated on the right. In lane 7, there is no Esa1 signal but rather a nonspecific keratin band (asterisk). (D) Tra1 requires the SANT domain region of Eaf1 for its association with NuA4. Purified material from strains indicated in the legend to panel C was loaded onto low-percentage gels and visualized by silver staining. The 400-kDa band visible on this part of the gel represents Tra1. (E) Requirement of the HSA and SANT regions of Eaf1 for proper growth. Tenfold dilutions of mid-log-phase Δeaf1, EAF1, Eaf1ΔHSA, and Eaf1ΔSANT yeast cultures were grown in the presence or absence of 0.03% MMS or 25 nM rapamycin. (F) Expression levels of episomal wild-type (WT) and mutant Eaf1 are similar to that of endogenous Eaf1. Whole-cell extracts (WCE) from the strains used for panel E were analyzed by Western blotting with anti-Eaf1. (G) Schematic representation of Eaf1 as the platform for the assembly of the different functional modules of NuA4 and the subunits shared with other chromatin modifying/remodeling complexes.
We then analyzed the role of specific regions of Eaf1 in these interactions. The deletion of the Eaf1 C-terminal glutamine-rich region creates no phenotype and has no effect on NuA4 assembly (data not shown). The deletion of a region encompassing the HSA and highly charged domains (aa 329 to 538) again separates picNuA4 from the rest of the complex, indicating that the Epl1 C terminus binds somewhere in this region (Fig. 3C, lanes 1 to 3). The deletion of a region containing the SANT domain (aa 604 to 815) leaves the NuA4 complex almost intact (Fig. 3C, compare lanes 2 and 4). Upon closer examination, we can see that the deletion of the SANT domain region provokes the loss of a single but large subunit, Tra1 (Fig. 3D, compare lanes 1 and 4) (because of its size and the difficulty of transferring for Western blotting, conclusions about Tra1's presence primarily are based on the detection of a 400-kDa band by silver staining). We then repeated the affinity purifications in the same mutant cells but with a tagged Eaf5 subunit. These experiments confirmed that the Eaf1 SANT domain region is responsible for the association of Tra1 with NuA4 (Fig. 3C, compare lanes 6 and 8, and D, compare lanes 7 and 8). When Eaf5 was purified from cells lacking the HSA or charged region of Eaf1, a partial NuA4 complex was obtained, lacking, as expected, picNuA4 subunits and the subunits shared with ATP-dependent chromatin-remodeling complexes INO80 and SWR1, i.e., Arp4/Act1, Yaf9, and Eaf2 (Swc4) (Fig. 3C, compare lanes 6 and 7). Taken together, these results indicate that different regions of Eaf1 are responsible for the ordered assembly of functional subcomplexes into NuA4. Accordingly, the regions deleted in Eaf1 create strong growth phenotypes on different media, reflecting their importance in NuA4 assembly and, thus, function (Fig. 3E). Preliminary results suggest that the binding of picNuA4(Epl1) is located in the highly charged domain of Eaf1, while the association with Eaf5/7/3 occurs at its N terminus (data not shown).
Our results position the Eaf1 subunit at the center of the NuA4 complex, making distinct physical interactions with four different functional modules within NuA4 (Fig. 3G). These interactions lead to the assembly of the native NuA4 complex. Eaf1 coordinates the association of the HAT module (Esa1/Epl1/Yng2) (9, 60), the activator recruitment module (Tra1) (11), the histone methylation/transcription elongation module (Eaf5/7/3) (Lacoste and Côté, unpublished), and finally the DNA repair/telomere boundary module (Arp4/Act1/Eaf2/Yaf9) (17, 35, 38, 46, 48, 64, 69). The fact that Eaf1 is the platform upon which the NuA4 complex is assembled goes well with the fact that it is the only subunit restricted to NuA4 in vivo. Thus, it is the only tool that can be used to specifically analyze NuA4 function, distinguishing targeted Esa1-dependent acetylation from global action by the picNuA4 complex.
Functional implications of shared subunits between NuA4 and SWR1 complexes.
As mentioned above, the purification of the SWI2/SNF2-related ATPase Swr1 identified a multisubunit complex that shares four subunits with NuA4, i.e., Arp4 and Act1 (also shared with the INO80 complex), Eaf2 (Swc4), and Yaf9 (Fig. 4A) (17, 35, 37, 38, 46, and data not shown). A characteristic of the SWR1 complex is the presence of Rvb1/2 helicases related to bacterial ruvB, which allows the movement of the Holiday junctions during DNA recombination (Fig. 4A). Confirming its presence in both complexes, TAP of Yaf9 yields proteins specifically found in NuA4 (Esa1 and Yng2) as well as those found only in SWR1 (Rvb1) (Fig. 4B). More Arp4 is recovered in Yaf9-TAP than in Epl1-TAP, since it also is present in both complexes. TAP of Eaf2 also yields NuA4 and Swr1 components (Fig. 4C, lane 2) (data not shown). Interestingly, when we purified a truncated version of Eaf2 lacking its C terminus, all NuA4 and SWR1 components still were obtained, except for Yaf9 (Fig. 4C, compare lanes 2 and 3). These data, along with previous work, indicate that Yaf9 associates with NuA4 and Swr1 complexes through direct interactions between Eaf2 and Yaf9 C termini (8, 69).
