Abstract
Previously, two large-scale mutagenic analyses showed that mutations in the human cytomegalovirus (HCMV) gene UL117 resulted in a defect in virus growth in fibroblasts. Early transcriptional analyses have revealed several mRNAs from the UL119-UL115 region; however, specific transcripts encoding UL117-related proteins have not been identified. In this study, we identified two novel transcripts arising from the UL117 gene locus, and we reported that the UL117 open reading frame encoded the full-length protein pUL117 (45 kDa) and the shorter isoform pUL117.5 (35 kDa) as the result of translation initiation at alternative in-frame ATGs. Both proteins were expressed with early kinetics, but pUL117 accumulated at a lower abundance relative to that of pUL117.5. During HCMV infection, both proteins localized predominantly to the nucleus, and the major fraction of pUL117 localized in viral nuclear replication compartments. We constructed mutant HCMV viruses in which the entire UL117 coding sequence was deleted or the expression of pUL117 was specifically abrogated. The growth of mutant viruses was significantly attenuated, indicating that pUL117 was required for efficient virus infection in fibroblasts. Cells infected with the pUL117-deficient mutant virus accumulated representative viral immediate-early proteins and early proteins normally. In the absence of pUL117, the accumulation of replicating viral DNA was reduced by no more than twofold at early times and was indistinguishable from that of the wild type at 72 h postinfection. Strikingly, there was a 12- to 24-h delay in the development of nuclear replication compartments and a marked delay in the expression of late viral proteins. We conclude that pUL117 acts to promote the development of nuclear replication compartments to facilitate viral growth.
Human cytomegalovirus (HCMV) is the prototypical betaherpesvirus and a ubiquitous opportunistic pathogen infecting the majority of the world's population. HCMV infection is usually asymptomatic in healthy individuals, but the viral infection causes severe disease in immunocompromised adults and birth defects in newborns (reviewed in reference 3). Additionally, HCMV has been implicated as a possible cofactor in the development of vascular diseases such as atherosclerosis, transplant vascular sclerosis, and coronary restenosis after angioplasty surgery (17, 21, 30, 32, 44, 46, 56).
The ∼240-kb double-stranded DNA genome of HCMV has the potential to encode more than 160 putative open reading frames (ORFs) (10, 33). Fewer than 80 ORFs encode proteins that have been experimentally characterized or are homologous to viral proteins of other herpesviruses with known functions (reviewed in reference 31). The protein products of the remainder have not been identified experimentally, and their functions remain elusive. Recently, using global mutagenesis approaches, we and others initiated genome-scale studies to delineate the functions of the genes carried by HCMV (14, 53). Such studies make it possible to systematically identify viral genes that are required or are important for HCMV to establish infection in a particular cell culture system.
The UL119-UL115 region of the HCMV genome encodes a complex transcription unit (Fig. 1A). Transcriptional analysis of the HCMV laboratory strains AD169 and Towne indicates that this region gives rise to at least four transcripts that coterminate at the polyadenylation site downstream of UL115 (23, 39). The late 2.1-kb and 1.2-kb transcripts encode viral proteins pUL116 and gL (i.e., the product of UL115), respectively (23). Splicing between the UL119 and the UL118 coding sequences results in the 4.1-kb transcript encoding a 68-kDa glycoprotein termed gpUL119-UL118. This protein is an HCMV-encoded receptor for the Fc domain of immunoglobulin G (vFcγR) (2). Additional splicing between UL118 and UL117 results in a 3.1-kb transcript. Its protein product has not been identified, but the transcript is predicted to encode a spliced protein (termed pUL119-UL117) that is composed of UL119, UL118, and C-terminal 96 amino acids (aa) of UL117 (23). Importantly, despite thorough efforts to map the transcripts arising from the UL119-UL115 region, the transcript encoding the entire UL117 ORF has not been detected (2, 23). Current knowledge suggests that the coding capacity of the UL117 ORF lies within the 96 aa of its C terminus, which constitutes a component of the spliced UL119-UL117 protein.
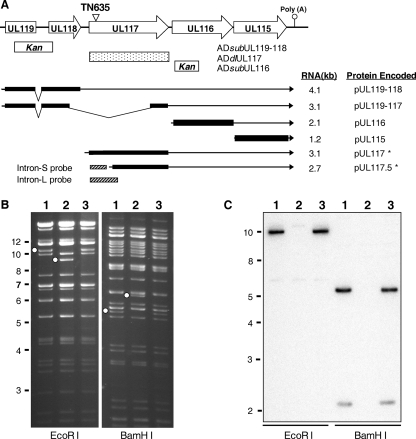
FIG. 1.
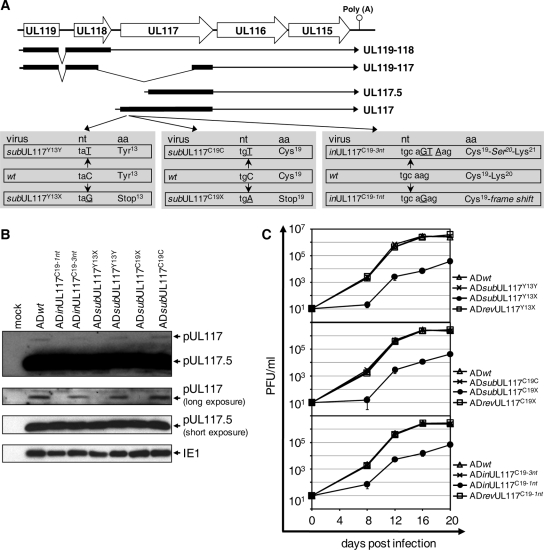
Construction of UL119-UL117 recombinant BAC-HCMV clones. (A) Viral genomic region encoding UL119-UL115. The first line represents the schematic structure of the viral genomic region. Viral ORFs are indicated by boxed arrows. Also indicated are the locations of the poly(A) signal downstream of UL115 and the transposon insertion in the recombinant BAC-HCMV clone TN635 (termed pADinUL117 in this study). The boxes below the first line represent the locations of the viral sequences within the UL119-UL116 region that are deleted in the substitution or deletion mutants, as indicated. Previously reported transcripts derived from the UL119-UL115 region (2, 23, 39) and the UL117-specific transcripts identified in this study (indicated by asterisks) are shown as lines with arrows representing the 3′ ends of the transcripts. Also indicated are the positions of two DNA probes used in Northern blotting analysis. Kan, kanamycin resistance gene cassette. (B) EcoRI and BamHI restriction digestion analysis and (C) Southern blotting analysis of UL117 recombinant BAC-HCMV clones. Open dots indicate restriction fragments unique to a recombinant BAC clone due to engineered sequence alteration. For Southern blotting analysis, a 32P-labeled probe of the UL117 ORF was used to hybridize EcoRI- or BamHI-digested BAC-HCMV DNA. Markers of molecular size (in kb) are indicated. Lane 1, the wild-type BAC pAD-GFP; lane 2, the UL117-deletion mutant BAC pADdlUL117; lane 3, the marker-rescued BAC pADrevUL117-1.
UL117 is a viral gene specific to betaherpesviruses of primates, and sequence homologues of UL117 are not found in cytomegaloviruses of distantly related species such as those from mice or rats. The 1,275-nucleotide full-length UL117 gene is predicted to encode a 425-aa protein that is classified as one of the noncore proteins and does not belong to any known viral protein family (13). However, UL117 is highly conserved among various HCMV strains such as laboratory strains AD169, Towne, and Toledo, as well as the clinical isolates TR, Fix, and Merlin (6, 10, 13, 14, 33). Forty out of 1,275 nucleotides of the UL117 gene vary among these HCMV strains and are randomly dispersed throughout the gene but result in only 9 aa changes, suggesting that most of the nucleotide variations are silent, due potentially to the requirement to maintain the coding capacity of UL117 during HCMV growth in cell culture. Recently, two large-scale mutagenic analyses revealed that mutations in UL117 resulted in a severe attenuation of HCMV growth in human fibroblasts, indicating that the UL117 gene locus is critical to the virus for establishing efficient infection (14, 53). In this study, we identified and characterized two proteins expressed from the UL117 gene, pUL117 and pUL117.5. We constructed and analyzed recombinant viruses lacking in the expression of the UL117-related proteins, and we report that pUL117 is expressed at the early stages of the virus infection cycle, it promotes the development of viral replication compartments, and it facilitates HCMV infection in fibroblasts.
MATERIALS AND METHODS
Plasmids, retroviral vectors, and antibodies.
The pGS284-based shuttle vectors pYD-C241 and pYD-C267 were used in allelic exchange to create recombinant infectious bacterial artificial chromosome (BAC) clones (53, 54). pYD-C241 contained a total of 1.9-kb viral genomic sequences immediately upstream and downstream of the UL117 ORF. pYD-C267 contained a 3.2-kb viral genomic fragment spanning the entire UL117 ORF and its flanking sequences. pYD-C245 and pYD-C266 are pRetro-EBNA-based retroviral overexpression vectors (19). pYD-C245 contained a DsRed gene whose expression was driven by an internal ribosome entry site (IRES). pYD-C266 was created by cloning the UL117 ORF upstream of the IRES site of pYD-C245. Finally, pYD-C255 was derived from pGalK (50), containing a GalK/kanamycin dual-expression cassette that was used for the first step of linear recombination (see below).
