Abstract
Variola virus, the causative agent of smallpox, encodes a soluble complement regulator named SPICE. Previously, SPICE has been shown to be much more potent in inactivating human complement than the vaccinia virus complement control protein (VCP), although they differ only in 11 amino acid residues. In the present study, we have expressed SPICE, VCP, and mutants of VCP by substituting each or more of the 11 non-variant VCP residues with the corresponding residue of SPICE to identify hot spots that impart functional advantage to SPICE over VCP. Our data indicate that (i) SPICE is ∼90-fold more potent than VCP in inactivating human C3b, and the residues Y98, Y103, K108 and K120 are predominantly responsible for its enhanced activity; (ii) SPICE is 5.4-fold more potent in inactivating human C4b, and residues Y98, Y103, K108, K120 and L193 mainly dictate this increase; (iii) the classical pathway decay-accelerating activity of activity is only twofold higher than that of VCP, and the 11 mutations in SPICE do not significantly affect this activity; (iv) SPICE possesses significantly greater binding ability to human C3b compared to VCP, although its binding to human C4b is lower than that of VCP; (v) residue N144 is largely responsible for the increased binding of SPICE to human C3b; and (vi) the human specificity of SPICE is dictated primarily by residues Y98, Y103, K108, and K120 since these are enough to formulate VCP as potent as SPICE. Together, these results suggest that principally 4 of the 11 residues that differ between SPICE and VCP partake in its enhanced function against human complement.
Variola virus, the causative agent of smallpox, continues to be a cause of concern among virologists, in spite of the successful eradication of the disease about 27 years ago (3). This is primarily due to the looming threat of a reemergence of this disease owing to the potential usage of variola virus as a biological weapon (http://www.bt.cdc.gov). Variola virus is a member of the poxvirus family and belongs to the genus Orthopoxviruses (28, 30). It is an exceedingly contagious virus associated with a high mortality rate, ranging between 20 and 30% (10). The recent global interest in understanding the variola virus pathogenesis has increased due to the notable upsurge in terrorism. This has rekindled the variola virus research to be au fait with the factors influencing its virulence, pathogenesis, and control (2, 7, 15, 17, 41, 56).
The outcome and severity of viral diseases depends on the intricate balance and interplay between the host's immune response and the viral mechanisms to evade these responses. It is evident from various studies that viruses, in particular viruses with large viral genomes such as poxviruses and herpesviruses, have framed diverse mechanisms to mask themselves from the host's immune assault; these mechanisms include humoral, cellular, and effector immune responses (9, 11, 23, 35, 46). Because the complement system is an important humoral effector mechanism, it has also become a target for immune evasion by various viruses (8, 16, 29, 32, 51, 54). The different strategies adapted by viruses to shield themselves from the host complement system include encoding structural and/or functional homologs of complement regulators belonging to the regulators of complement activation (RCA) family, capturing of host membrane complement regulators, and the usage of host complement receptors for cellular entry (5, 11, 19, 21, 33). The RCA family proteins are composed of tandemly repeating ∼60-amino-acid modules termed short consensus repeats or complement control protein (CCP) domains that fold into compact bead-like structures separated by two- to seven-residue linkers (13, 53, 57). The RCA proteins regulate complement by two distinct mechanisms: (i) acceleration of irreversible dissociation of C3 and C5 convertases, which cleave complement proteins C3 and C5 (termed decay-accelerating activity), and (ii) inactivation of C3b and C4b, the subunits of the convertases, by serving as cofactors for serine protease factor I (termed cofactor activity) (22, 39, 45).
Variola virus is known to encode within its genome a homologue of RCA named SPICE (for smallpox inhibitor of complement enzyme) (43). Like other viral complement regulators, it is formed by four units of CCP domains joined together by linkers of four amino acid residues. Interestingly, SPICE differs from the vaccinia virus complement control protein (VCP) by only 11 amino acids (27, 49), which are scattered throughout CCP 2, CCP 3, and CCP 4 domains of the molecule. Despite the minimum difference between the two proteins, SPICE was found to be significantly more active than VCP in inactivating the human complement (43), a finding which is intriguing because variola virus is much more virulent than vaccinia virus. In an elegant study, Rosengard et al. (43) demonstrated SPICE to be 100- and 6-fold more potent, respectively, than VCP in inactivating human C3b and C4b. The study also demonstrated that SPICE is more human complement specific than VCP. Later, Sfyroera et al. (48) showed that SPICE is 75- and 1,000-fold more efficient, respectively, than VCP in inhibiting the classical and alternative pathways of complement. In a recent study, Liszewski et al. (25) compared the decay-accelerating activities of SPICE and VCP and demonstrated that SPICE decays classical pathway (CP) C3 and C5 convertases 5- to 10-fold more efficiently than VCP. These studies suggest that one or more of the 11 residue variances between the two proteins, VCP and SPICE, account for the robust activity of SPICE (33, 43, 48).
Although considerable progress has been made in understanding the functional advantage of SPICE over VCP (25, 43, 48), what remained unclear was the contribution of each of the 11 amino acids that differ between SPICE and VCP toward the higher complement regulatory activities of SPICE. The only effort made to date in this direction was a study by Sfyroera et al. (48), who utilized an electrostatic modeling approach to look into the effect of positive charge on the C3b cofactor activity of SPICE. These authors predicted and showed that a two-residue substitution (E108K/E120K) in VCP enhanced its C3b cofactor activity. However, Sfyroera et al. did not look into the contributions of other individual residues toward C3b cofactor activity or the role of any of the 11 residues toward C4b cofactor activity or decay-accelerating activity. In the present study, we conducted a systematic analysis of the contribution of each of the 11 variant amino acids of SPICE toward various complement regulatory activities. Our results, obtained using single-point mutants, suggest that Y98, Y103, K108, and K120 significantly augment the C3b cofactor activity of SPICE, whereas Y98, Y103, K108, K120, and L193 mainly contribute toward increased C4b cofactor activity. Additional experiments performed using tetra- and pentamutants of VCP indicated that the residues Y98, Y103, K108, and K120 are sufficient to make VCP as potent as SPICE and confer enhanced human specificity. Unlike the earlier study (25), we failed to observe any difference in the CP decay-accelerating activity of SPICE and VCP, and none of the 11 residues affected the decay activity.
(This study was done by V. N. Yadav in partial fulfillment of the requirements for a Ph.D. from the University of Pune, Pune, India, 2007.)
MATERIALS AND METHODS
Purified proteins, reagents, and buffers.
The human complement proteins C1, C4, and C2 were purchased from Calbiochem (Calbiochem, La Jolla, CA) and human factor I was a generous gift from Michael K. Pangburn (University of Texas Health Centre at Tyler). Human C3b was generated by limited tryptic cleavage of human C3 and purified on a Mono Q 5/5 column (Amersham Pharmacia Biotech, Uppsala, Sweden) (44). Human C4b was obtained from Calbiochem. The purity of all of the proteins exceeded 95% as judged by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis. Antibody-coated sheep erythrocytes (EA) were made by incubating the sheep erythrocytes with anti-sheep erythrocyte antibody purchased from ICN Biomedical, Inc. (Irvine, CA). Gelatin Veronal-buffered saline (GVB) contained 5 mM barbital, 145 mM NaCl, and 0.1% gelatin (pH 7.4). GVBE was GVB with 10 mM EDTA. Magnesium EGTA contained 0.1 M MgCl2 and 0.1 M EGTA. Dextrose gelatin Veronal-buffered saline (DGVB) contained 2.5 mM barbital, 72.5 mM NaCl, 0.1% gelatin, and 2.5% dextrose (pH 7.4), and DGVB++ was DGVB with 0.5 mM MgCl2 and 0.15 mM CaCl2. Phosphate-buffered saline contained 10 mM sodium phosphate and 145 mM NaCl (pH 7.4).