A role of the SWR1-dependent incorporation of the H2AZ variant in chromatin is to block the heterochromatin-stabilizing silent information regulator (SIR) complex from spreading into euchromatin regions near telomeres (35, 38, 44, 46), and Yaf9 plays an essential role in this process (69). This is clearly depicted in an assay of telomere position effect, in which a URA3 gene is located next to the telomere repeats of chromosome VII (Fig. 4D) (27). In this assay, the URA3 gene normally is silenced by the local high concentration of the SIR complex. The spreading of the SIR complex into the euchromatin regions decreases its local concentration, allowing the expression of the URA3 gene, which is detected by a loss of growth on 5-FOA-containing media. Figure 4D demonstrates that the deletion of YAF9 provokes a complete loss of telomere silencing. Interestingly, this effect is stronger than that of other SWR1/Htz1 mutants, which could reflect Yaf9 function in both SWR1 and NuA4 complexes near telomere regions, as suggested by our previous work (69). The mutation of the NuA4 subunit Epl1 also affects telomere silencing (9). Accordingly, Eaf1, which physically links Epl1- and Yaf9-containing modules in NuA4 (Fig. 3G), also affects telomere silencing (Fig. 4D, lower gels).
These findings, as well as those of several reports in the literature, suggest that NuA4 and SWR1 cooperate in vivo (2, 5, 18, 20, 34, 37, 41, 45, 52, 69; Lacoste and Côté, unpublished). A nice parallel can be made to the transcription of the PHO5 gene. The promoter region of this gene has been shown to be highly enriched in nucleosomes containing H2AZ (named Htz1 in yeast) in repressed conditions, a chromatin state that prepares the promoter for chromatin remodeling and transcription activation (59). The chromatin over the PHO5 promoter also is enriched in acetylated H4 (66), and we have shown that NuA4 is recruited there by the Pho2 homeodomain transcription factor (50). Preacetylation by NuA4 is essential to prepare the promoter for chromatin remodeling and transcription activation. Thus, it is tempting to speculate that NuA4 and SWR1 could collaborate in presetting the PHO5 promoter for induction. This also is supported by the fact that a significant fraction of the SWR1 complexes also contains the bromodomain-containing protein Bdf1 (35, 38). Bdf1 has been shown to genetically interact with Esa1 and to physically bind acetylated histone H4 (43). Local chromatin acetylation by NuA4 may potentiate SWR1 complex binding through Bdf1, which would allow the exchange of H2A for Htz1 in local chromatin. We first performed ChIP analysis by real-time PCR to measure histone H4 acetylation over the PHO5 promoter region after correction for nucleosome occupancy (total histone H3). As shown in Fig. 4E, the deletion of EAF1 has a strong effect on the level of histone H4 acetylation at the PHO5 promoter. In comparison, the loss of Swr1 has little effect. We then analyzed the presence of Htz1 in our different mutant strains (Fig. 4F). As expected, the knockout of SWR1 completely depletes Htz1 from the PHO5 promoter. The same effect is seen in a Δyaf9 strain. When EAF1 is deleted, Htz1 still is detected in the promoter region but is diminished compared to its level under wild-type conditions. These data indicate that the NuA4-dependent acetylation of the PHO5 promoter participates in, but is not essential for, the incorporation of Htz1 by SWR1, in agreement with recent genome-wide analyses (20, 52, 70). This effect of NuA4 on SWR1 function is not reciprocal, since the loss of Htz1, Yaf9, or Swr1 has little effect on the histone H4 acetylation level at the PHO5 promoter (Fig. 3E and G). Taken together, these results demonstrate that SWR1/Htz1 can function independently of NuA4, but the two activities collaborate in preparing the PHO5 gene for induction.
Functional and physical interactions between SWR1 and NuA4 complexes.
The sharing of some subunits between NuA4 and Swr1 protein complexes suggests that these two activities cooperate. We have shown that they act together to antagonize heterochromatin spreading/silencing from telomere regions (69) and to preset chromatin over the PHO5 promoter for gene induction. Our data also indicate that these two activities can function independently of each other. Their independent functions are portrayed by certain distinct phenotypes between NuA4 and SWR1 mutants. As seen in Fig. 4H, both eaf1 and swr1 deletion mutants are highly sensitive to DNA breaks introduced by MMS, supporting their functional interaction in double-strand break repair (17). In contrast, the deletion of a key subunit of the INO80 ATP-dependent complex shows no sensitivity (Δarp8). The relative phenotypes are clearly different on rapamycin, for which the deletion of SWR1 has no effect while eaf1 and arp8 mutants are unable to grow. This supports our previous results that suggested that the Eaf2-containing module is not required for the biosynthesis of ribosomes (Fig. 2C). While NuA4 and SWR1 can act independently of each other, their combined action clearly is essential for cell viability. Genetic interactions between the two complexes have been reported (5, 34, 35, 37), but we wanted to address specifically that these interactions involved the functions of the full native NuA4 complex and the incorporation of Htz1 by SWR1. Since Δeaf1 cells show genome instability and high incidences of sporulation defects and aneuploidy (22, 30, 37, and data not shown), it is technically difficult to characterize EAF1 genetic interactions. Instead, we used the NuA4 mutant strain that separates picNuA4 from the rest of the NuA4 complex, i.e., the C-terminal truncation of Epl1 (Fig. 3). Figure 4I and J show that cells that are still able to globally acetylate histone H4/H2A but lack both the native NuA4 complex and either Swr1 or Htz1 are synthetic lethal. Since the epl1 truncation mutant separates NuA4 into two stable entities, this indicates that the functional interaction between SWR1/Htz1 and NuA4 depends on the targeted HAT function of NuA4.
