Abstract
Group A rotavirus classification is currently based on the molecular properties of the two outer layer proteins, VP7 and VP4, and the middle layer protein, VP6. As reassortment of all the 11 rotavirus gene segments plays a key role in generating rotavirus diversity in nature, a classification system that is based on all the rotavirus gene segments is desirable for determining which genes influence rotavirus host range restriction, replication, and virulence, as well as for studying rotavirus epidemiology and evolution. Toward establishing such a classification system, gene sequences encoding VP1 to VP3, VP6, and NSP1 to NSP5 were determined for human and animal rotavirus strains belonging to different G and P genotypes in addition to those available in databases, and they were used to define phylogenetic relationships among all rotavirus genes. Based on these phylogenetic analyses, appropriate identity cutoff values were determined for each gene. For the VP4 gene, a nucleotide identity cutoff value of 80% completely correlated with the 27 established P genotypes. For the VP7 gene, a nucleotide identity cutoff value of 80% largely coincided with the established G genotypes but identified four additional distinct genotypes comprised of murine or avian rotavirus strains. Phylogenetic analyses of the VP1 to VP3, VP6, and NSP1 to NSP5 genes showed the existence of 4, 5, 6, 11, 14, 5, 7, 11, and 6 genotypes, respectively, based on nucleotide identity cutoff values of 83%, 84%, 81%, 85%, 79%, 85%, 85%, 85%, and 91%, respectively. In accordance with these data, a revised nomenclature of rotavirus strains is proposed. The novel classification system allows the identification of (i) distinct genotypes, which probably followed separate evolutionary paths; (ii) interspecies transmissions and a plethora of reassortment events; and (iii) certain gene constellations that revealed (a) a common origin between human Wa-like rotavirus strains and porcine rotavirus strains and (b) a common origin between human DS-1-like rotavirus strains and bovine rotaviruses. These close evolutionary links between human and animal rotaviruses emphasize the need for close simultaneous monitoring of rotaviruses in animals and humans.
Group A rotaviruses are major pathogens associated with acute gastroenteritis in humans and animals. Rotaviruses form a genus in the Reoviridae family and are characterized by a segmented double-stranded RNA genome (3, 16). The rotavirus genome is enclosed in a triple-layered icosahedral capsid and consists of 11 segments, encoding six viral structural proteins (VP1 to VP4, VP6, and VP7) and six nonstructural proteins (NSP1 to NSP6) (16). Each genome segment, with the exception of gene 11 that encodes two proteins (NSP5 and NSP6), codes for a single viral protein. The inner layer of the rotavirus virion is mainly composed of VP2, which encases VP1, the viral RNA-dependent RNA polymerase, and VP3, the viral capping enzyme (16). The middle layer of the virion is composed entirely of VP6 trimers, which determine rotavirus groups. For group A rotaviruses four different subgroups (SGs) have been recognized, based on the immunoreactivity of SG-specific monoclonal antibodies (MAbs) (19). VP7 and VP4 comprise the outer layer of the virion and are the basis of a binary classification system defining G types (glycoprotein) and P types (protease sensitive), respectively. VP7 and VP4 are capable of independently eliciting neutralizing antibodies, initially used to define rotavirus G and P serotypes (16). The nonstructural proteins are variously involved in replication (NSP1 to NSP3, NSP5, and NSP6) and morphogenesis (NSP4), and with the exception of NSP4, all interact with nucleic acids (16).
Due to the lack of proper immunological reagents and the increasing ease of sequencing, serotyping is being complemented with genotyping, which is based on identities between sequences of cognate rotavirus gene segments. So far, 15 G genotypes (14 G serotypes) have been identified, and out of 27 different P genotypes, 14 P serotypes (1A, 1B, and 2 to 14) have been identified with available VP4-specific antibodies (5, 30, 34-37, 42, 62, 68). Traditionally, a cutoff value of 89% VP7 amino acid sequence identity has been used to classify G genotypes, yielding a nearly complete concordance with the different G serotypes (16, 57). In contrast, the 89% amino acid identity cutoff value for VP4, established by Gorziglia and colleagues (18), does not result in an absolute concordance between different P genotypes and P serotypes. Specifically, P serotypes have not been defined for approximately half of the P genotypes, which are designated by an Arabic numeral between square brackets (16). Molecular analyses of VP6 is limited to only a 379-bp fragment of VP6, which results in two broad genogroups that do not correlate with the SG specificities (27). The classification of rotavirus nonstructural proteins is limited to NSP4, and six genotypes (A to F) have been recognized based on clustering patterns in amino-acid-based phylogenetic dendrograms (9, 22, 26, 46). To date, no classification for VP1 to VP3, NSP1 to NSP3, or NSP5 has been described.
RNA-RNA hybridization assays have been used to analyze and compare complete genomes of group A rotaviruses. For human strains, three genogroups have been established: two major genogroups represented by the reference strains Wa and DS-1 and one minor genogroup represented by reference strain AU-1 (51). Similar genogroups also have been established for several animal rotavirus strains although they are more complex (50). The hybridization technique has proven to be useful to investigate possible reassortment events between human strains belonging to different genogroups (49, 76) or between human and animal strains (48). Currently, rotavirus strains are being analyzed and compared to one another by partial or complete sequencing of all 11 gene segments as this approach allows direct determination of genetic relationships (38-40, 63, 67). In addition, sequencing of rotavirus genomes is critical to the understanding of phylogenetic analyses and to the elucidation of the patterns of virus evolution. One method that is used to study typical evolutionary distances between virus strains is pairwise sequence identity profiles (2). Specifically, this method illustrates virus genotypes as well-resolved peaks, providing the basis for classification systems. To properly study the evolution of rotaviruses, the establishment of a classification system in which individual genes fall into defined clusters/genotypes based on reliable percentage identity cutoff values is fundamental. Such a classification system could be an important tool to elucidate how rotaviruses evolve over time. In addition, for viruses with segmented genomes, this type of classification system could also be used to determine whether or not certain rotavirus genes cosegregate during reassortment events (gene linkage) or whether certain gene constellations play a role in rotavirus host range restriction or virulence.
In this study, phylogenetic analyses and pairwise sequence identity profiles were constructed for each of the gene segments of group A rotaviruses to develop a uniform classification and nomenclature system for all 11 rotavirus genome segments in a similar fashion to that established for VP4 and VP7. Based on percentage identity cutoff values, this novel classification system illustrates phylogenetic relationships of all 11 rotavirus genome segments and allows the identification of distinct genotypes (which likely followed separate evolutionary paths) and reassortment events. In addition, the comprehensive classification system revealed genetic relationships among rotaviruses from different host species, including evidence that human rotaviruses belonging to the Wa-like genogroup have a common origin with porcine rotaviruses while those belonging to the DS-1-like genogroup have a common origin with bovine rotaviruses.
MATERIALS AND METHODS
Rotavirus strains.
The human rotavirus strains Wa (G1P1A[8]), D (G1P1A[8]), DS-1 (G2P1B[4]), S2 (G2P1B[4]), AU-1 (G3P3[9]), YO (G3P1A[8]), P (G3P1A[8]), Hosokawa (G4P1A[8]), ST3 (G4P2A[6]), IAL28 (G5P1A[8]), SE584 (G6P3[9]), 69 M (G8P4[10]), WI61 (G9P1A[8]), A64 (G10P11[14]), and L26 (G12P1B[4]); the bovine strains NCDV (G6P6[1]), BRV033 (G6P6[1]), and WC3 (G6P7[5]); and the porcine strains A131 (G3P9[7]), A253 (G11P9[7]), and A411 (G3P9[7]) are standard strains that have been adapted to growth in cell culture (serotypes are given in parentheses). The human rotavirus strain B3458 (G9P1A[8]) was isolated in Belgium during the 2003 to 2004 season (unpublished data).
RNA extraction, reverse transcription-PCR (RT-PCR), and sequencing of human rotavirus strains Wa, DS-1, S2, AU-1, YO, Hosokawa, and B3458 and bovine rotavirus strain NCDV.
For each strain 140 μl of cell culture supernatant (or fecal suspension in the case of strain B3458) was used to extract viral RNA using a QIAamp Viral RNA mini kit (Qiagen/Westburg, Leusden, The Netherlands) according to the manufacturer's instructions.
Ten microliters of extracted RNA was denatured at 97°C for 3 min and RT-PCR was carried out using a Qiagen One Step RT-PCR kit (Qiagen/Westburg). The primers used are shown in Data S1 in the supplemental material. The RT-PCR was carried out with an initial reverse transcription step at 45°C for 30 min; PCR activation was at 95°C for 15 min, followed by 40 cycles of amplification (45 s at 94°C, 45 s at 45°C, and 6 min at 68°C), with a final extension of 7 min at 72°C in a GeneAmp PCR System 9700 thermal cycler (Applied Biosystems, Foster City, CA).
The PCR amplicons were purified with a QIA Quick PCR purification kit (Qiagen/Westburg) and sequenced using the dideoxynucleotide chain termination method with an ABI Prism BigDye Terminator Cycle Sequencing Reaction kit (Perkin-Elmer) on an ABI Prism 3100 automated sequencer (Perkin-Elmer). The sequencing was performed with the forward and reverse primers used for the RT-PCR. In addition, primer walking sequencing was performed to cover the complete sequence of a segment on both strands.
Determination of the 5′ and 3′ terminal sequences of human rotavirus strains Wa, DS-1, S2, AU-1, YO, Hosokawa, and B3458 and bovine rotavirus strain NCDV.
To obtain the complete nucleotide sequence of each segment, the 5′ and 3′ terminal sequences of the 11 gene segments were determined using a modified version of the single-primer amplification method, as described previously (38).
RNA and protein sequence analyses of human rotavirus strains Wa, DS-1, S2, AU-1, YO, Hosokawa, and B3458 and bovine rotavirus strain NCDV.
The chromatogram sequencing files were analyzed using Chromas 2.23 (Technelysium, Queensland, Australia), and contigs were prepared using SeqMan II (DNAstar, Madison, WI). Nucleotide and protein sequence identity searches were performed using the National Center for Biotechnology Information (National Institutes of Health, Bethesda, MD) BLAST (Basic Local Alignment Search Tool) server on GenBank database release 154.0 (1). Multiple sequence alignments were calculated using ClustalX, version 1.81 (71). Sequences were manually edited in the GeneDoc, version 2.6.002, alignment editor (55).
In vitro transcription, PCR, cloning, and sequencing of the bovine rotavirus strains BRV033 and WC3 and porcine rotavirus strains A131, A411, and A253.