Molecular engineering of SPICE and the substitution mutants of VCP by site-directed mutagenesis.
The VCP gene (CCPs 1 to 4) was PCR amplified from the pHIL-S1-VCP clone (44) with the specific primers 5′-GAATTCCATATGTGCTGTCTATTCCGTCACGACC-3′ (the NdeI site is underlined) and 5′-GGGTTCGAAGCGTACACATTTTGGAAGTTCCGG-3′ (the HindIII site is underlined) using Proofstart DNA polymerase (Qiagen, Hilden, Germany) and cloned into the bacterial expression vector pET29 at the NdeI and HindIII sites. The PCR-amplified VCP1-4 was also cloned into the pGEM-T Easy vector (Promega) for use as a template for site-directed mutagenesis. The fidelity of these clones was confirmed by DNA sequencing using an automated ABI 3730 DNA analyzer.
SPICE and the 11 single-amino-acid substitution mutants of VCP were generated by introducing mutations in the pGEMT-VCP clone using the commercially available QuikChange II and QuikChange multi-site-directed mutagenesis kits (Stratagene, La Jolla, CA) according to the manufacturer's instructions. The single-amino-acid substitution mutants were Q77H, H98Y, S103Y, E108K, E120K, S131L, E144N, D178N, S193L, K214T, and K236Q, wherein the number denotes the location of the amino acid in the mature VCP protein and the letters before and after the number represent the amino acid in the VCP and its corresponding amino acid in SPICE to which it was mutated, respectively. The tetra- and penta-amino acid substitution mutants of VCP were generated by changing four (H98Y, S103Y, E108K, and E120K) and five (H98Y, S103Y, E108K, E120K, and S193L) residues, respectively. After these mutants were constructed they were validated by automated DNA sequencing and then transformed into Escherichia coli BL21 cells for expression.
Expression, purification, and refolding of SPICE, VCP, and the substitution mutants of VCP.
Expression of SPICE, VCP, and the respective mutants was performed as described below. Single colonies of bacterial clones expressing VCP, SPICE, or the mutants were inoculated into 50 ml of Luria-Bertani medium with kanamycin (25 μg/ml, final concentration) and grown overnight at 37°C. Thereafter, 5 ml of the grown culture was inoculated into 500 ml of Luria-Bertani medium plus kanamycin and grown at 37°C till the optical density of the culture reached 0.6 at A600. Induction of the protein was performed by addition of 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside), and the induced culture was further grown for 4 h. The cells were then harvested by centrifugation at 7,000 rpm at 4°C.
For protein purification, the harvested cell pellet was dissolved in 125 ml of lysis buffer (50 mM Tris [pH 8.0], 5 mM EDTA, 100 mM NaCl, 0.1% NaN3, 0.1 mM phenylmethylsulfonyl fluoride), and the cell suspension was sonicated with 15 pulses of 15 s each. Thereafter, it was treated with 10 mM MgSO4 to chelate EDTA and incubated with 0.1 mg of lysozyme/ml at 37°C for 30 min. The cell lysate was then centrifuged at 10,000 rpm for 10 min at 4°C, and the pellet containing the inclusion body was washed twice with the lysis buffer containing 0.5% Triton X-100. The inclusion body obtained as described above was solubilized in 100 mM Tris (pH 8.0), 50 mM glycine, and 8.0 M urea and centrifuged at 8,000 rpm for 10 min at 4°C. The supernatant thus obtained was loaded onto nickel nitrilotriacetic acid-agarose column (Qiagen) pre-equilibrated with binding buffer (100 mM NaH2PO4, 10 mM Tris [pH 8.0], 8.0 M urea), and the column was washed with binding buffer containing 20 mM imidazole. The bound protein was eluted in the same buffer containing 200 mM imidazole.
Refolding of the purified proteins was performed by using the rapid dilution method as previously described (50). The refolded proteins were further purified by using a Superose-12 gel filtration column (Pharmacia) in phosphate-buffered saline (pH 7.4) to obtain a monodispersed population of the expressed proteins. The protein samples obtained after gel filtration were concentrated and subjected to SDS-PAGE and circular dichroism (CD) analysis (18, 50).
Measurement of factor I cofactor activity for human C3b and C4b.
Quantitative analyses of factor I cofactor activities of SPICE, VCP, and the mutants were performed in 10 mM phosphate buffer (pH 7.4) containing 145 mM NaCl as described previously (4). In brief, 3 μg of human C3b or C4b was mixed with 250 ng of SPICE, VCP, or the mutants and 15 ng of human factor I in a total volume of 15 μl. The samples were then incubated at 37°C for various time periods as indicated, and the reactions were stopped by adding the sample buffer containing dithiothreitol. The cleavage fragments of C3b and C4b were separated by subjecting the samples to SDS-PAGE and visualized by staining with Coomassie blue. For quantitation of α′-chain, the gels were scanned for densitometric analysis by using the VersaDoc XRS system (Bio-Rad, Segrate, Italy). The data obtained were normalized by considering 100% of the α′-chain to be equal to the α′-chain intensity obtained in the absence of factor I.
Measurement of the CP for C3 convertase decay-accelerating activity.
The CP decay-accelerating activity of SPICE, VCP, and the mutants was determined by forming EAC142 as described earlier (31, 38) with minor modifications. In brief, EAC142 was formed by incubating 100 μl of EA (3.75 × 109/ml) in DGVB++ with 3.8 μg of human C1 for 20 min at 30°C in a total volume of 120 μl. The cells were then washed with ice-cold DGVB++, resuspended in 225 μl of DGVB++ containing 11 μg of human C4, and incubated for 20 min at 30°C. After incubation, the cells were mixed with 3.8 μg of human C2 and further incubated for 4 min at 30°C to form EAC142. The convertase formation was stopped by adding EDTA to a final concentration of 10 mM, and the cells were adjusted to 0.5 × 109/ml. To determine the effect of SPICE, VCP, and the mutants on decay of the CP C3 convertase, various concentrations of each protein were mixed with 10 μl of EAC142 in a total volume of 25 μl and allowed to decay for 5 min at 22°C. The reaction mixtures were then mixed with 100 μl of guinea pig serum diluted 1:100 in DGVB containing 40 mM EDTA in a final volume of 250 μl. After incubation for 30 min at 37°C, the reaction mixtures were centrifuged, and the percentage of lysis was determined by measuring the absorbance at 405 nM.
SPR measurements.
Binding of SPICE, VCP and the substitution mutants of VCP to human C3b and C4b was determined on the surface plasmon resonance (SPR)-based biosensor Biacore 2000 (Biacore AB, Uppsala, Sweden). Binding analyses of all of the proteins were performed in HBS-EP (0.01 M HEPES, 0.15 M NaCl, 50 μM EDTA, 0.05% surfactant P20) at 25°C using an Ni2+-nitrilotriacetic acid (NTA) sensor chip (37). The addition of 0.05% P-20 blocked the nonspecific adsorption of analytes to the sensor chips (4). An NTA chip was coated with nickel by injecting 40 μl of 500 μM NiCl2 at a flow rate of 20 μl/min. SPICE, VCP, and the mutants containing a C-terminal His6 tag were then immobilized (approximately 500 response units [RU]) on the test flow cells (FC-2, FC-3, and FC-4), and the control flow cell was formed by immobilizing the equivalent RU of an inactive Kaposica mutant. To measure the binding of C3b and C4b to the immobilized ligands, 1 μM C3b or 250 nM C4b was injected for 180 s at 10 μl/min, and the dissociation was measured for 180 s. The sensor chip was regenerated by brief pulses of 0.2 M sodium carbonate (pH 9.5) and regeneration buffer (0.01 M HEPES, 0.15 M NaCl, 0.35 M EDTA, 0.005% surfactant P20). Sensograms obtained for the control flow cell (FC-1) were subtracted from the data for the flow cell immobilized with test ligands to obtain the specific binding response.