Based on the homology between 12 yeast NuA4 subunits and components of the human Tip60 complex, we proposed that these two HAT complexes are functional orthologs (19). On the other hand, the human complex has additional subunits not present in yeast NuA4. It is striking to see that each of these additional human components corresponds precisely to homologs of yeast SWR1-specific subunits, including Bdf1, Rvb1/2, Vps72 (Swc2), and Swr1 (2, 12, 18). We then proposed that the human Tip60 complex represents an exact physical merge of yeast NuA4 and SWR1 complexes (18). This model has been supported by results with the Drosophila melanogaster TIP60 complex (39). This report clearly shows that the TIP60 complex can incorporate H2Av-H2B dimers into chromatin in an ATP-dependent fashion in a reaction stimulated by the prior acetylation of chromatin by Tip60 (H2Av is equivalent to both H2AZ and H2AX histone variants). The TIP60 subunit harboring the homology with Swr1 ATPase is p400/Domino, a protein required for oncogenic transformation by E1A and development in Drosophila and Caenorhabditis elegans (14, 23, 57). p400/Domino also has been shown recently to be responsible for the ATP-dependent incorporation of histone H2AZ variant in chromatin in vitro and in vivo (26). Interestingly, this large protein also has distinct structures homologous to those of yeast Eaf1, including HSA, SANT, glutamine-rich, and highly charged domains (Fig. 5A). We speculated that the single human complex corresponding to two distinct yeast complexes could be explained by the fact that yeast Swr1 and Eaf1 are merged into a single human protein, p400/Domino. To test this hypothesis, we created an artificial p400/Domino-like protein in yeast cells by inserting the SWI2/SNF2-related ATPase domain of Swr1 in the middle of Eaf1 (Fig. 5A). The expression of this synthetic yDomino protein demonstrates that it is functional, since it can suppress the growth defects of eaf1 deletion cells on rich media in the presence of rapamycin and also can do so partly in the presence of MMS (Fig. 5B and C). TAP of Epl1 in cells lacking Eaf1 but expressing the fusion protein indicates that it is efficiently integrated into the NuA4 complex and functionally replaces Eaf1 in the assembly of the full set of NuA4 subunits (Fig. 5D and E, compare lanes 1, 2, and 3). In addition, the association of yDomino in NuA4 leads to the presence of Rvb1 and Rvb2 helicases in the purified complex (Fig. 5E, lane 3). Unfortunately, we could not detect the suppression of swr1 mutant phenotypes by yDomino in growth assays (data not shown). On the other hand, while ChIP analysis of Htz1 at the PHO5 promoter confirms the very poor incorporation of Htz1 in swr1 mutant cells, it is reproducibly increased in yDomino-expressing cells (Fig. 5F). Nevertheless, this increase does not seem sufficient to restore Swr1 function. In contrast, the complementation of Eaf1 function by yDomino is significant in analyses of the NuA4-dependent acetylation of Htz1 at the PHO5 promoter. NuA4 has been shown to acetylate Htz1 at lysine 14 in vivo, a modification important for Htz1 function at promoters and heterochromatin boundaries (5, 34, 45, and data not shown). As shown in Fig. 5G, the expression of yDomino in eaf1 mutant cells restores the acetylation of Htz1 at lysine 14 to more than 65% of the wild-type level.
Taken together, these data validate our model of TIP60 being a physical merge of NuA4 and SWR1 complexes through an evolutionary fusion of Swr1 and Eaf1 into p400/Domino. This model predicts that the human TIP60 complex has at least three interrelated enzymatic activities: histone H4/H2A acetyltransferase, ATP-dependent H2AZ-H2B histone dimer exchange, and DNA helicase. It will be interesting to test these activities in vitro with the purified human and the synthetic yeast complexes.
DISCUSSION
In this study, we analyzed the structure and function of two stable subunits of the yeast NuA4 HAT complex. Eaf1 and Eaf2 both harbor a SANT domain, are required for normal cell cycle progression through mitosis, and do not affect the global nontargeted Esa1-dependent acetylation of chromatin but affect the cellular response to DNA damage and telomere silencing. In addition, we showed that Eaf1 is required for gene-specific transcription, including that of the ribosomal protein genes. More importantly, we demonstrated that Eaf1 is found exclusively in the NuA4 complex in vivo, which makes it the only subunit that is not also found in a distinct protein complex. This is an important finding, since Eaf1 becomes the only molecular tool that can be used to unequivocally study native NuA4 function in vivo. ChIP with Eaf1 identifies loci bound by NuA4, while experiments with antibodies targeting other subunits could reflect the association of a mixture of complexes or distinct complexes. It is an important development, since published ChIP experiments with NuA4 mostly have used tags on the Esa1 HAT protein, while it is known that Esa1 also is part of picNuA4, which is responsible for global acetylation (54). In contrast, the association of Eaf1 reflects targeted Esa1-dependent acetylation, as shown by the loss of H4 acetylation at the PHO5 promoter in eaf1 mutant cells while bulk acetylation levels are unchanged (Fig. 1F and 4E). The specificity of Eaf1 to NuA4 is explained by its central key role in the complex assembly. Our experiments have shown that Eaf1 makes independent interactions with distinct functional subcomplexes within NuA4, coordinating the assembly of the full native complex. It will be very interesting to further delineate the specific domains in Eaf1 responsible for each particular interaction. The information gathered here will increase the speed and significance of our upcoming experimental characterization of NuA4 function in transcription regulation and DNA repair and its cross-talk with other chromatin modifiers/remodelers.
