Abstract
The herpes simplex virus (HSV) ICP27 immediate-early protein plays an essential role in the expression of viral late genes. ICP27 is a multifunctional protein and has been reported to regulate multiple steps of mRNA synthesis and processing, including transcription, splicing, and nuclear export. Recently, ICP27 was reported to interact with translation factors and to stimulate translation of the viral late mRNA encoding VP16. We examined the effects of ICP27 on accumulation, nuclear export, and translation of HSV 1 (HSV-1) late mRNAs encoding VP16, ICP5, and gD. We confirm here that ICP27 stimulates translation of VP16 mRNA as well as an additional HSV-1 late ICP5 mRNA. The data presented here demonstrate that translation levels of both VP16 and ICP5 mRNA is reduced during infections with the ICP27-null virus mutant d27-1, and with ICP27 C-terminal deletion mutant viruses n406 and n504, compared to wild-type virus. In contrast, the translation of gD mRNA is not affected by the presence of ICP27 during infection. These data demonstrate that ICP27 functions to increase the translation levels of a subset of HSV-1 late genes, and this function requires the C terminus of ICP27.
Herpes simplex virus 1 (HSV-1) gene expression is regulated in a temporal pattern, where immediate-early, early, and late genes are expressed sequentially, and each group is dependent upon the expression of the preceding group of proteins. HSV-1 late genes can be further subdivided into two classes: γ1 leaky-late genes, and γ2 true-late genes. True-late genes require immediate-early and early proteins, as well as viral DNA replication for efficient expression, while leaky-late expression occurs at low levels prior to DNA replication (reviewed in reference 35). ICP27 is required for efficient expression of a subset of early genes involved in viral DNA replication, as well as for efficient late gene expression (34, 36, 47). Furthermore, we have shown that ICP27 is required for the transcriptional activation of viral late genes, including gC, UL44, and UL47 (17).
It is well documented that in conjunction with ICP4 and ICP0, ICP27 acts as both a positive and a negative regulator of viral transcription, and this generates a switch in viral gene expression from early to late genes (26, 32-34, 36). ICP27 has been shown to interact with a variety of cellular proteins involved in both transcriptional and posttranscriptional processes, including serine/arginine-rich proteins, casein kinase 2, heterogeneous nuclear ribonucleoprotein K, RNA polymerase II, Aly-REF, and TAP, and it is thought that its functions are mediated through interactions with these proteins (4, 5, 7, 38, 39, 48, 51). However, the exact mechanism by which ICP27 regulates mRNA synthesis and processing remains unclear. ICP27 acts posttranscriptionally to inhibit mRNA splicing, in part by redistributing splicing proteins (12, 31). In addition, ICP27 has been reported to increase poly(A) processing, as well as viral mRNA export from the nucleus to the cytoplasm (24, 25, 37).
ICP27 contains both a nuclear export and a nuclear localization signal, which allow it to shuttle between the nucleus and the cytoplasm (13, 27, 44). ICP27 also contains an arginine- and glycine-rich motif, the RGG-box, which allows it to bind to RNA (28). In addition, ICP27 is known to interact with members of the cellular RNA export machinery Aly/REF and TAP and was shown to stimulate the nuclear export of mRNAs via the REF/TAP pathway in Xenopus laevis oocytes (5, 20, 28). However, more recent studies have shown that ICP27 is not required in HSV-infected cells for efficient cytoplasmic accumulation of at least certain HSV-1 mRNAs, including the late VP16, gB, and gC mRNAs (8, 29).
Recent studies have also predicted a role for ICP27 in regulating translation of viral mRNAs. We showed that ICP27 interacts with translation factors (10), and Ellison et al. (9) showed that ICP27 increases translation of VP16 mRNA. ICP27 cosediments with polyribosomes (21), suggesting that ICP27 associates with polyribosomes. Finally, when ICP27 was tethered to an mRNA, it increased translation of the mRNA after injection into Xenopus oocytes (21).
MATERIALS AND METHODS
Cells and viruses.
Vero cells obtained from the American Type Culture Collection were maintained in Dulbecco's modified Eagle medium (DMEM) (Gibco) supplemented with heat-inactivated 5% fetal calf serum and 5% bovine calf serum (Invitrogen) in a humidified 5% CO2 atmosphere at 37°C.
The HSV-1 wild-type (WT) KOS1.1 strain (15), originally obtained from M. Levine (University of Michigan, Ann Arbor), was propagated and titrated on Vero cells. The ICP27 mutant viruses d27-1, n504, and n406 were derived from KOS1.1 and were titrated on V27 cell line as described elsewhere (33).
Antibodies.
The ICP5 and gD mouse monoclonal antibodies were purchased from EastCoast Biol. Anti-TIF (VP16) rabbit serum was provided by T. Kristie, National Institute of Allergy and Infectious Diseases (49).
Viral infections, metabolic labeling, and immunoprecipitations.
Vero cells were grown in 100-mm2 tissue culture dishes to 90% confluence at the time of infection and infected at a multiplicity of infection (MOI) of 20 PFU per cell with WT, d27-1, n504, or n406 virus diluted in cold phosphate-buffered saline containing 0.1% glucose. After 1 h adsorption at 37°C, cells were switched into DMEM containing 1% heat-inactivated bovine calf serum. At 5.5 h postinfection (hpi) the medium was removed and replaced with 3 ml of methionine-cysteine-free DMEM (Gibco) containing 100 μCi of [35S]methionine-cysteine protein labeling mix (Perkin-Elmer)/ml. Cells were harvested at 6 hpi by scraping them into the medium, and cells from four dishes containing the same virus infections were pooled. After one wash in cold PBS, cells were divided into aliquots in four 1.5-ml microcentrifuge tubes. One set of samples was set aside for RNA isolation. The remaining samples were incubated on ice for 30 min in 1 ml of immunoprecipitation buffer (140 mM Tris-HCl [pH 7.5], 20 mM NaCl, 0.5% sodium deoxycholate, 1 mg of bovine serum albumin/ml, 1% NP-40, 10 mM β-glycerophosphate, 5 mM NaF, 1 mM phenylmethylsulfonyl fluoride, 1 Complete mini protease inhibitor cocktail tablet [Roche Applied Science, Indianapolis, IN] per 10 ml). Cell lysates were clarified by centrifugation at 10,000 × g at 4°C for 10 min and precleared overnight by incubation with protein A-agarose beads at 4°C. The appropriate antibody was added at a concentration of 2.5 μg per 1-ml reaction and incubated at 4°C for 2 h. After four washes in wash buffer (140 mM Tris-HCl [pH 7.5], 20 mM NaCl, 10% NP-40, 1 mM phenylmethylsulfonyl fluoride), the immunoprecipitates were dissolved in gel sample buffer for sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
RNA isolation.