Single-stranded RNA transcripts were prepared from purified double-layered BRV033, WC3, A131, A411, or A253 rotavirus particles as described previously (10). Briefly, reverse transcription was used to generate gene 1, 2, 3, 5, 7, 8, or 11 complementary cDNA(s) using the corresponding primers complementary to the 3′ end as indicated in Data S1 in the supplemental material. Immediately after synthesis of the first cDNA strand, samples were heated to 95°C for 10 min, and PCR was carried out with the corresponding 3′ end primer and a primer complementary to the 5′ end (see Data S1 in the supplemental material); PCR was carried out with 40 cycles of amplification (55 s at 94°C, 1 min at 50°C, and 7 min at 68°C) and a final extension of 10 min at 72°C. Amplified DNA was immediately cloned into the TA3pCR2.1 vector (Invitrogen Corp., San Diego, CA) according to the manufacturer's instructions. For accuracy in sequence determination, two to four independent clones of VP1 to VP3, NSP1 to NSP3, or NSP5/NSP6 from individual PCRs using strains BRV033, WC3, A131, A411, and A253 were sequenced by the dideoxynucleotide chain termination method. Confirmation of the DNA sequence was performed by sequencing both DNA strands of each of the different clones using the M13 sense and antisense standard primers and additional primers (available upon request). The 5′ and 3′ end sequences of the clones are identical or complementary to the corresponding 5′ or 3′ end PCR primers, respectively.
RNA extraction, RT-PCR, and sequencing of the genomes of human rotavirus strains D, DS-1, P, ST3, IAL28, SE584, 69 M, W161, A64, and L26 and primate rotavirus strains TUCH and RRV were performed as described elsewhere (E. Heiman, S. M. McDonald, M. Barro, Z. F. Taraporewala, T. Bar-Magen, and J. T. Patton, unpublished data; also H. Yang, Z. F. Taraporewala, K. Kerstak, and J. T. Patton, unpublished data).
Phylogenetic analysis.
Phylogenetic and molecular evolutionary analyses were conducted both at the nucleotide and amino acid levels using MEGA, version 2.1, software. Genetic distances were calculated using the Poisson correction parameter at the amino acid level and the Kimura-2 correction parameter at the nucleotide level. The dendrograms were constructed using the neighbor-joining method (33).
Construction of pairwise identity frequency graphs.
To obtain suitable cutoff values for evolution-based classification of each rotavirus genome segment, the percentages of nucleotide and amino acid identities between the complete open reading frames of a large number of completely sequenced rotavirus genome segments in GenBank as well as the sequences determined in this study were calculated using the pairwise distances program of the MEGA program, version 2.1 (33). The use of pairwise identity frequency graphs for the classification of viruses has been approved by the International Committee on Taxonomy of Viruses (2), and the pairwise identity frequency graphs were constructed by plotting all the calculated pairwise identities in a graph with the percentage identities in the abscissa (x axis) and the frequency of each of the calculated pairwise identities in the ordinate (y axis).
Determination of the most appropriate nucleotide/amino acid identity cutoff percentages.
For each phylogenetic dendrogram, there were several alternatives to assign certain clusters as genotypes. To determine the most appropriate cutoff value for classification, several possible alternatives were investigated. For each alternative, the nucleotide/amino acid identities between strains belonging to the same cluster/genotype were designated the intragenotype identities, while the nucleotide/amino acid identities between strains belonging to different clusters/genotypes were designated the intergenotype identities. In the ideal case, the inter- and intragenotype identities should not overlap, and the cutoff percentage is defined as the percentage separating the inter- and the intragenotype identities. In practice, the inter- and intragenotype identities do partially overlap in some cases, and the most appropriate percentage cutoff value was chosen as the percentage at which the ratio of the intergenotype identity and the intragenotype identity (intergenotype identity/intragenotype identity) dropped below 1.
Nucleotide sequence accession numbers.
The nucleotide sequence data reported in this paper were deposited in GenBank under the accession numbers found in Data S2 in the supplemental material.
RESULTS
The nucleotide and deduced amino acid sequences of gene segments of several human rotavirus strains (Wa, DS-1, AU-1, YO, S2, Hosokawa, B3458, D, P, ST3, IAL28, SE584, 69 M, WI61, A64, and L26); the bovine rotavirus strains NCDV, WC3, and BRV033; and the porcine rotavirus strains A131, A253, and A411 were determined (see Data S2 in the supplemental material) (Heiman et al., unpublished; also Yang et al., unpublished). Multiple sequence alignments for the VP1 to 4, VP6, VP7, and NSP1 to NSP5 cognate gene segments were conducted using both the sequences available in GenBank databases and the sequences determined in this study. Nucleotide and amino acid sequence-based identity frequency graphs were constructed according to the different options to determine the most appropriate percentage identity cutoff values. Subsequently, the deduced complete amino acid sequences and the coding regions of the nucleotide alignments were used for further analyses. Based on the alignments, several phylogenetic dendrograms were constructed. For VP4, VP7, and NSP4, the established genotyping system was reevaluated, and for VP1 to VP3, VP6, NSP1 to NSP3, and NSP5, several alternative options to classify them into appropriate clusters/genotypes were investigated.
The results of the analyses conducted both at the nucleotide and amino acid levels were in complete accordance, resulting in identical genotypes/clusters (data not shown). However, there were several reasons favoring the use of the nucleotide-based classification over the amino acid-based classification. (i) The resolution of the different “peaks” in the nucleotide-based frequency-identity graphs was generally much better than that in the amino acid-based frequency-identity graphs, resulting in little or no overlap between the peaks of the inter- and intragenotype identities, which resulted in a limited number of deviations from the proposed cutoff values. (ii) Since several of the rotavirus proteins are rather conserved (VP1, VP2, VP6, and NSP5), the resulting cutoff values based on amino acid identity were very high (up to 96%). The high level of relatedness of these proteins also makes it difficult to classify and differentiate new strains based on (partial) amino acid sequences, since there is only a very limited amount of sequence diversity. (iii) The phylogenetic dendrograms and the different genetic clusters which were used to define the different genotypes were supported by higher bootstrap values in the nucleotide-based phylogenetic dendrograms than in the amino acid-based phylogenetic dendrograms. (iv) The genetic diversity observed between different rotavirus strains was more evenly distributed across the gene at the nucleotide level than at the amino acid level. This implies that genotyping of a partial gene sequence, using the proposed cutoff values, would be much more reliable at the nucleotide level than at the amino acid level.
In addition to the use of pairwise distances for the construction of the frequency identity graphs, several other more sophisticated models to estimate the genetic distance between homologous sequences, such as Jukes-Cantor, Kimura-2, and Tamura-Nei (54), were evaluated. These different models yielded slightly lower cutoff percentage values, but, most importantly, the resulting phylogenetic dendrograms demonstrated identical sets of genotypes for all 11 genes, regardless of which of the mathematical genetic distance models was used (data not shown).
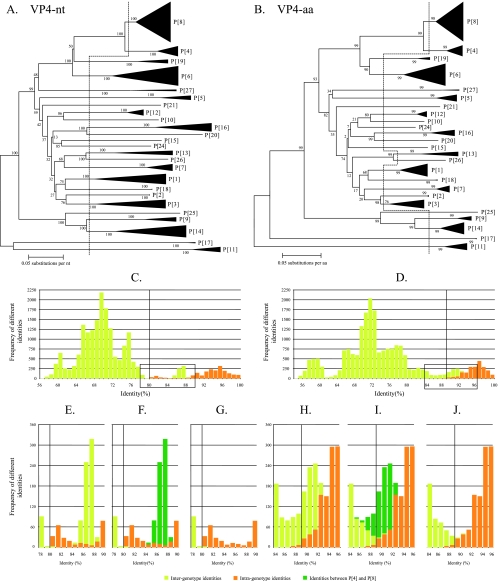
VP4.
More than 190 complete nucleotide and deduced amino acid sequences for VP4 were used in the analysis. The dendrograms based on the nucleotide and amino acid sequences are shown in Fig. 1A and B, respectively. The vertical dashed lines represent the division into different P genotypes as currently established, and the two resulting identity frequency graphs are shown in Fig. 1C and D. From these graphs, it was not obvious how to determine accurate nucleotide cutoff values or how to confirm that the established cutoff value of 89% at the amino acid level was appropriate since there was considerable overlap between the inter- and intragenotype identities (Fig. 1C and D). To further analyze the overlap between the inter- and intragenotypic identities, details of the boxed areas in Fig. 1C and D are shown magnified in Fig. 1E and H. In addition, Fig. 1F and I show the intergenotype identities between rotaviruses belonging to genotypes P[4] and P[8]; then in Fig. 1G and J, the identities between P[4] and P[8] were omitted to fully interpret the analyses. Figure 1G and J clearly show that an 80% cutoff value at the nucleotide level and the 89% cutoff value at the amino acid level, as currently established, were accurately suited to distinguish between different VP4 genotypes as they may have evolved. Among all P genotypes evaluated, only the identities between the P[4] and P[8] genotypes were not separated by the 80% nucleotide cutoff value (range, 84 to 89% nucleotide identity) or the 89% amino acid cutoff values (range, 85 to 93% amino acid identity), confirming the observation that these P genotypes are closely related. This close relationship between P[4] and P[8] should be kept in mind when newly sequenced VP4 gene segments are analyzed for their P genotype.
FIG. 1.
Phylogenetic dendrograms of VP4 at the nucleotide (nt) level (A) and the amino acid (aa) level (B) Bootstrap values (1,000 replicates) are shown. Certain clusters are replaced by triangles, in which the height of the triangle represents the number of sequences, and the width represents the genetic diversity inside that cluster. The dashed lines indicate the current division into P genotypes. Panels C and D show the respective identity frequency graphs. Panels E, F and G show details of the boxed regions of panel C, and panels H, I, and J show details of the boxed regions of panel D. In panels F and I, the identities between rotavirus strains belonging to the P[4] and P[8] genotypes are shown. In panels G and J, the identities between rotavirus strains belonging to the P[4] and P[8] genotypes are omitted to show that the 80% nucleotide and 89% amino acid cutoff values are the most suited cutoff values.
When the analyses were repeated for the VP8* region only (nucleotides 10 to 887 of VP4), a nearly identical result was found, with a 79% amino acid cutoff percentage (instead of 80%) completely separating the established 27 P genotypes, except P[4] and P[8], for which the identities ranged from 83% to 88% (data not shown).