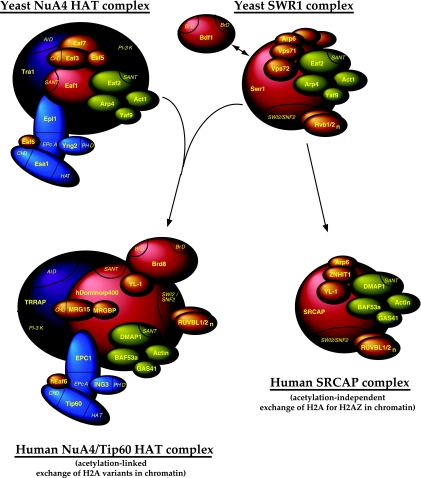
The present work also characterized the functional interactions between NuA4 and SWR1 complexes. Histone H4/H2A acetylation and H2AZ histone variant acetylation/incorporation into chromatin have been demonstrated to play roles in blocking the SIR complex from spreading out of telomere regions (5, 35, 38, 43, 46, 69), in presetting the PHO5 promoter for chromatin remodeling and transcription activation (45, 50, 59), and in the repair of DNA double-strand breaks (7, 17, 39). We showed that NuA4-dependent acetylation influences, but is not essential for, H2AZ incorporation at PHO5 (Fig. 4). Also, the presence of H2AZ does not greatly affect H4 acetylation by NuA4 in vivo. While these data show some functional cross-talk between NuA4 and SWR1/Htz1, they also show that each one can work independently of the other toward a common goal. Finding this limited cross-talk was somewhat surprising, since the double bromodomain factor Bdf1, which is associated with the SWR1 complex (35, 38), has been shown to bind NuA4-dependent acetylated histone H4 and functionally interacts with NuA4 (20, 43). On the other hand, we have not detected stoichiometric amounts of Bdf1 in our purifications of the SWR1 complex, similarly to what was reported by another group (46). These data suggest that there could be two different classes of SWR1 complexes: with Bdf1 and without Bdf1. We propose that the detected effect of EAF1 deletion on Htz1 incorporation at PHO5 corresponds to the activity of the subset of SWR1 complexes containing Bdf1. The presence of two functionally different forms of SWR1 complexes recently has been supported by the identification of the human SNP2-related CBP activator protein (SRCAP) complex. The SRCAP protein is highly related to yeast Swr1 and human p400/Domino, and it has been purified as a protein complex sharing many subunits with TIP60 that are homologous to yeast SWR1 components (12). In addition, the complex is able to load H2AZ-H2B dimers onto chromatin in vitro and in vivo (58, 67). On the other hand, in contrast to TIP60/p400/Domino complexes, the SRCAP complex does not contain Brd8, the homolog of Bdf1 in higher eukaryotes (12, 19, 39). We suggest that human SRCAP corresponds to a yeast SWR1 complex without Bdf1 that functions independently of NuA4, while p400/Domino corresponds to a Bdf1-associated SWR1 complex, which collaborates and functionally interacts with NuA4-dependent acetylated histones (Fig. 6). This important functional interaction would be merged into a single molecular entity in higher eukaryotes, i.e., the TIP60 complex, while being between two distinct partners in yeast (18).
FIG. 6.
Model for the evolution of NuA4 HAT and SWR1 ATP-dependent remodeling complexes from yeast to human cells. The yeast NuA4 and SWR1 complexes are depicted. SWR1 could be present in two forms, with or without a bromodomain-containing Bdf1 subunit. These distinct complexes regulating histone H2AZ incorporation in chromatin would differentially reflect the link to NuA4-dependent chromatin acetylation. In human cells, SWR1 plus a Bdf1 equivalent is physically merged with NuA4 into the TIP60/hNuA4 complex (through the fusion of yeast Eaf1 and Swr1 platform proteins into human p400/Domino). The yeast SWR1 lacking Bdf1 corresponds to the human SRCAP complex, which should not be linked to chromatin acetylation.
Finally, we tested the model of a physical merge between yeast NuA4 and SWR1 complexes to create a single complex in higher eukaryotes, TIP60. The protein p400/Domino has the striking feature of harboring strong and distinct homologies to both Swr1 and Eaf1 proteins, suggesting the evolutionary merging of the two yeast proteins into a single protein in higher eukaryotes. We tested that hypothesis by creating a synthetic p400/Domino-like protein in yeast, merging together parts of Eaf1 and Swr1 to create a domain structure similar to the human protein. The expression of this chimeric protein in yeast supported our theory, since a single complex containing both NuA4 and SWR1 components was obtained (Fig. 5). These data are in agreement with recent work showing that the Swr1 ATPase domain is responsible for the association of six critical SWR1 complex subunits and Htz1-H2B dimers (68). While the fusion protein can replace Eaf1 function in vivo, it also allows some incorporation of the H2AZ variant into chromatin in vivo, but not in amounts sufficient to suppress swr1 mutant phenotypes. This is likely explained by the absence of Swc5, which was shown to associate with the Swr1 N-terminal region and to be required for the efficient swapping of histone dimers in vitro (68). On the other hand, nucleosomal H2AZ recently has been shown to be acetylated by the NuA4 complex in vitro and in vivo (5, 34, 45; A. Auger, J. Brodeur, L. Gaudreau, and J. Côté, unpublished observations), and the Swr1-Eaf1 fusion protein is proficient at suppressing the H2AZ acetylation defect on the PHO5 promoter (Fig. 5G).