Nuclear and cytoplasmic RNA fractions were isolated from infected Vero cells (as described above) by using the RNeasy purification kit (Qiagen) according to the manufacturer's instructions for adherent cells. Cytoplasmic RNA fractions were isolated according to the Qiagen RNeasy protocol for the isolation of cytoplasmic RNA. Subsequently, RNA was isolated from nuclear pellets according to the Qiagen RNeasy protocol for the isolation of total RNA. Total RNA for analysis of RNA levels was isolated by using the RNeasy purification kit for isolation of total RNA or using TRIzol (Gibco) according to the manufacturer's instructions for the isolation of RNA from cells grown in a monolayer.
Northern blot hybridization and analysis.
[32P]dCTP- and [32P]dATP-labeled DNA probes (Perkin-Elmer) were generated by using the random primer DNA labeling system (Gibco) according to the manufacturer's instructions or T4 polynucleotide kinase (New England Biolabs) according to the manufacturer's instructions. The DNA templates used to make various DNA probes were as follows: for the VP16 (UL48) gene, the 437-bp template was amplified by PCR from viral DNA by using the primers 5′-TGGGCAGCGTTGATAGGAAT-3′ and 5′-GTTTGGGGGTTTTCTCTTCC-3′; for the ICP5 (UL19) gene, the 203-bp probe was amplified by PCR from viral DNA by using the primers 5′-CTTAGCACGATCGAGGT-3′ and 5′-GTTCATGTAGGCCAAGCT-3′; and for the gD (US6) gene, the 342-bp probe was amplified by PCR from viral DNA by using the primers 5′-CCGTGATTTTGTTTGTCGTCATAGTGGGC-3′ and 5′-CAAGCGATGGTCAGGTTGTAGGGTTGTTTC-3′. To probe for the U3 snRNA, radiolabeled oligonucleotide 5′-ACCACTCAGACCGCGTTCTCTCCCTCTCAC-3′ was used as a probe (6). All oligonucleotides were obtained from Invitrogen. GAPDH (glyceraldehyde-3-phosphate dehydrogenase) mRNA levels were determined by using 1.3-kb PstI fragment of pUC19-GAPDH as a probe (2). RNA samples were resolved in a 1% denaturing agarose (SeaKem) gel containing formaldehyde in 3[N-morpholino]propanesulfonic acid. Hybridizations with double-stranded DNA probes were carried out in QuickHyb hybridization buffer (Stratagene) at 68°C for 1 h. Hybridizations with oligonucleotides were carried out in ULTRAhyb-Oligo (Ambion) at 42°C for 16 h, and the filters were exposed to a phosphorimager screen (Amersham Biosciences) and then to Biomax XAR film (Kodak). Quantification of bands on all Northern blots was carried out by using a Typhoon PhosphorImager (GE Healthcare) with ImageQuant software. Levels of viral mRNAs were normalized to GAPDH RNA levels to correct for recovery, because GAPDH levels at 6 hpi have been shown to be similar to levels in mock-infected cells (40). To ensure that the assay yielded accurate quantification, a 10-fold dilution of total RNA from WT-infected cells was resolved, along with WT and mutant samples, and the values obtained represented a 10-fold decrease in total WT RNA for the diluted samples (results not shown).
SDS-PAGE and phosphorimager analysis.
Proteins in the immunoprecipitates were resolved by SDS-PAGE in bis-acrylamide cross-linked 10% polyacrylamide gels at 100 V. Gels were fixed in 10% acetic acid-20% methanol and dried under vacuum for 2 h at 65°C. Dried gels were exposed to a phosphorimager screen (Amersham Biosciences) and then to Biomax XAR film (Kodak). Quantification of protein bands was carried out by using a Typhoon PhosphorImager (GE Healthcare) with ImageQuant software. To ensure that the assay yielded accurate quantification, a dilution series of immunoprecipitate from WT-infected cells (30, 15, 7.5, and 3.75 μl) was used to construct a standard curve, which showed a linear response.
RESULTS
ICP27 is required for the efficient expression of viral proteins VP16, ICP5, and gD.
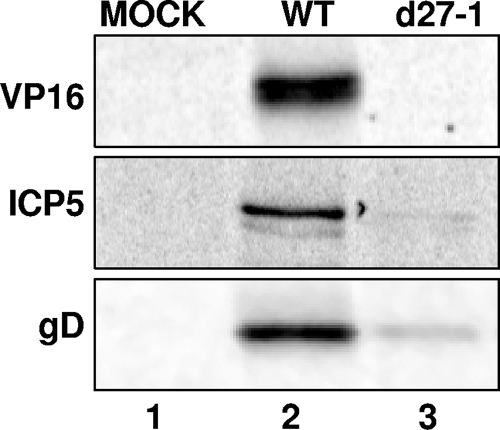
A recent report (8) demonstrated that ICP27 increases the translation of the HSV-1 late mRNA encoding VP16. We wanted to determine whether ICP27 was specifically stimulating VP16 mRNA translation or whether ICP27 can stimulate translation of additional HSV-1 late mRNAs. We therefore examined expression of two additional late proteins, ICP5 and gD, as well as VP16. We mock infected or infected Vero cells with HSV-1 WT or d27-1 (an ICP27 deletion mutant virus) at an MOI of 20. Cells were pulse-labeled with [35S]methionine-cysteine from 5.5 to 6 hpi and harvested. Cells were lysed in immunoprecipitation buffer, and viral proteins were immunoprecipitated from lysates using antibodies specific for VP16, ICP5, or gD. Immunoprecipitates were resolved on an SDS-polyacrylamide gel, and exposed to a phosphorimager screen. Protein synthesis levels were dramatically reduced for VP16, ICP5, and gD proteins during d27-1 infection compared to WT infection (Fig. 1, lane 3). Therefore, all three proteins were expressed at greatly reduced levels in the absence of ICP27.