Measurement of inhibition of human and dog complement.
Inhibition of human and dog complement by SPICE, VCP, and tetra- and pentamutants of VCP was studied by measuring their effect on the alternative-pathway-mediated lysis of rabbit erythrocytes (32, 44). Briefly, 10 μl of rabbit erythrocytes (109/ml) in GVB was mixed with increasing concentrations of the viral regulators in the presence of 5 μl of 0.1 M magnesium EGTA and 6 μl of human or 4.5 μl of dog sera. The final volume of the reaction mixture was brought to 100 μl by adding GVB. The reaction mixtures were then incubated for 20 min at 37°C, stopped by adding 200 μl of GVBE, and centrifuged. The absorbance of the supernatants was measured at 405 nm to determine the percentage of lysis. The data obtained were normalized by considering lysis in the absence of the regulators as 100% lysis. The data were fit by using nonlinear regression analysis (Grafit; Erithacus Software, London, United Kingdom), and a four-parameter logistic analysis was performed to identify the best-fit 50% inhibitory concentration value (IC50).
RESULTS
Cloning, expression, purification, and characterization of recombinant SPICE, VCP and the substitution mutants of VCP.
Both SPICE and VCP are soluble proteins composed entirely of four CCP domains without any membrane attachments sites. Therefore, we expressed these proteins as soluble proteins and not as membrane bound proteins as described earlier for VCP (42). SPICE was generated by molecular engineering using VCP as a template, while the 11 single-amino-acid substitution mutants of VCP were generated by mutating the individual residues that differ between VCP and SPICE (Fig. 1). The tetra- and pentamutants of VCP were generated by mutating the H98Y, S103Y, E108K, and E120K residues and the H98Y, S103Y, E108K, E120K, and S193L residues, respectively. All of the proteins were expressed in E. coli using the pET expression system, since it provided a large quantity of protein for performing multiple assays. The expressed proteins were purified to homogeneity by histidine affinity chromatography and refolded, and homogeneous protein fractions devoid of aggregates were obtained by loading the samples onto a gel filtration column. All of the purified recombinant proteins ran as a single band on SDS-PAGE and were estimated to be >95% pure (Fig. 1). SPICE, E120K, and the tetra- and pentamutants migrated slightly faster than VCP and the other mutants on the gel. The purified proteins were also validated for proper refolding by CD analysis. They yielded a typical peak at 230 nm, which is a characteristic of CCP-containing proteins (18; data not shown). The CD data, along with the conservation of CP decay-accelerating activity but selective gain or loss in cofactor activity (described below) in various expressed proteins, suggest that these proteins have maintained proper conformation.
FIG. 1.
Space-filling model of SPICE and sequence comparison of SPICE and VCP depicting the 11 mutations in SPICE compared to VCP, and SDS-PAGE analysis of purified recombinant SPICE, VCP, and various substitution mutants of VCP. (A) Front and back faces of SPICE model built by utilizing crystal structure of VCP (Ig40) (36) as the template using the program SWISS-MODEL (12, 40, 47). The 11 residues that differ between SPICE and VCP are labeled and depicted in red. (B) Sequence alignment of VCP and SPICE identifying CCP domains and linkers. Residues highlighted in red and indicated by asterisks represent those that differ between SPICE and VCP. These residues were mutated in VCP to generate SPICE, 11 single-amino-acid substitution mutants, and tetra- and pentamutants. (C) SDS-PAGE analysis of purified proteins. Purified VCP, SPICE, single-point, and tetra- and pentamutants of VCP were analyzed by SDS-12% PAGE under reducing conditions and stained with Coomassie blue. Molecular weight (MW) was determined by SDS-PAGE: VCP, 29,000; SPICE, 27,000; Q77H, 29,000; H98Y, 28,500; S103Y, 29,000; E108K, 28,500; E120K, 27,500; S131L, 28,000; E144N, 28,000; D178N, 29,000; S193L, 29,000; K214T, 29,000; K236Q, 28,000; tetramutant, 27,500; pentamutant, 27,500. Molecular mass is expressed in kilodaltons in the figure.
Characterization of factor I cofactor activity of SPICE, VCP, and single-amino-acid substitution mutants of VCP for human C3b and C4b.
Earlier reports have demonstrated that SPICE is ∼100-fold more potent than VCP in inactivating human C3b (25, 43, 48) and 6-fold more potent in inactivating human C4b (25, 43) due to its increased factor I cofactor activity. In the present study, we generated various single-point mutants to identify the determinant(s) responsible for this increased cofactor activity. As in the earlier studies, we also used a fluid-phase assay to characterize the cofactor activities of the various mutants. Using this assay, human complement protein C3b or C4b was incubated with equimolar concentrations of SPICE, VCP, or the mutants and factor I for various time periods, and the inactivation of C3b and C4b was assessed by quantitating the cleavage of their α′-chains. Inactivation of C3b was indicated by cleavage of C3b α′-chain into 68- and 46-kDa fragments, whereas inactivation of C4b was indicated by cleavage of C4b α′-chain into N-terminal 27-kDa, C-terminal 16-kDa, and central C4d fragments.
The data presented in Fig. 2 clearly show that the C3b cofactor activity of SPICE was vigorous compared to VCP, which supported the factor I-mediated cleavage of α′-chain of C3b at a much slower rate. The time required for 50% cleavage of α′-chain of C3b by VCP was 81 min, whereas that for SPICE was 0.9 min, indicating that SPICE is ∼90-fold more efficient cofactor for C3b cleavage than VCP. These data are in line with those of earlier studies (43, 48). Among the VCP single-amino-acid substitution mutants, H98Y, S103Y, E108K, and E120K exhibited significantly higher cofactor activity for human C3b than did VCP. The order of activity was E120K > S103Y > H98Y > E108K (Fig. 2 and Table 1) . These results indicate that the above-mentioned four residues are primarily responsible for the augmented cofactor activity of SPICE compared to VCP.
FIG. 2.
Time course analysis of factor I cofactor activity of SPICE, VCP, and the single-amino-acid substitution mutants of VCP for human complement protein C3b. Cofactor activity was determined by incubating human C3b with VCP, SPICE, or the mutants in the presence or absence of human factor I. The reactions were stopped after the indicated time periods by adding sample buffer containing dithiothreitol, and C3b cleavages were visualized by subjecting the samples to SDS-PAGE analysis on a 9% gel. During C3b cleavage, the α′-chain was cleaved into N-terminal 68-kDa and C-terminal 46-kDa fragments. The C-terminal fragment is subsequently cleaved into a 43-kDa fragment. Generation of these fragments indicates the inactivation of C3b. The amount of α′-chain remaining was assessed by measuring its intensity by densitometric analysis and is represented graphically in the lower panel.
TABLE 1.