The functional cross-talk between different histone modifications is part of the process of establishing a local chromatin identity/signature. The additional implication in this cross-talk of histone variants and ATP-dependent chromatin remodelers further highlights the complexity and fine-tuning of chromatin dynamics in regulating diverse nuclear functions. The ongoing structural and functional analysis of the NuA4 HAT complex in eukaryotes proves to lead us to the heart of chromatin biology.
Acknowledgments
We thank Nicolas Lacoste for technical help, advice, and useful discussions during the course of this work. We are grateful to Michael Grunstein, David Stillman, Song Tan, Y. Makino, Jerry Workman, and John Lucchesi for antibodies and Dan Gottschling and Nicolas Lacoste for yeast strains. We also thank Kristin Baetz, Michael Kobor, and Nevan Krogan for sharing results prior to publication.
This work was supported by a grant from the Canadian Institutes of Health Research (CIHR) to J.C. A.A. held a National Science and Engineering Research Council doctoral studentship. A.N. and R.T.U. were CIHR postdoctoral fellows. Y.D. held a CIHR/Canada graduate scholarship. J.C. is a CIHR Investigator.
Footnotes
Published ahead of print on 22 January 2008.
REFERENCES
- 1.Allard, S., R. T. Utley, J. Savard, A. Clarke, P. Grant, C. J. Brandl, L. Pillus, J. L. Workman, and J. Cote. 1999. NuA4, an essential transcription adaptor/histone H4 acetyltransferase complex containing Esa1p and the ATM-related cofactor Tra1p. EMBO J. 185108-5119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altaf, M., N. Saksouk, and J. Cote. 2007. Histone modifications in response to DNA damage. Mutat. Res. 61881-90. [DOI] [PubMed] [Google Scholar]
- 3.Avvakumov, N., and J. Cote. 2007a. Functions of myst family histone acetyltransferases and their link to disease. Subcell. Biochem. 41295-317. [PubMed] [Google Scholar]
- 4.Avvakumov, N., and J. Cote. 2007b. The MYST family of histone acetyltransferases and their intimate links to cancer. Oncogene 265395-5407. [DOI] [PubMed] [Google Scholar]
- 5.Babiarz, J. E., J. E. Halley, and J. Rine. 2006. Telomeric heterochromatin boundaries require NuA4-dependent acetylation of histone variant H2A.Z in Saccharomyces cerevisiae. Genes Dev. 20700-710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bennett, C. B., L. K. Lewis, G. Karthikeyan, K. S. Lobachev, Y. H. Jin, J. F. Sterling, J. R. Snipe, and M. A. Resnick. 2001. Genes required for ionizing radiation resistance in yeast. Nat. Genet. 29426-434. [DOI] [PubMed] [Google Scholar]
- 7.Bird, A. W., D. Y. Yu, M. G. Pray-Grant, Q. Qiu, K. E. Harmon, P. C. Megee, P. A. Grant, M. M. Smith, and M. F. Christman. 2002. Acetylation of histone H4 by Esa1 is required for DNA double-strand break repair. Nature 419411-415. [DOI] [PubMed] [Google Scholar]
- 8.Bittner, C. B., D. T. Zeisig, B. B. Zeisig, and R. K. Slany. 2004. Direct physical and functional interaction of the NuA4 complex components Yaf9p and Swc4p. Eukaryot. Cell 3976-983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boudreault, A. A., D. Cronier, W. Selleck, N. Lacoste, R. T. Utley, S. Allard, J. Savard, W. S. Lane, S. Tan, and J. Cote. 2003. Yeast enhancer of polycomb defines global Esa1-dependent acetylation of chromatin. Genes Dev. 171415-1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boyer, L. A., R. R. Latek, and C. L. Peterson. 2004. The SANT domain: a unique histone-tail-binding module? Nat. Rev. Mol. Cell Biol. 5158-163. [DOI] [PubMed] [Google Scholar]
- 11.Brown, C. E., L. Howe, K. Sousa, S. C. Alley, M. J. Carrozza, S. Tan, and J. L. Workman. 2001. Recruitment of HAT complexes by direct activator interactions with the ATM-related Tra1 subunit. Science 2922333-2337. [DOI] [PubMed] [Google Scholar]
- 12.Cai, Y., J. Jin, L. Florens, S. K. Swanson, T. Kusch, B. Li, J. L. Workman, M. P. Washburn, R. C. Conaway, and J. W. Conaway. 2005. The mammalian YL1 protein is a shared subunit of the TRRAP/TIP60 histone acetyltransferase and SRCAP complexes. J. Biol. Chem. 28013665-13670. [DOI] [PubMed] [Google Scholar]
- 13.Carrozza, M. J., B. Li, L. Florens, T. Suganuma, S. K. Swanson, K. K. Lee, W. J. Shia, S. Anderson, J. Yates, M. P. Washburn, and J. L. Workman. 2005. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell 123581-592. [DOI] [PubMed] [Google Scholar]