FIG. 1.
The expression of HSV-1 VP16, ICP5, and gD late proteins is dramatically decreased in the absence of ICP27. Vero cells were either mock infected (lane 1) or infected at an MOI of 20 with HSV-1 WT (lane 2) or d27-1 virus (lane 3). Cells were pulse-labeled with [35S]methionine-cysteine at 5.5 hpi and harvested at 6 hpi. Lysates were immunoprecipitated by using an antibody specific for VP16, ICP5, or gD, and the immunoprecipitates were resolved on a SDS polyacrylamide gel. The resulting autoradiogram is shown.
ICP27 is not required for cytoplasmic accumulation of ICP5 or gD mRNAs.
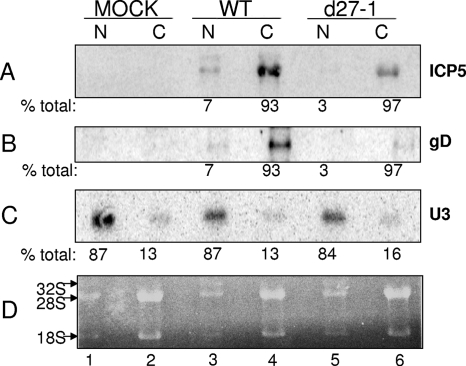
To determine whether the decrease in protein expression observed during d27-1 infection was due to a defect in mRNA export, we examined cytoplasmic accumulation of viral mRNAs during infection in the presence or absence of ICP27. We mock infected or infected Vero cells with HSV-1 WT or d27-1 at an MOI of 20. Cells were harvested at 6 hpi, and the total RNA was isolated from nuclear and cytoplasmic fractions. Equal amounts of total nuclear and cytoplasmic RNA were resolved in a denaturing formaldehyde gel (Fig. 2A, B, and C). Consistent with effective separation of nuclei and cytoplasm (30), the 32S rRNA precursor was observed in the nuclear RNA fractions and was absent from the cytoplasmic fractions, and the mature 28S and 18S rRNAs were located predominantly in the cytoplasm (Fig. 2D). This demonstrated that our isolation of RNA from separate nuclear or cytoplasmic cellular fractions was successful. Nucleic acids from this gel were transferred to a nylon membrane for Northern analysis. The membrane was first probed with an oligonucleotide specific for U3 snRNA (6, 46) to quantify the amount of nuclear RNA contamination in the cytoplasm (Fig. 2C). As expected from the RNA gel, the levels of U3 nuclear RNA contamination in the cytoplasm were very low for mock, WT, and d27-1 infections at 13 to 16% (Fig. 2C, lanes 2, 4, and 6), whereas the nucleus contained 84 to 87% of the total cellular U3 RNA (Fig. 2C, lanes 1, 3, and 5). This provides further evidence that fractionation of the nuclei and cytoplasm was successful. The membrane was stripped and reprobed for ICP5 or gD mRNAs. The total amount of ICP5 mRNA from d27-1 and the WT nuclear and cytoplasmic fractions was quantitated. The total ICP5 mRNA from the d27-1 infection expressed as a percentage of the total from the WT infection was ca. 26%, and the total gD mRNA was ca. 16% (data not shown), which are values consistent with subsequent studies (see Fig. 4 and 5). In WT infections, ICP5 and gD cytoplasmic mRNA accumulation was similar to d27-1 infections, with 93% of the total ICP5 or gD mRNA located in the cytoplasm (Fig. 2A and B, lanes 3 and 4). In d27-1 infections the majority (97%) of ICP5 and gD mRNAs was located in the cytoplasmic fraction, whereas the nuclear fraction contains only 3% of the total (Fig. 2A and B, lanes 5 and 6). Thus, in the presence or absence of ICP27 during infection, ICP5 and gD mRNAs were exported from the nucleus to the cytoplasm with equal efficiency. The membrane was also stripped and reprobed for VP16 mRNA, and similar results were obtained (data not shown). Consistent with observations from other laboratories for other HSV-1 mRNAs (8, 29), we concluded from these data that ICP27 is not required for efficient export of VP16, ICP5, or gD mRNAs into the cytoplasm.
FIG. 2.
ICP5 and gD mRNAs accumulate in the cytoplasm in the absence of ICP27. Vero cells were either mock infected (lanes 1 and 2) or infected at an MOI of 20 with HSV-1 WT (lanes 3 and 4) or d27-1 (lanes 5 and 6). Cells were harvested at 6 h postinfection, and RNA was isolated from nuclear (N) and cytoplasmic (C) fractions. Equal amounts of total RNA were resolved on a denaturing formaldehyde agarose gel stained with ethidium bromide. The positions of 28S and 18S rRNAs are indicated on the left side in panel D. The membrane was hybridized with a probe specific for U3 snRNA (C), stripped, and rehybridized with a probe specific for ICP5 (A) or gD (B). The percent total values represent the percentage for each fraction (N or C) relative to the total for the sample.
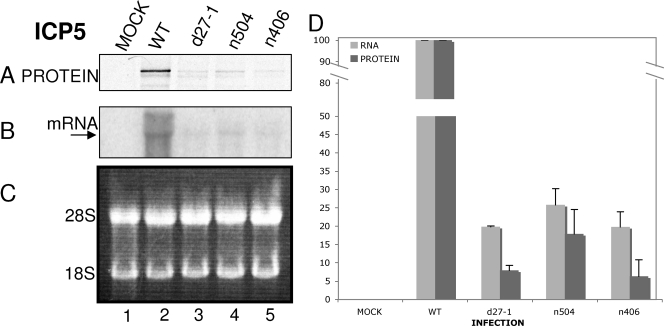
FIG. 4.