Summary of functional activities of SPICE, VCP, and the single-amino-acid substitution mutants of VCPa
| Wild type or mutant | Time (min) for 50% cleavage of human C3b α′-chain | Relative C3b cofactor activity | Time (min) for 50% cleavage of human C4b α′-chain | Relative C4b cofactor activity | CP-DAA IC50 (μM) | Relative CP-DAA |
|---|---|---|---|---|---|---|
| VCP | 81 | 1.0 | 96.6 | 1.0 | 0.078 | 1.0 |
| S131L | 162 | 0.5 | 135 | 0.72 | 0.056 | 1.4 |
| Q77H | 126 | 0.6 | 128.4 | 0.75 | 0.05 | 1.6 |
| K236Q | 56 | 1.4 | 46.6 | 2.0 | 0.026 | 3.0 |
| K214T | 54 | 1.5 | 44.4 | 2.2 | 0.035 | 2.2 |
| S193L | 49 | 1.6 | 28.5 | 3.4 | 0.068 | 1.1 |
| D178N | 44 | 1.8 | 60 | 1.6 | 0.043 | 1.8 |
| E144N | 34 | 2.3 | 160 | 0.6 | 0.11 | 0.7 |
| E108K | 26 | 3.1 | 37.5 | 2.6 | 0.078 | 1.0 |
| H98Y | 12 | 6.7 | 30 | 3.2 | 0.038 | 2.0 |
| S103Y | 6 | 13.5 | 23 | 4.2 | 0.079 | 0.99 |
| E120K | 2.5 | 32.4 | 13 | 7.5 | 0.034 | 2.3 |
| SPICE | 0.9 | 90 | 18 | 5.4 | 0.039 | 2.0 |
Relative cofactor activity refers to the relative activity compared to VCP. CP-DAA, CP decay-accelerating activity for human C3 convertase.
The time required for 50% cleavage of α′-chain of human C3b and the relative cofactor activity of SPICE and the 11 single-amino-acid substitution mutants with respect to VCP are tabulated in Table 1. It is pertinent from the data that, apart from the four mutants mentioned above, all of the other mutants except two demonstrated C3b cofactor activity comparable to VCP; S131L and Q77H displayed less C3b cofactor activity than did VCP (Fig. 1 and Table 1). The C3b cleavage pattern supported by SPICE, VCP, and the mutants were the same. All of them supported the cleavage of α′-chain into 68- and 46-kDa fragments. These cleavages occurred immediately in the presence of SPICE, E120K, S103Y, H98Y, and E108K, whereas with VCP and rest of the mutants the cleavages occurred at a much slower rate.
Analysis of factor I cofactor activity of SPICE and VCP for human C4b showed that SPICE is 5.4-fold more efficient than VCP in inactivating C4b (Fig. 3 and Table 1), a finding which is in accordance with the earlier data (43). Further analysis of C4b cofactor activity of the various mutants showed that H98Y, S103Y, E108K, E120K, and S193L mutations resulted in significant increase in activity in comparison to VCP (E120K > S103Y > S193L > H98Y > E108K), indicating that the increase in C4b cofactor activity of SPICE in relation to VCP is because of these mutations. It should be highlighted here that four of these five mutations also increased the C3b cofactor activity (Fig. 2 and Table 1). As with the C3b cofactor activity, 3 of the 11 mutations (Q77H, S131L, and E144N) marginally decreased the C4b cofactor activity. The cleavage patterns of C4b supported by SPICE, VCP, and the mutants were similar.
FIG. 3.
Time course analysis of factor I cofactor activity of SPICE, VCP, and the single-amino-acid substitution mutants of VCP for human complement protein C4b. Cofactor activity was determined by incubating human C4b with VCP, SPICE, or the mutants in the presence or absence of human factor I. The reactions were stopped after the indicated time periods by adding sample buffer containing dithiothreitol, and C4b cleavages were visualized by subjecting the samples to SDS-PAGE analysis on an 11% gel. During C4b cleavage, the α′-chain is cleaved into N-terminal 27-kDa, C-terminal 16-kDa, and central C4d fragments; generation of these fragments indicates the inactivation of C4b. The amount of α′-chain left was assessed by measuring its intensity by densitometric analysis and is represented graphically in the lower panel.
Characterization of CP decay-accelerating activity of SPICE, VCP, and the single-amino-acid substitution mutants of VCP.
Previously, SPICE has been shown to possess 10-fold-greater CP decay-accelerating activity than VCP (25). In order to determine the relative CP decay-accelerating activity of SPICE and VCP and identify the amino acid residue(s) responsible for the increased activity of SPICE, if any, we utilized a hemolytic assay. In this assay, the CP C3 convertase C4b,2a was formed on sensitized sheep erythrocytes by using purified human complement components, and then the enzyme was allowed to decay in the presence of various concentrations of SPICE, VCP, or the mutant proteins. The activity of the remaining C3 convertase enzyme was quantitated by measuring hemolysis after the addition of EDTA sera (source of C3 to C9). Surprisingly, we found that SPICE was only twofold more active than VCP (Fig. 4 and Table 1). Because our results were not in agreement with the recent study of Liszewski et al. (25), which reported SPICE to be 10-fold more potent than VCP in decaying CP C3 convertase, we verified whether this discrepancy was due to minor differences in the hemolytic assays used. We repeated the CP decay-accelerating activity assay according to their protocol but failed to observe the stated 10-fold difference in activity between VCP and SPICE and consistently found the same twofold difference observed earlier (data not shown).
FIG. 4.
Analysis of CP C3-convertase decay-accelerating activity of SPICE, VCP, and the single-amino-acid substitution mutants of VCP. The CP C3 convertase C4b,2a was assembled on EA by sequentially incubating the cells with human C1, C4 and C2 (Calbiochem). The C3 convertase was allowed to decay by incubating EA-C4b,2a with various concentrations of VCP, SPICE, or the mutants for 5 min at 22°C, and the remaining enzyme activity was assessed by measuring cell lysis after incubation of the cells with guinea pig sera diluted 1:100 in DGVB containing 40 mM EDTA for 30 min at 37°C. The data obtained were normalized by setting lysis that occurred in the absence of an inhibitor (VCP or SPICE or the mutants) as 100% lysis.
The molar concentrations of single-amino-acid substitution mutants required for 50% decay of the convertase enzyme are shown in Table 1. It is evident from the data that none of the mutants showed considerable increments in the decay of the CP C3-convertase compared to VCP or SPICE (Fig. 4 and Table 1). Thus, our results suggest that none of these residues in SPICE contribute to the decay-accelerating activity of SPICE.
Binding of SPICE, VCP and the single-amino-acid substitution mutants of VCP to human C3b and C4b.
Binding of complement regulators to C3b and C4b is an indispensable step for imparting the cofactor and decay-accelerating activities; however, higher binding does not always correlate with higher activity (1, 6, 20, 24, 26, 31, 34, 50). In order to probe into the correlation between binding and functional activities, we analyzed the binding of SPICE, VCP, and the various mutants of VCP by the SPR-based assay using NTA sensor chip, since all of the proteins have been expressed to carry a His tag. In this assay, we first captured the His-tag-containing expressed proteins onto the NTA sensor chip to about 500 RU. Immobilization at this level provided a stable baseline. After ligand immobilization, human C3b (1 μM) or C4b (250 nM) were flown at 10 μl/min over the chip to measure binding.
The data presented in Fig. 5A show that binding response of SPICE to C3b was 130-fold higher than for VCP. The binding response of the single-amino-acid substitution mutants to C3b varied from equal to moderately higher than VCP, except for one mutant, E144N. The binding response of E144N to C3b was 108-fold higher than that of VCP, suggesting that Asn at position 144 in SPICE is primarily responsible for the remarkably higher binding of SPICE to C3b. This mutation, however, did not result in a significant increase in C3b cofactor activity (Table 1). Mutations that resulted in considerable enhancement in C3b cofactor activity included E120K, S103Y, H98Y, and E108K. These mutants demonstrated moderately higher binding response to C3b compared to VCP, suggesting that increased cofactor activity in these mutants could be in part due to their increased binding ability to C3b.
FIG. 5.