To generate the mouse monoclonal antibody to viral proteins originating from UL117, the viral sequence corresponding to aa 131 to 425 of the UL117 ORF was cloned upstream of a six-His tag in the expression vector pET-22b (Novagen). The His-tagged peptide was produced in Escherichia coli, purified using Ni-agarose beads, and used as an immunogen to generate the mouse hybridomas that were screened for specific interaction with the immunogen and with the native UL117 proteins made in virus infection. Additional primary antibodies used in this study include anti-β-actin (clone AC15; Abcam), anti-green fluorescent protein (GFP) (clone ab6556; Abcam), anti-pUL44 (Virusys), anti-major capsid protein MCP (a gift from Wade Gibson, John Hopkins University), anti-immediate-early protein type 2 (IE2) (a gift from Jay Nelson, Oregon Health and Science University), and anti-IE1 (48) and anti-pp28 (43) (gifts from Thomas Shenk, Princeton University).
Cells and viruses.
Primary human foreskin fibroblasts (HF) were propagated in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum. To create HF cells expressing UL117-related proteins (HF-UL117) or DsRed (HF-DsRed), retrovirus stocks were made by transfecting the retroviral vector pYD-C266 or pYD-C245 into Phoenix Ampho cells (19), respectively. HF cells were transduced with retrovirus three times to generate a pool of cells expressing the protein of interest.
Various BAC-HCMV clones were constructed and used to reconstitute recombinant HCMV viruses. Two BAC-HCMV clones (pAD-GFP and pAD/Cre) were used to produce wild-type virus (ADwt and ADwt-2, respectively). pAD/Cre carries the full-length genome of HCMV strain AD169, and pAD-GFP is derived from pAD/Cre but contains a simian virus 40 (SV40) early promoter-driven GFP gene in place of the viral US4-US6 region (48, 54). The BAC clone that lacks the entire UL117 coding sequence (pADdlUL117) was created by performing an allelic exchange with the shuttle vector pYD-C241 with pAD-GFP. The marker-rescued BAC clone pADrevUL117-1, in which the UL117 allele was repaired, was created by performing an allelic exchange with the shuttle vector pYD-C267 with pADdlUL117. Other recombinant BAC clones were constructed by a two-step linear recombination in the bacterial strain SW102, with modifications (50). Briefly, a kanamycin cassette in pYD-C191, or a GalK/kanamycin dual marker cassette in pYD-C255, was amplified by PCR with a pair of 70-bp primers that had 5′-terminal 50-bp sequences homologous to the viral genomic sequences of the targeted sites and subsequently recombined into a BAC-HCMV clone at the locus of interest by the first step of linear recombination to generate the deletion or the insertional mutant BAC-HCMV clones. Resulting transformants were selected on kanamycin-containing LB plates to identify clones carrying the marker cassette, and the correct integration of the marker cassette was verified by restriction digestion, Southern blotting, PCR, and direct sequencing analyses. To introduce point mutations in UL117, the mutations were first generated by using a QuickChange XL kit (Stratagene) with primers containing the desired mutations and the template plasmid pYD-C259 carrying the HCMV UL118-UL116 sequence, resulting in the carrier plasmids carrying these mutations (Table 1). The mutant viral DNA fragments were then amplified by PCR from the carrier plasmids and subsequently recombined into the BAC clones to replace the GalK/kanamycin marker cassette by the second step of linear recombination. Similarly, to introduce the GFP-tagged UL117, the GFP coding sequence was amplified by PCR with 70-bp primer pairs from the plasmid pIC113 (7) and recombined into the viral genome in frame at the N terminus of the UL117 ORF by the second step of linear recombination. The resulting recombinants from the second step of the linear recombination were selected on 2-deoxy-galactose (2-DOG)-containing minimal medium plates for the loss of GalK/kanamycin (50) and verified by restriction digestion, Southern blotting, PCR, and direct sequencing analyses. Finally, the recombinant BAC-HCMV clone carrying the transposon insertion in UL117 (termed pADinUL117 in this study) and the marker-rescued clone (pADrevUL117-2) were described previously (53).
TABLE 1.
Primers used to introduce mutations into the UL117 coding sequence
| Targeted amino acid | Mutation type | Primers used to introduce mutationsa | Resulting constructb | Resulting BAC clone |
|---|---|---|---|---|
| Cys19 | Silent | 5′-ggctccacacgcatctgtaagtccctggccccg-3′ | pYD-C294 | pADsubUL117C19C |
| 5′-cggggccagggacttacagatgcgtgtggagcc-3′ | ||||
| Cys19 | Nonsense | 5′-ggctccacacgcatctgaaagtccctggccccg-3′ | pYD-C295 | pADsubUL117C19X |
| 5′-cggggccagggactttcagatgcgtgtggagcc-3′ | ||||
| Tyr13 | Silent | 5′-cacgtccagatcgtctatggctccacacgcatc-3′ | pYD-C297 | pADsubUL117Y13Y |
| 5′-gatgcgtgtggagccatagacgatctggacgtg-3′ | ||||
| Tyr13 | Nonsense | 5′-cacgtccagatcgtctagggctccacacgcatc-3′ | pYD-C296 | pADsubUL117Y13X |
| 5′-gatgcgtgtggagccctagacgatctggacgtg-3′ | ||||
| Cys19 | Insertion | 5′-gggccagggactctgcaGatgcgtgtg-3′ | pYD-C265 | pADinUL117C19-1nt |
| 5′-cacacgcatCtgcagagtccctggccc-3′ | ||||
| Cys19 | Insertion | 5′-cacacgcatctgcaGTAagtccctggccc-3′ | pAD-C298 | pADinUL117C19-3nt |
| 5′-gggccagggactTACtgcagatgcgtgtg-3′ |
Underlined lowercase or uppercase letters represent nucleotide substitutions or insertions engineered in the UL117 coding sequence, respectively.
These constructs were derived from pYD-C259 but carried the mutated UL117 coding sequences that were subsequently used as templates to introduce mutations into the BAC-HCMV by the second step of linear recombination, as described in Materials and Methods.
To reconstitute the virus, 2 μg of the BAC-HCMV DNA and 1 μg of the pp71 expression plasmid were transfected into HF cells by electroporation as described previously (54). Culture medium was changed 24 h later, and the virus stock was prepared by harvesting cell-free culture supernatant when the entire monolayer of cells was lysed. Alternatively, virus stocks were produced by collecting cell-free culture medium from infection at the multiplicity of infection of 0.05 PFU/cell.
Analysis of intracellular DNA, RNA, and proteins.
The DNA levels of infected cells were measured by real-time PCR as previously described (48). HF cells were infected with wild-type virus or pUL117-deficient virus at the multiplicity of infection of 0.1 PFU/cell, collected at various times postinfection, resuspended in lysis buffer (10 mM Tris-HCl [pH 7.8], 10 mM EDTA, 400 mM NaCl, 50 mg/ml proteinase K, 0.2% sodium dodecyl sulfate [SDS]), and lysed by incubation at 37°C overnight. DNA was extracted with phenol-chloroform, treated with RNase A, extracted again with phenol-chloroform, precipitated with ethanol, and resuspended in water. Viral DNA was quantified by real-time PCR analysis using a TaqMan probe (Applied Biosystems) and primers specific for the HCMV UL54 gene (35). Cellular DNA was quantified with SYBR Green PCR Master Mix (Applied Biosystems) and a primer pair specific for the human β-actin gene (5′-CTC CAT CCT GGC CTC GCT GT-3′ and 5′-GCT GTC ACC TTC ACC GTT CC-3′). The accumulation of viral DNA was normalized by dividing UL54 gene equivalents by β-actin equivalents.
RNA transcripts expressed during HCMV infection were analyzed by Northern blotting and rapid amplification of cDNA ends (RACE), as previously described (54). For the Northern blotting analysis, probes were prepared by using PCR-generated templates and a Strip-EZ PCR kit (Ambion) according to the manufacturer's instructions. Primer pairs used to generate the templates were 5′-AGA GCG TCG CCC AGA CAG ACT-3′ and 5′-ATG TTC TCC CAG GAC CAC GTC-3′ (for the intron-L probe [Fig. 1 and 3]), 5′-TGG AAG ACG ATT AGC TTG GAG C-3′ and 5′-ATG TTC TCC CAG GAC CAC GTC-3′ (for the intron-S probe [Fig. 1 and 3]), and 5′-GTA GCC TAC ACT TTG GCC ACC-3′ and 5′-TTA CTG GTC AGC CTT GCT TCT A-3′ (for the UL123 probe [Fig. 3]). RACE analysis was performed by using a SMART PCR cDNA synthesis kit (Clontech), and the gene-specific primer (5′-AGA GCG TCG CCC AGA CAG ACT-3′) that was located 500 bp downstream of the initiation codon of the UL117 ORF was used for 5′-end RACE.
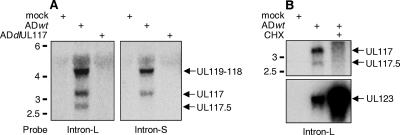
FIG. 3.
Northern blotting analysis of transcripts specifically arising from the UL117 gene locus. (A) HF were either mock-infected or infected with wild-type virus (ADwt) or UL117 deletion virus (ADdlUL117) at a multiplicity of infection of 1 PFU/cell. Cells were harvested at 24 h postinfection, total RNA was isolated, and UL117-specific transcripts were analyzed by Northern blotting using two specific probes of the 5′ terminus of the UL117 ORF (as shown in Fig. 1). (B) HF were either mock infected or infected with wild-type HCMV at a multiplicity of infection of 1 PFU/cell in the presence or absence of 100 μg/ml CHX. Cells were harvested at 8 h postinfection, total RNA was isolated, and the UL117-specific transcripts and the control UL123 transcript were analyzed by Northern blotting using the intron-L probe and the UL123 specific probe. Molecular size markers (kb) are indicated.