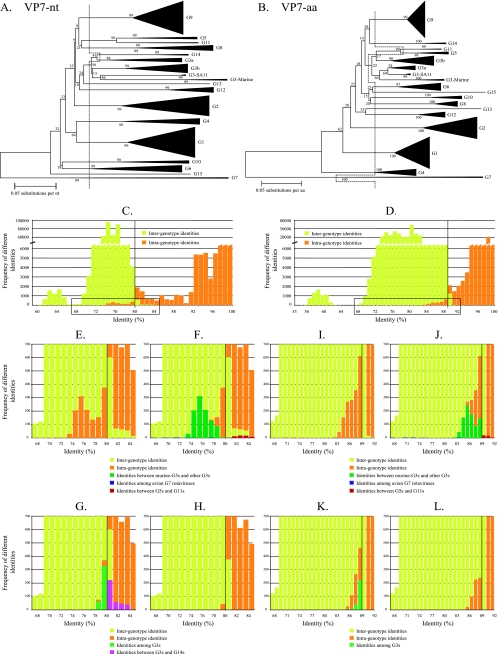
VP7.
More than 1,000 VP7-encoding nucleotide and deduced amino acid sequences, representing the 15 established G genotypes, were used for the analysis. The recently described equine strain ERV99, identified as a new G genotype (21), was revealed to be a G6 rotavirus strain with a high level of identity to NCDV (data not shown). The nucleotide- and amino acid-based phylogenetic dendrograms are shown in Fig. 2A and B, respectively. Identity frequency graphs were constructed according to the 15 G genotypes, as they are currently defined, and the 89% amino acid cutoff value is shown as a vertical line in Fig. 2B. At the nucleotide level, an 80% cutoff value seemed the most appropriate based on the data shown in Fig. 2C. Details of Fig. 2C and D (boxed regions) are depicted in Fig. 2E and I and show that a considerable degree of intraspecies diversity was found below the nucleotide and amino acid cutoff percentages, and a lower degree of interspecies identity was observed above the indicated cutoff percentages. Figure 2F and J show that a large proportion of the intragenotype identities, below the proposed cutoff values, were the result of identities between murine rotaviruses and G3 rotaviruses isolated from other species. Their sequence identities ranged from 73% to 79% (Fig. 2F, dark green bars) at the nucleotide level, completely below the 80% nucleotide identity cutoff value, and ranged from 80% to 92% at the amino acid level, which is almost entirely below the 89% amino acid identity cutoff value. This result strongly suggests that the murine rotavirus strains should be assigned to a different G genotype, tentatively designated G16. The detailed phylogenetic relationship between G3 rotavirus strains and the murine rotavirus strains is available in Data S3 in the supplemental material.
FIG. 2.
Phylogenetic dendrograms of VP7 at the nucleotide (nt) level (A) and the amino acid (aa) level (B). Bootstrap values (1,000 replicates) are shown. Certain clusters are replaced by triangles, in which the height of the triangle represents the number of sequences, and the width represents the genetic diversity inside that cluster. The dashed lines indicate the current division into G genotypes. Panels C and D show the respective identity frequency graphs. Panels E, F, G, and H show details of the boxed regions of panel C, and panels I, J, K, and L show details of the boxed regions of panel D. In panels F and J, identities between rotavirus strains belonging to the murine G3 strains and the remaining G3 strains (dark green), between rotavirus strains belonging to the G5 and G11 strains (red), and among rotavirus strains belonging to the G7 genotype (dark blue) are shown. In panels G and K identities among rotavirus strains belonging to the G3 genotype (light green) and between rotavirus strains belonging to the G3 and G14 genotypes (turquoise) are shown.
Likewise, it is also clear in Fig. 2F and J (dark blue bars) that the identities among the avian rotavirus strains, all previously classified as genotype G7, ranged from 67% to 99% at both nucleotide and amino acid levels. According to the newly proposed 80% nucleotide and 89% amino acid identity cutoff values, there should be four different G genotypes comprised of avian or avian-like rotavirus strains. Since the avian rotavirus strain Ch-2 (56) was the first G7 rotavirus to be characterized and sequenced, it should remain as the prototype genotype G7 avian strain, along with the avian strain Ty-3 (>99% nucleotide identity with Ch-2). Avian rotavirus strain Ty-1 (69 to 78% nucleotide identity with other avian strain) can tentatively be assigned to a new genotype, G17 (32). The pigeon rotavirus strain PO-13 and the avian-like bovine rotavirus strain 993/83 (65) (>95% nucleotide identity with PO-13) may be assigned to new genotype G18, while avian strain Ch-1 (47) (67 to 74% nucleotide identity with other avian strain) may be assigned to new genotype G19. The detailed phylogenetic relationship between the avian rotavirus strains is available in Data S4 in the supplemental material.
Some of the intergenotype identities above the cutoff values (Fig. 2F and J) were caused by identities between G5 and G11 rotavirus strains (red bars), ranging from 80% to 85% at the nucleotide level (completely above the 80% cutoff value) and from 82% to 92% at the amino acid level (partially above the 89% cutoff value). This suggests that strains assigned to G5 and G11 could be considered a single genotype, especially when analyzed at the nucleotide level. The detailed phylogenetic relationship between G5 and G11 rotavirus strains is available in Data S5 in the supplemental material. Figure 2G and K show that most of the remaining intragenotype identities below the cutoff values were due to identities between rotaviruses belonging to the G3 genotype (light green bars). Also the remaining intergenotype identities above the cutoff values were mostly caused by identities between G3 and G14 rotaviruses (turquoise bars). Figure 2H and L show that, after the omission of the above-mentioned identities, an 80% identity cutoff value at the nucleotide level and an 89% identity cutoff value at the amino acid level were the most appropriate. These values allowed the identities between strains belonging to the same genotypes to be mostly above the proposed cutoff values and identities between strains that belong to different genotypes to be mostly below the proposed cutoff values. The very last remaining intergenotype identities above 89% (Fig. 2L) were due to identities between rotavirus strains belonging to the G3 and G9 genotypes. Finally, the remaining intragenotype identities below 89% (Fig. 2L) were due to a high level of intragenotype diversity among rotavirus strains belonging to genotypes G1, G2, G4, and G6.
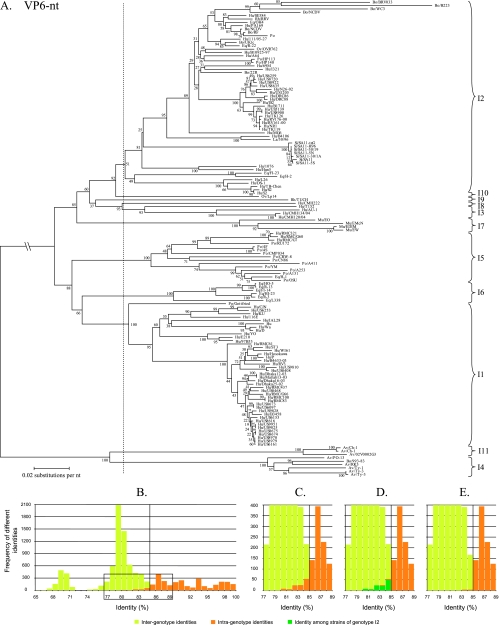
VP6.
A phylogenetic dendrogram constructed with 142 VP6 nucleotide sequences is shown in Fig. 3A. Several alternatives were tested to divide the dendrogram into appropriate genotypes, but only the best alternative is shown in Fig. 3A (vertical dashed line). The corresponding identity frequency graph (Fig. 3B) suggested that 85% was the most appropriate percentage identity cutoff value. A detailed view of the boxed region of Fig. 3B is shown in Fig. 3C. A total of 10 VP6 genotypes were determined and tentatively designated I (for intermediate capsid shell) genotypes, which contrasts with the two broad genogroups identified when only a 379-bp fragment of VP6 was analyzed (27). Further analyses of the origin of the intragenotype identities below the proposed cutoff value demonstrated that the greatest contribution to these identities (Fig. 3D, dark green bars) was the identity among strains assigned to genotype I2. The few remaining intragenotype identities below the 85% cutoff value (Fig. 3E) were caused by identities between the porcine rotavirus strain A411 and other rotavirus strains belonging to the VP6 genotype I5.
FIG. 3.
Phylogenetic dendrogram of VP6 at the nucleotide (nt) level (A). Bootstrap values (2,000 replicates) are shown. Designations of species of origin are as follows: Bo, bovine; Hu, human; Rh, rhesus; Eq, equine; Po, porcine; Ov, ovine; La, lapine; Si, simian; Mu, murine; Av, avian. The dashed line indicates the best option to divide the dendrogram into appropriate genotypes. The closing braces on the right side of the dendrograms depict the genotypes as they are proposed in this study. Panel B shows the identity frequency graph. Panels C, D, and E show details of the boxed region of panel B. In panel D, the identities among rotavirus strains belonging to the I2 genotype are shown. VP6 nucleotide accession numbers collected from GenBank are as follows for the indicated strains: BRV033, AF317126; B223, AF317128; WC3, AF411322; NCDV, AF317127; RRV, EF583009; OH4, D82975; PA169, EF554130; RF, K02254; Po, DQ119822; 111/05-27, EF554141; UKtc, X53667; R-22, D82977; OVR762, EF554152; B10925-97, EF554119; HP113, DQ003294; HP140, DQ003295; I321, X94618; 22R, AB040055; US6259, EF426123; US8720, EF426124; US8922, EF426132; US8635, EF426133; N26-02, DQ146686; US1205, AF079357; DRC86, DQ005121; DRC88, DQ005110; IS2, X94617; B1711, EF554086; US5139, EF426130; US8908, EF426140; TK126, AY456528; RV176-00, DQ490555; RV161-00, DQ490549; NR1, AF309652; TK119, AY456527; MG6, EF554097; AY456527; B4106, AY740737; 30/96, DQ205226; SA11-tsG, L15384; SA11-H96, DQ838650; SA11-30/19, DQ838648; SA11-5N, DQ838646; SA11-30/1A, DQ838649; SA11, AY187029; SA11-5S, DQ838647; 1076, D00325; Hun5, EF554108; FI-23, D82971; H-2, D00324; L26, DQ146695; TB-Chen, AY787645; S2, Y00437; Lp14, L11595; TUCH, AY594670; CMH222, ABC41660; T152, DQ146702; CMH134/04, DQ923800; CMH120/04, DQ923796; EO, AY947543; EMcN, AY267007; EDIM, U65988; EW, U36474; RMC321, AF531913; RMC/G60, AAT99008; RMC/G7, AY601551; RU172, DQ204741; 4F, L29184; 4S, L29186; CMP034, DQ534018; CRW-8, U82971; CN86, U10031; A411, AF317125; YM, X69487; A253, AF317122; A131, AF317124; H-1, AF242394; OSU, AF317123; HO-5, D82973; R-13, D82976; FI-14, D00323; HI-23, D82972; R-3, D82978; L338, D82974; Gottfried, D00326; CJN, AF461757; US6253, EF426131; KU, AB022768; 116E, U85998; Hu, X57943; Wa, K02086; E210, U36240; 97B53, AF260931; RMC61, AY601549; B4633-03, DQ146642; RV3, U04741; US9810, EF426127; US0408, EF426119; Dhaka12-03, DQ146664; Matlab13-03, DQ146675; Dhaka16-03, DQ492673; Dhaka25-02, DQ146653; RMC437, AY601554; US0468, EF426120; RMC/G66, AY601553; RMC100, AF531912; RMC83, AY601550; US8673, EF426135; US6097, EF426134; US9828, EF426139; US6153, EF426121; US8616, EF426138; US9951, EF426129; US9825, EF426128; US9875, EF426137; US9874, EF426136; US8960, EF426125; US8979, EF426126; US6161, EF426122; Ch-1, D82970; Ch-1, X98870; 02V0002G3, DQ096805; PO-13, D16329; 993-83, L13765; RK3, BAA22523; Ty-1, D82980; Ty-3, D82981; Ty-3, X98872.