SPR analysis of SPICE, VCP, and the single-amino-acid substitution mutants of VCP. (A) Binding of SPICE, VCP, and the mutants to human C3b. (B) Binding of SPICE, VCP, and the mutants to human C4b. SPICE, VCP, and the mutants were oriented onto the NTA sensor chip, and C3b (1 μM) or C4b (250 nM) was flown over the chip to measure binding.
The binding response pattern for C4b shown by the proteins was much different from that for C3b. Interestingly, binding response of SPICE to C4b was dramatically lower than that of VCP (Fig. 5B). As a matter of fact, SPICE showed the least binding to C4b among all of the proteins. The substitution mutants displayed various degrees of binding to C4b, but the binding responses of all of the mutants were less than those of VCP. Thus, it is clear from the data that the binding ability to C4b does not correlate with the C4b cofactor activity data. Further, although there were considerable differences in the C4b binding response of SPICE, VCP, and the mutants, these differences were not reflected in their CP decay-accelerating activities.
Characterization of factor I cofactor activities of the tetra- and pentamutants of VCP and their binding to human C3b and C4b.
The data presented on the C3b and C4b cofactor activities of the 11 mutants suggest that the residues Y98, Y103, K108, and K120 are important for C3b inactivation (Fig. 2 and Table 1) and that the residues Y98, Y103, K108, K120, and L193 are important for C4b inactivation (Fig. 3 and Table 1). Since none of these mutants showed full activity as that of SPICE, we sought to determine, whether simultaneous mutations of the above four or five residues would result in the generation of mutants with activity equivalent to that of SPICE. We thus created a tetramutant (H98Y, S103Y, E108K, and E120K) and a pentamutant (H98Y, S103Y, E108K, E120K, and S193L) and assessed their factor I cofactor activities against human C3b and C4b. The data depicted in Fig. 6 show that the tetra- and pentamutants are as potent as SPICE in inactivating C3b and C4b, indicating that just four residues—Y98, Y103, K108, and K120—are enough to formulate VCP as potent as SPICE in the inactivation of C3b and C4b, and the addition of S193L to these mutations does not contribute any additional increase in the cofactor activities.
FIG. 6.
Time course analysis of factor I cofactor activity of SPICE and the tetra- and pentamutants of VCP for human complement proteins C3b and C4b. The cofactor activity was determined by incubating human C3b or C4b with SPICE, tetramutant (H98Y, S103Y, E108K, and E120K), or pentamutant (H98Y, S103Y, E108K, E120K, and S193L) of VCP in the presence or absence of human factor I. The reactions were stopped after the indicated time periods by adding sample buffer containing dithiothreitol. C3b and C4b cleavages were visualized by subjecting the samples to SDS-PAGE analysis on 9 and 11% gels, respectively. The amount of α′-chain remaining was assessed by measuring its intensity by densitometric analysis and is represented graphically. The upper panels show the cleavage of C3b by tetramutant and pentamutant, and the lower panels depict the cleavage of C4b.
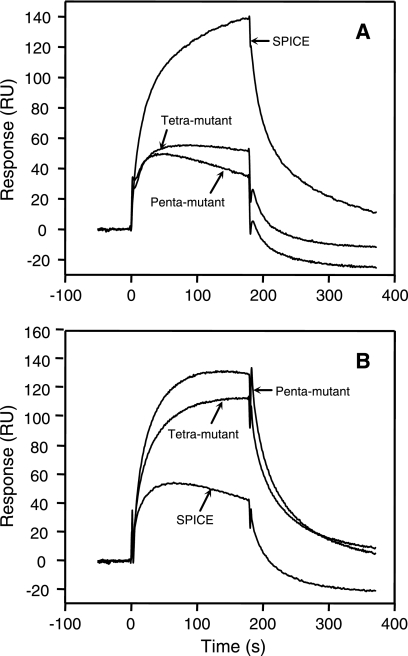
Next, we analyzed binding of the tetra- and the pentamutants to human C3b and C4b using an SPR assay. The binding response of these mutants to C3b was much lower than that of SPICE (Fig. 7A), whereas the response to C4b was appreciably greater (Fig. 7B). Thus, the binding ability of these mutants to C3b and C4b did not correlate with their cofactor activities.
FIG. 7.
SPR analysis of SPICE and the tetra- and pentamutants of VCP. (A) Binding of SPICE, tetramutant (H98Y, S103Y, E108K, and E120K), and pentamutant (H98Y, S103Y, E108K, E120K, and S193L) to human C3b. (B) Binding of SPICE, tetramutant, and pentamutant to human C4b. SPICE and the mutants were oriented onto the NTA sensor chip, and C3b (1 μM) or C4b (250 nM) was flown over the chip to measure binding.
Amino acid variations between SPICE and VCP dictate human specificity.
It has been shown that SPICE is more human specific than VCP (43). SPICE showed better inhibition of human complement than did VCP, whereas VCP demonstrated a preference for dog and guinea pig complement compared to SPICE. We therefore wondered whether the increase in activity seen in the mutants is by means of the functional forte provided by the mutations or due to an increase in the specificity toward human complement. We determined the inhibitory activities of SPICE, VCP, and the tetra- and pentamutants toward human and dog complement by measuring their effects on the alternative pathway-mediated lysis of rabbit erythrocytes. SPICE showed ∼25-fold more potent activity toward human complement than did VCP (SPICE, IC50 = 0.23 μM; VCP, IC50 = 5.8 μM) and, conversely, VCP displayed ∼12-fold more activity against dog complement (VCP, IC50 = 0.025 μM; SPICE, IC50 = 0.3 μM) (Fig. 8). The tetra- and the pentamutants that displayed activities equivalent to SPICE in C3b and C4b inactivation assays (Fig. 6 and Table 1) demonstrated a gain in activity toward human complement (tetramutant, IC50 = 0.29 μM; pentamutant, IC50 = 0.35 μM) and a loss in activity toward dog complement (tetramutant, IC50 = 0.11 μM; pentamutant, IC50 = 0.13 μM) compared to VCP (Fig. 8). Thus, the data indicate that amino acid variations in SPICE increase its specificity toward human complement.
FIG. 8.
Inhibition of human and dog complement by SPICE, VCP, and the tetra- and pentamutants of VCP. Inhibition of human (A) and dog (B) complement by SPICE, VCP, or tetramutant (H98Y, S103Y, E108K, and E120K), and pentamutant (H98Y, S103Y, E108K, E120K, and S193L) of VCP was studied my measuring their effect on the alternative pathway-mediated lysis of rabbit erythrocytes (ER).
DISCUSSION
Earlier studies have established VCP as an immune evasion molecule (14). Because genome analyses showed that a structurally similar molecule is also encoded and conserved in position and sequence in all of the strains of variola virus, efforts were also made to characterize this molecule. SPICE, the variola virus complement inhibitor, displayed 100-fold more potent activity than VCP in inactivating human C3b and 6-fold more activity in inactivating human C4b (43). These results were later corroborated by other studies (25, 48). In a further study, it was shown that SPICE also possesses significantly higher CP C3 convertase decay-accelerating activity (25). Because SPICE and VCP vary in only 11 amino acid residues, the functional differences between these two molecules can be attributed to these amino acids. Although previous studies characterized the functional activities of SPICE (25, 43, 48) and looked into the domains responsible for the enhanced activity of SPICE (48), no efforts have yet been made to characterize the role of each of the 11 amino acid differences in C3b and C4b inactivation (C3b and C4b cofactor activities) and CP C3 convertase decay-accelerating activity. Our objective here was to decipher the role of these nonvariant residues of SPICE in enhancing its activity to inactivate human C3b and C4b and the decay of CP C3 convertase.