Proteins were analyzed by Western blotting or immunofluorescence as previously described (48). To analyze proteins by Western blotting, infected cells were collected, washed, and lysed in the SDS sample buffer. Proteins from equal cell numbers were resolved by electrophoresis on an SDS-containing 10% polyacrylamide gel, transferred to a polyvinylidene difluoride membrane, hybridized with primary antibodies, reacted with horseradish peroxidase-conjugated secondary antibodies, and visualized by ECL-plus (Amersham) detection. To analyze proteins by immunofluorescence, cells grown on glass coverslips were fixed in 2% paraformaldehyde, permeabilized with 1 mg/ml Zwittergent for 1 min or with 0.1% Triton X-100 for 15 min, incubated with the primary antibody, and subsequently labeled with the secondary antibody. To label viral replication compartments with the anti-IE2 antibody, cells grown on coverslips were washed with phosphate-buffered saline and incubated with CSK buffer (10 mM piperazine-N, N′-bis(2-ethanesulfonic acid) [pH 6.8], 100 mM NaCl, 300 mM sucrose, 3 mM MgCl2) containing 0.5% Triton X-100 at 4°C for 10 min before fixation (29). Finally, labeled cells were counterstained with TO-PRO-3 (Molecular Probes) to visualize the nuclei and then mounted on slides with Prolong Gold antifade reagent (Molecular Probes). Images were captured using Zeiss LSM Image software with a Zeiss LSM 510 META confocal laser scanning microscope.
RESULTS
The UL117 gene is required for efficient virus replication in fibroblasts.
The UL119-UL115 region of the HCMV genome encodes a complex transcription unit (Fig. 1A). Transcripts encoding pUL116, gL (i.e., pUL115), pUL119-UL118, and pUL119-UL117 have been identified, of which the latter two resulted from alternative splicing (2, 23). Previously, two large-scale mutagenic analyses tentatively classified UL117 as an augmenting gene because mutations in UL117 resulted in attenuated growth of the virus in fibroblasts (14, 53). As the specific transcripts and the protein products arising from the UL117 gene have not been identified, we first wanted to determine whether the defective growth of the UL117 mutant was caused by an alteration in UL117 expression or by an alteration in neighboring gene expression as an inadvertent result of mutations introduced in UL117. Recombinant HCMV viruses used in this study were generated from the transfection of BAC-HCMV clones derived from parental clones pAD/Cre and pAD-GFP, both of which carried the genome of the HCMV AD169 strain (48, 54).
We constructed three recombinant viruses (Fig. 1A). ADdlUL117 lacked the entire UL117 ORF. ADsubUL116 carried a deletion at the N-terminal half of the UL116 ORF (i.e., aa 1 to 158). ADsubUL119-UL118 lost the substantial portion of the UL119-UL118 coding sequence (i.e., from aa 41 of the UL119 ORF to the aa 2 of the UL118 ORF). These viruses were reconstituted from recombinant BAC-AD169 clones (pADdlUL117, pADsubUL116, and pADsubUL119-UL118, respectively) that were constructed by allelic exchange or linear recombination. Virus ADwt reconstituted from the parental BAC clone pAD-GFP carrying the genome of the HCMV strain AD169 and expressing an SV40 early promoter-driven GFP gene was used as the wild-type control (48). Moreover, the UL117 gene in pADdlUL117 was subsequently repaired, producing the marker-rescued BAC clone (pADrevUL117-1) that was used to generate the marker-rescued virus (ADrevUL117-1). Recombinant BAC clones were examined for integrity, using EcoRI and BamHI restriction endonuclease digestions, and for the intended mutation at the specific locus, by Southern blotting analysis. Figure 1B and C show the analyses for pADdlUL117 and pADrevUL117-1. Restriction digestion patterns and Southern blotting analysis were consistent with the predictions, indicating that the recombinant BAC clones carried the intact viral genome and contained the precise modification at the correct locus. Finally, modifications introduced in the BAC-HCMV clones were also confirmed by PCR and direct sequencing analysis (data not shown). All other recombinant BAC clones constructed in this study were analyzed with the same level of scrutiny (data not shown).
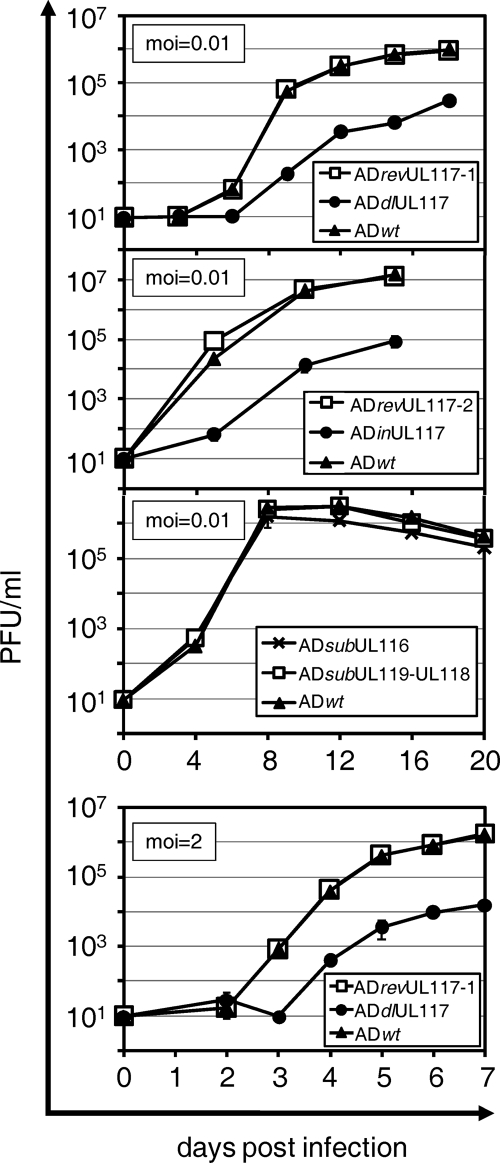
We first examined the impact of the neighboring genes, UL119-UL118 and UL116, on HCMV growth in HF. Under the conditions of multistep growth, the cell-free recombinant viruses lacking UL119-UL118 (ADsubUL119-UL118) or UL116 (ADsubUL116) were produced at wild-type levels, indicating that these genes are dispensable for HCMV growth in fibroblasts (Fig. 2). We then examined the role of the UL117 gene in HCMV growth by measuring the production of cell-free virus over the course of the UL117 deletion mutant infection. Both the single-step and the multistep growth levels of the UL117 deletion mutant relative to that of the wild-type virus and the marker-rescued virus were examined in fibroblasts (Fig. 2). In drastic contrast to the growth levels of the UL119-UL118 mutant and the UL116 mutant, the growth of the UL117-deficient virus was severely reduced under both conditions, whereas marker-rescued virus and wild-type virus replicated indistinguishably. Compared to wild-type virus, the UL117 deletion mutant produced 300- and 110-fold less infectious virus at 9 days postinfection at a multiplicity of infection of 0.01 PFU/cell and at 5 days postinfection at 2 PFU/cell, respectively. In an independent, single-step growth analysis experiment where the accumulation of cell-associated virus was measured, we observed that the UL117 deletion mutant also produced reduced levels of intracellular virus compared to that of the wild-type HCMV, suggesting that one defect of the UL117 deletion mutant remains prior to the release of virus from infected cells (data not shown). In addition, a mutant virus (ADinUL117) that carried a transposon insertion in the UL117 gene (53) was similarly attenuated; it produced 310-fold less infectious virus than the wild-type (ADwt) or the marker-rescued virus (ADrevUL117-2) at 10 days postinfection at a multiplicity of infection of 0.01 PFU/cell. Thus, we conclude that UL117 encodes a gene product independent of UL116 and UL119-UL118 and the UL117-specific product is required for efficient virus growth in fibroblasts.
FIG. 2.
Deletion of the UL117 gene locus resulted in severely attenuated growth of HCMV. HF were infected at a multiplicity of infection (moi) of either 0.01 PFU/cell (for multistep growth analysis) or at 2 PFU/cell (for single-step growth analysis) with various recombinant HCMV viruses as indicated. Culture medium was collected on different days postinfection, and yields of cell-free virus were determined by plaque assay. ADrevUL117-1 and ADrevUL117-2 are marker-rescued viruses of ADdlUL117 and ADinUL117, respectively.
Two novel transcripts are expressed from the UL117 gene during virus infection.
Previously, the only identified transcript predicted to encode a part of the UL117 ORF was the 3.1-kb spliced mRNA transcribed from the promoter upstream of UL119 (Fig. 1A). Because of the different growth phenotypes of ADsubUL119-UL118 and ADdlUL117, we hypothesized that another transcript(s) independent of UL119-UL118 must be expressed to encode UL117-specific products. Consequently, we carried out Northern blotting analyses to identify novel UL117-specific transcripts using UL117-specific probes (Fig. 3A). We detected three transcripts (4.1, 3.1, and 2.7 kb) at 24 h postinfection, using the DNA (intron-L) probe that was located at the intron region of the spliced UL119-UL117 transcript but within the 5′-terminal 500 bp of the UL117 ORF (Fig. 1A and 3A). While the 4.1-kb transcript corresponded to the known UL119-UL118 mRNA, the 3.1-kb and the 2.7-kb transcripts detected by the intron-L probe have not been previously reported. To map the 5′ ends of the 3.1-kb and the 2.7-kb transcripts more precisely, we performed Northern blotting analysis using a shorter DNA probe (intron-S probe) corresponding to the 5′-terminal 329 bp of the UL117 ORF (Fig. 1A and 3A). The intron-S probe detected only the 4.1-kb and the 3.1-kb transcripts but not the 2.7-kb transcript. Sequence analysis revealed that a putative TATA box was located 329 bp downstream of the initiation codon of the UL117 ORF and that methionine 131 was within the Kozak translation initiation consensus sequence. RACE analysis confirmed that the 2.7-kb transcript initiates 31 bp upstream of methionine 131, which was 30 bp downstream of the intron-S probe (data not shown). Thus, both the 3.1-kb and the 2.7-kb transcripts are specifically expressed from the UL117 gene. The 3.1-kb mRNA is expressed from a promoter upstream of UL117 and has the capacity to encode the full-length UL117 protein, termed pUL117. The 2.7-kb transcript is expressed from an alternative promoter within the UL117 coding sequence and has the capacity to use the internal ATG (Met 131) of the UL117 ORF as the initiation codon to translate a short isoform of the UL117 protein (termed pUL117.5). Although we did not directly examine other HCMV strains for their ability to express pUL117 and pUL117.5, both Met 1 and Met 131 of the UL117 ORF as well as the TATA box upstream of Met 131 are conserved among HCMV strains that have been sequenced, such as the laboratory strains Towne and Toledo, as well as the clinical strains TR, Fix, and Merlin, suggesting that the expression of both proteins is likely to be a common theme in HCMV (date not shown).