ICP5 synthesis rates are decreased in the absence of ICP27, and this effect requires the C terminus of ICP27. Vero cells were either mock infected (lane 1) or infected at an MOI of 20 with HSV-1 WT (lane 2), d27-1 (lane 3), n504 (lane 4), or n406 (lane 5) viruses. Cells were pulse-labeled with [35S]methionine-cysteine (100 μCi/ml) for 30 min at 5.5 hpi and harvested at 6 hpi. Aliquots of cells were lysed or used for RNA isolation as described in Materials and Methods. (A) For protein synthesis analysis, lysates were immunoprecipitated by using a monoclonal antibody to ICP5. Immunoprecipitates were resolved on an SDS-polyacrylamide gel. (B) For mRNA accumulation, the membrane was incubated with a probe specific for ICP5 mRNA, stripped, and rehybridized with a probe specific for GAPDH mRNA (data not shown). (C) Equivalent concentrations of total RNA were resolved on a denaturing formaldehyde agarose gel stained with ethidium bromide. The positions of 28S and 18S rRNAs are indicated on the left of the panel. (D) The levels of ICP5 protein and RNA for mock, HSV-1 WT, d27-1, n504, and n406 infections were quantified relative to the HSV-1 WT infection levels and are represented here as a percentage. The data shown are representative of three experiments.
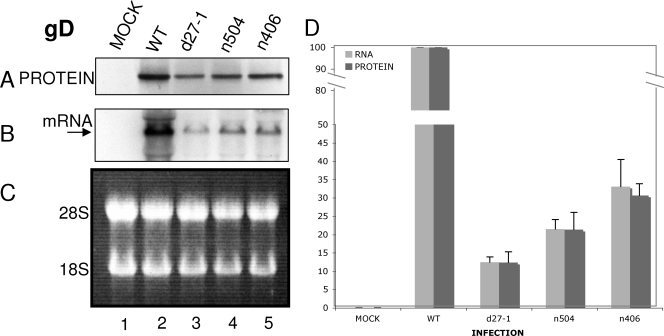
FIG. 5.
gD synthesis rates are not affected by ICP27. Vero cells were either mock infected (lane 1) or infected at an MOI of 20 with HSV-1 WT (lane 2), d27-1 (lane 3), n504 (lane 4), or n406 (lane 5) viruses. Cells were pulse-labeled with [35S]methionine-cysteine (100 μCi/ml) for 30 min at 5.5 hpi and harvested at 6 hpi. Aliquots of cells were lysed or used for RNA isolation as described in Materials and Methods. (A) For protein synthesis analysis, lysates were immunoprecipitated using a monoclonal antibody to gD. Immunoprecipitates were resolved on an SDS-polyacrylamide gel. (B) For mRNA accumulation, the membrane was incubated with a probe specific for gD, stripped, and rehybridized with a probe specific for GAPDH (data not shown). (C) Equivalent amounts of total RNA were resolved on a denaturing formaldehyde agarose gel stained with ethidium bromide. The positions of 28S and 18S rRNAs are indicated to the left of the panel. (D) The levels of VP16 protein and RNA for mock, HSV-1 WT, d27-1, n504, and n406 infections were quantified relative to WT levels and normalized to GAPDH levels. The levels are represented here as a percentage of the WT levels. The data shown are representative of three experiments.
ICP27 increased translation of VP16 and ICP5 mRNAs during HSV-1 infection, but not of gD mRNA.
It had been documented that ICP27 affects the accumulation of mRNA of HSV-1 late genes; therefore, to examine translation of these mRNAs it was necessary to quantify the amount of mRNA present to account for effects on the mRNA prior to the initiation of translation (17). For measurement of translation efficiencies, we compared the steady-state levels of mRNA to the levels of protein synthesis and the normalized levels during infections with mutant ICP27 viruses to the levels obtained during infection with WT virus. Vero cells were either mock infected or infected at an MOI of 20 with HSV-1 WT, d27-1, n504, or n406 viruses (Fig. 3A, 4A, and 5A, lanes 1 to 5). Cells were pulse-labeled with [35S]methionine-cysteine at 5.5 hpi and harvested at 6 hpi. Aliquots of the cells were either lysed or set aside for RNA isolation. Viral proteins were immunoprecipitated from lysates using antibodies specific for VP16, ICP5, or gD, and immunoprecipitates were resolved by SDS-PAGE (Fig. 3A, 4A, and 5A, respectively). For Northern analysis, equivalent amounts of total RNA were resolved on a denaturing formaldehyde agarose gel stained with ethidium bromide. The positions of 28S and 18S rRNAs are indicated in the figures (Fig. 3C, 4C, and 5C, the left side of each panel). The nucleic acids were transferred onto a nylon membrane, and the membrane was hybridized with a probe specific for VP16, ICP5, or gD mRNAs (Fig. 3B, 4B, and 5B, respectively), stripped, and rehybridized with a probe specific for GAPDH (data not shown). All viral mRNA values were normalized to GAPDH mRNA values, and no statistically significant changes in levels of GAPDH were observed between mock and infected samples in these samples (data not shown).
FIG. 3.
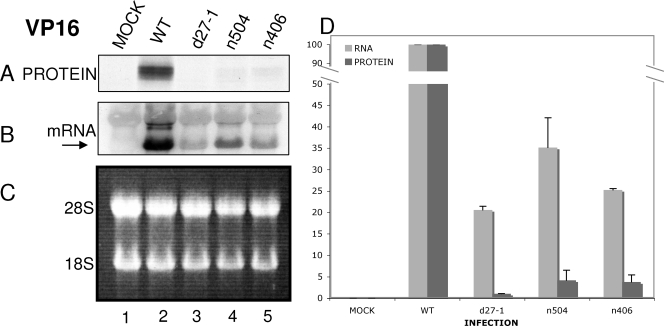
VP16 synthesis rates are decreased in the absence of ICP27, and this effect requires the C terminus of ICP27. Vero cells were either mock infected (lane 1) or infected at an MOI of 20 with HSV-1 WT (lane 2), d27-1 (lane 3), n504 (lane 4), or n406 (lane 5) viruses. Cells were pulse-labeled with [35S]methionine-cysteine (100 μCi/ml) for 30 min at 5.5 hpi and harvested at 6 hpi. Aliquots of cells were lysed or used for RNA isolation as described in Materials and Methods. (A) For protein synthesis analysis, lysates were immunoprecipitated using an antibody to VP16. Immunoprecipitates were resolved on an SDS-polyacrylamide gel. (B) For mRNA accumulation, the membrane was incubated with a probe specific for VP16 mRNA, stripped, and rehybridized with a probe specific for GAPDH mRNA (results not shown). (C) Equivalent concentrations of total RNA were resolved on a denaturing formaldehyde agarose gel stained with ethidium bromide. The positions of 28S and 18S rRNAs are indicated to the left of the panel. (D) The levels of VP16 protein and RNA for mock and HSV-1 WT, d27-1, n504, and n406 infections were quantified relative to the WT levels and normalized to the GAPDH mRNA levels. The levels are represented here as a percentage of the HSV-1 WT levels. The data shown are representative of three experiments.