We expressed the molecularly engineered SPICE, VCP, and single-point and multiresidue mutants of VCP by using a bacterial expression system and verified them for proper refolding (Fig. 1). Our data on human C3b inactivation by SPICE and VCP showed that SPICE is 90-fold more potent than VCP (Fig. 2 and Table 1). Analysis of C3b cofactor activities of the various mutants showed that H98Y, S103Y, E108K, and E120K possess significantly greater activity compared to VCP (Table 1). These results point out that only 4 of the 11 residues are primarily responsible for the robust C3b inactivation by SPICE. It is interesting that all of these residues are located within the CCP 2 domain of SPICE. This conclusion is also favored by an earlier report, which showed that substitution of CCP 2 domain of SPICE in VCP resulted in 48-fold enhancement in cofactor activity against human C3b (48).
Our data on the C4b cofactor activity demonstrated that SPICE is 5.4-fold more efficient than VCP in inactivating human C4b, and analysis of the various single-point mutants showed that the H98Y, S103Y, E108K, E120K, and S193L mutations led to a significant gain in C4b cofactor activity compared to VCP (Fig. 3 and Table 1). Since none of the single-point mutants possessed activities similar to SPICE, we also generated a tetramutant (H98Y, S103Y, E108K, and E120K) and a pentamutant (H98Y, S103Y, E108K, E120K, and S193L) of VCP and measured their cofactor activities against C3b and C4b to verify whether concurrent substitution of these residues is enough to achieve full activity. The data presented in Fig. 6 show that the substitution of just four residues (H98Y, S103Y, E108K and E120K) in VCP is enough to achieve an activity equivalent to SPICE. It should be noted that these residues are harbored in the CCP 2 domain. Thus, it seems that the CCP 2 domain in SPICE plays a predominant role in enhancing its C3b and C4b cofactor activities.
It is thus clear that Y98, Y103, K108, and K120 are primarily responsible for the increase in cofactor activities of SPICE (Fig. 6). In order to examine whether similar residues are conserved in position in other complement regulators, we aligned the sequence of SPICE with various other viral and human complement regulators. We observed that a positively charged amino acid at a position comparable to K120, but not K108, of SPICE is conserved in complement regulators of herpesvirus saimiri (HVS CCPH; R118), herpesvirus ateline (R118), rhesus rhadinovirus 17577 (RCP-1; K421), complement receptor 1 (CR1, CD35; K566 and K1466), and membrane cofactor protein (MCP, CD46; K119). Among these proteins, the importance of the charged residue has been demonstrated by us in HVS CCPH (50) and others in CR1 (20) and MCP (24). We noticed that the mutation of R118 to Ala in HVS CCPH resulted in a 50-fold decrease in C3b cofactor activity and a 12-fold decrease in C4b cofactor activity, whereas the mutation of K566 (in CCP 9) to Glu in CR1 nearly abrogated the C3b and C4b cofactor activities. The mutation of K119 to Ala in MCP, however, resulted in a moderate decrease in C4b cofactor activity. These data suggest that the positive charge at position comparable to K120 of SPICE plays a paramount role in the cofactor activities of both viral and human complement regulators. While looking at the conservation of Y/F residues at positions comparable to Y98 and/or Y103 of SPICE, we noticed that these are conserved in Kaposica (Kaposi's sarcoma-associated herpesvirus complement regulator; F102 and Y108), MCP (Y98), and C4b-binding protein (F97). A single-point mutation in MCP showed that substitution of Y98 to Ala substantially reduced its C4b cofactor activity (24); the importance of conserved Y/F in Kaposica and C4b-binding protein has not been studied. Thus, it seems that both viral and human complement regulators use a common mechanism to enhance their innate ability to inactivate C3b and C4b.
According to a prevailing model (52), the process of C3b and C4b inactivation entails two steps: (i) binding of the complement regulator (viral or human) to complement proteins C3b or C4b and (ii) interaction of factor I with C3b or C4b and the complement regulator. Since the single-point mutants H98Y, S103Y, E108K, E120K, and S193L displayed a significant increase in cofactor activity, we examined whether these mutations also resulted in an enhancement in binding to human C3b and C4b. Point mutations that enhanced the C3b cofactor activity (H98Y, S103Y, E108K, and E120K) showed a moderate increase in C3b binding, but mutations that resulted in a gain in C4b cofactor activity (H98Y, S103Y, E108K, E120K, and S193L) did not correlate with C4b binding (Fig. 5 and Table 1). Similarly, binding of the tetra- and pentamutants to C3b and C4b did not correlate well with their cofactor activities (Fig. 6 and 7). We therefore suggest that the increased cofactor activities of these mutants are likely due to their better interaction with factor I. Earlier mutational studies of MCP (1, 24), CR1 (20), C4BP (6), Kaposica (26, 34), and HVS-CCPH (50) have also indicated that binding and cofactor activities are not always parallel.
Because previously Liszewski, et al. (25) showed that SPICE is 10-fold more potent than VCP in decaying the CP C3 convertase, we also looked into the decay-accelerating activities of SPICE and the mutants to identify the residues responsible for this increase. Our data, however, showed only a twofold increase in CP decay-accelerating activity of SPICE compared to VCP (Fig. 4 and Table 1). To rule out whether minor differences in the assays between our study and the previous study were responsible for this difference we performed the assays again according to the method utilized by Liszewski et al. (25), but even then we detected only a twofold difference (data not shown). Whether the variation in our data and the previous data is due to difference in the expression systems utilized (E. coli versus mammalian cells) is not clear and requires further investigation. However, the contribution of glycosylation to these differences can be ruled out since both SPICE and VCP lack putative glycosylation sites. Examination of CP decay-accelerating activities of the single-amino-acid substitution mutants exhibited activities comparable to that of VCP, except for the K236Q mutant, which showed threefold increase in activity. The present data gain some support from our previous observation in HVS CCPH, with which we had demonstrated that mutation of R118 to Ala drastically reduced the C3b and C4b cofactor activities but not the decay-accelerating activities (50). Based on these data we suggest that the amino acid differences in SPICE compared to VCP result only in the enhancement of its cofactor activities and not of its decay-accelerating activity.
Earlier, Rosengard et al. (43) demonstrated that SPICE and VCP exhibited a preference for complement from different species. For example, SPICE showed a preference for human complement, whereas VCP showed a preference for dog complement. We therefore looked into the influence of the amino acid variations in SPICE on species specificity. We tested the tetra- and the pentamutants for the inhibition of human and dog complement and compared their activities to that of SPICE and VCP. Both mutants behaved in a manner similar to SPICE by inhibiting human complement more preferentially than dog complement (Fig. 8). The inhibitory activity of both mutants against human complement was comparable to that of SPICE (Fig. 8A), suggesting that just four residues (Y98, Y103, K108, and K120) are enough to provide human specificity to SPICE.
In summary, our data suggest that the increased effectiveness of SPICE against human complement compared to VCP is mainly due to its increased ability to inactivate C3b and C4b and that Y98, Y103, K108, and K120 are the hot spots within SPICE that are largely responsible for imparting this robust activity to the molecule. It has been argued earlier that because complement effectively neutralizes poxviruses (14, 55), targeting SPICE could form an alternative strategy to control variola virus infections (43). Our data suggest that for designing such therapeutics, targeting the CCP2 domain of SPICE would be fundamental since residues that enhance its activity reside within CCP2 domain.
Acknowledgments
We thank John D. Lambris (Department of Pathology and Laboratory Medicine, University of Pennsylvania, Philadelphia) and Michael K. Pangburn (Department of Biochemistry, University of Texas Health Center, Tyler) for their continuous support and Dulal Panda (School of Biosciences and Bioengineering, Indian Institute of Technology, Mumbai, India) for access to his spectropolarimeter. We also express appreciation to Yogesh Panse and Sarang Satoor for excellent technical assistance.