Expression and localization of UL117 proteins during virus infection.
We used two approaches to directly detect UL117-related protein products. First, we made a monoclonal mouse antibody against a recombinant peptide derived from the pUL117.5 coding sequence and purified from E. coli. This antibody was anticipated to recognize the common sequence of the gene products of both UL117 and UL117.5, and it was used to examine the expression of UL117-related proteins by Western blotting analysis (Fig. 4A). We detected two proteins of 45 and 35 kDa in the lysate of ADwt-infected cells but not in mock-infected or ADdlUL117-infected cells, indicating that both pUL117 and pUL117.5 were expressed at the predicted sizes and that the deletion mutant has lost the ability to produce both proteins. Moreover, both pUL117 and pUL117.5 were expressed from fibroblasts transduced with the retroviral vector carrying the UL117 ORF (Fig. 4A), thereby providing additional proof that pUL117 and pUL117.5 are translated from the UL117 ORF.
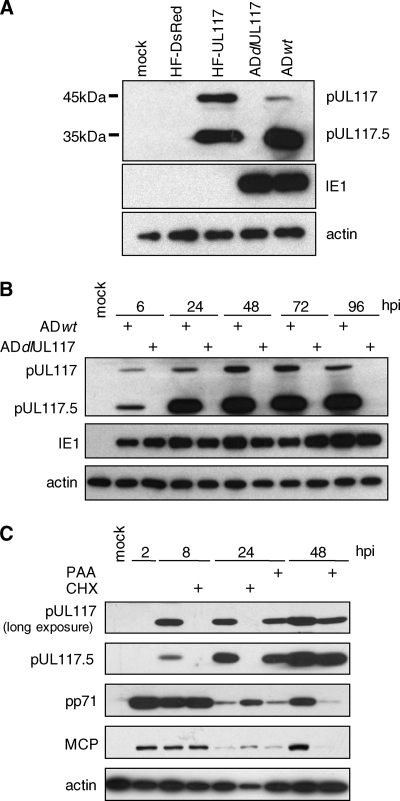
FIG. 4.
Western blotting analysis of proteins arising from the UL117 gene locus. (A) HF were transduced with retrovirus expressing DsRed (HF-DsRed) or UL117 (HF-UL117) or were infected with wild-type HCMV (ADwt) or UL117 deletion mutant virus (ADdlUL117) at a multiplicity of infection of 10 PFU/cell. At 96 h postinfection (hpi), cell lysates were prepared and analyzed by Western blotting using the mouse antibody raised against the peptide derived from UL117. (B) HF were infected with ADwt or ADdlUL117 at a multiplicity of infection of 10 PFU/cell, cell lysates were prepared at different times postinfection, and the accumulation of pUL117 and pUL117.5 was analyzed by Western blotting. The antibodies to the viral protein IE1 and cellular protein β-actin were used as the infection control and the loading control, respectively. (C) HF were infected with ADwt at a multiplicity of infection of 10 PFU/cell in the presence or absence of 100 μg/ml CHX or 200 μg/ml PAA; cell lysates were prepared at different times as indicated; and the accumulation of pUL117, pUL117.5, the tegument protein pp71, and the major capsid protein MCP was analyzed by Western blotting. Longer exposure of the ECL Western blot was used to reveal the accumulation of pUL117.
To determine the expression kinetics of UL117-related proteins, we first analyzed the accumulation of pUL117 and pUL117.5 at different times postinfection (Fig. 4B). The expression level of pUL117 peaked at 48 to 72 h, while the expression level of pUL117.5 increased drastically from 6 h to 24 h and then remained relatively stable throughout the remainder of the experiment. Nevertheless, both pUL117 and pUL117.5 were detected as early as 6 h postinfection, a time point prior to viral DNA replication. When infected cells were treated with cycloheximide (CHX), the accumulation of UL117 and the UL117.5 mRNAs was blocked, whereas the immediate-early transcript of UL123 accumulated at abundant levels (Fig. 3B), indicating that UL117 and UL117.5 are not immediate-early genes. To further define the expression kinetics of pUL117 and pUL117.5, we investigated the accumulation of viral proteins during HCMV infection in the presence of CHX or of the viral DNA synthesis inhibitor phosphonoacetic acid (PAA) (Fig. 4C). Tegument proteins (pp71) and capsid proteins (MCP) that were delivered into cells by input virus were evident at 2 to 8 h when the multiplicity of infection of 10 PFU/cell was used, but their levels were markedly reduced at 24 h, likely due to protein degradation. Neither pUL117 nor pUL117.5 was detectable at 2 h or 8 to 24 h in the presence of CHX, but they were readily detectable at 8 to 48 h in the absence of CHX, indicating that pUL117 and pUL117.5 accumulated under this experimental condition represented newly synthesized proteins. Importantly, at 24 to 48 h, both pUL117 and pUL117.5 accumulated at significant levels in the presence of PAA, whereas de novo synthesis of late proteins pp71 and MCP was inhibited. Together, these data indicate that pUL117 and pUL117.5 are expressed with early kinetics.
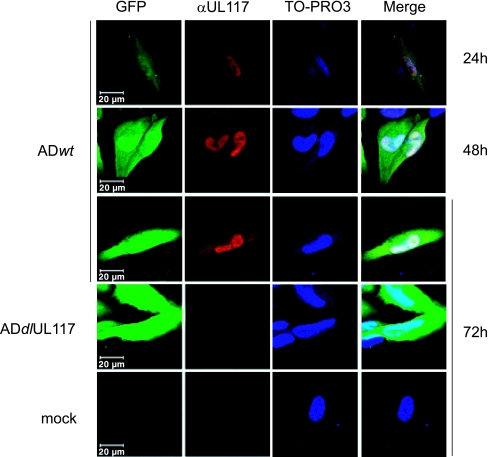
To define the cellular distribution of UL117-related proteins, we performed immunofluorescence analysis using the anti-UL117 antibody (Fig. 5). No appreciable UL117 staining was present in cells mock infected or infected with the UL117 deletion mutant, indicating the stringent specificity of the antibody to UL117-related proteins. On the other hand, we detected the strong UL117 staining that was predominantly localized within the nuclei of cells infected with wild-type virus, indicating that pUL117 and pUL117.5 are primarily nuclear proteins during the course of infection.
FIG. 5.
UL117 proteins were localized in the nuclei of infected cells. HF were infected with either ADwt or ADdlUL117 at a multiplicity of infection of 1 PFU/cell, and at different times, cells were examined for the GFP signal that indicated infection of cells and for the localization of UL117 proteins by using confocal immunofluorescence microscopy with the anti-UL117 (αUL117) antibody. Cells were also counterstained with TO-PRO3 to visualize the nuclei of the cells. Scale bars are indicated.
pUL117 is required for the efficient growth of HCMV in fibroblasts.
Our analysis of viral protein expression revealed that the full-length pUL117 protein was expressed at much reduced levels compared to that of the shorter isoform pUL117.5 (Fig. 4). This interesting observation raised the question of whether the expression of pUL117 was relevant to virus infection. The growth of mutant virus ADdlUL117, deficient in the expression of both pUL117 and pUL117.5, was severely attenuated, as was the transposon insertion mutant ADinUL117, which still expressed pUL117.5 (Fig. 2 and data not shown). We hypothesized that pUL117 is required for efficient virus infection in fibroblasts despite its low expression levels, and we set out to investigate the role of pUL117 in HCMV replication. Attempts to complement the growth of ADdlUL117 with fibroblasts expressing UL117 proteins, however, were unsuccessful (data not shown). Using an alternative approach, we constructed three pairs of recombinant HCMV viruses that carried point mutations in UL117 (Fig. 6A). For every pair, one mutation was designed to disrupt the translation of pUL117, whereas the other was introduced at the same position but preserved the coding capacity of UL117 (i.e., silent mutation). Silent mutations were included in this experiment to test the formal possibility that the cis elements (i.e., DNA sequences) within the UL117 ORF rather than its protein-coding capacity was required for the effective growth of the virus. Moreover, both types of mutations were designed to maintain the expression of pUL117.5. In the first pair, the mutant virus ADsubUL117Y13X carried a (C to G) mutation at nucleotide 39 of the UL117 gene that would replace aa Tyr13 with a stop codon, and ADsub silent UL117Y13Y carried a (C to T) mutation at the same position, thus maintaining the UL117 coding sequence. Similarly, in the second pair, ADsubUL117C19X carried a (C to A) change at nucleotide 57 to terminate the translation of pUL117 at aa 19, whereas ADsubUL117C19C carried a (C to T) silent mutation at nucleotide 57. The third pair included the mutant ADinUL117C19-1nt that carried a single-nucleotide insertion at nucleotide 58, resulting in a frameshift of the UL117 gene; and ADinUL117C19-3nt carried a three-nucleotide insertion at nucleotide 58. In ADinUL117C19-3nt, the translation frame of the UL117 gene was preserved despite the insertion of an extra serine at aa 19.
