Abstract
We have initiated a screen for cellular factors that can induce human papillomavirus type 16 (HPV-16) late gene expression in human cancer cells. We report that the overexpression of polypyrimidine tract binding protein (PTB), also known as heterologous nuclear ribonucleoprotein I (hnRNP I), induces HPV-16 late gene expression in cells transfected with subgenomic HPV-16 plasmids or with full-length HPV-16 genomes and in persistently HPV-16-infected cells. In contrast, other hnRNPs such as hnRNP B1/A2, hnRNP F, and hnRNP Q do not induce HPV-16 late gene expression. PTB activates SD3632, the only 5′ splice site on the HPV-16 genome that is used exclusively by late mRNAs. PTB interferes with splicing inhibitory sequences located immediately upstream and downstream of SD3632, thereby activating late gene expression. One AU-rich PTB-responsive element was mapped to a 198-nucleotide sequence located downstream of SD3632. The deletion of this element induced HPV-16 late gene expression in the absence of PTB. Our results suggest that the overexpression of PTB interferes with cellular factors that interact with the inhibitory sequences. One may speculate that an increase in PTB levels or a reduction in the concentration of a PTB antagonist is required for the activation of HPV-16 late gene expression during the viral life cycle.
Persistent human papillomavirus type 16 (HPV-16) infection is a risk factor for several anogenital cancers, primarily cancer of the uterine cervix (22, 64). These infections are characterized by the presence of HPV-16 DNA and the production of viral mRNAs encoding early proteins needed for the replication of the viral DNA (16, 21). The HPV-16 late genes are not expressed in mitotic cell populations but are expressed in nonmitotic but differentiating cells. For example, late HPV-16 genes are not expressed in cervical cancer cells containing HPV-16 DNA (14). This property of HPV-16 probably aids in the establishment of viral persistence and in escape from the immune response of the infected host. It is therefore of interest to investigate how HPV-16 late gene expression is regulated.
The HPV-16 life cycle is strictly linked to the differentiation stage of the infected cell, and late viral mRNAs are expressed only in differentiated cells (14, 37, 43). Since all known HPV-16 promoters are located in one end of the viral genome, late gene expression is strongly influenced by posttranscriptional gene regulation (62). Viral RNA elements and cellular RNA binding factors are therefore important players in HPV-16 gene regulation (39, 45). The activities of these RNA elements and factors are responsible for the inhibition of HPV-16 late gene expression in proliferating cells and contribute to the induction of late gene expression in differentiating cells (45). RNA elements in the late untranslated region (UTR) of HPV-1 (50, 54, 56), HPV-16 (28), and HPV-31 (12) have been shown to inhibit late gene expression. These elements appear to be conserved among HPVs (60) and were originally identified in bovine papillomavirus (17). Late UTRs from different HPVs interact with the elav-like HuR protein (3, 48). Other RNA elements that suppress HPV-16 late gene expression have been identified previously. For example, the L1 and L2 coding regions contain inhibitory RNA elements that prevent the expression of L1 and L2 from subgenomic expression plasmids (10, 11, 42, 49, 53). The RNA elements in the L1 coding region were later shown to coincide with splicing silencers that suppress SA5639 (61), the only 3′ splice site used exclusively by late mRNAs, whereas the RNA elements in the L2 coding region are active primarily as positive regulators of the early poly(A) signal pAE (41), thereby indirectly inhibiting late gene expression (41). Polyadenylation elements in L2 were originally identified in HPV-31 (55). In addition, splicing inhibitory sequences surround SD3632, the only 5′ splice site used exclusively by late mRNAs, and therefore suppress late gene expression in proliferating cells (44). Finally, a major splicing enhancer downstream of the early 3′ splice site SA3358 directs splicing to the early region of the genome and promotes polyadenylation at pAE (44), indirectly inhibiting late gene expression (44). Together, cellular factors interacting with these elements regulate HPV-16 gene expression. While we have previously identified hnRNP A1 as a cellular factor that binds to the splicing silencers in the HPV-16 L1 coding region and inhibits late mRNA splicing (61), most of the transacting factors regulating HPV-16 late gene expression remain to be identified. We have therefore initiated a screen for cellular factors that can induce HPV-16 late gene expression in proliferating cells.
MATERIALS AND METHODS
Plasmid constructions.
pBEL, pBELM, pBSplice, and pBSpliceM have been described previously (pBSplice was referred to as pC16L1L2splice) (61). pT1, pT2, pT3, pT4, pT9, pT10, pT1OPSA, and pOPSDM have also been described previously (44), as have pBearly and pBELMDPU (59) and p1-22 M (58). pC16L1 and pC16L1M have been described previously as pC16L1MUT123 (11).
pMT1, pMT2, and pMT3 were constructed by the cleaving of pT1, pT2, and pT3 with SalI and BssHII, the filling in of overhangs, and religation. pMT22 was constructed by the cleaving of pT2 with SalI and MluI, the filling in of overhangs, and religation. In order to construct p2xpAL, PCR was first performed with oligonucleotides M7S and M8A (Table 1) on pBR-HPV-16. The PCR fragment was subcloned into pCR2.1-TOPO (Invitrogen) and then transferred as an ApaI-MluI fragment into pBELMDP (59), resulting in p2xpAL. pBSpliceD3703-4530 was created by first PCR amplifying a sequence from pBR-HPV-16 with oligonucleotides E4S (61) and M3A (Table 1). The PCR fragment was subcloned into pCR2.1-TOPO, released with BssHII and ApaI, and subcloned into pBSpliceM. pBSpMD4288-4530 was constructed in the same manner except that oligonucleotides E4S (61) and M6A (Table 1) were used for the generation of the PCR fragment. pBSpMDL1 was constructed by BamHI and XhoI digestion of pBSpliceM, followed by filling in and religation. To construct pFLwt and pFLmut, upstream HPV-16 sequences were first PCR amplified with primers M7STOPHXS and M1A (Table 1) and inserted into pCR2.1-TOPO. The HPV-16 sequences were excised with HindIII and NcoI and inserted into pBEX and pBEXM (44) to generate pFL16wt and pFL16mut, respectively. To construct pC97EL, HPV-16 sequences were first PCR amplified with primers M97S and M1A (Table 1) and cloned into pCR2.1-TOPO. The HPV-16 sequences were excised with HindII and SalI and inserted into pBEX (44) to generate pC97EL. To construct pBELDL2, a DNA fragment was PCR amplified with primers M4S (61) and 16F (Table 1) and cloned into TOPO. The fragment was excised with ApaI and BamHI and transferred into pBEL, generating pBELDL2.
TABLE 1.
Oligonucleotides used in this study
| Oligonucleotide name | Oligonucleotide sequence |
|---|---|
| M7S | ACGCGTTAACTATTGTGTCATGCAACATAAATAAAC |
| M8A | CCCGGGAGGCCTGGTTGAAGCTACAAAATGGGCCTGG |
| M3A | GGGCCCAGGCCTCGACACTGCAGTATACAATGTACAATGCT |
| M6A | GGGCCCAGGCCTAGGGTTGGGTAGCCGATGCACGTTTTGTGC |
| M4S | AGGCCTGGGCCCGCTCCTTCATTAATTCCTATAGTTCCAGGG |
| 16F | CCTCGAGCATTTTAATATAATCTGGATATTTGC |
| 880AS | TCTAGACGCGTCCACTACAGCCTCTACATAAAACCATCC |
| 3902ae | GGAATTCACGCGTATAGACATAAATCCAGTAG |
| PTBaS | GGATCCATGGACGGCATTGTCCCA |
| PTBaA | CTCGAGCATCGCCGCCGAGGGGATGGCCAG |
| PTBbS | GGATCCATGGCGGCAGGTCGGATCGCCATC |
| PTBbA | CTCGAGCATCTAGATGGTGGACTTGGCGAA |
| A2B1S | GGCGCGCACCATGGAGAGAGAAAAGGAACAGTTCC |
| A2B1A | CCTCGAGTCAGTATCGGCTCCTCCCACCATAACCC |
| QS | GGCGCGCACCATGGCTACAGAACATGTTAATGGA |
| QA | CCTCGAGCTACTTCCACTGTTGCCCAAAAGTATCCTGATAAAA |
| M7STOPHXS | AAGCTTCTCGAGGTATTGTATGTATGTTGAATTAGTGTGTTG |
| M1A | ACGCGTTCTAGATCTCCATCCATTACATCCCGTACCCTCTTC |
| M97S | GAAGCTTGTTAACGTCGACAACTGCAATGTTTCAGGACCCACAGG |
| HCMVS | GTCAGATCGCCTGGAGACGCC |
| SD3632S | GGTACCCCAATCCTCACTGCATT |
| FS | GCGGGCGAAATGATGCTGGGCCCTGAGG |
| FA | CTCGAGCTAGTCATAGCCACCCATGCTG |
To construct pPTBA, the 5′ half of the gene encoding polypyrimidine tract binding protein (PTB) was first PCR amplified with oligonucleotides PTBaS and PTBaA (Table 1) and cloned into pCDNA3.1/V5-His (Invitrogen), generating pPTBA. pPTBB was constructed in the same manner except that primers PTBbS and PTBbA (Table 1) were used. PTB1 expression plasmid pCMVPTB1 was generously provided by Clare Gooding, and plasmids encoding Raver1 or Raver2 under the control of the cytomegalovirus (CMV) promoter were generously provided by Susanne Illenberger. hnRNP Q, hnRNP B1/A2, and hnRNP F were generously provided by Natalja Funk, Oriol Bachs, and Douglas Black, respectively. To construct CMV promoter-driven expression plasmids, the open reading frames of each cDNA were PCR amplified using primers QS and QA and A2B1S and A2B1A and FS and FA (Table 1) and the products were subcloned into pCR2.1-TOPO. The protein-encoding regions were excised with BssHII and XhoI and individually subcloned into plasmid pCL086 (11), resulting in pCMVhnRNPQ, pCMVhnRNPB1/A2, and pCMVhnRNPF.