VP1.
Fifty-eight complete VP1 nucleotide sequences were used to construct the phylogenetic dendrogram (see Data S6A in the supplemental material). As with the VP6 analyses, only the best alternative to divide the dendrogram into genotypes is shown. The results show the existence of four VP1 genotypes, tentatively designated R (for RNA-dependent RNA-polymerase) genotypes. An 83% nucleotide identity cutoff value perfectly reflected the classification as shown in the corresponding identity frequency graph (see Data S6B in the supplemental material).
VP2.
For the gene encoding VP2, no unambiguous nucleotide alignment could be obtained for the 5′ end region (nucleotides 1 to 134) since numerous insertion/deletions are present in this region. At present, the reason or function of this sequence heterogeneity is unknown. For this reason, the gene 2 alignments preceding nucleotide 134 (amino acid 40) with respect to human rotavirus reference strain Wa (accession numbers X14942 and CAA33074) were omitted from the analyses. The resulting phylogenetic dendrogram (see Data S7A in the supplemental material) was based on 58 complete VP2 nucleotide sequences. The most appropriate classification of VP2 into five genotypes, tentatively designated C (for core shell protein) genotypes, is shown by the vertical dashed line in Data S7A in the supplemental material. This classification was best reflected by a nucleotide identity cutoff value of 84%, as shown in the identity frequency graph in Data S7B in the supplemental material. The few intragenotype identities below the proposed 84% cutoff value (see Data S7C, D, and E in the supplemental material) are entirely due to the identities between the three strains A131 (porcine), A253 (porcine), and BRV033 (bovine) and the other members of the C2 genotype.
VP3.
Sixty-seven VP3 nucleotide sequences were used to construct the phylogenetic dendrogram shown in Data S8A in the supplemental material. The best alternative to classify the dendrogram into six suitable VP3 genotypes, tentatively designated M (for methyltransferase) genotypes, is shown as a vertical dashed line. The corresponding identity frequency graph (see Data S8B in the supplemental material) showed that an 81% nucleotide identity cutoff value was a perfect reflection of these proposed genotypes. Interestingly, the VP3-encoding gene segment of L26 determined in this study belonged to genotype M2. In contrast, the VP3-encoding gene of L26 as described previously by Cook and McCrae belonged to the M1 genotype (11). The reason for this discrepancy is not known.
NSP1.
One hundred complete NSP1 nucleotide sequences were used to construct the phylogenetic dendrogram shown in Data S9A in the supplemental material. The most suited alternative to divide the dendrogram into appropriate genotypes revealed 14 NSP1 genotypes, tentatively designated A (for interferon antagonist) genotypes (indicated by the vertical dashed line in Data S9A in the supplemental material). A 79% nucleotide identity cutoff value accurately reflects the classification as shown in the corresponding identity frequency graph (see Data S9B in the supplemental material). The few intragenotype identities below the 79% cutoff value were all due to identities among strains belonging to the A1 genotype.
NSP2.
Seventy-one NSP2 nucleotide sequences were used to construct the phylogenetic dendrogram in shown in Data S10A in the supplemental material. The best alternative to classify the dendrogram into five suitable NSP2 genotypes, tentatively designated N (for NTPase activity) genotypes, is shown as a vertical dashed line (see Data S10A in the supplemental material). The corresponding identity frequency graph (see Data S10B in the supplemental material) showed that an 85% nucleotide identity cutoff value was a good reflection of these proposed genotypes. The few intragenotype identities below the 85% the cutoff value were mainly due to identities among strains belonging to the N1 genotype, and the few intergenotype identities above the cutoff value are due to identities between N1 and N2 (see Data S10B in the supplemental material).
NSP3.
Seventy-seven NSP3 nucleotide sequences were used to construct the phylogenetic dendrogram in shown in Data S11A in the supplemental material. The best alternative to classify the dendrogram into seven suitable NSP3 genotypes, tentatively designated T (for translation enhancer) genotypes, is shown as a vertical dashed line (see Data S11A in the supplemental material), and the corresponding identity frequency graph (see Data S11B in the supplemental material) showed that an 85% nucleotide identity cutoff value was the most appropriate for these proposed genotypes. The few intragenotype identities below the 85% cutoff value are due to identities among strains belonging to the T1 genotype.
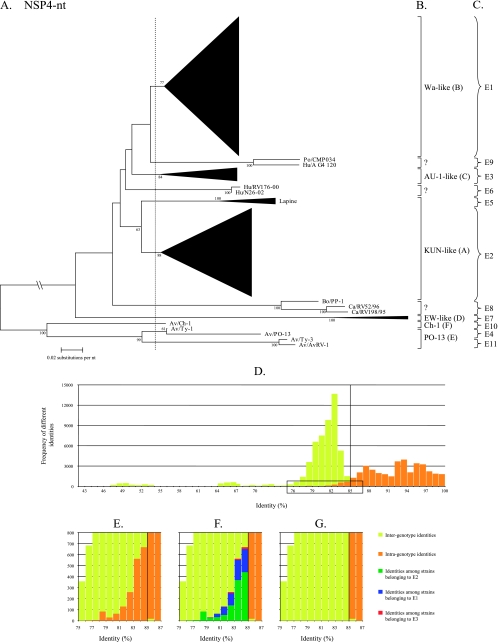
NSP4.
For NSP4, a classification into six genotypes (A to F) is currently used, based on the clustering into amino acid-based phylogenetic dendrograms using approximately 100 sequences (9, 22, 26, 46). In our analysis, a phylogenetic dendrogram was constructed with 430 available NSP4 nucleotide sequences (Fig. 4A). Large clusters have been replaced by triangles, but the full dendrogram is shown in Data S12 in the supplemental material. Several alternatives to divide the dendrogram into appropriate genotypes were analyzed, but only the best alternative is shown in Fig. 4A (vertical dashed line). The corresponding identity frequency graph (Fig. 4D) suggested that 85% was the most appropriate percentage identity cutoff value although a considerable number of intragenotype identities were found below this cutoff value. Although six NSP4 genotypes have been described to date, a total of 11 NSP4 genotypes, largely corresponding with the existing classification, were determined and tentatively designated E (for enterotoxin) genotypes when all available sequences were utilized. NSP4 genotype E1 corresponded completely with the Wa-like (genotype B) NSP4 genotype; genotype E2 largely corresponded to the KUN-like (genotype A) NSP4 genotype and was the most diverse genotype, as the majority of the rotavirus strains isolated from several species of origin (human, bovine, equine, simian, and ovine) clustered together, although the lapine or lapine-like human rotavirus strains were not included; genotype E3 corresponded completely to the AU-1-like (genotype C) NSP4 genotype; genotype E4 (previously NSP4 genotype E) was restricted to avian rotavirus strains (PO-13 and Ty-1); genotype E5 was composed of lapine and a human lapine-like rotavirus strains, previously belonging to the KUN-like (genotype A) NSP4 genotype. The assignment of the lapine NSP4 sequences into a single distinct genotype is also supported by previous observations showing a rather low degree of identity between lapine rotavirus strains and the remaining NSP4 genotype A strains (9); genotype E6 was restricted to newly described unusual human G12 strains (N26-02 and RV176-00) (63); genotype E7 (previously NSP4 genotype D) was composed entirely of murine rotavirus strains; genotype E8 contained a bovine strain (PP-1, which was originally proposed to belong to the NSP4 genotype B) (15) and two canine strains (RV52/96 and RV198/95); genotype E9 contained a human (A_G4_120) and a porcine (CMP034) strain that were described recently (30), while genotype E10 (previously NSP4 genotype F) contained the avian strain Ch-1 only; and genotype E11 was composed of the avian rotavirus strains Ty-3 and AvRV-1. The majority of the newly added NSP4 genotypes are due to the inclusion of new NSP4 sequences found in the GenBank database. A detail of the boxed region of Fig. 4D is shown in Fig. 4E and reveals that the greatest contribution to the intragenotype identities below the proposed cutoff value was generated by the identities among strains assigned to the three largest genotypes, E2, E1, and E3 (Fig. 4F), in descending order.
FIG. 4.
Phylogenetic dendrogram of NSP4 at the nucleotide (nt) level (A). Bootstrap values (2,000 replicates) above 50 are shown. Designations of species of origin are as follows: Bo, bovine; Hu, human; Po, porcine; Ca, canine; Av, avian. The dashed line indicates the best option to divide the dendrogram into appropriate genotypes. Certain clusters are replaced by triangles, in which the height of the triangle represents the number of sequences, and the width represents the genetic diversity inside that cluster. The full NSP4 phylogenetic dendrogram is provided in Data S6 in the supplemental material as well as the accession numbers used to construct the dendrogram. The closing braces on the right side of the dendrograms depict the 11 E genotypes as they are proposed in this study (C). Panel B depicts the genotypes as they were previously used. Panel D shows the identity frequency graph. Panels E, F, and G show details of the boxed region of panel D. In panel F, the identities among rotavirus strains belonging to the E3, E1, and E2 genotypes are shown. In panel G, the above-mentioned bars are omitted.
NSP5.