FIG. 6.
pUL117 was required for efficient growth of HCMV in fibroblasts. (A) The diagram illustrates changes introduced into the viral genome that specifically inactivated the expression of pUL117 without altering the expression of pUL117.5. The three gray boxes represent three groups of recombinant HCMV viruses constructed for this study. Each group consists of a parental wild-type virus and two recombinant viruses with single- or three-nucleotide alterations in the gene UL117. For each virus, the targeted nucleotide codon and its encoded aa (whose position within the UL117 ORF is also shown) as the result of the engineered nucleotide change (underlined uppercase letters) are indicated. In ADsubUL117Y13Y or ADsubUL117C19C, a single-nucleotide silent change of (C to T) was introduced in the codon of Tyr13 or Cys19, respectively. In ADsubUL117Y13X or ADsubUL117C19X, a single-nucleotide nonsense mutation of (C to G) or (C to A) was introduced at the same nucleotide position as that in ADsubUL117Y13Y or in ADsubUL117C19C, respectively, resulting in the premature termination of the pUL117 translation. In ADinUL117C19-1nt or in ADinUL117C19-3nt, a single- or a three-nucleotide insertion was introduced after the codon of Cys19 to generate a frameshift mutation or a single-serine insertion in the UL117 ORF, respectively. (B) HF were infected with the indicated HCMV recombinant viruses at a multiplicity of infection of 10 PFU/cell, cells were harvested at 24 h postinfection, and expressions of pUL117 and pUL117.5 were analyzed by Western blotting using the anti-UL117 antibody. Shown are both the long and the short exposures made to visualize protein bands. The viral protein IE1 was used as the infection control. (C) HF were infected with indicated recombinant HCMV viruses at a multiplicity of infection of 0.001 PFU/cell, culture medium was collected at the indicated times, and the yields of cell-free virus were determined by plaque assay.
To confirm the effect of the mutations on the expression levels of pUL117 and pUL117.5, we performed Western blotting analysis of lysates of cells infected with various mutants. As expected, recombinant viruses carrying the silent mutations (ADsubUL117Y13Y and ADsubUL117C19C) or the three-nucleotide insertion (ADinUL117C19-3nt) expressed both pUL117 and pUL117.5 (Fig. 6B). On the contrary, recombinant viruses carrying nonsense mutations (ADsubUL117Y13X and ADsubUL117C19X) or the frameshift mutation (ADinUL117C19-1nt) lost the ability to express pUL117 but accumulated pUL117.5 at wild-type levels (Fig. 6B). These results validated our mutagenesis strategy for differentiating the expression of pUL117 from that of pUL117.5 in the context of virus infection.
To determine the requirement of pUL117 expression for HCMV infection in fibroblasts, we performed a multistep growth analysis of the UL117 mutant viruses (Fig. 6C). All of the mutants deficient in pUL117 expression (ADsubUL117Y13X, ADsubUL117C19X, and ADinUL117C19-1nt) were severely growth attenuated, whereas all of the recombinant viruses that retained the ability to express pUL117 (ADsubUL117Y13Y, ADsubUL117C19C, and ADinUL117C19-3nt) replicated at wild-type levels. Finally, the mutant alleles in pUL117-deficient viruses were also repaired, and marker-rescued viruses (ADrevUL117Y13X, ADrevUL117C19X, and ADrevUL117C19-1nt) grew indistinguishably from wild-type virus. Thus, the defective growth of the pUL117 mutant viruses was due to the abrogation of pUL117 expression but not to the alteration in any potential critical cis elements within the UL117 ORF or in pUL117.5 expression. We conclude that the expression of full-length pUL117 is required for HCMV to establish efficient infection in HF.
The biologically functional GFP-tagged full-length UL117 protein localizes in replication compartments during virus infection.
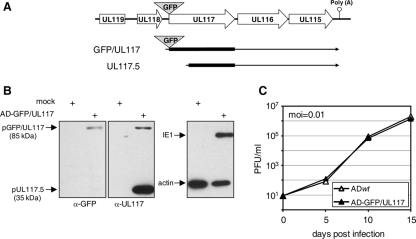
In order to specifically detect and analyze pUL117 independent of pUL117.5, we constructed the recombinant HCMV virus (AD-GFP/UL117), which was identical to ADwt, except that the GFP coding sequence that the virus carried was fused in frame to the 5′ terminus of the UL117 ORF (Fig. 7A). We performed Western blotting analysis to validate the expression of the fusion protein pGFP/UL117 in AD-GFP/UL117 infection (Fig. 7B). Cells infected with AD-GFP/UL117 produced two proteins of 85 kDa and 35 kDa that were recognized by the anti-UL117 antibody. These proteins were absent in mock-infected cells, and their apparent molecular masses were consistent with the sizes predicted for the GFP-tagged pUL117 and the native pUL117.5, respectively. Moreover, the anti-GFP polyclonal antibody recognized only the 85-kDa protein, indicating that we specifically tagged the full-length pUL117 but not pUL117.5 with GFP. Finally, AD-GFP/UL117 replicated at wild-type levels in multistep growth analysis (Fig. 7C), indicating that the GFP-tagged pUL117 was fully functional during virus infection and was likely to retain the biological properties of the native pUL117.
FIG. 7.
Construction of the recombinant HCMV (AD-GFP/UL117) expressing the functional GFP-tagged pUL117. (A) The diagram illustrates the UL119-UL115 genomic region of AD-GFP/UL117. The GFP coding sequence (gray triangle) was fused in frame to the N terminus of the UL117 ORF. As a result, the transcript encoding the GFP/UL117 fusion protein (indicated as the second line) was expressed under the control of the endogenous UL117 promoter, whereas the UL117.5 transcript (indicated as the third line) remains unaltered. (B) Detection of the GFP/UL117 fusion protein. HF were either mock infected or infected with AD-GFP/UL117 at a multiplicity of infection (moi) of 10 PFU/cell. At 96 h postinfection, cell lysates were analyzed by Western blotting using antibodies (α-) specific to GFP or UL117. The antibody to the viral IE1 protein or β-actin was used as the infection control or the loading control, respectively. (C) AD-GFP/UL117 replicated indistinguishably from wild-type virus. HF were infected with AD-GFP/UL117 or wild-type virus at a multiplicity of infection of 0.01 PFU/cell, culture medium was collected at indicated times, and yields of cell-free virus were determined by plaque assay.
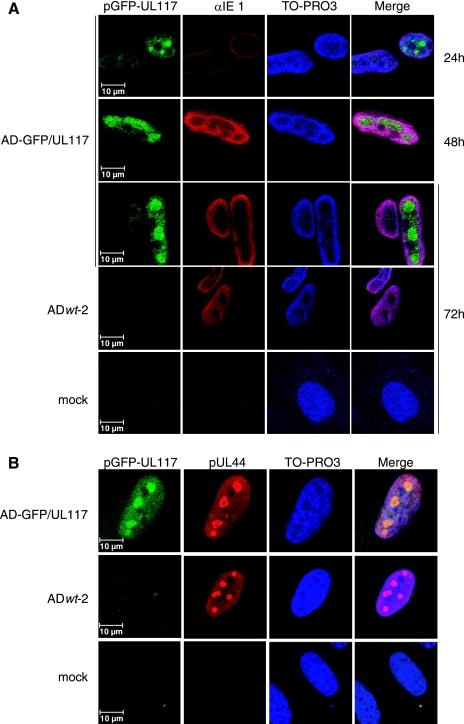
The UL117 proteins recognized by the anti-UL117 antibody were localized predominantly within the nuclei of infected cells (Fig. 5); however, this was likely to represent the localization pattern of pUL117.5, because it accumulated at a much higher level than pUL117 (Fig. 4). To determine the subcellular localization of pUL117 specifically, we performed fluorescence microscopy analysis of cells infected with AD-GFP/UL117 (Fig. 8A). The GFP fluorescence was observed predominantly within the nucleus of the cell infected with AD-GFP/UL117, during the entire course of infection. This represented the intracellular distribution of the GFP-tagged pUL117 because no appreciable green fluorescence was seen in the cells that were mock infected or those infected with the wild-type virus that did not express GFP (ADwt-2). Interestingly, the nuclear GFP/UL117 signal was not evenly distributed; on the diffuse background of nuclear fluorescence, we observed the accumulation of a strong GFP signal at global intranuclear structures, reminiscent of viral replication compartments (Fig. 8A).
FIG. 8.
A major fraction of pUL117 was localized in replication compartments. (A) pUL117 was localized in the nucleus during HCMV infection. HF were infected with the non-GFPtagged wild-type virus (ADwt-2) or AD-GFP/UL117 at a multiplicity of infection of 1 PFU/cell. At different times postinfection, cells were examined for infection by using indirect confocal immunofluorescence microscopy using the anti-IE1 (αIE1) antibody and for the pUL117 distribution by direct GFP fluorescence microscopy. (B) pUL117 was colocalized with pUL44 in replication compartments. HF were infected with AD-GFP/UL117 at a multiplicity of infection of 1 PFU/cell. Cells were harvested at 48 h postinfection and examined by direct GFP confocal fluorescence microscopy and indirect confocal immunofluorescence microscopy with anti-pUL44. Scale bars are indicated.