Transfection and cell culture.
HeLa cells were cultured in Dulbecco's modified Eagle medium containing 10% heat-inactivated fetal bovine calf serum and penicillin-streptomycin. Transfections were carried out by using FuGENE 6 according to the protocol of the manufacturer (Roche Molecular Biochemicals). In short, 3 μl of FuGENE 6 and 200 μl of serum-free Dulbecco's modified Eagle medium were mixed with 1 μg of DNA, and the mixture was incubated for 15 min. Thereafter, the mixture was added dropwise to 60-mm plates of HeLa cells at approximately 60% confluence. Cells were harvested 24 h posttransfection. Cells were transfected with each plasmid in a minimum of three independent experiments with similar results.
W12E cell culture, transfection, and RNA preparation.
The cervical cell line W12E (subclone 20850), containing extrachromosomal and replicating HPV-16 genomes, was obtained from Paul Lambert (University of Wisconsin). Cells were grown on mitomycin C-treated J2-3T3 fibroblast feeder cells (also from Paul Lambert) in F medium as described previously (25). Cells were harvested after 3 days (±1 day) of incubation after the removal of the feeder layer, spread onto 30-mm-well plates, and grown overnight in keratinocyte serum-free medium with epidermal growth factor (5 ng/ml; Invitrogen) and bovine pituitary extract (60 μg/ml) (Invitrogen). The medium was changed the next day, and cells were transfected with 1 μg of plasmid DNA and FuGENE 6 according to the instructions of the manufacturer (Roche Molecular Biochemicals). Cells were harvested 24 h posttransfection, and RNA was prepared with a QIAamp viral RNA mini kit (Qiagen) and reverse transcribed with Moloney murine leukemia virus reverse transcriptase (Invitrogen) according to the instructions of the manufacturers.
RNA extraction, Northern blotting, and synthesis of radiolabeled DNA probes.
Cytoplasmic RNA was extracted using NP-40 buffer as described previously (56). The RNA was treated with DNase I prior to Northern blotting. The Northern blot analysis was carried out by size separation of 10 μg of cytoplasmic RNA on a 1.2% agarose gel containing 2.2 M formaldehyde. The RNA was transferred overnight onto a nitrocellulose filter and hybridized (11) with either an L1, E4, or GAPDH (glyceraldehyde-3-phosphate dehydrogenase) probe as described previously (61). The L1 probe was excised from pBEL with BamHI and XhoI, whereas the E4 probe was generated by PCR using the following primers: E4S (61) and K1 (44). DNA probes were radiolabeled with [α-32P]dCTP by using a Decaprime kit (Ambion).
Reverse transcription (RT)-PCR.
Two hundred nanograms of cytoplasmic RNA was reverse transcribed at 42°C by using Superscript II and random hexamers according to the protocol of the manufacturer (Invitrogen). Two microliters of cDNA in a 100-μl reaction mixture was amplified by PCR using the oligonucleotides indicated in the figures, as described previously (61). GAPDH cDNA was amplified as a control by using previously described primers (27).
Real-time RT-PCR.
RNA was reverse transcribed with Moloney murine leukemia virus reverse transcriptase according to the instructions of the manufacturer (Invitrogen). To detect the absolute number of RNA molecules, a copy number standard was made by cloning a PCR fragment of the HPV-16 L1 coding region into a TOPO expression vector, RNA was expressed with a MEGAscript high-yield transcription kit (Ambion), and the concentration of RNA molecules was determined with a NanoDrop ND-1000 spectrophotometer. A standard curve of absolute molecule numbers was created for each for real-time reaction, and samples were analyzed with the software of ABI Prism 7700 (Applied Biosystems). The primers (forward primer TGATACTACACGCAGTACAAATATGTCAT and reverse primer CCCCATGTCGTAGGTACTCCT) and major groove binding probe (TGCTGCCATATCTAC-6-carboxyfluorescein) were designed with Primer Express 3.0 (Applied Biosystems), and reactions were performed with TaqMan 1000 RXN PCR core reagents by using the ABI Prism 7700 thermocycler (Applied Biosystems).
RESULTS
The overexpression of PTB induces the production of partially spliced HPV-16 L2/L1 mRNAs.
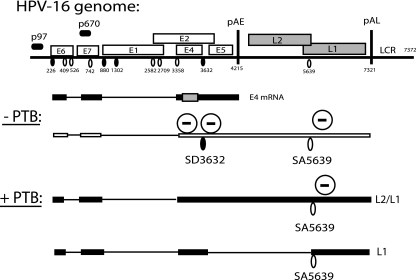
The HPV-16 genome can be divided into early and late regions (Fig. 1). HPV-16 late genes are not expressed in dividing cells, including cancer cells. One may speculate that this pattern contributes to the virus's ability to establish persistent infections in the presence of a functional immune system. Late gene expression is actively suppressed by the presence of splicing silencers that inhibit late mRNA splicing (44, 58, 61) or RNA elements that indirectly inhibit late gene expression by promoting early mRNA splicing (44) and polyadenylation (41, 42, 55, 59) (Fig. 1). To identify cellular factors that induce late gene expression, and presumably are present in suboptimal concentrations in dividing cells and cancer cells, we have initiated a screen for such cellular factors. Since the presently available evidence suggests that HPV-16 late gene expression is regulated primarily at the level of RNA processing, RNA binding factors are likely to regulate HPV-16 late gene expression. Two big families of RNA binding proteins regulate RNA splicing: the hnRNP proteins (15) and the SR proteins (4). Here, we have investigated the hnRNP proteins. We have previously identified several hnRNP proteins that bind to HPV-16 mRNAs and presumably inhibit late gene expression by stimulating early polyadenylation or inhibiting late mRNA splicing or translation, including hnRNP A1, hnRNP E1 and hnRNP E2 [also known as poly(rC) binding proteins 1 and 2], hnRNP H, and hnRNP K (10, 41, 61). Here, we wished to identify factors that could potentially activate the expression of late HPV-16 mRNAs and focused on other hnRNPs, primarily those with documented functions in RNA splicing, such as hnRNP I (PTB), hnRNP F, hnRNP A2/B1, and hnRNP Q (15).
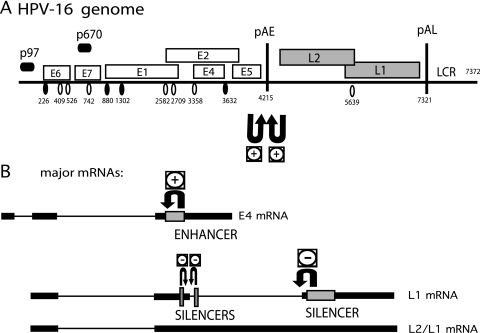
FIG. 1.
(A) Schematic diagram of the HPV-16 genome. Early and late viral promoters p97 and p670 are indicated. Numbers indicate the nucleotide positions of 5′ splice sites (filled circles), 3′ splice sites (empty circles), or early and late poly(A) signals pAE and pAL, respectively. (B) Potential HPV-16 mRNAs representing major early mRNA (E4 mRNA) and late mRNAs (L1 and L2/L1 mRNAs). Previously identified splicing regulatory elements are indicated, including a major splicing enhancer downstream of SA3358 (44), splicing suppressor sequences at SD3632 (44), and multiple splicing silencers in the L1 coding region that suppress the use of SA5639 (58, 61). Polyadenylation regulatory elements in the early UTR (59) and in the L2 region (41) are indicated. U-shaped arrows indicate positive (+) or negative (−) effects of previously identified regulatory RNA elements on viral splice sites (44, 61). LCR, long control region.
We used primarily a subgenomic HPV-16 plasmid named pBEL (61), which carries viral early and late genes after a human CMV immediate-early promoter placed at the position of the proposed late viral promoter p670 (Fig. 2A). This plasmid produces only early mRNAs upon the transfection of cervical cancer cells (Fig. 2B, E4 and L1 probes) (61) and can therefore be used to identify cellular factors that activate HPV-16 late gene expression.
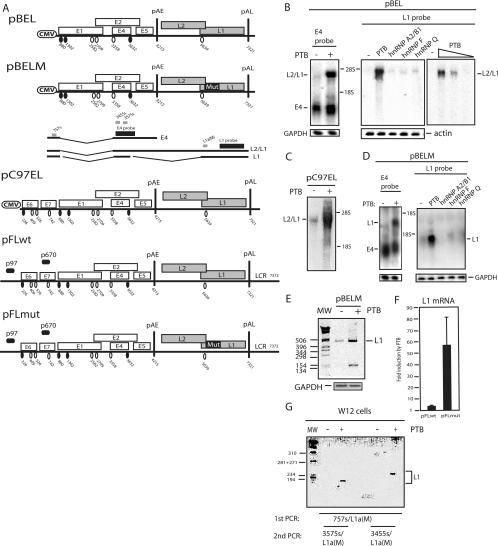
FIG. 2.
