Abstract
Zhang et al. (G. Zhang, D. Shoham, D. Gilichinsky, S. Davydov, J. D. Castello, and S. O. Rogers, J. Virol. 80:12229-12235, 2006) have claimed to have recovered influenza A virus RNA from Siberian lake ice, postulating that ice might represent an important abiotic reservoir for the persistence and reemergence of this medically important pathogen. A rigorous phylogenetic analysis of these influenza A virus hemagglutinin gene sequences, however, indicates that they originated from a laboratory reference strain derived from the earliest human influenza A virus isolate, WS/33. Contrary to Zhang et al.'s assertions that the Siberian “ice viruses” are most closely related either to avian influenza virus or to human influenza virus strains from Asia from the 1960s (Zhang et al., J. Virol. 81:2538 [erratum], 2007), they are clearly contaminants from the WS/33 positive control used in their laboratory. There is thus no credible evidence that environmental ice acts as a biologically relevant reservoir for influenza viruses. Several additional cases with findings that seem at odds with the biology of influenza virus, including modern-looking avian influenza virus RNA sequences from an archival goose specimen collected in 1917 (T. G. Fanning, R. D. Slemons, A. H. Reid, T. A. Janczewski, J. Dean, and J. K. Taubenberger, J. Virol. 76:7860-7862, 2002), can also be explained by laboratory contamination or other experimental errors. Many putative examples of evolutionary stasis in influenza A virus appear to be due to laboratory artifacts.
Zhang et al. (20) recently reported the recovery of influenza virus RNA from ice sampled from a Siberian lake. If correct, this finding could have far-reaching implications for understanding the emergence and evolutionary dynamics of one of the most important human pathogens. Those authors reported reverse transcription-PCR (RT-PCR) amplification of an RNA fragment of the hemagglutinin (HA) gene from 20 out of 373 amplification attempts with ice sampled from Lake Park, as well as 1 out of 161 attempts with water sampled from Lake Edoma, both in northeastern Siberia. An additional 40 samples from a third lake tested negative (20).
If influenza viruses can become trapped in suspended animation in environmental ice, their subsequent reawakening might explain some perplexing cases in which what look like relics from the evolutionary past of influenza A virus have appeared at later time points. By far the most important example is the reemergence, in 1977, of the human lineage of the H1N1 subtype, relatives of the 1918 “Spanish flu” (14). An extraordinary event—Zhang et al. postulate viral escape from ice—evidently allowed the human H1N1 influenza A virus lineage, which had gone extinct after the H2N2 pandemic in 1957, to reestablish itself in humans after a 20-year absence. Intriguingly, comparative analyses have shown that the virus that reemerged in 1977 was not directly related to the H1N1 strains that had been displaced by H2N2 viruses in 1957; rather, it was virtually identical to a strain from 1950, resulting in an effective 27-year gap in H1N1 evolution between 1950 and 1977 (14).
In addition to the reemergence of the H1N1 subtype, there are several other apparent exceptions to the rule of relatively constant and clock-like evolutionary change in influenza A virus. Taken together, these anomalies seem to present a challenge to current understanding of how this virus evolves. Involving either genetically primitive viruses reportedly circulating in contemporary human or swine populations (1, 2, 7, 8) or a genetically modern virus reportedly recovered from a bird specimen from 1917 (10), these cases appear to violate the ground rules of molecular evolution for a virus with high rates of replication and mutation: they imply that extreme evolutionary stasis can occur such that some influenza A virus strains remain virtually unchanged across many decades.
As Zhang et al. argued (20), a natural abiotic reservoir could potentially explain these anomalies and would have serious implications for the emergence of future pandemic influenza virus strains in humans, constituting an “abiotic reservoir of prime importance over short and long periods of time.” In order to evaluate the strength of the evidence for these claims, we reanalyzed the relevant sequences using up-to-date Bayesian Markov chain Monte Carlo (BMCMC) and maximum likelihood (ML) phylogenetic methods. The results argue strongly against any of these cases representing real instances of influenza viruses naturally reemerging from an abiotic reservoir or exhibiting true evolutionary stasis.
MATERIALS AND METHODS
The partial (∼460-nucleotide) “ice virus” HA sequences reported by Zhang et al. (20) were aligned with the homologous region of a panel of human, swine, and avian H1N1 HA sequences spanning several decades. The accession number for each sequence in the alignment is provided in Fig. 1, with the exception of the sequence from the positive control reference strain used by Zhang et al. (20), WS/33 clone p1.9, which was provided by S. O. Rogers upon request. In addition to the “ice virus” sequences, the other potentially problematic sequences included in the analysis were Mongolia/153/88 and Mongolia/111/91 (1), swine/St-Hyacinthe/148/90 (2), Alma Ata/1417/84 (7), and Brant goose/Alaska/1/17 (10). The Brant goose HA1 sequence (AY095226) was only 166 nucleotides in length, of which 139 overlapped with the larger HA region analyzed here. The sequences were aligned by eye using SE-AL (A. Rambaut; http://tree.bio.ed.ac.uk.software/seal/). Aligning the sequences was straightforward, with few insertions or deletions required.