Replication compartments are intranuclear structures formed during herpesvirus infection (1, 11, 20, 26, 34, 36, 37). A set of viral proteins (e.g., pUL44, pUL57, and IE2) and cellular proteins (e.g., p53, Nbs1, and Rad50) are recruited to these sites during infection. Localization of these proteins to replication compartments is thought to facilitate viral DNA synthesis (11, 34, 36, 38, 47), gene transcription (20, 24, 36, 38, 40), and DNA packaging (12, 22, 34, 49, 55), which occur within or juxtaposed to these organized structures, as well as suppress the host responses (e.g., DNA damage responses) that are detrimental to HCMV infection (15, 29). To directly test the hypothesis that a subpopulation of pUL117 was distributed to replication compartments, we stained cells with a monoclonal antibody to pUL44, the virus-encoded DNA polymerase accessory protein that has been used as a marker for replication compartments (1, 34). pUL44 localized mainly to replication compartments, with diffuse background staining as previously reported (Fig. 8B) (1, 34). Furthermore, the pGFP/UL117 fluorescence signals that formed intranuclear large domains colocalized with pUL44, clearly indicating that a major fraction of the GFP-tagged pUL117 is localized in replication compartments (Fig. 8B).
Development of viral replication compartments is delayed in the absence of pUL117.
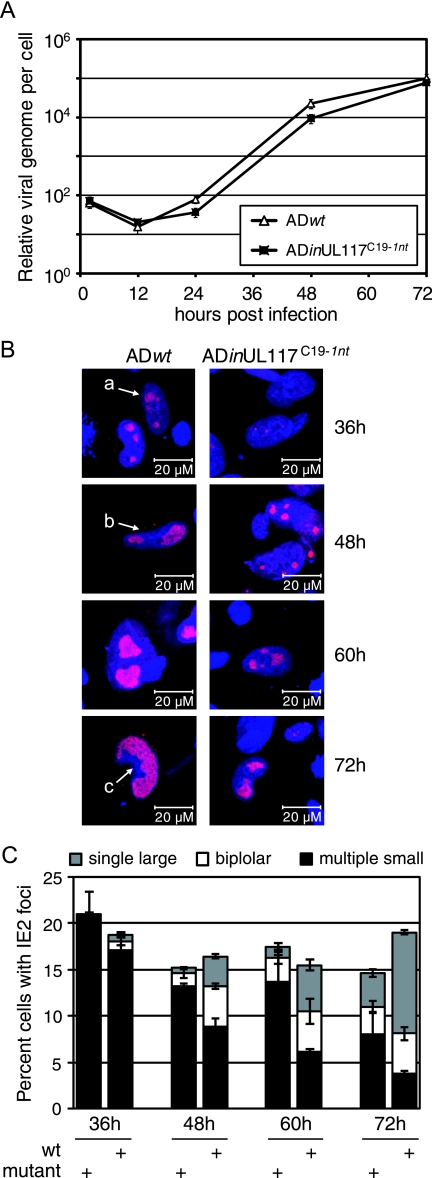
The localization of pUL117 in replication compartments led us to hypothesize that pUL117 might play a role in the replication of viral DNA or in the development of DNA replication compartments. We first examined the potential involvement of pUL117 in viral DNA replication. Cells were infected with the wild-type virus (ADwt) or the pUL117-deficient virus (ADinUL117C19-1nt), and the amounts of viral DNA accumulated within the infected cells were monitored by real-time PCR (Fig. 9A). Consistent with early reports (1, 18, 45), viral DNA replication was initiated at approximately 12 to 24 h, and at 72 h, both viruses accumulated DNA at the level that represented more than 1,000-fold increase compared to the amount of the input DNA. At any given time point during the course of infection (2 to 72 h), the pUL117-deficient virus replicated its DNA at the level comparable to that of the wild-type virus. The difference detected by real-time PCR was twofold at most, which approached the resolution limit of the assay. Therefore, the reduction of viral DNA replication in the absence of pUL117 was at most twofold during the early stages of infection, and the mutant accumulated viral DNA at a level that was indistinguishable from that of the wild-type virus, at 72 h postinfection.
FIG. 9.
The development of viral replication compartments was delayed in cells infected with pUL117-deficient virus. (A) Viral DNA replication in cells infected with pUL117-deficient virus was largely unaltered. HF were infected with wild-type virus (ADwt) or the pUL117-deficient mutant (ADinUL117C19-1nt) at a multiplicity of infection of 0.1 PFU/cell, the total cell-associated DNA was isolated at different times postinfection, and the accumulation of viral genomes was examined by real-time PCR, as described in Material and Methods. (B) Representative confocal images and (C) quantitative analysis of IE2 focus-positive cells. HF were infected with wild-type virus or pUL117-deficient virus, as described in the legend to panel A, and cells were treated with CSK extraction and stained with anti-IE2 antibody at the indicated times postinfection. IE2 focus-positive cells were counted and scored according to the stage of development, as described in the text. At least three fields and 900 cells were counted blind for each infection per time point. Multiple small foci (a), bipolar foci (b), and single large foci (c) are indicated in panel B by arrows.
We then examined the development of replication compartments in cells infected with pUL117-deficient virus. Replication compartments are initiated from the periphery of the promyelocytic leukemia protein-associated nuclear bodies (PODs or ND10) as puncture prereplicative sites at very early times in HCMV infection. These structures become larger in size after viral DNA synthesis begins and finally grow into large globular replication compartments occupying most of the nucleus (1, 27, 34, 41). We performed confocal immunofluorescence using the anti-IE2 antibody to visualize replication compartments because IE2 is known to localize to replication compartments (1) and is produced at wild-type levels in cells infected with pUL117-deficient virus (Fig. 10). Most of the HCMV proteins used to mark replication compartments, such as pUL44 and IE2, also have diffuse background staining that would make their localization in replication compartments less visible (Fig. 8A and B, and data not shown) (1, 34). In order to better reveal viral replication compartments, we treated infected cells with the CSK buffer containing nonionic detergent Triton X-100, which maintains the nuclear structures of treated cells but removes viral and cellular proteins that are not tightly associated with DNA (4, 9, 16). CSK extraction is a very useful method for specifically examining functional fractions of viral replication proteins morphologically and biochemically, and it has been used previously by other investigators for analyzing viral replication compartments in cells infected with herpesvirus such as HCMV (29), herpes simplex virus (42), and Epstein-Barr virus (8, 9). CSK extraction reveals IE2-positive replication compartments clearly, with little diffuse background staining in HCMV-infected cells (Fig. 9B). As a control, it completely removes IE1 that is a diffuse nuclear protein and is known not to localize within replication compartments in infected cells (data not shown) (1).
FIG. 10.
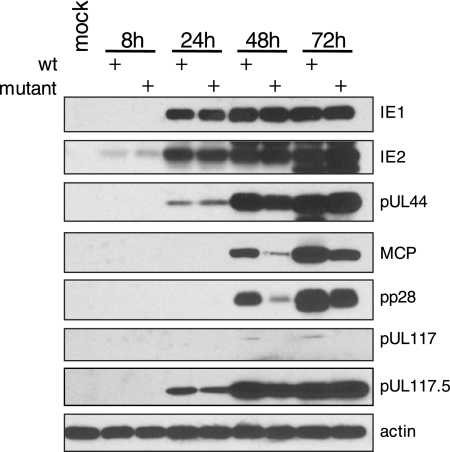
pUL117 was required for efficient accumulation of representative late viral proteins during HCMV infection. HF were infected with ADwt or ADinUL117C19-1nt at a multiplicity of infection of 0.1 PFU/cell; cells were harvested at different times postinfection; and accumulations of immediate-early proteins (IE1 and IE2), early protein (pUL44), and late proteins (MCP and pp28) were determined by Western blotting analysis. The antibody to actin was used as a loading control.
Consistent with previous reports (28, 34), we observed that during wild-type HCMV infection, viral replication compartments stained by anti-IE2 appeared initially as multiple small foci (termed RC I in this study), grew into large bipolar foci with the progression of the infection (RC II), and eventually formed a single global compartment at the advanced stage of the infection (RC III) (Fig. 9B). Strikingly, the progression of replication compartments in cells infected with pUL117-deficient virus was markedly delayed. At any time postinfection (36 h, 48 h, 60 h, and 72 h), while cells containing RC I in mutant infection were more than those in wild-type infection, the accumulation of cells containing RC III was delayed in infection of the mutant virus. At 36 h, focus-positive cells with RC III were visible in wild-type infection, whereas focus-positive cells in the mutant infection consisted almost exclusively of cells with RC I (Fig. 9C). At 48 h, 60 h, and 72 h, the numbers of the cells with RC III in mutant infection were 18%, 24%, and 33% of those in wild-type infection, respectively (Fig. 9C). Similar results were also obtained when anti-pUL44 was used for marking and analyzing the maturation of replication compartments in mutant infection (data not shown). Therefore, we conclude that pUL117 plays an important role in the development of replication compartments during HCMV infection.
Synthesis of late viral gene products is delayed in the absence of pUL117.
To determine whether the delay of replication compartment development was related to the potential alteration in viral protein synthesis, we monitored the accumulation of several representative viral genes with immediate-early (IE1 and IE2), early (pUL44), and late (MCP and pp28) expression kinetics in cells infected with pUL117-deficient virus (Fig. 10). At all examined times postinfection (8 to 72 h), the mutant virus accumulated the immediate-early proteins IE1 and IE2 at a level comparable to that of the wild-type virus. The level of pUL44 expressed by the mutant virus was also similar to that expressed by the wild-type virus (Fig. 10). Remarkably, the accumulation of late proteins (MCP and pp28) by the mutant was significantly delayed compared to that by the wild-type virus at 48 h and 72 h postinfection.
Taken together, our results indicate that two isoforms of UL117 proteins are expressed during HCMV infection and that the full-length protein pUL117 is required for viral growth, efficient development of viral replication compartments, and expression of late viral gene products.