(A) Schematic representation of subgenomic HPV-16 expression plasmids used to study the induction of late gene expression by PTB. Numbers indicate the nucleotide positions of 5′ splice sites (filled circles) and 3′ splice sites (empty circles) or poly(A) sites pAE and pAL. Major potential mRNAs produced by pBEL and pBELM are indicated below the plasmids. Gray bars represent RT-PCR primers, and black bars represent E4 and L1 probes used for Northern blotting. Mut, a mutant L1 gene in which splicing silencers have been inactivated (11, 61); LCR, long control region. (B, C, and D) Northern blots of cytoplasmic RNA extracted from HeLa cells transfected with pBEL, pC97EL, or pBELM in the absence or presence of plasmids expressing PTB, hnRNP A2/B1, hnRNP F, or hnRNP Q. Blots were probed with the E4 or L1 probe as indicated. Gels were also probed for GAPDH or actin expression. The positions of the E4, L1, and L2/L1 mRNAs are indicated. +, present; −, absent. (E) RT-PCR with primers 757s and L1a(M) (see panel A) (61) on the same cytoplasmic RNA that was analyzed in the blot in panel D. GAPDH cDNA was amplified as a control. MW, molecular weight marker. (F) Real-time PCR was performed with RNA extracted from cells transfected with plasmid pFLwt or pFLmut in the absence or presence of PTB-expressing plasmid pCMVPTB1, as described in Materials and Methods. The levels of induction (n-fold) by PTB were calculated for pFLwt and pFLmut and are shown in the graph. (G) Results from nested RT-PCRs to detect HPV-16 L1 mRNA in HPV-16-containing W12 cells transfected with an empty vector (−) or with the pCMVPTB1 plasmid (+). First, PCR was performed with primers 757s and L1a(WT). This reaction was nested with either 3455s and L1a(WT) or 3575s and L1a(WT), as indicated in the figure.
Cotransfection with pCMVPTB1, which expresses PTB1/hnRNP I (hereinafter called PTB), and pBEL induced HPV-16 late gene expression in a dose-dependent manner (Fig. 2B), whereas early E4 mRNAs were largely unaffected (Fig. 2B, left panel). The late mRNAs detected were primarily L2/L1 mRNAs (Fig. 2B), although longer exposures of the gels revealed that spliced L1 mRNAs were induced as well (data not shown). In contrast, the overexpression of hnRNP A2/B1, hnRNP F, or hnRNP Q had no detectable effect on HPV-16 late gene expression (Fig. 2B, middle panel). We also constructed and tested plasmid pC97EL (Fig. 2A), in which the CMV promoter was placed at nucleotide position 97 in the HPV-16 genome, thereby replacing the viral early promoter. PTB induced primarily L2/L1 mRNAs from pC97EL (Fig. 2C), although L1 mRNAs could be detected by RT-PCR (data not shown).
The overexpression of PTB induces the production of spliced HPV-16 L1 mRNAs in the absence of splicing silencer elements in the L1 coding region.
To investigate if PTB induced L2/L1 mRNAs specifically or if PTB could induce the expression of the spliced L1 mRNAs also (Fig. 2A), cells were cotransfected with pCMVPTB1 and a mutant version of pBEL named pBELM (61) in which splicing silencers in the L1 coding region had been inactivated (Fig. 2A) (61). PTB induced high levels of primarily spliced L1 mRNAs from pBELM (Fig. 2D), while hnRNPs A2/B1, F, and Q had no detectable effect, as expected (Fig. 2D). The cloning and sequencing of RT-PCR products confirmed that the late splice sites SD3632 and SA5639 were used (Fig. 2E). The quantitation of the L1 mRNA levels in multiple, independent Northern blot experiments revealed an average induction of L1 mRNA from pBELM of 42-fold by PTB, whereas the level of induction of L2/L1 mRNAs from pBEL was 15-fold. We concluded that PTB induced L2/L1 mRNAs when the late 3′ splice site at genomic position 5639 was suppressed by splicing silencers in the L1 coding region but induced spliced L1 mRNAs when splicing suppression was relieved by splicing silencer-inactivating mutations in the L1 coding region (as in pBELM). In general, the level of induction was greater when PTB induced spliced L1 mRNAs.
PTB induces HPV-16 late gene expression from full-length HPV-16 genomes and in HPV-16-infected cells.
The subgenomic HPV-16 plasmids described above were all driven by a CMV promoter. To confirm that full-length HPV-16 genomes also respond to PTB, pFLwt and pFLmut (Fig. 2A) were constructed and cells were cotransfected with these plasmids and a plasmid expressing PTB. Viral gene expression is driven by the weak HPV-16 early promoter, and the effect of PTB was therefore monitored by real-time PCR rather than Northern blotting. The results revealed a high level of induction of HPV-16 late gene expression from both plasmids by PTB (Fig. 2F). The level of induction was higher with pFLmut than with pFLwt. Real-time PCR analyses were designed to detect spliced L1 mRNAs. Since PTB induces primarily unspliced L2/L1 mRNAs when silencers in the L1 gene are intact, as in pFLwt, the level of induction of late gene expression from pFLwt by PTB (2.5-fold) (Fig. 2F) was probably underestimated in the real-time PCR analyses, whereas the 58-fold induction of expression from pFLmut (Fig. 2F) is similar to the 42-fold induction seen with pBELM (Fig. 2D).
To investigate if PTB could induce HPV-16 late gene expression in chronically HPV-16-infected cells also, W12E (20850) cells (61) that contained episomal HPV-16 genomic DNA were transfected with pCMVPTB1. An analysis of RNA by RT-PCR revealed that PTB induced late gene expression in persistently infected cells also in multiple independent transfection experiments (Fig. 2G). We concluded that the increase in PTB levels in HeLa cells and in W12E (20850) cells (61) could induce HPV-16 late gene expression.
The induction of HPV-16 late gene expression by PTB occurs independently of the HPV-16 early poly(A) signal (pAE), early splice sites, and the L1 sequence.
The activation of HPV-16 late gene expression by PTB may be caused by an inhibition of early splice sites or the early poly(A) signal, which would redirect splicing or polyadenylation to the late region, or by a direct interaction of PTB with the L1 coding region, selectively stabilizing L2/L1 mRNAs and/or L1 mRNAs or exporting them to the cytoplasm. To investigate if PTB induces HPV-16 late mRNA production by inhibiting polyadenylation at pAE, we cotransfected cells with a PTB-expressing plasmid and pBELMDPU (59), a pBELM-derived plasmid in which the early poly(A) signal at position 4215 had been mutationally inactivated by replacing the AATAAA sequence and the U-rich region in the early UTR with an MluI site (Fig. 3A) (59). This mutation activated an efficiently used cryptic polyadenylation site (CpA) at position 3820 in the early region (Fig. 3A), as described previously (59). The major mRNA species produced from this plasmid is an mRNA that is spliced from SD880 to SA3358 and polyadenylated at 3820 (59). The overexpression of PTB induced late mRNA production from pBELMDPU (Fig. 3B), demonstrating that PTB did not target pAE specifically and that the early UTR was not required. To confirm that PTB did not act through the early UTR, a plasmid with a deletion that removes the entire UTR but without affecting the pAE itself (pBELMDU) (61) (Fig. 3A) was analyzed in parallel with pBELM. PTB induced L1 mRNAs from the two plasmids to the same extent (Fig. 3C), demonstrating that this effect of PTB was not mediated through the U-rich region in the early UTR.
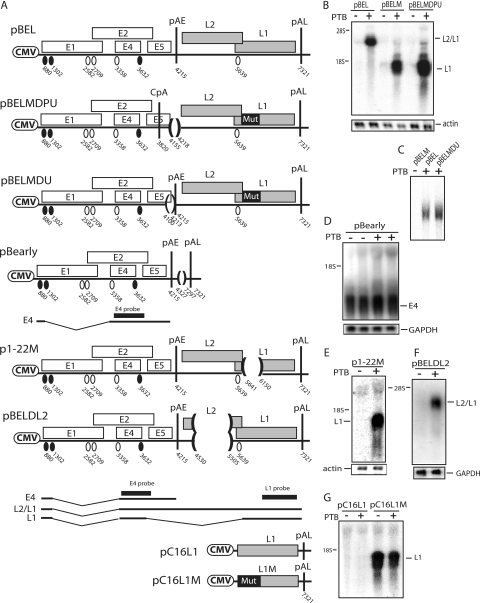
FIG. 3.
(A) Schematic representation of subgenomic HPV-16 expression plasmids used to study the induction of late gene expression by PTB. Numbers indicate the nucleotide positions of 5′ splice sites (filled circles) and 3′ splice sites (empty circles) or the positions of pAE and pAL or mark the borders of deletions. Major potential mRNAs produced by pBEL and pBELM are indicated below the plasmids. Black bars represent the E4 and L1 probes used for Northern blotting. Mut, a mutant L1 gene in which splicing silencers have been inactivated (11, 61). (B, C, D, E, and F) Northern blots of cytoplasmic RNA extracted from HeLa cells transfected with pBEL, pBELM, pBELMDU, pBELMDPU, pBearly, p1-22 M, pBELDL2, pC16L1, or pC16L1M in the absence or presence of a PTB-expressing plasmid. Blots were probed with the E4 probe (D) or the L1 probe (B, C, E, F, and G). Gels were also probed for GAPDH or actin expression. Positions of the E4, L1, and L2/L1 mRNAs are indicated. +, present; −, absent.
We next investigated if PTB affected the processing of early mRNAs in the absence of the late region. Cells were transfected with plasmid pBearly (Fig. 3A) in the absence or presence of PTB. Results revealed that PTB did not have a detectable effect on early mRNA splicing or polyadenylation (Fig. 3D). The lack of effect on the major early 3′ splice site at position 3358 is reasonable since the E4 3′ splice site is suboptimal (44) and the polypyrimidine tract lacks continuous pyrimidine stretches of significance, as well as predicted PTB binding sites (44).