The data shown in Fig. 3, 4, and 5 are the averages of results from three separate experiments for mRNA and protein levels. The VP16 mRNA levels in d27-1-infected cells were ca. 20% of levels in WT virus-infected cells, and the VP16 protein levels in d27-1-infected cells were ca. 1% of levels in WT virus-infected cells (Fig. 3D), which is an ∼20-fold decrease in protein expression. ICP5 mRNA levels in d27-1-infected cells were ca. 20% of levels in WT virus-infected cells, and ICP5 protein levels in d27-1-infected cells were ca. 8% of levels in WT virus-infected cells (Fig. 4D), which is an approximately two- to threefold decrease in protein expression. It appeared that ICP27 can increase protein expression of ICP5 mRNA, but the effect was not as dramatic as that observed with VP16. gD mRNA levels in d27-1-infected cells were ca. 14% of levels in WT virus-infected cells, and gD proteins levels in d27-1-infected cells were ca. 13% of levels in WT virus-infected cells (Fig. 5D). Thus, gD mRNA translation was equivalent in the presence or absence of ICP27. Therefore, these data suggested that ICP27 can increase the translation of specific late mRNAs.
The C terminus of ICP27 is required for increased translation of ICP5 and VP16 mRNAs.
Because the C terminus of ICP27 has been shown to be important for stimulating translation in an mRNA reporter assay system, we determined the effect on protein synthesis during infection with the two C-terminal deletion viruses n504 and n406, which lack the C-terminal 8 and 106 amino acid residues of ICP27, respectively (Fig. 3, 4, and 5) (21). Experiments were performed as described above. VP16 mRNA levels in n504-infected cells were ca. 35% of the levels in WT virus-infected cells, and VP16 proteins levels in n504-infected cells were ca. 4% of the levels in WT virus-infected cells (Fig. 3D), which is an ∼9-fold decrease in protein expression. VP16 mRNA levels in n406-infected cells were ca. 25% of the levels in WT virus-infected cells, and VP16 proteins levels in n406-infected cells were ca. 4% of the levels in WT virus-infected cells (Fig. 3D), which is an ∼6-fold decrease in protein expression. These data indicated that the C terminus of ICP27 was required for efficient translation of VP16 mRNA.
ICP5 mRNA levels in n504-infected cells were ca. 25% of the levels in WT virus-infected cells, and ICP5 proteins levels in n504-infected cells were ca. 18% of the levels in WT virus-infected cells (Fig. 4D), so reduction in mRNA and expression were similar. However, ICP5 mRNA levels in n406-infected cells were ca. 20% of levels in WT virus-infected cells, and ICP5 proteins levels in n406-infected cells were ca. 6% of levels in WT virus-infected cells (Fig. 4D), which is an ∼3-fold decrease in protein expression. These data suggested that the C terminus of ICP27 was required for the effect observed on ICP5 protein expression, because the effect was similar to that observed during infection in the absence of ICP27. However, unlike what we observed for VP16, ICP5 protein expression did not appear to be affected by the last eight amino acid residues of the C terminus. As observed for gD during infections with d27-1, we also observed that infections with n504 and n406 generated an mRNA/protein ratio equivalent to that for WT virus (Fig. 5D). Accordingly, gD protein expression did not appear to be affected by the C terminus of ICP27. We can conclude from the data presented here that ICP27 increases the translation of certain late mRNAs but exhibits no effect on the translation of at least one other late mRNA.
DISCUSSION
In this study we present evidence that ICP27 increases the translation of a subset of HSV mRNAs and that the C terminus of ICP27 is important for this function. We found that ICP27 increased ICP5 mRNA translation, in addition to VP16 mRNA translation, but had no effect on gD mRNA translation. We examined translation during infection with two ICP27 C terminus truncation mutant viruses, n504 and n406, from which the last 8 and 106 amino acids, respectively, are deleted. We found that for VP16 mRNA an intact C terminus is required for efficient translation because both the n504 and the n406 deletions showed decreases in translation similar to that observed for the ICP27-null virus. For ICP5 mRNA, the n406 deletion showed similar decreases in translation to the ICP27-null virus, while the n504 deletion showed a less dramatic decrease for translation. However, the n504 deletion did still appear to decrease translation of ICP5 mRNA, and it can be concluded that an intact C terminus is required for efficient translation of both VP16 and ICP5 mRNA.
Role of ICP27 in late mRNA translation.
The multifunctional ICP27 protein has been studied extensively and has been proposed to play multiple roles in mRNA metabolism from transcription to posttranscriptional processes and, more recently, to play a role in increasing translation (8, 21). However, the exact mechanisms of many of its functions remain unclear. We previously showed that ICP27 interacts with translation factors during HSV-1 infection (10), and in the present study we have shown that ICP27 also increases the translation rates of certain HSV-1 late genes. It may be that interactions with translation factors are required for efficient translation of specific viral mRNAs. ICP27 could be transporting viral mRNAs to sites of translation through these interactions. Another idea is that ICP27 could be loading translation factors onto viral mRNAs in the nucleus. It has been shown that translation factors PABP and eIF3, which have both been shown to associate with ICP27, are present in the nucleus (1, 10, 14, 41).