A phylogenetic dendrogram constructed with 113 NSP5 nucleotide sequences is shown in Data S13A in the supplemental material. Several alternatives to divide the dendrogram into appropriate genotypes were tested, but only the best alternative is shown (see Data S13A in the supplemental material; vertical dashed line). The corresponding identity frequency graph (see Data S13B in the supplemental material) suggested that 91% was the most appropriate percentage identity cutoff value. A total of six NSP5 genotypes were determined and tentatively designated H (for phosphoprotein) genotypes. A detail of the boxed region of Data S13B is shown in Data S13C in the supplemental material. Further analyses of the origin of the intragenotype identities below the proposed cutoff value (91%) demonstrated that the greatest contribution to these identities (see Data S13D in the supplemental material; green, blue, and red bars) was the identity among strains assigned to genotypes H2, H1, and H3, in descending order.
Recommendations for a rotavirus classification system of all 11 genome segments.
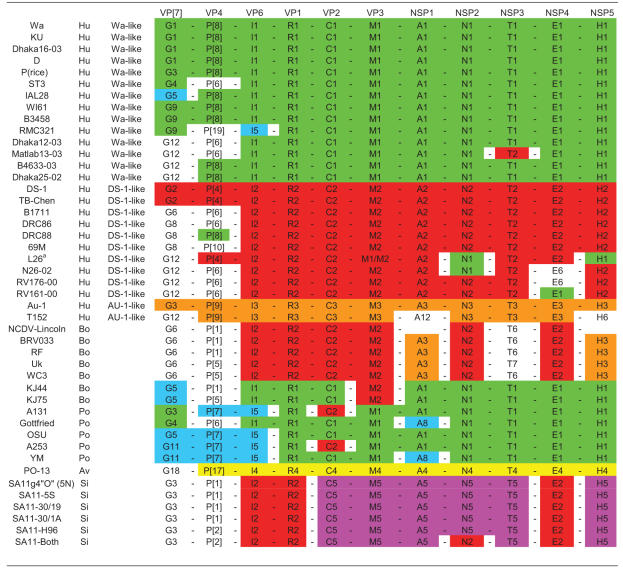
A summary of all 11 calculated cutoff values for the rotavirus genes segments is shown in Table 1. With these nucleotide cutoff percentages and the resulting appropriate genotypes for all the rotavirus proteins, a complete nomenclature and classification system can be proposed based on the evidence provided in this study. To designate the complete genetic constellation of a virus, the following schematic nomenclature is suggested: Gx-P[x]-Ix-Rx-Cx-Mx-Ax-Nx-Tx-Ex-Hx, representing the genotypes of, respectively, the VP7-VP4-VP6-VP1-VP2-VP3-NSP1-NSP2-NSP3-NSP4-NSP5 genes, with x indicating the numbers of the corresponding genotypes. In Table 2, this classification system has been applied to a number of human, bovine, porcine, avian, and simian rotavirus strains, demonstrating the practical use of the system in identifying reassortment events and examples of interspecies transmission and providing evidence on the role of animals as a source of rotavirus infection in humans. An extended table with more strains, including equine, murine, lapine, and ovine strains is available in Data S13 in the supplemental material.
TABLE 1.
Nucleotide (amino acid) percentage identity cutoff values defining genotypes for 11 rotavirus gene segments
| Gene product | Percentage identity cutoff value | Genotype (n)b | Description of gene product |
|---|---|---|---|
| VP7 | 80 (89)a | G (19) | Glycolylated |
| VP4 | 80 (89)a | P (27) | Protease sensitive |
| VP6 | 85 | I (11) | Intermediate capsid shell |
| VP1 | 83 | R (4) | RNA-dependent RNA polymerase |
| VP2 | 84 | C (5) | Core shell protein |
| VP3 | 81 | M (6) | Methyltransferase |
| NSP1 | 79 | A (14) | Interferon antagonist |
| NSP2 | 85 | N (5) | NTPase |
| NSP3 | 85 | T (7) | Translation enhancer |
| NSP4 | 85 | E (11) | Enterotoxin |
| NSP5 | 91 | H (6) | Phosphoprotein |
TABLE 2.
Application of the proposed classification and nomenclature to the structural and nonstructural protein-encoding genes for known human, bovine, porcine, simian, and avian rotavirus strainsb
The VP3 gene segment of L26 determined in this study was found to belong to a different genotype than the VP3 gene sequence determined by Cook (11). The reason for this is unknown. The Wa-, DS-1, or AU-1-like genogroup was assigned to human strains if at least seven gene segments belonged to the respective Wa-, DS-1, or AU-1-like genotype.
Colors were added to visualize certain patterns or gene constellations. Green, red, and orange indicate Wa-like, DS-1-like, and AU-like gene segments, respectively. Yellow, blue, and purple indicate the avian PO-13-like rotavirus gene segments; some typical porcine VP4, VP7, and VP6 genotypes; and the SA-11-like gene segments, respectively. For an extended table including equine, lapine, murine, and ovine rotavirus strains, see Data S14 in the supplemental material.
Identification of reassortment events and interspecies transmission.
In addition to identifying distinct genotypes for each rotavirus gene, this comprehensive sequence-based classification system also allowed complete comparison of the genome data of rotavirus strains, which in turn allowed the identification of reassortment events between strains belonging to different genogroups and confirmation of some cases of interspecies transmission. For example, the human rotavirus strain B4106 was speculated to have originated after interspecies transmission from a rabbit to a child (12, 38). Our analyses support this hypothesis because all the 11 rotavirus gene segments of B4106 and 30/96, a lapine rotavirus isolated in an Italian rabbit (37), were found to belong to the same genotype: G3-P[14]-I2-R2-C2-M3-A9-N2-T6-E5-H3 (see Data S13 in the supplemental material). A second example of an interspecies transmission is the equine rotavirus strain H-1, possessing genes belonging to typical porcine genotypes G5-P[7]-I5-A8-E1 (see Data S13 in the supplemental material), as was described previously (8). The hypothesis that DRC88, a G8P[8] rotavirus strain isolated in the Democratic Republic of Congo, was the result of a reassortment involving only the VP4 encoding gene segment between a G8P[6] human rotavirus strain and a strain with the P[8] specificity, is also strengthened by our analyses (Table 2; see also Data S13 in the supplemental material). The analyses of several SA11 strains, which are being used as rotavirus reference strains in different laboratories around the world, revealed that over the years reassortment events have occurred introducing different VP4 and NSP2 gene segments into these SA11 laboratory strains (67). These suggested reassortments are clearly visible in our classification system (Table 2; see also Data S13 in the supplemental material). Several reassortment events are suggested to be responsible for the large genetic diversity that is observed between different G12 rotavirus strains circulating in Bangladesh (63). Again, these reassortments are confirmed in our analyses as well (Table 2; see also Data S13 in the supplemental material). Recently, two bovine rotavirus strains, KJ44 and KJ75, were found to have porcine characteristics in all their gene segments except for VP3 and VP4, which have bovine characteristics (59), and our analyses confirmed this conclusion (Table 2; see also Data S13 in the supplemental material). These examples demonstrate that a classification including all 11 rotavirus gene segments can be very useful to study rotavirus gene reassortment and interspecies transmission.
Origin of rotaviruses infecting humans.
There has been speculation on the role of animals as a source of rotavirus infection in humans (5). As Table 2 and Data S13 in the supplemental material show, certain naturally occurring animal rotavirus strains may infect humans, but this type of interspecies transmission appears to be a rare event (5, 8, 15, 31, 38, 50, 52, 53).
Colors were added to Table 2 to visualize certain patterns or constellations of genes more easily: green, red, and orange were used for Wa-like, DS-1-like, and AU-1-like genogroup gene segments, respectively. Yellow, blue, and purple were used for the avian PO-13-like rotavirus gene segments; some typical porcine VP4, VP7, and VP6 genotypes; and the SA11-like gene segments, respectively. With the help of the color codes in Table 2, the source or origin of some human rotavirus strains, namely the Wa-like and DS-1-like genogroup human strains, became evident.
First, the most widespread human rotavirus strains belonging to the Wa-like genogroup (G1P[8], G3P[8], G4P[8], and G9P[8]) share the majority, if not all, of the genotypes with porcine rotavirus strains: R1-C1-M1-A1-N1-T1-E1-H1, corresponding to VP1 to VP3 and NSP1 to NSP5 genotypes (Table 2). In terms of the VP4, VP7, and VP6 genotypes, the human Wa-like rotavirus strains are mostly found to possess the G(1, 3, 4, or 9)-P[8]-I1 genotypes, while the porcine rotavirus strains mostly posses the G(5 or 11)-P[7]-I5 genotypes. Genotypes G1, G3, G4, G5, G9, G(5 or 11), and G12 have been also shown to infect pigs and humans (Table 2). The observation that the genes encoding VP4, VP7, and VP6 are partially distinct between the human Wa-like rotavirus strains and porcine rotavirus strains might be explained by the different selective pressures triggered in different host species. The close evolutionary relationship between NSP1, NSP2, and NSP4 of human Wa-like and porcine rotavirus strains has been described previously (9, 31, 61), and RNA-RNA hybridization studies showed at least five to six hybrid bands between porcine rotavirus strain YM and some Wa-like genogroup human rotavirus strains (31). Together, these data strongly suggest a common origin between human Wa-like strains and porcine rotaviruses.
Second, human rotavirus strains belonging to the DS-1-like genogroup shared the majority of their genotypes, namely VP6, VP1, VP2, VP3, NSP2, and NSP4 (I2-R2-C2-M2-N2-E2), with thoseof bovine rotavirus strains (Table 2). The different G and P genotypes commonly found associated with human DS-1 genogroup rotaviruses (G2 and P[4]) and bovine rotaviruses (G6, G8, and G10 and P[5] and P[11]) might again be due to different selective pressures in these two host species. However, the genotypes of NSP1 (A2 and A3), NSP3 (T2 and T6/T7), and NSP5 (H2 and H3), which are commonly found in human DS-1-like and bovine rotavirus strains, respectively, are not phylogenetically closely related (see Data S9, S11, and S13 in the supplemental material). A close evolutionary relationship between human DS-1-like and bovine rotaviruses has been previously described for several gene segments such as NSP2, NSP4, and VP3 (9, 11, 61). Again, these data point toward a common origin between the human DS-1-like rotavirus strains and bovine rotaviruses although a few gene segments might have been replaced by reassortment. The third, but minor, human AU-1-like genogroup (G3-P[9]-I3-R3-C3-M3-A3-N3-T3-E3-H3), is believed to have a close evolutionary relationship with canine and feline rotavirus strains, based on RNA-RNA hybridization studies (53). The very few sequence data that are available for feline rotaviruses (Table 2; see also Data S13 in the supplemental material) support this hypothesis, but additional sequence data of feline, and possibly canine, strains are necessary to properly answer this question. These results suggest that human and animal rotavirus strains are linked very closely and emphasize the importance of monitoring rotaviruses in animal populations, as well as following uncommon rotavirus strains in the human population very closely.