Since both L2/L1 and L1 mRNAs could be induced by PTB, the PTB-responsive sequence may be located in the L1 coding region that is present on all late mRNAs. The first 514 nucleotides of the L1 exon that contains previously identified hnRNP A1 binding splicing silencers (61) were deleted in pBELM, resulting in p1-22 M (Fig. 3A). In a second plasmid, the remaining part of the L1 coding region was deleted (data not shown). Both plasmids responded well to PTB, indicating that PTB did not target sequences in the L1 coding region (Fig. 3E and data not shown). In addition, cotransfection with a PTB-expressing plasmid and the HPV-16 L1 expression plasmid pC16L1 (Fig. 3A) (11), which encodes only L1 but does not express detectable levels of L1 mRNA as a result of inhibitory RNA elements in the L1 gene, did not activate L1 expression (Fig. 3G). A genetically modified HPV-16 L1 gene that expressed high levels of L1, carried on pC16L1M (11), was also unaffected by PTB (Fig. 3G). Finally, we also introduced a deletion in the L2 region into pBEL, resulting in pBELDL2 (Fig. 3A). This deletion did not affect the ability of PTB to induce late gene expression either (Fig. 3F). Taken together, our results failed to support a model in which PTB acts directly on sequences in the late region or indirectly on early processing signals of HPV-16. This finding suggested that PTB may act on late RNA processing signals located in the early region.
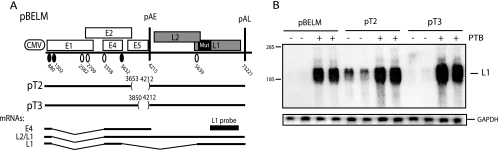
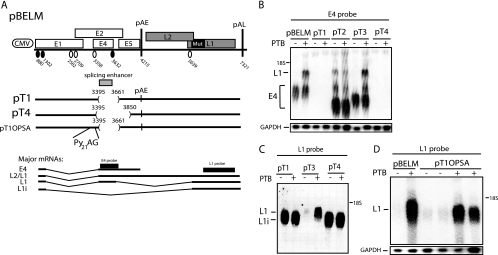
The deletion of sequences immediately downstream of the HPV-16 major late 5′ splice site SD3632 reduces the effect of PTB.
One of the splice sites in the early region, SD3632 (Fig. 4A), is used exclusively by late mRNAs (1). It is suboptimal, and we have shown previously that upstream and downstream RNA sequences suppress SD3632 (44). Sequences located downstream of SD3632, between SD3632 and pAE, were deleted in pT2 and pT3 (Fig. 4A) (44). The deletion of the entire sequence between SD3632 and pAE, as in pT2, reduced the effect of PTB (Fig. 4B), demonstrating that this sequence inhibited late gene expression in the absence of PTB. However, PTB did exert a positive effect on pT2, although to a lower extent than on pBELM, suggesting that other sequences also inhibited late gene expression in the absence of PTB. The pT3 deletion includes a U-rich element that we have previously shown binds PTB (44). pT3 displayed the same phenotype as pBELM (Fig. 4B), suggesting that PTB targeted sequences upstream of nucleotide position 3850.
FIG. 4.
(A) Schematic representation of subgenomic HPV-16 expression plasmids pBELM, pT2, and pT3. Numbers indicate the nucleotide positions of 5′ splice sites (filled circles) and 3′ splice sites (empty circles) or the positions of pAE and pAL or mark the borders of deletions. Major potential mRNAs produced by these plasmids are indicated below. The L1 probe used for Northern blotting is indicated. Mut, a mutant L1 gene in which splicing silencers have been inactivated (11, 61). (B) Northern blot of cytoplasmic RNA extracted from HeLa cells transfected with pBELM, pT2, or pT3 in the absence or presence of a PTB-expressing plasmid. The blot was probed with the L1 probe. Gels were also probed for GAPDH expression. The position of the L1 mRNA is indicated. +, present; −, absent.
PTB induces the production of HPV-16 mRNAs that are spliced from SD3632 to SA5639 but not from SD880 to SA5639.
Sequences upstream of SD3632, as well as SD3632 itself, were deleted in pT1 and pT4 (Fig. 5A) (44). These deletions also removed an essential splicing enhancer of the upstream-located 3′ splice site SA3358, and splicing was therefore totally redirected from SD880 to SA5639, resulting in the production of L1i mRNAs that were not detected by the E4 probe (Fig. 5B) but were detected by the L1 probe (Fig. 5C). The overexpression of PTB did not affect mRNAs that were spliced from SD880 to SA5639 (Fig. 5C). Plasmids pT2 and pT3 served as controls, and as expected, PTB had a small effect on pT2, whereas pT3 responded well to PTB (Fig. 5B and C). In conclusion, mRNAs spliced directly from SD880 to SA5639 did not respond to PTB, demonstrating that sequences in the central, early region of the genome were required for the effect of PTB on late gene expression.
FIG. 5.
(A) Schematic representation of subgenomic HPV-16 expression plasmids pBELM, pT1, pT4, and pT1OPSA. Numbers indicate the nucleotide positions of 5′ splice sites (filled circles) and 3′ splice sites (empty circles) or the positions of pAE and pAL or mark the borders of deletions. Major potential mRNAs produced by these plasmids are indicated below. The optimized 3′ splice site with 21 consecutive pyrimidines (Py21) upstream of the invariable AG dinucleotide of the 3′ splice site is indicated. The L1i mRNA is a previously described mRNA that is spliced directly from SD880 to SA5639 when splicing silencers at SA5639 are inactivated by mutations (61). Splicing directly from SD880 to SA5639 in differentiated W12 epithelial cells has also been observed previously (38). The E4 and L1 probes used for Northern blotting are indicated. Mut, a mutant L1 gene in which splicing silencers have been inactivated (11, 61). (B, C, and D) Northern blots of cytoplasmic RNA extracted from HeLa cells transfected with pBELM, pT1, pT2, pT3, pT4, or pT1OPSA in the absence or presence of a PTB-expressing plasmid. Blots were probed with the E4 probe (B) or the L1 probe (C and D). Gels were also probed for GAPDH expression. The positions of the E4, L1, and L1i mRNAs are indicated. +, present; −, absent.
Restoring splicing to SA3358 in pT1 (Fig. 5A), in the absence of the downstream enhancer at SA3358, by optimizing SA3358 with an extended polypyrimidine tract (in plasmid pT1OPSA [Fig. 5A] [44]) restored high E4 mRNA levels and undetectable L1 mRNA expression (Fig. 5D and data not shown). The overexpression of PTB induced L1 mRNA production from pT1OPSA (Fig. 5D), which confirmed that the major sequence elements that inhibit late gene expression in the absence of PTB, as well as sequences that respond to PTB, are located downstream of the deletion in pT1, that is, downstream of nucleotide position 3661 and downstream of SD3632.
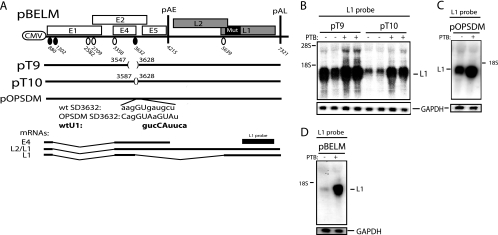
The deletion of sequences immediately upstream of HPV-16 SD3632 reduces the effect of PTB.
To establish if sequences upstream of SD3632 also affected the PTB response, smaller deletions that did not affect the splicing enhancer at SA3358 were introduced upstream of SD3632 (Fig. 6A). Plasmid pT10 contains a deletion that does not affect the efficiency of either SA3358 or SD3632 (44), whereas the pT9 deletion activates SD3632 as a result of the removal of splicing inhibitory sequences (44). As can be seen in Fig. 6B, the enhancing effect of PTB was reduced by the pT9 deletion but not by the pT10 deletion. These results suggested that PTB also targeted sequences located immediately upstream of SD3632 that acted by suppressing SD3632. Alternatively, the PTB response element may have been lost as a result of increased splicing. We concluded that PTB alleviated the effects of splicing inhibitory RNA elements at SD3632. If these conclusions are correct, the optimization of SD3632 in pBELM should reduce the requirement for induction by PTB as a result of the increased splicing of L1 mRNAs. Figure 6C shows that a pBELM derivative with an optimized SD3632 (pOPSDM) (44) responded less well to PTB than pBELM (Fig. 6D), as expected. However, PTB exerted a small stimulatory effect on L1 expression from pOPSDM. We have previously shown that the optimization of SD3632 does not overcome the inhibitory effect of the splicing inhibitory elements, although it does enhance splicing from SD3632 (44). Combined, our results indicate that PTB acts on HPV-16 sequences that suppress SD3632.
FIG. 6.
(A) Schematic representation of subgenomic HPV-16 expression plasmids pBELM, pT9, pT10, and pOPSDM. Numbers indicate the nucleotide positions of 5′ splice sites (filled circles) and 3′ splice sites (empty circles) or the positions of pAE and pAL or mark the borders of deletions. The exact sequences of wild-type (wt) and mutant SD3632 are indicated. Major potential mRNAs produced by these plasmids are indicated below. The L1 probe used for Northern blotting is indicated. Mut, a mutant L1 gene in which splicing silencers have been inactivated (11, 61). (B, C, and D) Northern blots of cytoplasmic RNA extracted from HeLa cells transfected with pBELM, pT9, pT10, or pOPSDM in the absence or presence of a PTB-expressing plasmid. Blots were probed with the L1 probe and a GAPDH probe. The position of the L1 mRNA is indicated. +, present; −, absent.