This study was supported by a project grant from the Department of Biotechnology of India to A.S. We also acknowledge financial assistance to V.N.Y. and K.P. from the Council of Scientific and Industrial Research, New Delhi, India, and to J.M. from the Department of Science and Technology, New Delhi, India.
Footnotes
Published ahead of print on 23 January 2008.
REFERENCES
- 1.Adams, E. M., M. C. Brown, M. Nunge, M. Krych, and J. P. Atkinson. 1991. Contribution of the repeating domains of membrane cofactor protein (CD46) of the complement system to ligand binding and cofactor activity. J. Immunol. 1473005-3011. [PubMed] [Google Scholar]
- 2.Aldaz-Carroll, L., J. C. Whitbeck, D. L. Ponce, H. Lou, L. Hirao, S. N. Isaacs, B. Moss, R. J. Eisenberg, and G. H. Cohen. 2005. Epitope-mapping studies define two major neutralization sites on the vaccinia virus extracellular enveloped virus glycoprotein B5R. J. Virol. 796260-6271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anonymous. 1980. Declaration of global eradication of smallpox. Wkly. Epidemiol. Rec. 55145-152. [Google Scholar]
- 4.Bernet, J., J. Mullick, Y. Panse, P. B. Parab, and A. Sahu. 2004. Kinetic analysis of the interactions between vaccinia virus complement control protein and human complement proteins C3b and C4b. J. Virol. 789446-9457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bernet, J., J. Mullick, A. K. Singh, and A. Sahu. 2003. Viral mimicry of the complement system. J. Biosci. 28249-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blom, A. M., B. O. Villoutreix, and B. Dahlback. 2003. Mutations in alpha-chain of C4BP that selectively affect its factor I cofactor function. J. Biol. Chem. 27843437-43442. [DOI] [PubMed] [Google Scholar]
- 7.Chen, Z., P. Earl, J. Americo, I. Damon, S. K. Smith, Y. H. Zhou, F. Yu, A. Sebrell, S. Emerson, G. Cohen, R. J. Eisenberg, J. Svitel, P. Schuck, W. Satterfield, B. Moss, and R. Purcell. 2006. Chimpanzee/human mAbs to vaccinia virus B5 protein neutralize vaccinia and smallpox viruses and protect mice against vaccinia virus. Proc. Natl. Acad. Sci. USA 1031882-1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cooper, N. R. 1998. Complement and viruses, p. 393-407. In J. E. Volanakis and M. M. Frank (ed.), The human complement system in health and disease. Marcel Dekker, Inc., New York, NY.
- 9.Coscoy, L. 2007. Immune evasion by Kaposi's sarcoma-associated herpesvirus. Nat. Rev. Immunol. 7391-401. [DOI] [PubMed] [Google Scholar]
- 10.Fenner, F. 2005. Smallpox and its eradication. World Health Organization, Geneva, Switzerland.
- 11.Finlay, B. B., and G. McFadden. 2006. Anti-immunology: evasion of the host immune system by bacterial and viral pathogens. Cell 124767-782. [DOI] [PubMed] [Google Scholar]
- 12.Guex, N., and M. C. Peitsch. 1997. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 182714-2723. [DOI] [PubMed] [Google Scholar]
- 13.Hourcade, D., V. M. Holers, and J. P. Atkinson. 1989. The regulators of complement activation (RCA) gene cluster. Adv. Immunol. 45381-416. [DOI] [PubMed] [Google Scholar]
- 14.Isaacs, S. N., G. J. Kotwal, and B. Moss. 1992. Vaccinia virus complement-control protein prevents antibody-dependent complement-enhanced neutralization of infectivity and contributes to virulence. Proc. Natl. Acad. Sci. USA 89628-632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jahrling, P. B., L. E. Hensley, M. J. Martinez, J. W. Leduc, K. H. Rubins, D. A. Relman, and J. W. Huggins. 2004. Exploring the potential of variola virus infection of cynomolgus macaques as a model for human smallpox. Proc. Natl. Acad. Sci. USA 10115196-15200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jayasekera, J. P., E. A. Moseman, and M. C. Carroll. 2007. Natural antibody and complement mediate neutralization of influenza virus in the absence of prior immunity. J. Virol. 813487-3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kidokoro, M., M. Tashiro, and H. Shida. 2005. Genetically stable and fully effective smallpox vaccine strain constructed from highly attenuated vaccinia LC16m8. Proc. Natl. Acad. Sci. USA 1024152-4157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kirkitadze, M. D., M. Krych, D. Uhrin, D. T. Dryden, B. O. Smith, A. Cooper, X. Wang, R. Hauhart, J. P. Atkinson, and P. N. Barlow. 1999. Independently melting modules and highly structured intermodular junctions within complement receptor type 1. Biochemistry 387019-7031. [DOI] [PubMed] [Google Scholar]
- 19.Kotwal, G. J., and B. Moss. 1988. Vaccinia virus encodes a secretory polypeptide structurally related to complement control proteins. Nature 335176-178. [DOI] [PubMed] [Google Scholar]
- 20.Krych, M., L. Clemenza, D. Howdeshell, R. Hauhart, D. Hourcade, and J. P. Atkinson. 1994. Analysis of the functional domains of complement receptor type 1 (C3b/C4b receptor, CD35) by substitution mutagenesis. J. Biol. Chem. 26913273-13278. [PubMed] [Google Scholar]
- 21.Lachmann, P. J. 2002. Microbial subversion of the immune response. Proc. Natl. Acad. Sci. USA 998461-8462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lambris, J. D., A. Sahu, and R. Wetsel. 1998. The chemistry and biology of C3, C4, and C5, p. 83-118. In J. E. Volanakis and M. Frank (ed.), The human complement system in health and disease. Marcel Dekker, Inc., New York, NY.
- 23.Lilley, B. N., and H. L. Ploegh. 2005. Viral modulation of antigen presentation: manipulation of cellular targets in the ER and beyond. Immunol. Rev. 207126-144. [DOI] [PubMed] [Google Scholar]
- 24.Liszewski, M. K., M. Leung, W. Cui, V. B. Subramanian, J. Parkinson, P. N. Barlow, M. Manchester, and J. P. Atkinson. 2000. Dissecting sites important for complement regulatory activity in membrane cofactor protein (MCP; CD46). J. Biol. Chem. 27537692-37701. [DOI] [PubMed] [Google Scholar]
- 25.Liszewski, M. K., M. K. Leung, R. Hauhart, R. M. Buller, P. Bertram, X. Wang, A. M. Rosengard, G. J. Kotwal, and J. P. Atkinson. 2006. Structure and regulatory profile of the monkeypox inhibitor of complement: comparison to homologs in vaccinia and variola and evidence for dimer formation. J. Immunol. 1763725-3734. [DOI] [PubMed] [Google Scholar]
- 26.Mark, L., W. H. Lee, O. B. Spiller, D. Proctor, D. J. Blackbourn, B. O. Villoutreix, and A. M. Blom. 2004. The Kaposi's sarcoma-associated herpesvirus complement control protein mimics human molecular mechanisms for inhibition of the complement system. J. Biol. Chem. 27945093-45101. [DOI] [PubMed] [Google Scholar]
- 27.Massung, R. F., V. N. Loparev, J. C. Knight, A. V. Totmenin, V. E. Chizhikov, J. M. Parsons, P. F. Safronov, V. V. Gutorov, S. N. Shchelkunov, and J. J. Esposito. 1996. Terminal region sequence variations in variola virus DNA. Virology 221291-300. [DOI] [PubMed] [Google Scholar]
- 28.McFadden, G. 2005. Poxvirus tropism. Nat. Rev. Microbiol. 3201-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Means, R. E., J. K. Choi, H. Nakamura, Y. H. Chung, S. Ishido, and J. U. Jung. 2002. Immune evasion strategies of Kaposi's sarcoma-associated herpesvirus. Curr. Top. Microbiol. Immunol. 269187-201. [DOI] [PubMed] [Google Scholar]
- 30.Moss, B. 2001. Poxviridae: the viruses and their replication, p. 2849-2883. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology. Lippincott/Williams and Wilkins Co., Philadelphia, PA.