- 14.Ceol, C. J., and H. R. Horvitz. 2004. A new class of C. elegans synMuv genes implicates a Tip60/NuA4-like HAT complex as a negative regulator of Ras signaling. Dev. Cell 6563-576. [DOI] [PubMed] [Google Scholar]
- 15.Clarke, A. S., J. E. Lowell, S. J. Jacobson, and L. Pillus. 1999. Esa1p is an essential histone acetyltransferase required for cell cycle progression. Mol. Cell. Biol. 192515-2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Doerks, T., R. R. Copley, J. Schultz, C. P. Ponting, and P. Bork. 2002. Systematic identification of novel protein domain families associated with nuclear functions. Genome Res. 1247-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Downs, J. A., S. Allard, O. Jobin-Robitaille, A. Javaheri, A. Auger, N. Bouchard, S. J. Kron, S. P. Jackson, and J. Cote. 2004. Binding of chromatin-modifying activities to phosphorylated histone H2A at DNA damage sites. Mol. Cell 16979-990. [DOI] [PubMed] [Google Scholar]
- 18.Doyon, Y., and J. Cote. 2004. The highly conserved and multifunctional NuA4 HAT complex. Curr. Opin. Genet. Dev. 14147-154. [DOI] [PubMed] [Google Scholar]
- 19.Doyon, Y., W. Selleck, W. S. Lane, S. Tan, and J. Cote. 2004. Structural and functional conservation of the NuA4 histone acetyltransferase complex from yeast to humans. Mol. Cell. Biol. 241884-1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Durant, M., and B. F. Pugh. 2007. NuA4-directed chromatin transactions throughout the Saccharomyces cerevisiae genome. Mol. Cell. Biol. 275327-5335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eisen, A., R. T. Utley, A. Nourani, S. Allard, P. Schmidt, W. S. Lane, J. C. Lucchesi, and J. Cote. 2001. The yeast NuA4 and Drosophila MSL complexes contain homologous subunits important for transcription regulation. J. Biol. Chem. 2763484-3491. [DOI] [PubMed] [Google Scholar]
- 22.Enyenihi, A. H., and W. S. Saunders. 2003. Large-scale functional genomic analysis of sporulation and meiosis in Saccharomyces cerevisiae. Genetics 16347-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fuchs, M., J. Gerber, R. Drapkin, S. Sif, T. Ikura, V. Ogryzko, S. W. Lane, Y. Nakatani, and M. D. Livingston. 2001. The p400 complex is an essential E1A transformation target. Cell 106297-307. [DOI] [PubMed] [Google Scholar]
- 24.Galarneau, L., A. Nourani, A. A. Boudreault, Y. Zhang, L. Heliot, S. Allard, J. Savard, W. S. Lane, D. J. Stillman, and J. Cote. 2000. Multiple links between the NuA4 histone acetyltransferase complex and epigenetic control of transcription. Mol. Cell 5927-937. [DOI] [PubMed] [Google Scholar]
- 25.Gangaraju, V. K., and B. Bartholomew. 2007. Mechanisms of ATP dependent chromatin remodeling. Mutat. Res. 6183-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gévry, N., H. M. Chan, L. Laflamme, D. M. Livingston, and L. Gaudreau. 2007. p21 transcription is regulated by differential localization of histone H2A.Z. Genes Dev. 211869-1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gottschling, D. E., O. M. Aparicio, B. L. Billington, and V. A. Zakian. 1990. Position effect at S. cerevisiae telomeres: reversible repression of Pol II transcription. Cell 63751-762. [DOI] [PubMed] [Google Scholar]
- 28.Grant, P. A., L. Duggan, J. Cote, S. M. Roberts, J. E. Brownell, R. Candau, R. Ohba, T. Owen-Hughes, C. D. Allis, F. Winston, S. L. Berger, and J. L. Workman. 1997. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 111640-1650. [DOI] [PubMed] [Google Scholar]
- 29.Grant, P. A., D. Schieltz, M. G. Pray-Grant, J. R. Yates III, and J. L. Workman. 1998. The ATM-related cofactor Tra1 is a component of the purified SAGA complex. Mol. Cell 2863-867. [DOI] [PubMed] [Google Scholar]
- 30.Hughes, T. R., C. J. Roberts, H. Dai, A. R. Jones, M. R. Meyer, D. Slade, J. Burchard, S. Dow, T. R. Ward, M. J. Kidd, et al. 2000. Widespread aneuploidy revealed by DNA microarray expression profiling. Nat. Genet. 25333-337. [DOI] [PubMed] [Google Scholar]
- 31.Kamakaka, R. T., and S. Biggins. 2005. Histone variants: deviants? Genes Dev. 19295-310. [DOI] [PubMed] [Google Scholar]
- 32.Katan-Khaykovich, Y., and K. Struhl. 2002. Dynamics of global histone acetylation and deacetylation in vivo: rapid restoration of normal histone acetylation status upon removal of activators and repressors. Genes Dev. 16743-752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Keogh, M. C., S. K. Kurdistani, S. A. Morris, S. H. Ahn, V. Podolny, S. R. Collins, M. Schuldiner, K. Chin, T. Punna, N. J. Thompson, et al. 2005. Cotranscriptional set2 methylation of histone H3 lysine 36 recruits a repressive Rpd3 complex. Cell 123593-605. [DOI] [PubMed] [Google Scholar]