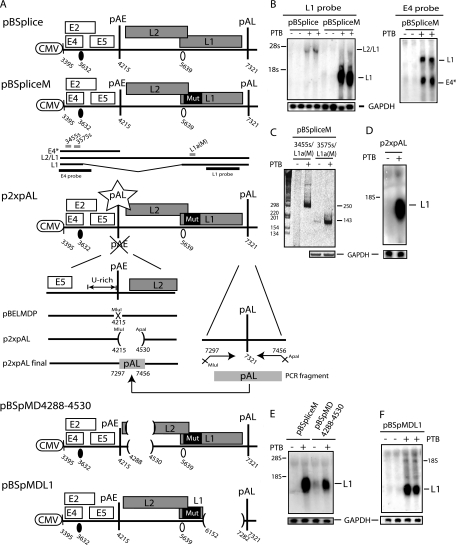
PTB induces splicing from the HPV-16 late 5′ splice site SD3632 by interfering with adjacent splicing inhibitory elements.
To provide evidence for a direct effect of PTB on splicing from SD3632 to SA5639, we investigated the effect of PTB on HPV-16 plasmids pBSplice and pBSpliceM (Fig. 7A). These plasmids contain a large deletion in the early region that removes all splice sites but the two late HPV-16 splice sites SD3632 and SA5639. The predicted mRNAs are depicted in Fig. 7A below the schematics of the plasmids. However, we have shown previously that these plasmids do not express detectable levels of any mRNAs (see also Northern blots in the absence of PTB in Fig. 7B) (44). This lack of detectable expression is presumably a direct effect of the presence on the mRNAs of unutilized splice sites SD3632 and SA5639 and/or inhibitory sequences that target the predicted mRNAs for rapid degradation. However, cotransfection with pBSplice and pBSpliceM in the presence of PTB activated the expression of the L2/L1 mRNAs from pBSplice (Fig. 7B) and spliced late L1 mRNAs from pBSpliceM (Fig. 7B). That PTB induced L1 mRNAs using SD3632 and SA5639 from pBSpliceM was confirmed by the cloning and sequencing of the RT-PCR products seen in Fig. 7C. As described above for pBEL and pBELM, the level of induction of L1 mRNAs from pBSpliceM was higher than the level of induction of L2/L1 mRNAs from pBSplice (58-fold induction from pBSpliceM versus 8-fold induction from pBSplice). These results established that PTB induced splicing from SD3632. In contrast, L1 mRNAs that were spliced from SD880 rather than SD3632, as those from pT1 and pT4 (Fig. 5A), were unaffected by PTB, as described above (Fig. 5B and C), confirming that PTB acted specifically on late mRNAs using SD3632.
FIG. 7.
(A) Schematic representation of subgenomic HPV-16 expression plasmids pBSplice, pBSpliceM, p2xpAL, pBSpMD4288-4530, and pBSpMDL1. Numbers indicate the nucleotide positions of 5′ splice sites (filled circles) and 3′ splice sites (empty circles) or the positions of pAE and pAL or mark the borders of deletions. Major potential mRNAs produced by these plasmids are indicated below pBSpliceM. E4* indicates a short mRNA encoding E4 and E5 that is unspliced due to the absence of SD880 and other 5′ splice sites upstream of the major E4 3′ splice site SA3358 in the pBSplice-derived plasmids. The E4 and L1 probes used for Northern blotting are indicated. Gray bars represent RT-PCR primers 3455s, 3575s, and L1a(M) (44, 61). Mut, a mutant L1 gene in which splicing silencers have been inactivated (11, 61). (B, D, E, and F) Northern blots of cytoplasmic RNA extracted from HeLa cells transfected with pBSplice, pBSpliceM, p2xpAL, pBSpMD4288-4530, and pBSpMDL1in the absence or presence of a PTB-expressing plasmid. Blots were probed with the E4 probe (B, right panel), the L1 probe (B, left panel, and D, E, and F), and a GAPDH probe. The positions of the alternative E4 mRNA named E4* and the L2/L1 and L1 mRNAs are indicated. +, present; −, absent. (C) Results from RT-PCR with primers 3455s or 3575s and L1a(M) (see panel A) (44, 61) on the same cytoplasmic RNA that was analyzed in the blot in panel B. GAPDH cDNA was amplified as a control.
To confirm that pAE was not targeted by PTB, pAE was replaced by the HPV-16 late polyadenylation signal pAL in pBSpliceM. pAL is not negatively affected by PTB, as it is used by all HPV-16 mRNAs that are induced by PTB. These cloning steps resulted in plasmid p2xpAL (Fig. 7A). PTB induced high levels of late mRNAs from p2xpAL (Fig. 7D), confirming that PTB does not induce late gene expression by inhibiting HPV-16 polyadenylation. Sequences in L2 or L1 coding regions were not required either, as two large deletions in either the HPV-16 L2 or L1 coding region (in plasmids pBSpMD4288-4530 and pBSpMDL1) (Fig. 7A) failed to reduce the response to PTB (Fig. 7E and F), as expected.
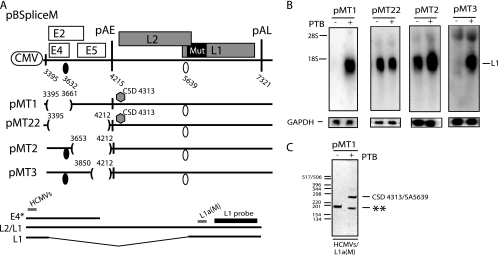
To test if sequences adjacent to SD3632, rather than SD3632 itself, were required for the PTB response, we deleted SD3632 and the sequence upstream of SD3632 in pBSpliceM, resulting in pMT1 (Fig. 8A). As expected, this plasmid did not produce HPV-16 mRNAs (Fig. 8B), since inhibitory sequences downstream of SD3632 were still present on the plasmid (Fig. 8A), and late gene expression was strongly induced by PTB (Fig. 8B). Surprisingly, the induced late mRNAs were spliced, although SD3632 had been deleted. The cloning and sequencing of RT-PCR amplification products identified a cryptic 5′ splice site (Fig. 8A and C) at position 4313 downstream of pAE (Fig. 8A). These results demonstrated that SD3632 was dispensable for the effect of PTB and further indicated that PTB acted on sequences adjacent to SD3632. However, we could not formally exclude the possibility that PTB acted on the cryptic 5′ splice site in pMT1. We therefore deleted the entire sequence upstream of the cryptic 5′ splice site, without deleting the cryptic 5′ splice site itself, resulting in pMT22 (Fig. 8A). This plasmid produced high levels of L1 mRNAs in the absence or presence of PTB and did not respond to PTB (Fig. 8B), demonstrating that sequences upstream of the cryptic 5′ splice site, not the cryptic 5′ splice site itself, were targeted by PTB. The mRNAs produced from pMT22 were spliced. Taken together, these results established that a sequence located between SD3632 and pAE inhibited late gene expression in the absence of PTB and that PTB induced late gene expression by targeting this sequence. In contrast, SD3632 and the cryptic 5′ splice site were dispensable for PTB induction.
FIG. 8.
(A) Schematic representation of subgenomic HPV-16 expression plasmids pBSpliceM, pBMT1, pBMT22, pBMT2, and pBMT3. Numbers indicate the nucleotide positions of 5′ splice sites (filled circles) and 3′ splice sites (empty circles) or the positions of pAE and pAL or mark the borders of deletions. Major potential mRNAs produced by these plasmids are indicated below pBSpliceM. E4* indicates a short mRNA encoding E4 and E5 that is unspliced due to the absence of SD880 and other 5′ splice sites upstream of the major E4 3′ splice site SA3358 in the pBSplice-derived plasmids. The E4 and L1 probes used for Northern blotting are indicated. Gray bars represent RT-PCR primers HCMVS (Table 1) and L1a(M) (61). CSD indicates a cryptic 5′ splice site that was identified by the cloning and sequencing of the RT-PCR products seen in panel C. Mut, a mutant L1 gene in which splicing silencers have been inactivated (11, 61). (B) Northern blots of cytoplasmic RNA extracted from HeLa cells transfected with pMT1, pMT22, pMT2, or pMT3 in the absence or presence of a PTB-expressing plasmid. Blots were probed with the L1 probe and a GAPDH probe. The position of the L1 mRNA is indicated. +, present; −, absent. (C) Results from RT-PCR with primers HCMVS and L1a(M) (see panel A) on cytoplasmic RNA extracted from HeLa cells transfected with pMT22 in the absence or presence of a PTB-expressing plasmid. All bands were cloned and sequenced. The band representing HPV-16 mRNA spliced from CSD4313 to SA5639 is indicated. **, PCR artifact due to mispriming.
Mapping of a PTB-responsive sequence to a 198-nucleotide region downstream of the HPV-16 5′ splice site SD3632.
To map the major PTB response element downstream of SD3632, two deletions were introduced into this region, resulting in plasmids pMT2 and pMT3 (Fig. 8A). While pMT3 was fully responsive to PTB (Fig. 8B), pMT2 produced high L1 mRNA levels in the absence and presence of PTB and did not respond to PTB (Fig. 8B). The mRNAs produced from pMT2 were spliced. These experiments established that one PTB element is 198 nucleotides and is located between nucleotide positions 3653 and 3850 in the early region of the HPV-16 genome. This sequence is AU rich (AU content, 69%) and contains two penta-A sequences and one penta-U sequence. While the U nucleotides are more dispersed, several tracts of three or more A nucleotides are present in the sequence. High-affinity PTB binding sites (UCUUU) were not present. These results do not exclude the possibility that sequences downstream of nucleotide position 3850 may also respond to PTB.