DISCUSSION
The betaherpesvirus-specific gene UL117 is conserved in human herpesvirus type 6 (HHV-6) and HHV-7 and cytomegaloviruses of primates, but its homologue appears absent from cytomegaloviruses from lower species. Early studies have identified two major spliced mRNAs transcribed through the UL117 region, but none is predicted to encode the UL117 protein (Fig. 1) (2, 23, 39). The failure to detect additional UL117-specific transcripts may be due to their low abundance or to the fact that the UL119-UL117 transcript and the UL117 transcript have a similar size. In this study we reported that two novel transcripts of 3.1 kb and 2.7 kb were expressed from the UL117 gene locus and they encoded the full-length protein pUL117 of 45 kDa and the short isoform pUL117.5 of 35 kDa (Fig. 3 to 4).
pUL117 is expressed at very low levels relative to that of pUL117.5 during virus infection (Fig. 4); however, an analysis of mutations that specifically disrupt the expression of pUL117, but not the expression of pUL117.5, demonstrates that pUL117 is required for the full growth of HCMV in fibroblasts (Fig. 6). The low abundance of pUL117 suggests that the protein may be tightly regulated during infection and that such regulation may be important to HCMV replication. It is conceivable that the proper regulation of the timing and/or the level of pUL117 is essential to promote the progression of the viral life cycle and, in the meantime, to minimize any potential detrimental effects that the protein might have on the host cellular environment. This may explain our unsuccessful attempts to rescue the growth of the mutant virus with fibroblast overexpressing UL117.
Currently, the role of pUL117.5 remains elusive. This protein was distributed in both the cytoplasm and the nucleus in the absence of pUL117, during virus infection (Z. Qian and D. Yu, unpublished results). However, during wild-type virus infection, proteins recognized by the anti-UL117 antibody were localized predominantly within the nucleus (Fig. 5). As pUL117.5 is much more abundant than pUL117, this suggests that pUL117.5 may represent a major portion of the nuclear staining, and thus, pUL117.5 may translocate to the nucleus in the presence of pUL117. It is intriguing to speculate that pUL117.5 interacts with pUL117 to enhance the function of the latter. Alternatively, pUL117.5 may act as an antagonist, thus constituting a regulatory loop to fine tune the activity of pUL117. Further genetic and biochemical analyses are needed in order to provide insights into the role of pUL117.5 in HCMV replication.
A major population of pUL117 was localized within replication compartments (Fig. 8B). Interestingly, the earliest major defect that was apparent during infection in the absence of pUL117 was the delay in the development of viral replication compartments (Fig. 9). However, the levels of replicating viral DNA made by the mutant virus were comparable to those made by the wild-type virus at any given time. The differences measured by real-time PCR were less than twofold, at most, between the infection of the wild type and the mutant, which was unlikely to be the reason for the differences that we observed in the progression of replication compartments (Fig. 9A). At 48 h and 72 h, while both the mutant virus and the wild-type virus actively replicated viral DNA, the number of cells containing large global foci in wild-type infection was 5.1- and 3.1-fold more than that in mutant infection (Fig. 9C). Moreover, the DNA levels in cells infected with the mutant virus at 72 h were 3.4-fold more than those in cells infected with the wild-type virus at 48 h, but the development of replication compartments in mutant-infected cells was, at most, comparable to, if not less than, that of the wild-type infected cells (Fig. 9B and 9C). Therefore, we propose that pUL117 facilitates the development of replication compartments independently of viral DNA synthesis. To our knowledge, this is the first example that a CMV protein is not required for viral DNA replication but is important for the maturation of replication compartments.
Viral DNA synthesis is essential for the maturation of replication compartments (34, 37); however, it is conceivable that it may not be the only factor driving this process. Replication compartments are the places where not only does viral DNA synthesis occur but other important events such as viral gene transcription and DNA packaging also take place. In addition to viral proteins required for DNA synthesis, an increasing number of cellular proteins, such as p53 and proteins involved in the host cell DNA damage response, are found to be recruited to replication compartments (5, 28, 47, 51, 52). It is possible that, in addition to their potential roles in viral DNA synthesis, these proteins are important for conferring the proper configuration of replicating viral DNA that might be instrumental to the progression of replication compartments and the transcription of viral late genes (5, 25). It is intriguing to speculate that pUL117 is required, directly or indirectly by interacting with cellular factors for the modeling of replicating viral DNA, the maturation of replication compartments, and the efficient expression of viral late genes (Fig. 10). We are currently investigating the potential interactions between pUL117 and cellular factors to elucidate its mechanism during HCMV infection.
Acknowledgments
We thank Herbert Virgin and the members of his laboratory for helpful discussions and invaluable advice, Anthony Fehr and Nathaniel Gualberto for their critical reading of the manuscript, and the Washington University School of Medicine Hybridoma Center and Department of Comparative Medicine for the excellent assistance in animal housing and antibody generation.
This work was supported by a Howard Temin Award (CA-101957) from the National Cancer Institute, a grant-in-aid from the American Heart Association (0555636Z), a Wyeth Young Investigator Award from the Infectious Diseases Society of America and National Foundation for Infectious Diseases, and a grant from the American Cancer Society (IRG-58-010-48).
Footnotes
Published ahead of print on 23 January 2008.
REFERENCES
- 1.Ahn, J. H., W. J. Jang, and G. S. Hayward. 1999. The human cytomegalovirus IE2 and UL112-113 proteins accumulate in viral DNA replication compartments that initiate from the periphery of promyelocytic leukemia protein-associated nuclear bodies (PODs or ND10). J. Virol. 7310458-10471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atalay, R., A. Zimmermann, M. Wagner, E. Borst, C. Benz, M. Messerle, and H. Hengel. 2002. Identification and expression of human cytomegalovirus transcription units coding for two distinct Fcγ receptor homologs. J. Virol. 768596-8608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Britt, W. J., and C. A. Alford. 1996. Cytomegalovirus, p. 2493-2523. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed. Lippincott-Raven, Philadelphia, PA.
- 4.Cardoso, M. C., H. Leonhardt, and B. Nadal-Ginard. 1993. Reversal of terminal differentiation and control of DNA replication: cyclin A and Cdk2 specifically localize at subnuclear sites of DNA replication. Cell 74979-992. [DOI] [PubMed] [Google Scholar]
- 5.Casavant, N. C., M. H. Luo, K. Rosenke, T. Winegardner, A. Zurawska, and E. A. Fortunato. 2006. Potential role for p53 in the permissive life cycle of human cytomegalovirus. J. Virol. 808390-8401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chee, M. S., A. T. Bankier, S. Beck, R. Bohni, C. M. Brown, R. Cerny, T. Horsnell, C. A. Hutchison III, T. Kouzarides, J. A. Martignetti, et al. 1990. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr. Top. Microbiol. Immunol. 154125-169. [DOI] [PubMed] [Google Scholar]
- 7.Cheeseman, I. M., and A. Desai. 2005. A combined approach for the localization and tandem affinity purification of protein complexes from metazoans. Sci. STKE 2005pl1. [DOI] [PubMed] [Google Scholar]
- 8.Daikoku, T., A. Kudoh, M. Fujita, Y. Sugaya, H. Isomura, N. Shirata, and T. Tsurumi. 2005. Architecture of replication compartments formed during Epstein-Barr virus lytic replication. J. Virol. 793409-3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Daikoku, T., A. Kudoh, M. Fujita, Y. Sugaya, H. Isomura, and T. Tsurumi. 2004. In vivo dynamics of EBNA1-oriP interaction during latent and lytic replication of Epstein-Barr virus. J. Biol. Chem. 27954817-54825. [DOI] [PubMed] [Google Scholar]
- 10.Davison, A. J., A. Dolan, P. Akter, C. Addison, D. J. Dargan, D. J. Alcendor, D. J. McGeoch, and G. S. Hayward. 2003. The human cytomegalovirus genome revisited: comparison with the chimpanzee cytomegalovirus genome. J. Gen. Virol. 8417-28. [DOI] [PubMed] [Google Scholar]
- 11.de Bruyn Kops, A., and D. M. Knipe. 1988. Formation of DNA replication structures in herpes virus-infected cells requires a viral DNA binding protein. Cell 55857-868. [DOI] [PubMed] [Google Scholar]
- 12.de Bruyn Kops, A., S. L. Uprichard, M. Chen, and D. M. Knipe. 1998. Comparison of the intranuclear distributions of herpes simplex virus proteins involved in various viral functions. Virology 252162-178. [DOI] [PubMed] [Google Scholar]
- 13.Dolan, A., C. Cunningham, R. D. Hector, A. F. Hassan-Walker, L. Lee, C. Addison, D. J. Dargan, D. J. McGeoch, D. Gatherer, V. C. Emery, P. D. Griffiths, C. Sinzger, B. P. McSharry, G. W. Wilkinson, and A. J. Davison. 2004. Genetic content of wild-type human cytomegalovirus. J. Gen. Virol. 851301-1312. [DOI] [PubMed] [Google Scholar]
- 14.Dunn, W., C. Chou, H. Li, R. Hai, D. Patterson, V. Stolc, H. Zhu, and F. Liu. 2003. Functional profiling of a human cytomegalovirus genome. Proc. Natl. Acad. Sci. USA 10014223-14228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fortunato, E. A., and D. H. Spector. 1998. p53 and RPA are sequestered in viral replication centers in the nuclei of cells infected with human cytomegalovirus. J. Virol. 722033-2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fujita, M., Y. Ishimi, H. Nakamura, T. Kiyono, and T. Tsurumi. 2002. Nuclear organization of DNA replication initiation proteins in mammalian cells. J. Biol. Chem. 27710354-10361. [DOI] [PubMed] [Google Scholar]