- 34.Keogh, M. C., T. A. Mennella, C. Sawa, S. Berthelet, N. J. Krogan, A. Wolek, V. Podolny, L. R. Carpenter, J. F. Greenblatt, K. Baetz, and S. Buratowski. 2006. The Saccharomyces cerevisiae histone H2A variant Htz1 is acetylated by NuA4. Genes Dev. 20660-665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kobor, M. S., S. Venkatasubrahmanyam, M. D. Meneghini, J. W. Gin, J. L. Jennings, A. J. Link, H. D. Madhani, and J. Rine. 2004. A protein complex containing the conserved Swi2/Snf2-related ATPase Swr1p deposits histone variant H2A.Z into euchromatin. PLoS Biol. 2E131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kouzarides, T. 2007. Chromatin modifications and their function. Cell 128693-705. [DOI] [PubMed] [Google Scholar]
- 37.Krogan, N. J., K. Baetz, M. C. Keogh, N. Datta, S. T. Cawa, C. Kwok, N. J. Thompson, M. G. Davey, J. Pootoolal, T. R. Hughes, et al. 2004. Regulation of chromosome stability by the histone H2A variant Htz1, the Swr1 chromatin remodeling complex, and the histone acetyltransferase NuA4. Proc. Natl. Acad. Sci. USA 10113513-13518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Krogan, N. J., M. C. Keogh, N. Datta, C. Sawa, O. W. Ryan, H. Ding, R. A. Haw, J. Pootoolal, A. Tong, V. Canadien, et al. 2003. A Snf2 family ATPase complex required for recruitment of the histone H2A variant Htz1. Mol. Cell 121565-1576. [DOI] [PubMed] [Google Scholar]
- 39.Kusch, T., L. Florens, W. H. Macdonald, S. K. Swanson, R. L. Glaser, J. R. Yates III, S. M. Abmayr, M. P. Washburn, and J. L. Workman. 2004. Acetylation by Tip60 is required for selective histone variant exchange at DNA lesions. Science 3062084-2087. [DOI] [PubMed] [Google Scholar]
- 40.Kusch, T., and J. L. Workman. 2007. Histone variants and complexes involved in their exchange. Subcell. Biochem. 4191-109. [PubMed] [Google Scholar]
- 41.Lindstrom, K. C., J. C. Vary, Jr., M. R. Parthun, J. Delrow, and T. Tsukiyama. 2006. Isw1 functions in parallel with the NuA4 and Swr1 complexes in stress-induced gene repression. Mol. Cell. Biol. 266117-6129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Longtine, M. S., A. McKenzie III, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14953-961. [DOI] [PubMed] [Google Scholar]
- 43.Matangkasombut, O., and S. Buratowski. 2003. Different sensitivities of bromodomain factors 1 and 2 to histone H4 acetylation. Mol. Cell 11353-363. [DOI] [PubMed] [Google Scholar]
- 44.Meneghini, M. D., M. Wu, and H. D. Madhani. 2003. Conserved histone variant H2A.Z protects euchromatin from the ectopic spread of silent heterochromatin. Cell 112725-736. [DOI] [PubMed] [Google Scholar]
- 45.Millar, C. B., F. Xu, K. Zhang, and M. Grunstein. 2006. Acetylation of H2AZ Lys 14 is associated with genome-wide gene activity in yeast. Genes Dev. 20711-722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mizuguchi, G., X. Shen, J. Landry, W. H. Wu, S. Sen, and C. Wu. 2004. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science 303343-348. [DOI] [PubMed] [Google Scholar]
- 47.Morillon, A., N. Karabetsou, A. Nair, and J. Mellor. 2005. Dynamic lysine methylation on histone H3 defines the regulatory phase of gene transcription. Mol. Cell 18723-734. [DOI] [PubMed] [Google Scholar]
- 48.Morrison, A. J., J. Highland, N. J. Krogan, A. Arbel-Eden, J. F. Greenblatt, J. E. Haber, and X. Shen. 2004. INO80 and gamma-H2AX interaction links ATP-dependent chromatin remodeling to DNA damage repair. Cell 119767-775. [DOI] [PubMed] [Google Scholar]
- 49.Nourani, A., Y. Doyon, R. T. Utley, S. Allard, W. S. Lane, and J. Cote. 2001. Role of an ING1 growth regulator in transcriptional activation and targeted histone acetylation by the NuA4 complex. Mol. Cell. Biol. 217629-7640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nourani, A., R. T. Utley, S. Allard, and J. Cote. 2004. Recruitment of the NuA4 complex poises the PHO5 promoter for chromatin remodeling and activation. EMBO J. 232597-2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Puig, O., F. Caspary, G. Rigaut, B. Rutz, E. Bouveret, E. Bragado-Nilsson, M. Wilm, and B. Seraphin. 2001. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods 24218-229. [DOI] [PubMed] [Google Scholar]