PTB alleviates the suppression of HPV-16 gene expression by the 198-nucleotide inhibitory element in the absence of splicing.
The induction of splicing from SD3632 by PTB caused high-level expression of spliced L1 mRNAs when the splicing silencers at SA5639 were destroyed and SA5639 could be used efficiently, for example, in pBSpliceM (Fig. 7A and B). In contrast, unspliced L2/L1 mRNAs were induced when SA5639 was suppressed (Fig. 2C and 7B). We proposed that PTB did not target SD3632 directly but targeted adjacent splicing suppressors and that these suppressors also inhibited the expression of unspliced L2/L1 mRNAs by their mere presence when late mRNA splicing was suppressed. Hence, PTB induced the expression of L2/L1 mRNAs rather than spliced L1 mRNAs when the 3′ splice site SA5639 was suppressed (compare pBEL and pBELM [Fig. 2B and D] or pBSplice and pBSpliceM [Fig. 7B]). If this line of reasoning was correct, it predicted that PTB should also induce the expression of the predicted short and unspliced E4* mRNA polyadenylated at pAE in the pBSplice plasmids (Fig. 7A). Similar to the L2/L1 mRNAs induced by PTB from these plasmids, the unspliced E4* mRNA contains an unutilized SD3632 with adjacent sequences and should therefore respond to PTB as the L2/L1 mRNAs. As predicted, the analysis of unspliced E4* mRNA expression from pBSplice with an E4 probe revealed that the unspliced E4* mRNAs were strongly induced by PTB (Fig. 7B, right panel). These results provided further evidence that PTB acts on sequences at SD3632 rather than on SD3632. These results are consistent with a model in which PTB interferes with the function of splicing silencer elements that inhibit splicing as well as the further processing of the unspliced mRNA, rather than targeting SD3632 directly (see Discussion and Fig. 10).
FIG. 10.
Schematic representation of the HPV-16 genome and potential viral mRNAs. With low levels of PTB, SD36332 is suppressed, presumably by cellular factors binding at SD3632. These unutilized splice sites may retain L2/L1 mRNAs in the nucleus. In the presence of high levels of PTB, the suppression of SD3632 is relieved. This effect results in the production of low levels of L2/L1 mRNAs if silencers at SA5639 (61) are active or high levels of L1 mRNAs if the splicing inhibition at SA5639 is alleviated. LCR, long control region; −PTB, without PTB; +PTB, with PTB. Minus signs in circles indicate cellular factors that bind to HPV-16 mRNAs and inhibit viral RNA splicing (44, 61).
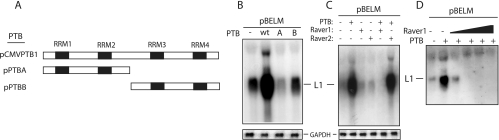
RRM1 and RRM2 of PTB are not sufficient for the induction of HPV-16 late gene expression.
PTB contains four independent RNA binding sites named RNA recognition motif 1 (RRM1) to RRM4 (Fig. 9A) (51). Two deletion mutant forms of PTB, one containing RRM1 and RRM2 and the other containing RRM3 and RRM4, both failed to induce HPV-16 late gene expression (Fig. 9B). We concluded that RRM1 and RRM2 could not induce HPV-16 late gene expression and that RRM3 and RRM4 were required as well.
FIG. 9.
(A) Schematic representation of PTB expressed from plasmids pCMVPTB1, pPTBA, and pPTBB. The RNA binding regions (RRMs) are indicated. (B, C, and D) Northern blots of cytoplasmic RNA extracted from HeLa cells transfected with pBELM in the absence or presence of various PTB-expressing plasmids or the Raver1 or Raver2 plasmid as indicated. Blots were probed with the L1 probe and a GAPDH probe. The position of the L1 mRNA is indicated. +, present; −, absent; wt, wild type.
The PTB binding protein Raver1 does not induce HPV-16 late gene expression.
PTB binds Raver1 and cooperates with Raver1 to inhibit the splicing of cellular mRNAs (19, 23). Raver1 can replace PTB (19, 23). However, in the experiments described here, PTB had a positive effect on HPV-16 late gene expression. We therefore wished to test if PTB acted together with Raver1, and we therefore overexpressed Raver1 in the absence or presence of overexpressed PTB. Raver2 served as a control, as it is expressed primarily in neurons (29). However, the expression of Raver1 alone had no effect on HPV-16 late gene expression from plasmid pBELM (Fig. 9C), and neither did that of Raver2. The coexpression of Raver1 and PTB did not enhance the effect of PTB (Fig. 9C and D). In contrast, Raver1 appeared to have a general inhibitory effect on HPV-16 gene expression (Fig. 9C and D and data not shown). We concluded that PTB enhanced HPV-16 late gene expression independently of Raver1.
DISCUSSION
It has been reported previously by several investigators that PTB binds to the 5′ and/or 3′ UTRs of different RNA viruses (2, 40, 46), of which only hepatitis C virus causes persistent infections that also increase the risk of developing cancer. In most of the RNA viruses, PTB appears to be involved in viral translation and/or replication and is therefore active primarily in the cytoplasm (2, 40). The hepatitis B virus (HBV), on the other hand, replicates in the cell nucleus. It encodes a posttranscriptional regulatory element (PRE) that increases the expression of unspliced HBV mRNAs by facilitating their export from the nucleus. The interaction of PTB with the HBV PRE suggested a role for PTB in the nuclear export of these viral mRNAs (31, 57). PTB has been shown previously to have a shuttling function, which appears to be Crm1 independent (31, 57). Cells that stably overexpress PTB show increased PRE-dependent gene expression (31, 57). HPV-16 and HBV are two examples of DNA tumor viruses that are positively regulated by human PTB. While PTB induces the nuclear export of HBV mRNAs, PTB apparently alleviates splicing suppression in HPV-16 and induces late mRNA splicing. This promotes nuclear export indirectly.
Although initially identified as a splicing factor binding to the polypyrimidine tracts at 3′ splice sites, PTB has also been shown to bind at U-rich elements present at a subset of polyadenylation signals (5, 30). This interaction inhibits polyadenylation. We have previously reported that PTB binds to a U-rich sequence upstream of the HPV-16 early polyadenylation signal (59). However, this U-rich element was not needed for the induction of late gene expression by PTB, nor was the HPV-16 early polyadenylation signal targeted by PTB.
In general, PTB affects gene expression by negatively interfering with exon definition to cause exon skipping or by physically preventing U2AF65 and U2AF35 from binding to RNA (52). PTB has been shown previously to regulate the splicing of several cellular mRNAs. PTB binds to intronic positions on each side of the N1 exon on the c-src mRNA, which results in the skipping of the N1 exon (6, 9, 35). On the Fas mRNA, PTB binds to exon 6 and causes the skipping of the same exon (24). The overexpression of PTB prevents the inclusion of exon 9 on the cystic fibrosis transmembrane conductance regulator mRNA (63), exon 12 on the chicken myosin phosphatase-targeting subunit 1 mRNA (13, 47), exon 9a9′ on the alpha-tropomyosin mRNA (18, 30, 32), and exon 4 on the calcitonin/calcitonin gene-regulated peptide mRNA (34). In contrast, PTB does not cause exon skipping in HPV-16. Here, we show that the overexpression of PTB activates a viral 5′ splice site by counteracting adjacent splicing inhibitory elements. This process is different from the exon-skipping mechanism described above for cellular mRNAs. In addition, previous results have shown that Raver1 (an hnRNP-like protein that interacts with PTB) (44) functions as a corepressor of alpha-tropomyosin splicing (44). As a matter of fact, the recruitment of Raver1 alone, in the absence of PTB, may induce exon skipping (19). However, Raver1 overexpression does not induce HPV-16 late gene expression, nor does Raver1 enhance the induction of HPV-16 late gene expression by PTB (Fig. 9B, C, and D). The mechanism of induction of HPV-16 late gene expression by PTB is different from the exon-skipping mechanism seen with many cellular mRNAs (Fig. 10).
We have not been able to show that PTB binds to the HPV-16 splicing inhibitory sequence directly. However, we have seen that a complex of cellular proteins forms on this viral RNA sequence, and we have detected a 55-kDa protein that binds directly to the RNA sequence (data not shown). Further work is needed to identify the cellular factors that form a complex with the viral splicing inhibitory element and to elucidate how PTB overexpression affects the composition of this complex.