DISCUSSION
VP7, VP4, and NSP4 are the only rotavirus proteins for which a genotype-based classification system currently exists (9, 16, 18, 22, 26, 46). A serological classification exists for VP6, based on SG I- and II-specific MAbs, but by molecular and phylogenetic analyses of a 379-bp fragment of VP6, two genogroups have been identified (27). Serological classification of the nonstructural proteins NSP1 to NSP3 and NSP5 and viral core proteins VP1 and VP3 have not been carried out, and while the reactivity pattern of a MAb (YO-60) directed against VP2 suggested the possible presence of VP2 subgroups in human rotavirus strains (70), the extent of this finding was never investigated further. Therefore, sequence analyses would provide useful information to identify major clusters or genotypes in each of the rotavirus proteins. A similar approach has already been used for the determination of appropriate cutoff percentages for the classification of astroviruses (58), sapoviruses (66), noroviruses (78), and papillomaviruses (13). Here, we propose a rotavirus classification system for all the rotavirus genes that recognizes phylogenetic relationships and defines genotypes based on percentage identity cutoff values for each of the genes, which may have followed separate evolutionary paths. Subsequently, the novel rotavirus classification system was used to identify reassortment events and gene constellations shared by human and animal rotaviruses.
Our phylogenetic classification of VP4 into different P genotypes showed that an 80% identity cutoff value was in accordance with the established P genotypes. However, identities between P[8] and P[4] ranged from 84% to 89%, completely above the 80% cutoff value, reinforcing the idea that P[8] and P[4] are not only subtypes P1A and P1B of one serotype (16, 18, 23) but also subtypes of one genotype. The three recently described novel porcine P genotypes, represented by strains 344/04-1, CMP034, and P21-5, isolated in Italy, Thailand, and Slovenia, respectively (30, 35, 68), were all classified as distant members of the P[27] genotype, as was noted before (60).
The phylogenetic analyses of the VP7 gene sequences revealed that 80% and 89% identity cutoff values at the nucleotide and amino acid levels, respectively, provided the most appropriate classification from an evolutionary perspective. However, the classification proposed herein does not fully concur with the current VP7 classification scheme, as outlined below.
(i) Until now, all the VP7 genes of rotaviruses isolated from mice have been designated G3 or G3-like (14, 41, 74). Our phylogenetic analyses indicate that the murine rotaviruses could be assigned to a new G genotype, tentatively designated G16. Serological analyses of five murine strains, EW (also known as EDIM), EB, EC, EL, and EHP, have revealed that strains EW and EHP are not recognized by G3-specific sera or G3-specific MAbs (6, 14, 20). Among the murine rotavirus strains, only strain EB meets traditional criteria as serotype G3, while murine strain EC, and probably EL, are recognized by most, but not all, G3 serotype-specific MAbs (14). The American murine rotavirus strain EMcN showed a diverse pattern of reactivity with G3-specific hyperimmune sera, and the Japanese murine rotavirus strain YR-1 did not react with G3 specific MAbs at all (41, 74). We recognize that some murine strains may exhibit a dual VP7 serotype similar to strains MDR-13 (G3 and G5) and IAL28 (G5 and G11) and escape mutants of strain A253 (G11 and G3) (7, 16, 72).
(ii) All avian rotaviruses have been classified as serotype G7 even though there are several conflicting serotyping reports in the literature (25, 43, 65). The traditional avian genotype G7 and the three proposed novel avian genotypes (G17, G18, and G19) show less than 78% and 84% identity at the nucleotide and amino acid levels, respectively, which are differences very similar to those observed between different mammalian rotavirus genotypes (data not shown).
(iii) As reported previously (7, 72), G5 and G11 rotavirus strains are genotypically closely related. A human rotavirus strain, IAL-28, exhibits both G5 and the G11 serotype specificities (72), further strengthening the observation that G5 and G11 are closely related. With a few exceptions (8, 64, 73), both G5 and G11 rotaviruses have been mainly isolated from pigs. Since genotype G11 is closer to genotype G5 than to genotype G3, it is interesting that escape mutants of porcine strain A253 (G11) exhibited both G11 and G3 serotype specificities instead of G11 and G5 serotype specificities (7). Until the antigenic relationships between G5 and G11 rotavirus strains are fully understood, we propose to keep these two genotypes separate as they are clearly antigenically distinct.
(iv) Finally, the large sequence identity range found for G3 rotaviruses (Fig. 2G, light green bars) is not to be ignored. Given that G3 rotaviruses have the largest spectrum of host species and a higher degree of intergenotype sequence diversity than other genotypes (57), it does raise the level of complexity of the phylogenetic analyses (see discussion of murine rotavirus strains above) (29). In addition G3 and G14 rotavirus strains are more closely related to each other serologically than different genotypes usually are (10).
The high level of intragenotype sequence diversity within the VP2 and VP6 genotype 2 (C2 and I2, respectively), together with the broad range of host species within these genotypes, resembles features observed in the VP7 serotype G3 (29, 57). A remarkable observation is that except for VP6 genotypes I2 and I5, all the remaining eight VP6 genotypes contain rotaviruses isolated from one or two host species. No correlation was found between our VP6 genotyping system and VP6 SG specificity (data not shown) as several of the VP6 genotypes contain strains with different SG specificities (17, 27, 41, 69).
The identification of distinct genotypes for each rotavirus genome segment yielded multiple examples of related strains (Wa and KU, DS-1 and TB-Chen, and Dhaka25-02 and B4633-03), which cluster closely together in all 11 phylogenetic dendrograms, suggesting similar patterns of evolution. Several of the phylogenetic dendrograms, such as those for VP1, VP2, VP3, VP6, NSP2, and NSP3, show similar branching patterns for most of the established genotypes.
Our classification system concurs with the main group A rotavirus genogroups (Wa, DS-1, and AU-1), as established through RNA-RNA hybridization analyses (51). In line with the established human genogroups Wa, DS-1, and AU-1, the genotypes for each genome segment of these reference rotavirus strains were assigned to genotypes 1, 2, and 3, respectively, for consistency. All genome segments of the avian rotavirus PO-13 strain were assigned to genotype 4. For the genome segments encoding NSP1, NSP2, NSP3, VP2, and VP3, genotype 5 was assigned to the cluster containing the SA11 clones. The remaining genotypes of the structural and nonstructural protein-encoding genes were assigned randomly, but systematically.
The demonstration that Wa-like human rotaviruses possess similar constellations of genes and, hence, a close genetic relationship to porcine rotaviruses while the DS-1-like human rotaviruses have a close relationship with bovine rotaviruses suggests provocative ideas about the role of animals as a source of rotavirus infection in humans. The fact that each of the three main human rotavirus families may have a different animal origin would be instrumental in obtaining a better understanding of rotavirus host-range restriction and rotavirus ecology in nature. It is of interest that the only animals that can be productively infected experimentally by human rotavirus strains are piglets and calves (4). Piglets are monogastric animals with intestinal physiology that resembles that of humans and are susceptible to infection and severe disease by the human rotavirus Wa strain (75) but not the DS-1 strain (24). On the other hand, human DS-1-like rotaviruses have been shown to successfully infect cattle (45, 77), and the human Wa-like strain D can also infect and induce mild disease in calves (44). So far, dogs have been experimentally infected only with canine rotavirus strains (28).
To date a total of 35 human rotavirus strains have been completely sequenced, while only five and four porcine and bovine strains, respectively, have been completely sequenced (16, 38, 39, 63; also this report). Additional porcine and bovine rotavirus strains will be fully sequenced, allowing a further investigation of whether pigs and cows act as reservoirs for Wa-like and DS-1-like human rotavirus strains, respectively. Likewise, we are currently sequencing all the genome segments of feline and canine strains to determine if these domestic animals act as reservoirs for AU-1-like rotavirus strains in humans.
A further advantage of the establishment of this novel genotyping classification system is that a more systematic approach can be used to investigate possible genetic linkages among rotavirus genome segments. A comprehensive evaluation to investigate the possible genetic linkage between (sets of) genes for all 11 genes is being currently conducted (unpublished data).
In summary, we calculated nucleotide percentage identity cutoff values to define genotypes for the gene segments encoding the 11 proteins of group A rotaviruses. Appropriate genotypes were defined, and a novel nomenclature system is being proposed, allowing a rational comparison between different rotavirus strains and international standardization. The application of this classification system to a large number of rotavirus strains identified distinct genotypes in all genes, which probably follow separate evolutionary paths, and allow the identification of multiple reassortment and interspecies transmission events. The system also revealed the possible animal origin of the most common human rotavirus strains. The data reemphasize the rationale for simultaneous analysis of animal and human rotavirus strains.
Supplementary Material
Acknowledgments
A special word of our most sincere gratitude goes to Ulrich Desselberger for insightful suggestions and critical revisions of the manuscript. We also thank all the colleagues of the Laboratory of Clinical and Epidemiological Virology, Department of Microbiology and Immunology, Rega Institute for Medical Research, University of Leuven, Belgium, for their helpful comments and discussions.
J.M. was supported by the Institute for the Promotion of Innovation through Science and Technology in Flanders (IWT Vlaanderen). J.T.P. and S.M.M. were supported by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases, NIH. E.H. was supported by an appointment to the Oak Ridge Associated Universities Research Associates/Specialists Program at the NIH.
Footnotes
Published ahead of print on 23 January 2008.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215403-410. [DOI] [PubMed] [Google Scholar]
- 2.Ball, L. A. 2005. The universal taxonomy of viruses in theory and practice, p. 3-8. In C. M. Fauquet, M. A. Mayo, J. Maniloff, U. Desselberger, and L. A. Ball (ed.), Virus taxonomy. Eighth report of the International Committee on Taxonomy of Viruses. Elsevier, Amsterdam, The Netherlands.
- 3.Both, G. W., A. R. Bellamy, and D. B. Mitchell. 1994. Rotavirus protein structure and function. Curr. Top. Microbiol. Immunol. 18567-105. [DOI] [PubMed] [Google Scholar]
- 4.Ciarlet, M., M. E. Conner, and M. K. Estes. 2003. The rat model of rotavirus infection, p. 291-306. In J. Gray and U. Desselberger (ed.), Perspectives in medical virology: viral gastroenteritis. Elsevier Science BV, Amsterdam, The Netherlands.