- 17.Grattan, M. T., C. E. Moreno-Cabral, V. A. Starnes, P. E. Oyer, E. B. Stinson, and N. E. Shumway. 1989. Cytomegalovirus infection is associated with cardiac allograft rejection and atherosclerosis. JAMA 2613561-3566. [PubMed] [Google Scholar]
- 18.Heider, J. A., W. A. Bresnahan, and T. E. Shenk. 2002. Construction of a rationally designed human cytomegalovirus variant encoding a temperature-sensitive immediate-early 2 protein. Proc. Natl. Acad. Sci. USA 993141-3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kinsella, T. M., and G. P. Nolan. 1996. Episomal vectors rapidly and stably produce high-titer recombinant retrovirus. Hum. Gene Ther. 71405-1413. [DOI] [PubMed] [Google Scholar]
- 20.Knipe, D. M., D. Senechek, S. A. Rice, and J. L. Smith. 1987. Stages in the nuclear association of the herpes simplex virus transcriptional activator protein ICP4. J. Virol. 61276-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuvin, J. T., and C. D. Kimmelstiel. 1999. Infectious causes of atherosclerosis. Am. Heart J. 137216-226. [DOI] [PubMed] [Google Scholar]
- 22.Lamberti, C., and S. K. Weller. 1998. The herpes simplex virus type 1 cleavage/packaging protein, UL32, is involved in efficient localization of capsids to replication compartments. J. Virol. 722463-2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leatham, M. P., P. R. Witte, and M. F. Stinski. 1991. Alternate promoter selection within a human cytomegalovirus immediate-early and early transcription unit (UL119-115) defines true late transcripts containing open reading frames for putative viral glycoproteins. J. Virol. 656144-6153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leopardi, R., P. L. Ward, W. O. Ogle, and B. Roizman. 1997. Association of herpes simplex virus regulatory protein ICP22 with transcriptional complexes containing EAP, ICP4, RNA polymerase II, and viral DNA requires posttranslational modification by the UL13 protein kinase. J. Virol. 711133-1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lilley, C. E., C. T. Carson, A. R. Muotri, F. H. Gage, and M. D. Weitzman. 2005. DNA repair proteins affect the lifecycle of herpes simplex virus 1. Proc. Natl. Acad. Sci. USA 1025844-5849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liptak, L. M., S. L. Uprichard, and D. M. Knipe. 1996. Functional order of assembly of herpes simplex virus DNA replication proteins into prereplicative site structures. J. Virol. 701759-1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lukonis, C. J., and S. K. Weller. 1997. Formation of herpes simplex virus type 1 replication compartments by transfection: requirements and localization to nuclear domain 10. J. Virol. 712390-2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luo, M. H., and E. A. Fortunato. 2007. Long-term infection and virus shedding of human cytomegalovirus, in T98G glioblastoma cells. J. Virol. 8110424-10436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luo, M. H., K. Rosenke, K. Czornak, and E. A. Fortunato. 2007. Human cytomegalovirus disrupts both ataxia telangiectasia mutated protein (ATM)- and ATM-Rad3-related kinase-mediated DNA damage responses during lytic infection. J. Virol. 811934-1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Melnick, J. L., E. Adam, and M. E. Debakey. 1993. Cytomegalovirus and atherosclerosis. Eur. Heart J. 14(Suppl. K)30-38. [PubMed] [Google Scholar]
- 31.Mocarski, E. S., and C. T. Courcelle. 2001. Cytomegaloviruses and their replication, p. 2629-2673. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed., vol. 2. Lippincott-Raven, Philadelphia, PA. [Google Scholar]
- 32.Muhlestein, J. B., B. D. Horne, J. F. Carlquist, T. E. Madsen, T. L. Bair, R. R. Pearson, and J. L. Anderson. 2000. Cytomegalovirus seropositivity and C-reactive protein have independent and combined predictive value for mortality in patients with angiographically demonstrated coronary artery disease. Circulation 1021917-1923. [DOI] [PubMed] [Google Scholar]
- 33.Murphy, E., D. Yu, J. Grimwood, J. Schmutz, M. Dickson, M. A. Jarvis, G. Hahn, J. A. Nelson, R. M. Myers, and T. E. Shenk. 2003. Coding potential of laboratory and clinical strains of human cytomegalovirus. Proc. Natl. Acad. Sci. USA 10014976-14981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Penfold, M. E., and E. S. Mocarski. 1997. Formation of cytomegalovirus DNA replication compartments defined by localization of viral proteins and DNA synthesis. Virology 23946-61. [DOI] [PubMed] [Google Scholar]
- 35.Petrik, D. T., K. P. Schmitt, and M. F. Stinski. 2006. Inhibition of cellular DNA synthesis by the human cytomegalovirus IE86 protein is necessary for efficient virus replication. J. Virol. 803872-3883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Phelan, A., J. Dunlop, A. H. Patel, N. D. Stow, and J. B. Clements. 1997. Nuclear sites of herpes simplex virus type 1 DNA replication and transcription colocalize at early times postinfection and are largely distinct from RNA processing factors. J. Virol. 711124-1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Quinlan, M. P., L. B. Chen, and D. M. Knipe. 1984. The intranuclear location of a herpes simplex virus DNA-binding protein is determined by the status of viral DNA replication. Cell 36857-868. [DOI] [PubMed] [Google Scholar]
- 38.Randall, R. E., and N. Dinwoodie. 1986. Intranuclear localization of herpes simplex virus immediate-early and delayed-early proteins: evidence that ICP 4 is associated with progeny virus DNA. J. Gen. Virol. 672163-2177. [DOI] [PubMed] [Google Scholar]
- 39.Rawlinson, W. D., and B. G. Barrell. 1993. Spliced transcripts of human cytomegalovirus. J. Virol. 675502-5513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rice, S. A., M. C. Long, V. Lam, and C. A. Spencer. 1994. RNA polymerase II is aberrantly phosphorylated and localized to viral replication compartments following herpes simplex virus infection. J. Virol. 68988-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sarisky, R. T., and G. S. Hayward. 1996. Evidence that the UL84 gene product of human cytomegalovirus is essential for promoting oriLyt-dependent DNA replication and formation of replication compartments in cotransfection assays. J. Virol. 707398-7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shirata, N., A. Kudoh, T. Daikoku, Y. Tatsumi, M. Fujita, T. Kiyono, Y. Sugaya, H. Isomura, K. Ishizaki, and T. Tsurumi. 2005. Activation of ataxia telangiectasia-mutated DNA damage checkpoint signal transduction elicited by herpes simplex virus infection. J. Biol. Chem. 28030336-30341. [DOI] [PubMed] [Google Scholar]
- 43.Silva, M. C., Q. C. Yu, L. Enquist, and T. Shenk. 2003. Human cytomegalovirus UL99-encoded pp28 is required for the cytoplasmic envelopment of tegument-associated capsids. J. Virol. 7710594-10605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Speir, E., R. Modali, E. S. Huang, M. B. Leon, F. Shawl, T. Finkel, and S. E. Epstein. 1994. Potential role of human cytomegalovirus and p53 interaction in coronary restenosis. Science 265391-394. [DOI] [PubMed] [Google Scholar]
- 45.Stinski, M. F. 1978. Sequence of protein synthesis in cells infected by human cytomegalovirus: early and late virus-induced polypeptides. J. Virol. 26686-701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Streblow, D. N., S. L. Orloff, and J. A. Nelson. 2001. Do pathogens accelerate atherosclerosis? J. Nutr. 1312798S-2804S. [DOI] [PubMed] [Google Scholar]
- 47.Taylor, T. J., and D. M. Knipe. 2004. Proteomics of herpes simplex virus replication compartments: association of cellular DNA replication, repair, recombination, and chromatin remodeling proteins with ICP8. J. Virol. 785856-5866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Terhune, S., E. Torigoi, N. Moorman, M. Silva, Z. Qian, T. Shenk, and D. Yu. 2007. Human cytomegalovirus UL38 protein blocks apoptosis. J. Virol. 813109-3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ward, P. L., W. O. Ogle, and B. Roizman. 1996. Assemblons: nuclear structures defined by aggregation of immature capsids and some tegument proteins of herpes simplex virus 1. J. Virol. 704623-4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Warming, S., N. Costantino, D. L. Court, N. A. Jenkins, and N. G. Copeland. 2005. Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Res. 33e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wilcock, D., and D. P. Lane. 1991. Localization of p53, retinoblastoma and host replication proteins at sites of viral replication in herpes-infected cells. Nature 349429-431. [DOI] [PubMed] [Google Scholar]
- 52.Wilkinson, D. E., and S. K. Weller. 2004. Recruitment of cellular recombination and repair proteins to sites of herpes simplex virus type 1 DNA replication is dependent on the composition of viral proteins within prereplicative sites and correlates with the induction of the DNA damage response. J. Virol. 784783-4796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yu, D., M. C. Silva, and T. Shenk. 2003. Functional map of human cytomegalovirus AD169 defined by global mutational analysis. Proc. Natl. Acad. Sci. USA 10012396-12401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yu, D., G. A. Smith, L. W. Enquist, and T. Shenk. 2002. Construction of a self-excisable bacterial artificial chromosome containing the human cytomegalovirus genome and mutagenesis of the diploid TRL/IRL13 gene. J. Virol. 762316-2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yu, D., and S. K. Weller. 1998. Genetic analysis of the UL 15 gene locus for the putative terminase of herpes simplex virus type 1. Virology 24332-44. [DOI] [PubMed] [Google Scholar]
- 56.Zhou, Y. F., M. B. Leon, M. A. Waclawiw, J. J. Popma, Z. X. Yu, T. Finkel, and S. E. Epstein. 1996. Association between prior cytomegalovirus infection and the risk of restenosis after coronary atherectomy. N. Engl. J. Med. 335624-630. [DOI] [PubMed] [Google Scholar]