The expression of PTB in relation to cell differentiation, as well as PTB expression in various cancers, has been investigated previously. For example, PTB is expressed in developing mammalian astrocytes, it is absent in mature adult astrocytes, and it is aberrantly elevated in gliomas (8) and neuronal tumors (36). Strong upregulation of PTB expression in tumor cells of glial or primitive neuroectodermal origin suggests the involvement of this protein in cellular transformation (36). The knockdown of PTB has been shown previously to suppress ovarian tumor cell growth and invasiveness in vitro (20). The results of these previous studies all suggest that high levels of PTB are associated with cell proliferation and correlate negatively with cell differentiation. PTB expression in the cervical epithelium as well as that in HPV-16-infected epithelial cells is therefore of interest in relation to the results reported in this study. The role of PTB in the HPV-16 life cycle has not been investigated previously. However, we have performed an immunohistochemical analysis of the expression profile of PTB in the cervical epithelium in the absence or presence of HPV that will be described elsewhere (J. Fay et al., submitted for publication). In general, these results demonstrate that PTB is downregulated in the superficial levels of the cervical epithelium and highly upregulated in cervical cancer cells. These results therefore suggest that HPV-16 late gene expression is induced by the downregulation of a PTB antagonist that inhibits HPV-16 late gene expression in nonterminally differentiated cells, rather than an increase in PTB levels. PTB antagonists may be induced as cervical epithelial cells differentiate. A few PTB antagonists, including RBM4, ETR-3, TIA-1, CELF, and Fox-1, have been identified previously (7, 24, 26, 33, 51). It would be of interest to investigate if these factors counteract the effect of PTB on HPV-16 late gene expression and/or if the expression of any of these factors is altered in response to the terminal differentiation of cervical keratinocytes. On the other hand, one cannot exclude the possibility that the relatively small number of HPV-16-infected cells that actually express viral capsid proteins and/or virions in the cervical epithelium may belong to a rare population of cells that maintain or increase the levels of PTB as they differentiate, thereby increasing the relative levels of PTB and activating HPV-16 late gene expression. A detailed immunohistochemical analysis of HPV-16 proteins and PTB in individual cells in infected cervical epithelia may clarify these connections. It would also be interesting to investigate the effects of PTB on late gene expression by other HPV types, in particular those that display high-level capsid protein expression in a high number of cells in vivo (44). However, in order to determine the exact mechanism of PTB induction of HPV-16 late gene expression, it is necessary to identify proteins in the complex that forms on the splicing element downstream of HPV-16 SD3632 (Fig. 10). It is presumably this complex that is targeted by PTB.
Acknowledgments
We thank Anna Tranell for providing plasmids and for discussion; Joanna Fay and Hilary Cassidy for discussion; the groups of Göran Magnusson, Göran Akusjärvi, and Catharina Svensson for comments and discussions at lab meetings; and project workers Eoin Sweeny, Oskar Skog, and Maria Malloy for contributing to some experiments. We also thank Susanne Illenberger for plasmids encoding Raver1 and Raver2, Natalja Funk for hnRNP Q, Clare Gooding for pCMVPTB1, Oriol Bachs for hnRNP B1/A2, and Douglas Black for hnRNP F.
This research was sponsored by the Swedish Cancer Society, by the Swedish Research Council/Medicine, and by Linneus support from the Swedish Research Council to the Uppsala RNA Research Center.
Footnotes
Published ahead of print on 23 January 2008.
REFERENCES
- 1.Baker, C., and C. Calef. 1997. Maps of papillomavirus mRNA transcripts. In S. R. Billakanti, C. E. Calef, A. D. Farmer, A. L. Halpern, and G. L. Myers (ed.), Human papillomaviruses: a compilation and analysis of nucleic acid and amino acid sequences. Theoretical Biology and Biophysics, Los Alamos National Laboratory, Los Almos, NM.
- 2.Belsham, G. J., and N. Sonenberg. 1996. RNA-protein interactions in regulation of picornavirus RNA translation. Microbiol. Rev. 60499-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carlsson, A., and S. Schwartz. 2000. Inhibitory activity of the human papillomavirus type 1 AU-rich element correlates inversely with the levels of the elav-like HuR protein in the cell cytoplasm. Arch. Virol. 145491-503. [DOI] [PubMed] [Google Scholar]
- 4.Cartegni, L., S. L. Chew, and A. R. Krainer. 2002. Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat. Rev. Genet. 3285-298. [DOI] [PubMed] [Google Scholar]
- 5.Castelo-Branco, P., A. Furger, M. Wollerton, C. Smith, A. Moreira, and N. J. Proudfoot. 2004. Polypyrimidine tract binding protein modulates efficiency of polyadenylation. Mol. Cell. Biol. 244174-4183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chan, R. C., and D. L. Black. 1997. The polypyrimidine tract binding protein binds upstream of neural cell-specific c-src exon N1 to repress the splicing of the intron downstream. Mol. Cell. Biol. 174667-4676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Charlet, B. N., P. Logan, G. Singh, and T. A. Cooper. 2002. Dynamic antagonism between ETR-3 and PTB regulates cell type-specific alternative splicing. Mol. Cell 9649-658. [DOI] [PubMed] [Google Scholar]
- 8.Cheung, H. C., L. J. Corley, G. N. Fuller, I. E. McCutcheon, and G. J. Cote. 2006. Polypyrimidine tract binding protein and Notch1 are independently re-expressed in glioma. Mod. Pathol. 191034-1041. [DOI] [PubMed] [Google Scholar]
- 9.Chou, M. Y., J. G. Underwood, J. Nikolic, M. H. Luu, and D. L. Black. 2000. Multisite RNA binding and release of polypyrimidine tract binding protein during the regulation of c-src neural-specific splicing. Mol. Cell 5949-957. [DOI] [PubMed] [Google Scholar]
- 10.Collier, B., L. Goobar-Larsson, M. Sokolowski, and S. Schwartz. 1998. Translational inhibition in vitro of human papillomavirus type 16 L2 mRNA mediated through interaction with heterogenous ribonucleoprotein K and poly(rC)-binding proteins 1 and 2. J. Biol. Chem. 27322648-22656. [DOI] [PubMed] [Google Scholar]
- 11.Collier, B., D. Öberg, X. Zhao, and S. Schwartz. 2002. Specific inactivation of inhibitory sequences in the 5′ end of the human papillomavirus type 16 L1 open reading frame results in production of high levels of L1 protein in human epithelial cells. J. Virol. 762739-2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cumming, S. A., C. E. Repellin, M. McPhilips, J. C. Redford, J. B. Clements, and S. V. Graham. 2002. The human papillomavirus type 31 untranslated region contains a complex bipartite negative regulatory element. J. Virol. 765993-6003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dirksen, W. P., S. A. Mohamed, and S. A. Fisher. 2003. Splicing of a myosin phosphatase targeting subunit 1 alternative exon is regulated by intronic cis-elements and a novel bipartite exonic enhancer/silencer element. J. Biol. Chem. 2789722-9732. [DOI] [PubMed] [Google Scholar]
- 14.Doorbar, J. 2005. The papillomavirus life cycle. J. Clin. Virol. 32(Suppl. 1)S7-S15. [DOI] [PubMed] [Google Scholar]
- 15.Dreyfuss, G., V. N. Kim, and N. Kataoka. 2002. Messenger-RNA-binding proteins and the messages they carry. Nat. Rev. Mol. Cell Biol. 3195-205. [DOI] [PubMed] [Google Scholar]
- 16.Fehrmann, F., and L. A. Laimins. 2003. Human papillomaviruses: targeting differentiating epithelial cells for malignant transformation. Oncogene 225201-5207. [DOI] [PubMed] [Google Scholar]
- 17.Furth, P. A., and C. C. Baker. 1991. An element in the bovine papillomavirus late 3′ untranslated region reduces polyadenylated cytoplasmic RNA levels. J. Virol. 655806-5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gooding, C., G. C. Roberts, and C. W. Smith. 1998. Role of an inhibitory pyrimidine element and polypyrimidine tract binding protein in repression of a regulated alpha-tropomyosin exon. RNA 485-100. [PMC free article] [PubMed] [Google Scholar]
- 19.Gromak, N., A. Rideau, J. Southby, A. D. Scadden, C. Gooding, S. Huttelmaier, R. H. Singer, and C. W. Smith. 2003. The PTB interacting protein raver1 regulates α-tropomyosin alternative splicing. EMBO J. 226356-6364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.He, X., M. Pool, K. M. Darcy, S. B. Lim, N. Auersperg, J. S. Coon, and W. T. Beck. 2007. Knockdown of polypyrimidine tract-binding protein suppresses ovarian tumor cell growth and invasiveness in vitro. Oncogene 264961-4968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hebner, C. M., and L. A. Laimins. 2006. Human papillomaviruses: basic mechanisms of pathogenesis and oncogenicity. Rev. Med. Virol. 1683-97. [DOI] [PubMed] [Google Scholar]
- 22.Howley, P. M., and D. R. Lowy. 2001. Papillomaviridae and their replication, p. 2197-2229. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Virology, vol. 2. Lippincott/The Williams & Wilkins Co., Philadelphia, PA. [Google Scholar]
- 23.Huttelmaier, S., S. Illenberger, I. Grosheva, M. Rudiger, R. H. Singer, and B. M. Jockusch. 2001. Raver1, a dual compartment protein, is a ligand for PTB/hnRNPI and microfilament attachment proteins. J. Cell Biol. 155775-786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Izquierdo, J. M., N. Majos, S. Bonnal, C. Martinez, R. Castelo, R. Guigo, D. Bilbao, and J. Valcarcel. 2005. Regulation of Fas alternative splicing by antagonistic effects of TIA-1 and PTB on exon definition. Mol. Cell 19475-484. [DOI] [PubMed] [Google Scholar]