- 31.Mullick, J., J. Bernet, Y. Panse, S. Hallihosur, A. K. Singh, and A. Sahu. 2005. Identification of complement regulatory domains in vaccinia virus complement control protein. J. Virol. 7912382-12393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mullick, J., J. Bernet, A. K. Singh, J. D. Lambris, and A. Sahu. 2003. Kaposi's sarcoma-associated herpesvirus (human herpesvirus-8) open reading frame 4 protein (Kaposica) is a functional homolog of complement control proteins. J. Virol. 773878-3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mullick, J., A. Kadam, and A. Sahu. 2003. Herpes and pox viral complement control proteins: ‘the mask of self.’ Trends Immunol. 24500-507. [DOI] [PubMed] [Google Scholar]
- 34.Mullick, J., A. K. Singh, Y. Panse, V. Yadav, J. Bernet, and A. Sahu. 2005. Identification of functional domains in Kaposica, the complement control protein homolog of Kaposi's sarcoma-associated herpesvirus (human herpesvirus-8). J. Virol. 795850-5856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murphy, P. M. 2001. Viral exploitation and subversion of the immune system through chemokine mimicry. Nat. Immunol. 2116-122. [DOI] [PubMed] [Google Scholar]
- 36.Murthy, K. H., S. A. Smith, V. K. Ganesh, K. W. Judge, N. Mullin, P. N. Barlow, C. M. Ogata, and G. J. Kotwal. 2001. Crystal structure of a complement control protein that regulates both pathways of complement activation and binds heparan sulfate proteoglycans. Cell 104301-311. [DOI] [PubMed] [Google Scholar]
- 37.Nieba, L., S. E. Nieba-Axmann, A. Persson, M. Hamalainen, F. Edebratt, A. Hansson, J. Lidholm, K. Magnusson, A. F. Karlsson, and A. Pluckthun. 1997. BIACORE analysis of histidine-tagged proteins using a chelating NTA sensor chip. Anal. Biochem. 252217-228. [DOI] [PubMed] [Google Scholar]
- 38.Pan, Q., R. O. Ebanks, and D. E. Isenman. 2000. Two clusters of acidic amino acids near the NH2 terminus of complement component C4 alpha'-chain are important for C2 binding. J. Immunol. 1652518-2527. [DOI] [PubMed] [Google Scholar]
- 39.Pangburn, M. K. 2000. Host recognition and target differentiation by factor H, a regulator of the alternative pathway of complement. Immunopharmacology 49149-157. [DOI] [PubMed] [Google Scholar]
- 40.Peitsch, M. C. 1995. Protein modeling by e-mail. Bio/Technology 13658-660. [Google Scholar]
- 41.Reeves, P. M., B. Bommarius, S. Lebeis, S. McNulty, J. Christensen, A. Swimm, A. Chahroudi, R. Chavan, M. B. Feinberg, D. Veach, W. Bornmann, M. Sherman, and D. Kalman. 2005. Disabling poxvirus pathogenesis by inhibition of Abl-family tyrosine kinases. Nat. Med. 11731-739. [DOI] [PubMed] [Google Scholar]
- 42.Rosengard, A. M., L. C. Alonso, L. C. Korb, W. M. Baldwin III, F. Sanfilippo, L. A. Turka, and J. M. Ahearn. 1999. Functional characterization of soluble and membrane-bound forms of vaccinia virus complement control protein (VCP). Mol. Immunol. 36685-697. [DOI] [PubMed] [Google Scholar]
- 43.Rosengard, A. M., Y. Liu, Z. Nie, and R. Jimenez. 2002. Variola virus immune evasion design: expression of a highly efficient inhibitor of human complement. Proc. Natl. Acad. Sci. USA 998808-8813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sahu, A., S. N. Isaacs, A. M. Soulika, and J. D. Lambris. 1998. Interaction of vaccinia virus complement control protein with human complement proteins: factor I-mediated degradation of C3b to iC3b1 inactivates the alternative complement pathway. J. Immunol. 1605596-5604. [PubMed] [Google Scholar]
- 45.Sahu, A., and J. D. Lambris. 2001. Structure and biology of complement protein C3, a connecting link between innate and acquired immunity. Immunol. Rev. 18035-48. [DOI] [PubMed] [Google Scholar]
- 46.Samuel, C. E. 2007. Innate immunity minireview series: making biochemical sense of nucleic acid sensors that trigger antiviral innate immunity. J. Biol. Chem. 28215313-15314. [DOI] [PubMed] [Google Scholar]
- 47.Schwede, T., J. Kopp, N. Guex, and M. C. Peitsch. 2003. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res. 313381-3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sfyroera, G., M. Katragadda, D. Morikis, S. N. Isaacs, and J. D. Lambris. 2005. Electrostatic modeling predicts the activities of orthopoxvirus complement control proteins. J. Immunol. 1742143-2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shchelkunov, S. N., V. M. Blinov, S. M. Resenchuk, A. V. Totmenin, L. V. Olenina, G. B. Chirikova, and L. S. Sandakhchiev. 1994. Analysis of the nucleotide sequence of 53 kbp from the right terminus of the genome of variola major virus strain India-1967. Virus Res. 34207-236. [DOI] [PubMed] [Google Scholar]
- 50.Singh, A. K., J. Mullick, J. Bernet, and A. Sahu. 2006. Functional characterization of the complement control protein homolog of herpesvirus saimiri: R118 is critical for factor I cofactor activities. J. Biol. Chem. 28123119-23128. [DOI] [PubMed] [Google Scholar]
- 51.Smith, G. L., J. A. Symons, A. Khanna, A. Vanderplasschen, and A. Alcami. 1997. Vaccinia virus immune evasion. Immunol. Rev. 159137-154. [DOI] [PubMed] [Google Scholar]
- 52.Soames, C. J., and R. B. Sim. 1997. Interactions between human complement components factor H, factor I and C3b. Biochem. J. 326553-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Soares, D. C., and P. N. Barlow. 2005. Complement control protein modules in the regulators of complement activation, p. 19-62. In D. Morikis and J. D. Lambris (ed.), Structural biology of the complement system. Taylor & Francis, New York, NY.
- 54.Stoiber, H., M. Pruenster, C. G. Ammann, and M. P. Dierich. 2005. Complement-opsonized HIV: the free rider on its way to infection. Mol. Immunol. 42153-160. [DOI] [PubMed] [Google Scholar]
- 55.Vanderplasschen, A., E. Mathew, M. Hollinshead, R. B. Sim, and G. L. Smith. 1998. Extracellular enveloped vaccinia virus is resistant to complement because of incorporation of host complement control proteins into its envelope. Proc. Natl. Acad. Sci. USA 957544-7549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wyatt, L. S., P. L. Earl, L. A. Eller, and B. Moss. 2004. Highly attenuated smallpox vaccine protects mice with or without immune deficiencies against pathogenic vaccinia virus challenge. Proc. Natl. Acad. Sci. USA 1014590-4595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang, L., and D. Morikis. 2006. Immunophysical properties and prediction of activities for VCP and SPICE using molecular dynamics and electrostatics. Biophys. J. 903106-3119. [DOI] [PMC free article] [PubMed] [Google Scholar]