In our experiments we observed that ICP27 is required for efficient translation of VP16 mRNA, which is consistent with observations made by another group (8), as well as having a moderate effect on translation of ICP5 mRNA. We did not observe any effect of ICP27 on translation of gD mRNA.
A previous study, which compared the rates of gD protein synthesis and mRNA accumulation at various times postinfection, suggested that gD expression was regulated translationally because these researchers found that gD protein levels began to decline even as mRNA accumulation increased during infection (19). However, another study found that the rates of synthesis for gD mRNA closely correlated with levels of polysome associated gD mRNA over the course of infection (42), and this is consistent with our results examining gD translation during WT and ICP27 mutant infections.
Laurent et al. (22) showed that over the course of HSV-1 infection for all viral mRNAs there is a gradual shutoff of viral mRNA translation caused by a shift from polyribosomes to monoribosomes, a finding similar to that observed for cellular mRNAs. VP16 mRNA was shown to be associated with polyribosomes at 12 hpi during WT infection of VP16 mRNA, and only in the absence of ICP27 shifted to monoribosomes (8). However, it may be more informative to examine ribosome association over a time course of infection, because any actual shift would not have been observed at only one time point. ICP27 has also been observed in association with polyribosomes during infection (21); thus, it is possible that if the shift is occurring for all viral and cellular mRNAs that ICP27 plays some role in delivering select mRNAs to newly made polyribosomes. Because ICP27 seems to act in a selective manner to increase VP16 and ICP5 mRNA translation, it would be informative to determine whether ICP27 also can increase the association of ICP5 mRNA with polyribosomes.
The selectivity exhibited by ICP27 does not appear to be sequence specific because a sequence comparison of VP16 and ICP5 5′ and 3′ untranslated regions showed no obvious similarity (data not shown). However, there may be a structural component of similarity that is not obvious by sequence comparison. In addition, ICP27 binds to multiple HSV-1 mRNAs, including VP16 and ICP5; however, no similar region or motif among the RNAs has been reported (37, 43).
ICP27 and mRNA Export.
Previously, ICP27 was reported to be required for efficient nuclear export of viral mRNAs (37). However, our results, as well as those of two previous studies (8, 29), showed that HSV-1 late mRNAs VP16, ICP5, gB, gC, and gD are all exported into the cytoplasm in the absence of ICP27. In the present study we showed that while the total abundance of ICP5 and gD mRNAs is reduced during infection in the absence of ICP27, the mRNAs that are present during d27-1 infection accumulate in the cytoplasm as efficiently as during HSV-1 WT infection. Taken together, these data demonstrate that ICP27 is not required for efficient nuclear export of these viral late mRNAs and possibly more viral late mRNAs.
ICP27 shuttles from the nucleus to the cytoplasm, binds to HSV-1 mRNAs, and interacts with members of the cellular mRNA export machinery Aly/REF and TAP (3, 5, 16, 27, 28, 44). It may be that the interactions of ICP27 with Aly/REF and TAP facilitate the export of viral mRNAs during WT infection, since Koffa et al. (20) demonstrated that ICP27 stimulates the export of viral intronless mRNAs, but not cellular mRNAs, after microinjection into X. laevis oocytes, although these interactions are not required for late mRNAs to be exported. It is possible that viral late mRNAs are exported via a different pathway in the absence of ICP27 or that viral late mRNAs continue to be exported via the REF/TAP pathway even in the absence of ICP27. In addition, TAP has been reported to play a role in promoting the translation of unspliced mRNA (18), and Aly/REF has been reported to enhance transcriptional promoter activity (45). It may be that ICP27 interactions with Aly/REF and TAP facilitate late mRNA transcription and translation, respectively, and that the role in late mRNA export is minimal. Chen et al. (4) showed that Aly/REF, but not TAP, is recruited by ICP27 to viral transcription sites during WT infection, which leads one to speculate that Aly/REF may play a role in activating viral transcription. It remains to be determined whether any of these effects are operative in HSV-infected cells.
Multifunctional C terminus of ICP27.
The C terminus of ICP27, which is involved in numerous protein-protein interactions, is also necessary for its effects on viral replication and transcription, alteration of cellular splicing machinery and, based on data generated in the present study and from a previous study, the stimulation of translation (11, 17, 21, 26, 33, 34, 39). It is unlikely that the C terminus of ICP27 by itself can carry out its multiple functions, given that a deletion of the entire N terminus (mutant virus d1-5) generates a virus that acts very much like the d27-1-null virus, although this N-terminal deletion may alter the structure and function of the C terminus (23). It is more likely that alterations in the C terminus of ICP27 cause misfolding of the protein or structural changes, which deleteriously affect its functions. ICP27 requires its C terminus for self-interaction, and it is interesting to speculate that this self-interaction may be required for ICP27 to function properly (50).
Based on these studies we conclude that ICP27 plays a role in stimulating the translation of certain viral mRNAs. Furthermore, ICP27's interactions with translation factors may provide a mechanism for this observed translational effect. Further studies are needed to define the protein-protein interactions needed for ICP27 to stimulate late mRNA translation and to define the effects of ICP27 on localization of late viral mRNA in infected cells in order to elucidate the mechanism of action of the ICP27 multifunctional regulatory protein.
Acknowledgments
This research was supported by NIH grants AI20530 and AI63106.
Footnotes
Published ahead of print on 23 January 2008.