- 25.Jeon, S., B. L. Allen-Hoffmann, and P. F. Lambert. 1995. Integration of human papillomavirus type 16 into the human genome correlates with a selective growth advantage of cells. J. Virol. 692989-2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jin, Y., H. Suzuki, S. Maegawa, H. Endo, S. Sugano, K. Hashimoto, K. Yasuda, and K. Inoue. 2003. A vertebrate RNA-binding protein Fox-1 regulates tissue-specific splicing via the pentanucleotide GCAUG. EMBO J. 22905-912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johansson, C., H. Zhao, E. Bajak, F. Granberg, U. Pettersson, and C. Svensson. 2005. Impact of the interaction between adenovirus E1A and CtBP on host cell gene expression. Virus Res. 11351-63. [DOI] [PubMed] [Google Scholar]
- 28.Kennedy, I. M., J. K. Haddow, and J. B. Clements. 1990. Analysis of human papillomavirus type 16 late mRNA 3′ processing signals in vitro and in vivo. J. Virol. 641825-1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kleinhenz, B., M. Fabienke, S. Swiniarski, N. Wittenmayer, J. Kirsch, B. M. Jockusch, H. H. Arnold, and S. Illenberger. 2005. Raver2, a new member of the hnRNP family. FEBS Lett. 5794254-4258. [DOI] [PubMed] [Google Scholar]
- 30.Le Sommer, C., M. Lesimple, A. Mereau, S. Menoret, M. R. Allo, and S. Hardy. 2005. PTB regulates the processing of a 3′-terminal exon by repressing both splicing and polyadenylation. Mol. Cell. Biol. 259595-9607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li, B., and T. S. Yen. 2002. Characterization of the nuclear export signal of polypyrimidine tract-binding protein. J. Biol. Chem. 27710306-10314. [DOI] [PubMed] [Google Scholar]
- 32.Lin, C. H., and J. G. Patton. 1995. Regulation of alternative 3′ splice site selection by constitutive splicing factors. RNA 1234-245. [PMC free article] [PubMed] [Google Scholar]
- 33.Lin, J. C., and W. Y. Tarn. 2005. Exon selection in α-tropomyosin mRNA is regulated by the antagonistic action of RBM4 and PTB. Mol. Cell. Biol. 2510111-10121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lou, H., D. M. Helfman, R. F. Gagel, and S. M. Berget. 1999. Polypyrimidine tract-binding protein positively regulates inclusion of an alternative 3′-terminal exon. Mol. Cell. Biol. 1978-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Markovtsov, V., J. M. Nikolic, J. A. Goldman, C. W. Turck, M. Y. Chou, and D. L. Black. 2000. Cooperative assembly of an hnRNP complex induced by a tissue-specific homolog of polypyrimidine tract binding protein. Mol. Cell. Biol. 207463-7479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McCutcheon, I. E., S. J. Hentschel, G. N. Fuller, W. Jin, and G. J. Cote. 2004. Expression of the splicing regulator polypyrimidine tract-binding protein in normal and neoplastic brain. Neuro-oncol. 69-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meyers, C., and L. A. Laimins. 1994. In vitro systems for the study and propagation of human papillomaviruses. Curr. Top. Microbiol. Immunol. 186199-215. [DOI] [PubMed] [Google Scholar]
- 38.Milligan, S. G., T. Veerapraditsin, B. Ahamet, S. Mole, and S. V. Graham. 2007. Analysis of novel human papillomavirus type 16 late mRNAs in differentiated W12 cervical epithelial cells. Virology 360172-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mole, S., T. Veerapraditsin, M. G. McPhilips, and S. V. Graham. 2006. Regulation of splicing-associated SR proteins by HPV-16. Biochem. Soc. Trans. 341145-1147. [DOI] [PubMed] [Google Scholar]
- 40.Nomoto, A., K. Tsukiyama-Kohara, and M. Kohara. 1995. Mechanism of translation initiation on hepatitis C virus RNA. Princess Takamatsu Symp. 25111-119. [PubMed] [Google Scholar]
- 41.Oberg, D., B. Collier, X. Zhao, and S. Schwartz. 2005. A downstream polyadenylation element in human papillomavirus type 16 L2 encodes multiple GGG motifs and interacts with hnRNP H. J. Virol. 799254-9269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Oberg, D., B. Collier, X. Zhao, and S. Schwartz. 2003. Mutational inactivation of two distinct negative RNA elements in the human papillomavirus type 16 L2 coding region induces production of high levels of L2 in human cells. J. Virol. 7711674-11684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ozbun, M., S. K. Campos, and J. L. Smith. 2007. The early events of human papillomavirus infections: implications for regulation of cell tropism and host range, p. 69-122. In B. Norrild (ed.), Human papillomavirus gene regulation and transformation. Transworld Research Network, Trivandrum, India.
- 44.Rush, M., X. Zhao, and S. Schwartz. 2005. A splicing enhancer in the E4 coding region of human papillomavirus type 16 is required for early mRNA splicing and polyadenylation as well as inhibition of premature late gene expression. J. Virol. 7912002-12015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schwartz, S., M. Somberg, X. Zhao, M. Rush, D. Öberg, H. Cassidy, J. Fay, and H. Lambkin. 2007. Regulation of HPV gene splicing and expression, p. 47-67. In B. Norrild (ed.), Human papillomavirus gene regulation and transformation. Transworld Research Network, Trivandrum, India.
- 46.Shi, S. T., and M. M. Lai. 2005. Viral and cellular proteins involved in coronavirus replication. Curr. Top. Microbiol. Immunol. 28795-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shukla, S., F. Del Gatto-Konczak, R. Breathnach, and S. A. Fisher. 2005. Competition of PTB with TIA proteins for binding to a U-rich cis-element determines tissue-specific splicing of the myosin phosphatase targeting subunit 1. RNA 111725-1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sokolowski, M., H. Furneaux, and S. Schwartz. 1999. The inhibitory activity of the AU-rich RNA element in the human papillomavirus type 1 late 3′ untranslated region correlates with its affinity for the elav-like HuR protein. J. Virol. 731080-1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sokolowski, M., W. Tan, M. Jellne, and S. Schwartz. 1998. mRNA instability elements in the human papillomavirus type 16 L2 coding region. J. Virol. 721504-1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sokolowski, M., C. Zhao, W. Tan, and S. Schwartz. 1997. AU-rich mRNA instability elements on human papillomavirus type 1 late mRNAs and c-fos mRNAs interact with the same cellular factors. Oncogene 152303-2319. [DOI] [PubMed] [Google Scholar]
- 51.Spellman, R., A. Rideau, A. Matlin, C. Gooding, F. Robinson, N. McGlincy, S. N. Grellscheid, J. Southby, M. Wollerton, and C. W. Smith. 2005. Regulation of alternative splicing by PTB and associated factors. Biochem. Soc. Trans. 33457-460. [DOI] [PubMed] [Google Scholar]
- 52.Spellman, R., and C. W. Smith. 2006. Novel modes of splicing repression by PTB. Trends Biochem. Sci. 3173-76. [DOI] [PubMed] [Google Scholar]
- 53.Tan, W., B. K. Felber, A. S. Zolotukhin, G. N. Pavlakis, and S. Schwartz. 1995. Efficient expression of the human papillomavirus type 16 L1 protein in epithelial cells by using Rev and the Rev-responsive element of human immunodeficiency virus or the cis-acting transactivation element of simian retrovirus type 1. J. Virol. 695607-5620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tan, W., and S. Schwartz. 1995. The Rev protein of human immunodeficiency virus type 1 counteracts the effect of an AU-rich negative element in the human papillomavirus type 1 late 3′ untranslated region. J. Virol. 692932-2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Terhune, S. S., C. Milcarek, and L. A. Laimins. 1999. Regulation of human papillomavirus 31 polyadenylation during the differentiation-dependent life cycle. J. Virol. 737185-7192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wiklund, L., M. Sokolowski, A. Carlsson, M. Rush, and S. Schwartz. 2002. Inhibition of translation by UAUUUAU and UAUUUUUAU motifs of the AU-rich RNA instability in the HPV-1 late 3′ untranslated region. J. Biol. Chem. 27740462-40471. [DOI] [PubMed] [Google Scholar]
- 57.Zang, W. Q., B. Li, P. Y. Huang, M. M. Lai, and T. S. Yen. 2001. Role of polypyrimidine tract binding protein in the function of the hepatitis B virus posttranscriptional regulatory element. J. Virol. 7510779-10786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhao, X., J. Fay, H. Lambkin, and S. Schwartz. 2007. Identification of a 17-nucleotide splicing enhancer in HPV-16 L1 that counteracts the effect of multiple hnRNP A1-binding splicing silencers. Virology 369351-363. [DOI] [PubMed] [Google Scholar]
- 59.Zhao, X., D. Öberg, M. Rush, J. Fay, H. Lambkin, and S. Schwartz. 2005. A 57-nucleotide upstream early polyadenylation element in human papillomavirus type 16 interacts with hFIP1, CstF-64, hnRNP C1/C2, and polypyrimidine tract binding protein. J. Virol. 794270-4288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhao, X., M. Rush, A. Carlsson, and S. Schwartz. 2007. The presence of inhibitory RNA elements in the late 3′-untranslated region is a conserved property of human papillomaviruses. Virus Res. 125135-144. [DOI] [PubMed] [Google Scholar]
- 61.Zhao, X., M. Rush, and S. Schwartz. 2004. Identification of an hnRNP A1-dependent splicing silencer in the human papillomavirus type 16 L1 coding region that prevents premature expression of the late L1 gene. J. Virol. 7810888-10905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zheng, Z. M., and C. C. Baker. 2006. Papillomavirus genome structure, expression, and posttranscriptional regulation. Front. Biosci. 112286-2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zuccato, E., E. Buratti, C. Stuani, F. E. Baralle, and F. Pagani. 2004. An intronic polypyrimidine-rich element downstream of the donor site modulates cystic fibrosis transmembrane conductance regulator exon 9 alternative splicing. J. Biol. Chem. 27916980-16988. [DOI] [PubMed] [Google Scholar]
- 64.zur Hausen, H. 2002. Papillomaviruses and cancer: from basic studies to clinical application. Nat. Rev. Cancer 2342-350. [DOI] [PubMed] [Google Scholar]