- 52.Raisner, R. M., P. D. Hartley, M. D. Meneghini, M. Z. Bao, C. L. Liu, S. L. Schreiber, O. J. Rando, and H. D. Madhani. 2005. Histone variant H2A.Z marks the 5′ ends of both active and inactive genes in euchromatin. Cell 123233-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Reid, J. L., V. R. Iyer, P. O. Brown, and K. Struhl. 2000. Coordinate regulation of yeast ribosomal protein genes is associated with targeted recruitment of Esa1 histone acetylase. Mol. Cell 61297-1307. [DOI] [PubMed] [Google Scholar]
- 54.Robert, F., D. K. Pokholok, N. M. Hannett, N. J. Rinaldi, M. Chandy, A. Rolfe, J. L. Workman, D. K. Gifford, and R. A. Young. 2004. Global position and recruitment of HATs and HDACs in the yeast genome. Mol. Cell 16199-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rohde, J. R., and M. E. Cardenas. 2003. The Tor pathway regulates gene expression by linking nutrient sensing to histone acetylation. Mol. Cell. Biol. 23629-635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rountree, M. R., K. E. Bachman, and S. B. Baylin. 2000. DNMT1 binds HDAC2 and a new co-repressor, DMAP1, to form a complex at replication foci. Nat. Genet. 25269-277. [DOI] [PubMed] [Google Scholar]
- 57.Ruhf, M. L., A. Braun, O. Papoulas, J. W. Tamkun, N. Randsholt, and M. Meister. 2001. The domino gene of Drosophila encodes novel members of the SWI2/SNF2 family of DNA-dependent ATPases, which contribute to the silencing of homeotic genes. Development 1281429-1441. [DOI] [PubMed] [Google Scholar]
- 58.Ruhl, D. D., J. Jin, Y. Cai, S. Swanson, L. Florens, M. P. Washburn, R. C. Conaway, J. W. Conaway, and J. C. Chrivia. 2006. Purification of a human SRCAP complex that remodels chromatin by incorporating the histone variant H2A.Z into nucleosomes. Biochemistry 455671-5677. [DOI] [PubMed] [Google Scholar]
- 59.Santisteban, M. S., T. Kalashnikova, and M. M. Smith. 2000. Histone H2A.Z regulates transcription and is partially redundant with nucleosome remodeling complexes. Cell 103411-422. [DOI] [PubMed] [Google Scholar]
- 60.Selleck, W., I. Fortin, D. Sermwittayawong, J. Cote, and S. Tan. 2005. The Saccharomyces cerevisiae Piccolo NuA4 histone acetyltransferase complex requires the Enhancer of Polycomb A domain and chromodomain to acetylate nucleosomes. Mol. Cell. Biol. 255535-5542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shahbazian, M. D., and M. Grunstein. 2007. Functions of site-specific histone acetylation and deacetylation. Annu. Rev. Biochem. 7675-100. [DOI] [PubMed] [Google Scholar]
- 62.Squatrito, M., C. Gorrini, and B. Amati. 2006. Tip60 in DNA damage response and growth control: many tricks in one HAT. Trends Cell Biol. 16433-442. [DOI] [PubMed] [Google Scholar]
- 63.Utley, R. T., N. Lacoste, O. Jobin-Robitaille, S. Allard, and J. Cote. 2005. Regulation of NuA4 histone acetyltransferase activity in transcription and DNA repair by phosphorylation of histone H4. Mol. Cell. Biol. 258179-8190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.van Attikum, H., O. Fritsch, B. Hohn, and S. M. Gasser. 2004. Recruitment of the INO80 complex by H2A phosphorylation links ATP-dependent chromatin remodeling with DNA double-strand break repair. Cell 119777-788. [DOI] [PubMed] [Google Scholar]
- 65.Vaquero, A., A. Loyola, and D. Reinberg. 2003. The constantly changing face of chromatin. Sci. Aging Knowledge Environ. 2003RE4. [DOI] [PubMed] [Google Scholar]
- 66.Vogelauer, M., J. Wu, N. Suka, and M. Grunstein. 2000. Global histone acetylation and deacetylation in yeast. Nature 408495-498. [DOI] [PubMed] [Google Scholar]
- 67.Wong, M. M., L. K. Cox, and J. C. Chrivia. 2007. The chromatin remodeling protein, SRCAP, is critical for deposition of the histone variant H2A.Z at promoters. J. Biol. Chem. 28226132-26139. [DOI] [PubMed] [Google Scholar]
- 68.Wu, W. H., S. Alami, E. Luk, C. H. Wu, S. Sen, G. Mizuguchi, D. Wei, and C. Wu. 2005. Swc2 is a widely conserved H2AZ-binding module essential for ATP-dependent histone exchange. Nat. Struct. Mol. Biol. 121064-1071. [DOI] [PubMed] [Google Scholar]
- 69.Zhang, H., D. O. Richardson, D. N. Roberts, R. Utley, H. Erdjument-Bromage, P. Tempst, J. Cote, and B. R. Cairns. 2004. The Yaf9 component of the SWR1 and NuA4 complexes is required for proper gene expression, histone H4 acetylation, and Htz1 replacement near telomeres. Mol. Cell. Biol. 249424-9436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang, H., D. N. Roberts, and R. B. Cairns. 2005. Genome-wide dynamics of Htz1, a histone H2A variant that poises repressed/basal promoters for activation through histone loss. Cell 123219-231. [DOI] [PMC free article] [PubMed] [Google Scholar]