- 5.Ciarlet, M., and M. K. Estes. 2002. Rotaviruses: basic biology, epidemiology and methodologies, p. 2573-2773. In G. Britton (ed.), Encyclopedia of environmental microbiology. John Wiley and Sons, New York, NY.
- 6.Ciarlet, M., M. Hidalgo, and F. Liprandi. 1996. Cross-reactive, serotype- and monotype-specific neutralization epitopes on VP7 of serotype G3 and G5 porcine rotavirus strains. Arch. Virol. 141601-614. [DOI] [PubMed] [Google Scholar]
- 7.Ciarlet, M., Y. Hoshino, and F. Liprandi. 1997. Single point mutations may affect the serotype reactivity of serotype G11 porcine rotavirus strains: a widening spectrum? J. Virol. 718213-8220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ciarlet, M., P. Iša, M. E. Conner, and F. Liprandi. 2001. Antigenic and molecular analyses reveal that the equine rotavirus strain H-1 is closely related to porcine, but not equine, rotaviruses: interspecies transmission from pigs to horses? Virus Genes 225-20. [DOI] [PubMed] [Google Scholar]
- 9.Ciarlet, M., F. Liprandi, M. E. Conner, and M. K. Estes. 2000. Species specificity and interspecies relatedness of NSP4 genetic groups by comparative NSP4 sequence analyses of animal rotaviruses. Arch. Virol. 145371-383. [DOI] [PubMed] [Google Scholar]
- 10.Ciarlet, M., F. Reggeti, C. I. Piña, and F. Liprandi. 1994. Equine rotaviruses with G14 serotype specificity circulate among Venezuelan horses. J. Clin. Microbiol. 322609-2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cook, J. P., and M. A. McCrae. 2004. Sequence analysis of the guanylyltransferase (VP3) of group A rotaviruses. J. Gen. Virol. 85929-932. [DOI] [PubMed] [Google Scholar]
- 12.De Leener, K., M. Rahman, J. Matthijnssens, L. Van Hoovels, T. Goegebuer, I. van der Donck, and M. Van Ranst. 2004. Human infection with a P[14], G3 lapine rotavirus. Virology 32511-17. [DOI] [PubMed] [Google Scholar]
- 13.de Villiers, E. M., C. Fauquet, T. R. Broker, H. U. Bernard, and H. zur Hausen. 2004. Classification of papillomaviruses. Virology 32417-27. [DOI] [PubMed] [Google Scholar]
- 14.Dunn, S. J., J. W. Burns, T. L. Cross, P. T. Vo, R. L. Ward, M. Bremont, and H. B. Greenberg. 1994. Comparison of VP4 and VP7 of five murine rotavirus strains. Virology 203250-259. [DOI] [PubMed] [Google Scholar]
- 15.El-Attar, L., W. Dhaliwal, C. R. Howard, and J. C. Bridger. 2001. Rotavirus cross-species pathogenicity: molecular characterization of a bovine rotavirus pathogenic for pigs. Virology 291172-182. [DOI] [PubMed] [Google Scholar]
- 16.Estes, M. K., and A. Z. Kapikian. 2007. Rotaviruses and their replication, p. 1917-1974. In B. N. Fields, D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 5th ed. Lippincott, Williams and Wilkins, Philadelphia, PA.
- 17.González, S. A., L. Tomasini, M. A. Tortorici, and J. L. Affranchino. 1995. VP6 from porcine rotavirus strain CN86: amino acid sequence divergence with conservation of subgroup II specificity. J. Gen. Virol. 76221-224. [DOI] [PubMed] [Google Scholar]
- 18.Gorziglia, M., G. Larralde, A. Z. Kapikian, and R. M. Chanock. 1990. Antigenic relationships among human rotaviruses as determined by outer capsid protein VP4. Proc. Natl. Acad. Sci. USA 877155-7159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Greenberg, H., V. McAuliffe, J. Valdesuso, R. Wyatt, J. Flores, A. Kalica, Y. Hoshino, and N. Singh. 1983. Serological analysis of the subgroup protein of rotavirus, using monoclonal antibodies. Infect. Immun. 3991-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Greenberg, H. B., P. T. Vo, and R. Jones. 1986. Cultivation and characterization of three strains of murine rotavirus. J. Virol. 57585-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gulati, B. R., R. Deepa, B. K. Singh, and C. D. Rao. 2007. Diversity in Indian equine rotaviruses: identification of genotype G10,P6[1] and G1 strains and a new VP7 genotype (G16) strain in specimens from diarrheic foals in India. J. Clin. Microbiol. 45972-978. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 22.Horie, Y., O. Masamune, and O. Nakagomi. 1997. Three major alleles of rotavirus NSP4 proteins identified by sequence analysis. J. Gen. Virol. 782341-2346. [DOI] [PubMed] [Google Scholar]
- 23.Hoshino, Y., R. W. Jones, R. M. Chanock, and A. Z. Kapikian. 2002. Generation and characterization of six single VP4 gene substitution reassortant rotavirus vaccine candidates: each bears a single human rotavirus VP4 gene encoding P serotype 1A[8] or 1B[4] and the remaining 10 genes of rhesus monkey rotavirus MMU18006 or bovine rotavirus UK. Vaccine 203576-3584. [DOI] [PubMed] [Google Scholar]
- 24.Hoshino, Y., L. J. Saif, S. Y. Kang, M. M. Sereno, W. K. Chen, and A. Z. Kapikian. 1995. Identification of group A rotavirus genes associated with virulence of a porcine rotavirus and host range restriction of a human rotavirus in the gnotobiotic piglet model. Virology 209274-280. [DOI] [PubMed] [Google Scholar]
- 25.Hoshino, Y., R. G. Wyatt, H. B. Greenberg, J. Flores, and A. Z. Kapikian. 1984. Serotypic similarity and diversity of rotaviruses of mammalian and avian origin as studied by plaque-reduction neutralization. J. Infect. Dis. 149694-702. [DOI] [PubMed] [Google Scholar]
- 26.Ito, H., M. Sugiyama, K. Masubuchi, Y. Mori, and N. Minamoto. 2001. Complete nucleotide sequence of a group A avian rotavirus genome and a comparison with its counterparts of mammalian rotaviruses. Virus Res. 75123-138. [DOI] [PubMed] [Google Scholar]
- 27.Iturriza-Gómara, M., C. Wong, S. Blome, U. Desselberger, and J. Gray. 2002. Molecular characterization of VP6 genes of human rotavirus isolates: correlation of genogroups with subgroups and evidence of independent segregation. J. Virol. 766596-6601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnson, C. A., R. W. Fulton, W. G. Henk, and T. G. Snider, III. 1983. Inoculation of neonatal gnotobiotic dogs with a canine rotavirus. Am. J. Vet. Res. 441682-1686. [PubMed] [Google Scholar]
- 29.Kapikian, A. Z., Y. Hoshino, and R. M. Chanock. 2001. Rotaviruses, p. 1787-1834. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott Williams and Wilkins, Philadelphia, PA.
- 30.Khamrin, P., N. Maneekarn, S. Peerakome, W. Chan-It, F. Yagyu, S. Okitsu, and H. Ushijima. 2007. Novel porcine rotavirus of genotype P[27] shares new phylogenetic lineage with G2 porcine rotavirus strain. Virology 361243-252. [DOI] [PubMed] [Google Scholar]
- 31.Kojima, K., K. Taniguchi, and N. Kobayashi. 1996. Species-specific and interspecies relatedness of NSP1 sequences in human, porcine, bovine, feline, and equine rotavirus strains. Arch. Virol. 1411-12. [DOI] [PubMed] [Google Scholar]
- 32.Kool, D. A., and I. H. Holmes. 1993. The avian rotavirus Ty-1 Vp7 nucleotide and deduced amino acid sequences differ significantly from those of Ch-2 rotavirus. Arch. Virol. 129227-234. [DOI] [PubMed] [Google Scholar]
- 33.Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei. 2001. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 171244-1245. [DOI] [PubMed] [Google Scholar]
- 34.Liprandi, F., M. Gerder, Z. Bastidas, J. A. López, F. H. Pujol, J. E. Ludert, D. B. Joelsson, and M. Ciarlet. 2003. A novel type of VP4 carried by a porcine rotavirus strain. Virology 315373-380. [DOI] [PubMed] [Google Scholar]
- 35.Martella, V., M. Ciarlet, K. Bányai, E. Lorusso, S. Arista, A. Lavazza, G. Pezzotti, N. Decaro, A. Cavalli, M. S. Lucente, M. Corrente, G. Elia, M. Camero, M. Tempesta, and C. Buonavoglia. 2007. Identification of group A porcine rotavirus strains bearing a novel VP4 (P) genotype in Italian swine herds. J. Clin. Microbiol. 45577-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martella, V., M. Ciarlet, K. Bányai, E. Lorusso, A. Cavalli, M. Corrente, G. Elia, S. Arista, M. Camero, C. Desario, N. Decaro, A. Lavazza, and C. Buonavoglia. 2006. Identification of a novel VP4 genotype carried by a serotype G5 porcine rotavirus strain. Virology 346301-311. [DOI] [PubMed] [Google Scholar]
- 37.Martella, V., M. Ciarlet, A. Camarda, A. Pratelli, M. Tempesta, G. Greco, A. Cavalli, G. Elia, N. Decaro, V. Terio, G. Bozzo, M. Camero, and C. Buonavoglia. 2003. Molecular characterization of the VP4, VP6, VP7, and NSP4 genes of lapine rotaviruses identified in Italy: emergence of a novel VP4 genotype. Virology 314358-370. [DOI] [PubMed] [Google Scholar]
- 38.Matthijnssens, J., M. Rahman, V. Martella, Y. Xuelei, S. De Vos, K. De Leener, M. Ciarlet, C. Buonavoglia, and M. Van Ranst. 2006. Full genomic analysis of human rotavirus strain B4106 and lapine rotavirus strain 30/96 provides evidence for interspecies transmission. J. Virol. 803801-3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Matthijnssens, J., M. Rahman, X. Yang, T. Delbeke, I. Arijs, J. P. Kabue, J. J. Muyembe, and M. Van Ranst. 2006. G8 rotavirus strains isolated in the Democratic Republic of Congo belong to the DS-1-like genogroup. J. Clin. Microbiol. 441801-1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maunula, L., and C. H. von Bonsdorff. 2002. Frequent reassortments may explain the genetic heterogeneity of rotaviruses: analysis of Finnish rotavirus strains. J. Virol. 7611793-11800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McNeal, M. M., J. Belli, M. Basu, A. H. Choi, and R. L. Ward. 2004. Discovery of a new strain of murine rotavirus that is consistently shed in large quantities after oral inoculation of adult mice. Virology 3201-11. [DOI] [PubMed] [Google Scholar]