REFERENCES
- 1.Afonina, E., R. Stauber, and G. N. Pavlakis. 1998. The human poly(A)-binding protein 1 shuttles between the nucleus and the cytoplasm. J. Biol. Chem. 27313015-13021. [DOI] [PubMed] [Google Scholar]
- 2.Bouton, C., and B. Demple. 2000. Nitric oxide-inducible expression of heme oxygenase-1 in human cells. Translation-independent stabilization of the mRNA and evidence for direct action of nitric oxide. J. Biol. Chem. 27532688-32693. [DOI] [PubMed] [Google Scholar]
- 3.Brown, C. R., M. S. Nakamura, J. D. Mosca, G. S. Hayward, S. E. Straus, and L. P. Perera. 1995. Herpes simplex virus trans-regulatory protein ICP27 stabilizes and binds to 3′ ends of labile mRNA. J. Virol. 697187-7195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen, I. H., L. Li, L. Silva, and R. M. Sandri-Goldin. 2005. ICP27 recruits Aly/REF but not TAP/NXF1 to herpes simplex virus type 1 transcription sites although TAP/NXF1 is required for ICP27 export. J. Virol. 793949-3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, I. H., K. S. Sciabica, and R. M. Sandri-Goldin. 2002. ICP27 interacts with the RNA export factor Aly/REF to direct herpes simplex virus type 1 intronless mRNAs to the TAP export pathway. J. Virol. 7612877-12889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheung, P., K. S. Ellison, R. Verity, and J. R. Smiley. 2000. Herpes simplex virus ICP27 induces cytoplasmic accumulation of unspliced polyadenylated alpha-globin pre-mRNA in infected HeLa cells. J. Virol. 742913-2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dai-Ju, J. Q., L. Li, L. A. Johnson, and R. M. Sandri-Goldin. 2006. ICP27 interacts with the C-terminal domain of RNA polymerase II and facilitates its recruitment to herpes simplex virus 1 transcription sites, where it undergoes proteasomal degradation during infection. J. Virol. 803567-3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ellison, K. S., R. A. Maranchuk, K. L. Mottet, and J. R. Smiley. 2005. Control of VP16 translation by the herpes simplex virus type 1 immediate-early protein ICP27. J. Virol. 794120-4131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ellison, K. S., S. A. Rice, R. Verity, and J. R. Smiley. 2000. Processing of alpha-globin and ICP0 nRNA in cells infected with herpes simplex virus type 1 ICP27 mutants. J. Virol. 747307-7319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fontaine-Rodriguez, E. C., T. J. Taylor, M. Olesky, and D. M. Knipe. 2004. Proteomics of herpes simplex virus infected cell protein 27: association with translation initiation factors. Virology 330487-492. [DOI] [PubMed] [Google Scholar]
- 11.Hardwicke, M. A., P. J. Vaughan, R. E. Sekulovich, R. O'Conner, and R. M. Sandri-Goldin. 1989. The regions important for the activator and repressor functions of herpes simplex virus type 1 alpha protein ICP27 map to the C-terminal half of the molecule. J. Virol. 634590-4602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hardy, W. R., and R. M. Sandri-Goldin. 1994. Herpes simplex virus inhibits host cell splicing, and regulatory protein ICP27 is required for this effect. J. Virol. 687790-7799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hibbard, M. K., and R. M. Sandri-Goldin. 1995. Arginine-rich regions succeeding the nuclear localization region of the herpes simplex virus type 1 regulatory protein ICP27 are required for efficient nuclear localization and late gene expression. J. Virol. 694656-4667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hosada, N., F. Lejeune, and L. E. Maquat. 2006. Evidence that poly(A) binding protein C1 binds nuclear pre-mRNA poly(A) tails. Mol. Cell. Biol. 263085-3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hughes, R. G., Jr., and W. H. Munyon. 1975. Temperature-sensitive mutants of herpes simplex virus type 1 defective in lysis but not in transformation. J. Virol. 16275-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ingram, A., A. Phelan, J. Dunlop, and J. B. Clements. 1996. Immediate early protein IE63 of herpes simplex virus type 1 binds RNA directly. J. Gen. Virol. 771847-1851. [DOI] [PubMed] [Google Scholar]
- 17.Jean, S., K. M. LeVan, B. Song, M. Levine, and D. M. Knipe. 2001. Herpes simplex virus 1 ICP27 is required for transcription of two viral late (gamma2) genes in infected cells. Virology 283273-284. [DOI] [PubMed] [Google Scholar]
- 18.Jin, L., G. B. W., Y. C. Bor, D. Rekosh, and M. L. Hammarskjold. 2003. Tap and NXT promote translation of unspliced RNA. Genes Dev. 173075-3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson, D. C., and P. G. Spear. 1984. Evidence for translational regulation of herpes simplex virus type 1 gD expression. J. Virol. 51389-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koffa, M. D., J. B. Clements, E. Izaurralde, S. Wadd, S. A. Wilson, I. W. Mattaj, and S. Kuersten. 2001. Herpes simplex virus ICP27 protein provides viral mRNAs with access to the cellular mRNA export pathway. EMBO J. 205769-5778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Larralde, O., R. W. Smith, G. S. Wilkie, P. Malik, N. K. Gray, and J. B. Clements. 2006. Direct stimulation of translation by the multifunctional herpesvirus ICP27 protein. J. Virol. 801588-1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laurent, A. M., J. J. Madjar, and A. Greco. 1998. Translational control of viral and host protein synthesis during the course of herpes simplex virus type 1 infection: evidence that initiation of translation is the limiting step. J. Gen. Virol. 792765-2775. [DOI] [PubMed] [Google Scholar]
- 23.Lengyel, J., C. Guy, V. Leong, S. Borge, and S. A. Rice. 2002. Mapping of functional regions in the amino-terminal portion of the herpes simplex virus ICP27 regulatory protein: importance of the leucine-rich nuclear export signal and RGG box RNA-binding domain. J. Virol. 7611866-11879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McGregor, F., A. Phelan, J. Dunlop, and J. B. Clements. 1996. Regulation of herpes simplex virus poly(A) site usage and the action of immediate-early protein IE63 in the early-late switch. J. Virol. 701931-1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McLauchlan, J., A. Phelan, C. Loney, R. M. Sandri-Goldin, and J. B. Clements. 1992. Herpes simplex virus IE63 acts at the posttranscriptional level to stimulate viral mRNA 3′ processing. J. Virol. 666939-6945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McMahan, L., and P. A. Schaffer. 1990. The repressing and enhancing functions of the herpes simplex virus regulatory protein ICP27 map to C-terminal regions and are required to modulate viral gene expression very early in infection. J. Virol. 643471-3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mears, W. E., V. Lam, and S. A. Rice. 1995. Identification of nuclear and nucleolar localization signals in the herpes simplex virus regulatory protein ICP27. J. Virol. 69935-947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mears, W. E., and S. A. Rice. 1996. The RGG box motif of the herpes simplex virus ICP27 protein mediates an RNA-binding activity and determines in vivo methylation. J. Virol. 707445-7453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pearson, A., D. M. Knipe, and D. M. Coen. 2004. ICP27 selectively regulates the cytoplasmic localization of a subset of viral transcripts in herpes simplex virus type 1-infected cells. J. Virol. 7823-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Penman, S., I. Smith, and E. Holtzman. 1966. Ribosomal RNA synthesis and processing in a particulate site in the HeLa cell nucleus. Science 154786-789. [DOI] [PubMed] [Google Scholar]
- 31.Phelan, A., M. Carmo-Fonseca, J. McLaughlan, A. I. Lamond, and J. B. Clements. 1993. A herpes simplex virus type 1 immediate-early gene product, IE63, regulates small nuclear ribonucleoprotein distribution. Proc. Natl. Acad. Sci. USA 909056-9060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rice, S. A., and D. M. Knipe. 1988. Gene-specific transactivation by herpes simplex virus type 1 alpha protein ICP27. J. Virol. 623814-3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rice, S. A., and D. M. Knipe. 1990. Genetic evidence for two distinct transactivation functions of the herpes simplex virus alpha protein ICP27. J. Virol. 641704-1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rice, S. A., L. S. Su, and D. M. Knipe. 1989. Herpes simplex virus alpha protein ICP27 possesses separable positive and negative regulatory activities. J. Virol. 633399-3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roizman, B., D. M. Knipe, and R. J. Whitley. 2007. Herpes simplex virus, p. 2501-2602. In D. M. Knipe and P. M. Howley (ed.), Fields Virology, 5th ed. Lippincott, Williams and Wilkins, Philadelphia, PA.
- 36.Sacks, W. R., C. C. Greene, D. P. Aschman, and P. A. Schaffer. 1985. Herpes simplex virus type 1 ICP27 is an essential regulatory protein. J. Virol. 55796-805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sandri-Goldin, R. M. 1998. ICP27 mediates HSV RNA export by shuttling through a leucine-rich nuclear export signal and binding viral intronless RNAs through an RGG motif. Genes Dev. 12868-879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sandri-Goldin, R. M., and M. K. Hibbard. 1996. The herpes simplex virus type 1 regulatory protein ICP27 coimmunoprecipitates with anti-Sm antiserum, and the C terminus appears to be required for this interaction. J. Virol. 70108-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sandri-Goldin, R. M., M. K. Hibbard, and M. A. Hardwicke. 1995. The C-terminal repressor region of herpes simplex virus type 1 ICP27 is required for the redistribution of small nuclear ribonucleoprotein particles and splicing factor SC35; however, these alterations are not sufficient to inhibit host cell splicing. J. Virol. 696063-6076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schang, L. M., J. Phillips, and P. A. Schaffer. 1998. Requirement for cellular cyclin-dependent kinases in herpes simplex virus replication and transcription. J. Virol. 725626-5637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shi, J., Y. Feng, A. C. Goulet, R. R. Vaillancourt, N. A. Sachs, J. W. Hershey, and M. A. Nelson. 2003. The p34cdc2-related cyclin-dependent kinase 11 interacts with the p47 subunit of eukaryotic initiation factor 3 during apoptosis. J. Biol. Chem. 2785062-5071. [DOI] [PubMed] [Google Scholar]
- 42.Smith, I. L., and R. M. Sandri-Goldin. 1988. Evidence that transcriptional control is the major mechanism of regulation for the glycoprotein D gene in herpes simplex virus type 1-infected cells. J. Virol. 621474-1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sokolowski, M., J. E. Scott, R. P. Heaney, A. H. Patel, and J. B. Clements. 2003. Identification of herpes simplex virus RNAs that interact specifically with regulatory protein ICP27 in vivo. J. Biol. Chem. 27833540-33549. [DOI] [PubMed] [Google Scholar]
- 44.Soliman, T. M., R. M. Sandri-Goldin, and S. J. Silverstein. 1997. Shuttling of the herpes simplex virus type 1 regulatory protein ICP27 between the nucleus and cytoplasm mediates the expression of late proteins. J. Virol. 719188-9197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Suganuma, H., M. Kumada, T. Omi, T. Gotoh, M. Lkhagvasuren, H. Okuda, T. Kamesaki, E. Kajii, and S. Iwamoto. 2005. Aly/REF, a factor for mRNA transport, activates RH gene promoter function. FEBS J. 2722696-2704. [DOI] [PubMed] [Google Scholar]
- 46.Suh, Y. L., H. Kim, J. G. Chi, H. R. Byun, and K. Lee. 1987. Disseminated neonatal herpes simplex virus infection with necrotizing encephalitis: an autopsy case. J. Korean Med. Sci. 2123-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Uprichard, S. L., and D. M. Knipe. 1996. Herpes simplex virus ICP27 mutant viruses exhibit reduced expression of specific DNA replication genes. J. Virol. 701969-1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wadd, S., H. Bryant, O. Filhol, J. E. Scott, T. Y. Hsieh, R. D. Everett, and J. B. Clements. 1999. The multifunctional herpes simplex virus IE63 protein interacts with heterogeneous ribonucleoprotein K and with casein kinase 2. J. Biol. Chem. 27428991-28998. [DOI] [PubMed] [Google Scholar]
- 49.Zhang, Y., D. A. Sirko, and J. L. McKnight. 1991. Role of herpes simplex virus type 1 UL46 and UL47 in alpha TIF-mediated transcriptional induction: characterization of three viral deletion mutants. J. Virol. 65829-841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhi, Y., K. S. Sciabica, and R. M. Sandri-Goldin. 1999. Self-interaction of the herpes simplex virus type 1 regulatory protein ICP27. Virology 257341-351. [DOI] [PubMed] [Google Scholar]
- 51.Zhou, C., and D. M. Knipe. 2002. Association of herpes simplex virus type 1 ICP8 and ICP27 proteins with cellular RNA polymerase II holoenzyme. J. Virol. 765893-5904. [DOI] [PMC free article] [PubMed] [Google Scholar]