- 42.McNeal, M. M., K. Sestak, A. H. Choi, M. Basu, M. J. Cole, P. P. Aye, R. P. Bohm, and R. L. Ward. 2005. Development of a rotavirus-shedding model in rhesus macaques, using a homologous wild-type rotavirus of a new P genotype. J. Virol. 79944-954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McNulty, M. S., G. M. Allan, D. Todd, J. B. McFerran, E. R. McKillop, D. S. Collins, and R. M. McCracken. 2006. Isolation of rotaviruses from turkeys and chickens: demonstration of distinct serotypes and RNA electropherotypes. Avian Pathol. 9363-375. [DOI] [PubMed] [Google Scholar]
- 44.Mebus, C., N. Stair, N. Underdahl, and M. Twiehaus. 1971. Pathology of neonatal calf diarrhea induced by a reo-like virus. Vet. Pathol. 8490-505. [DOI] [PubMed] [Google Scholar]
- 45.Mebus, C. A., R. G. Wyatt, R. L. Sharpee, M. M. Sereno, A. R. Kalica, A. Z. Kapikian, and M. J. Twiehaus. 1976. Diarrhea in gnotobiotic calves caused by the reovirus-like agent of human infantile gastroenteritis. Infect. Immun. 14471-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mori, Y., M. A. Borgan, N. Ito, M. Sugiyama, and N. Minamoto. 2002. Sequential analysis of nonstructural protein NSP4s derived from group A avian rotaviruses. Virus Res. 89145-151. [DOI] [PubMed] [Google Scholar]
- 47.Mori, Y., M. A. Borgan, M. Takayama, N. Ito, M. Sugiyama, and N. Minamoto. 2003. Roles of outer capsid proteins as determinants of pathogenicity and host range restriction of avian rotaviruses in a suckling mouse model. Virology 316126-134. [DOI] [PubMed] [Google Scholar]
- 48.Nakagomi, O., and T. Nakagomi. 2002. Genomic relationships among rotaviruses recovered from various animal species as revealed by RNA-RNA hybridization assays. Res. Vet. Sci. 73207-214. [DOI] [PubMed] [Google Scholar]
- 49.Nakagomi, O., and T. Nakagomi. 1991. Molecular evidence for naturally occurring single VP7 gene substitution reassortant between human rotaviruses belonging to two different genogroups. Arch. Virol. 11967-81. [DOI] [PubMed] [Google Scholar]
- 50.Nakagomi, O., and T. Nakagomi. 1991. Genetic diversity and similarity among mammalian rotaviruses in relation to interspecies transmission of rotavirus. Arch. Virol. 12043-55. [DOI] [PubMed] [Google Scholar]
- 51.Nakagomi, O., T. Nakagomi, K. Akatani, and N. Ikegami. 1989. Identification of rotavirus genogroups by RNA-RNA hybridization. Mol. Cell Probes 3251-261. [DOI] [PubMed] [Google Scholar]
- 52.Nakagomi, O., A. Ohshima, Y. Aboudy, I. Shif, M. Mochizuki, T. Nakagomi, and T. Gotlieb-Stematsky. 1990. Molecular identification by RNA-RNA hybridization of a human rotavirus that is closely related to rotaviruses of feline and canine origin. J. Clin. Microbiol. 281198-1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nakagomi, T., and O. Nakagomi. 1989. RNA-RNA hybridization identifies a human rotavirus that is genetically related to feline rotavirus. J. Virol. 631431-1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nei, M., and S. Kumar. 2000. Molecular evolution and phylogenetics, p. 33-50. Oxford University Press, New York, NY.
- 55.Nicholas, K. B., H. B. Nicholas, and D. W. Deerfield. 1997. GeneDoc: analysis and visualization of genetic variation. EMBnet News 414. [Google Scholar]
- 56.Nishikawa, K., Y. Hoshino, and M. Gorziglia. 1991. Sequence of the VP7 gene of chicken rotavirus Ch2 strain of serotype 7 rotavirus. Virology 185853-856. [DOI] [PubMed] [Google Scholar]
- 57.Nishikawa, K., Y. Hoshino, K. Taniguchi, K. Y. Green, H. B. Greenberg, A. Z. Kapikian, R. M. Chanock, and M. Gorziglia. 1989. Rotavirus VP7 neutralization epitopes of serotype 3 strains. Virology 171503-515. [DOI] [PubMed] [Google Scholar]
- 58.Noel, J. S., T. W. Lee, J. B. Kurtz, R. I. Glass, and S. S. Monroe. 1995. Typing of human astroviruses from clinical isolates by enzyme immunoassay and nucleotide sequencing. J. Clin. Microbiol. 33797-801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Park, S. H., L. J. Saif, C. Jeong, G. K. Lim, S. I. Park, H. H. Kim, S. J. Park, Y. J. Kim, J. H. Jeong, M. I. Kang, and K. O. Cho. 2006. Molecular characterization of novel G5 bovine rotavirus strains. J. Clin. Microbiol. 444101-4112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Parra, G. I., M. E. Galeano, and J. Arbiza. 2007. Genetic relationship between porcine rotavirus strains bearing a new P-type. Vet. Microbiol. 125193-195. [DOI] [PubMed] [Google Scholar]
- 61.Patton, J. T., L. Salter-Cid, A. Kalbach, E. A. Mansell, and M. Kattoura. 1993. Nucleotide and amino acid sequence analysis of the rotavirus nonstructural RNA-binding protein NS35. Virology 192438-446. [DOI] [PubMed] [Google Scholar]
- 62.Rahman, M., J. Matthijnssens, S. Nahar, G. Podder, D. A. Sack, T. Azim, and M. Van Ranst. 2005. Characterization of a novel P[25],G11 human group a rotavirus. J. Clin. Microbiol. 433208-3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rahman, M., J. Matthijnssens, X. Yang, T. Delbeke, I. Arijs, K. Taniguchi, M. Iturriza-Gómara, N. Iftekharuddin, T. Azim, and M. Van Ranst. 2007. Evolutionary history and global spread of the emerging G12 human rotaviruses. J. Virol. 812382-2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rahman, M., R. Sultana, G. Ahmed, S. Nahar, Z. M. Hassan, F. Saiada, G. Podder, A. S. Faruque, A. K. Siddique, D. A. Sack, J. Matthijnssens, M. Van Ranst, and T. Azim. 2007. Prevalence of G2P[4] and G12P[6] rotavirus, Bangladesh. Emerg. Infect. Dis. 1318-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rohwedder, A., H. Hotop, N. Minamoto, H. Ito, O. Nakagomi, and H. Brussow. 1997. Bovine rotavirus 993/83 shows a third subtype of avian VP7 protein. Virus Genes 14147-151. [DOI] [PubMed] [Google Scholar]
- 66.Schuffenecker, I., T. Ando, D. Thouvenot, B. Lina, and M. Aymard. 2001. Genetic classification of “Sapporo-like viruses.” Arch. Virol. 1462115-2132. [DOI] [PubMed] [Google Scholar]
- 67.Small, C., M. Barro, T. L. Brown, and J. T. Patton. 2007. Genome heterogeneity of SA11 rotavirus due to reassortment with “O” agent. Virology 359415-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Steyer, A., M. Poljsak-Prijatelj, D. Barlic-Maganja, U. Jamnikar, J. Z. Mijovski, and J. Marin. 2007. Molecular characterization of a new porcine rotavirus P genotype found in an asymptomatic pig in Slovenia. Virology 359275-282. [DOI] [PubMed] [Google Scholar]
- 69.Tang, B., J. M. Gilbert, S. M. Matsui, and H. B. Greenberg. 1997. Comparison of the rotavirus gene 6 from different species by sequence analysis and localization of subgroup-specific epitopes using site-directed mutagenesis. Virology 23789-96. [DOI] [PubMed] [Google Scholar]
- 70.Taniguchi, K., T. Urasawa, and S. Urasawa. 1986. Reactivity patterns to human rotavirus strains of a monoclonal antibody against VP2, a component of the inner capsid of rotavirus. Brief report. Arch. Virol. 87135-141. [DOI] [PubMed] [Google Scholar]
- 71.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 254876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Timenetsky, M. C., V. Gouvea, N. Santos, R. C. Carmona, and Y. Hoshino. 1997. A novel human rotavirus serotype with dual G5-G11 specificity. J. Gen. Virol. 781373-1378. [DOI] [PubMed] [Google Scholar]
- 73.Uchida, R., B. D. Pandey, J. B. Sherchand, K. Ahmed, M. Yokoo, T. Nakagomi, L. E. Cuevas, N. A. Cunliffe, C. A. Hart, and O. Nakagomi. 2006. Molecular epidemiology of rotavirus diarrhea among children and adults in Nepal: detection of G12 strains with P[6] or P[8] and a G11P[25] strain. J. Clin. Microbiol. 443499-3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ushijima, H., S. Morikawa, A. Mukoyama, and O. Nishio. 1995. Characterization of VP4 and VP7 of a murine rotavirus (YR-1) isolated in Japan. Jpn. J. Med. Sci. Biol. 48237-247. [DOI] [PubMed] [Google Scholar]
- 75.Ward, L. A., B. I. Rosen, L. Yuan, and L. J. Saif. 1996. Pathogenesis of an attenuated and a virulent strain of group A human rotavirus in neonatal gnotobiotic pigs. J. Gen. Virol. 771431-1441. [DOI] [PubMed] [Google Scholar]
- 76.Ward, R. L., O. Nakagomi, D. R. Knowlton, M. M. McNeal, T. Nakagomi, J. D. Clemens, D. A. Sack, and G. M. Schiff. 1990. Evidence for natural reassortants of human rotaviruses belonging to different genogroups. J. Virol. 643219-3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wyatt, R. G., C. A. Mebus, R. H. Yolken, A. R. Kalica, H. D. James, Jr., A. Z. Kapikian, and R. M. Chanock. 1979. Rotaviral immunity in gnotobiotic calves: heterologous resistance to human virus induced by bovine virus. Science 203548-550. [DOI] [PubMed] [Google Scholar]
- 78.Zheng, D. P., T. Ando, R. L. Fankhauser, R. S. Beard, R. I. Glass, and S. S. Monroe. 2006. Norovirus classification and proposed strain nomenclature. Virology 346312-323. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.