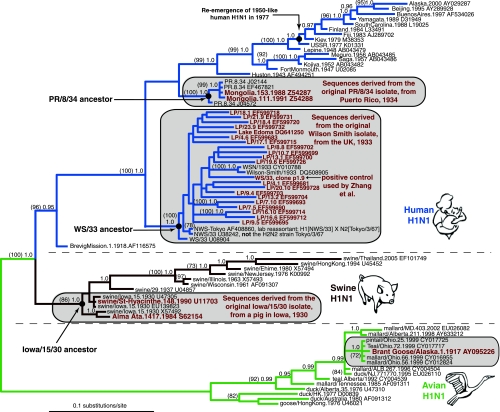
FIG. 1.
The majority-rule consensus tree summarizing the results from the BMCMC analysis of partial influenza A virus HA genes. The branch lengths are drawn to scale and represent the mean value observed for that branch among the post-burn-in sampled trees. Posterior probabilities greater than or equal to 0.95 are indicated for each node, and bootstrap percentages greater than or equal to 70 are given in parentheses. The potentially problematic sequences discussed in the main text are labeled in red. LP, Lake Park “ice virus.” The estimated maximum likelihood phylogeny was very consistent with the Bayesian results, though on the ML tree the clade including the Brant goose/Alaska 1917 and Ohio 1999 sequences was characterized by branch lengths equal to zero, reflecting the 100% identity between these sequences in the region of overlap. The slightly longer branch length of the Brant goose sequence on the Bayesian consensus tree is possibly due to the fact that only 139 nucleotides were available for analysis. As with the Bayesian result, the swine/Iowa/15/30 sequences were also paraphyletic in the ML tree, most likely representing a minor phylogenetic error in the placement of the branch leading to the remaining swine sequences; however, a significant proportion of bootstrap replicates (87%) supported monophyly for this group.
BMCMC phylogenetic inference was performed using MrBayes (13), under a general time-reversible (GTR) nucleotide substitution model. Rate heterogeneity among sites was modeled using a separate rate for each codon position, with heterogeneity within each codon position modeled with a gamma distribution. Substitution models incorporating codon-specific rate heterogeneity have previously been found to provide a good fit to influenza A virus hemagglutinin data (18). A GTR and gamma substitution model (without codon-specific rate heterogeneity) gave very similar results (not shown). Two independent runs of 10 million steps were performed, and examination of the MCMC samples with Tracer v1.3 (A. Rambaut and A. J. Drummond; http://beast.bio.ed.ac.uk) indicated adequate mixing of the Markov chains. The first 1 million steps from each run were discarded as burn-in, and the resulting MCMC samples from the independent runs were combined for subsequent estimation of posteriors.
A heuristic ML tree search with tree bisection reconnection branch swapping was performed using PAUP* (16), under a GTR and gamma substitution model. Rate matrix and gamma shape parameter estimates were generated using a neighbor-joining phylogeny and then fixed during the heuristic ML search. In order to assess the support for each node in the ML tree, a bootstrap analysis was subsequently performed using 1,000 bootstrap replicates. For each replicate, a neighbor-joining tree was inferred, with distances estimated under a GTR and gamma model with parameters estimated on the ML tree. The results of the ML search were very similar to the BMCMC results, with the ML tree showing no topological conflict with the (less-resolved) majority-rule consensus tree that summarized the BMCMC results (Fig. 1).
RESULTS AND DISCUSSION
The Siberian “ice viruses” are contaminants.
Before turning to the phylogenetic evidence presented in Fig. 1, some aspects of the results presented by Zhang et al. (20, 21) merit clarification. In their original paper (20), the authors misidentified the two published sequences that were most closely related to the sequences supposedly recovered from Siberian lake ice and water. Specifically, these two strains, labeled in Fig. 3 of Zhang et al. (20) as Av U38242 (Tokyo/3/67) and Av U08904 (A/WS/33), were described by the authors as avian in origin, when in fact they represent human influenza A virus strains. This was corrected in an erratum (21), which made clear that both these strains are human in origin. Incidentally, the WS/33 strain was derived from a strain of the first human influenza virus ever isolated, by (and from) Wilson Smith, in the United Kingdom, in 1933 (9).
The erratum, however, perpetuated an error in the GenBank entry of the sequence labeled by Zhang et al. (20, 21) as Tokyo/3/67 (accession number U38242). Sequence U38242 is not from Tokyo or from 1967 (Fig. 1). Indeed, the H1N1 subtype did not circulate in humans between 1957 and 1977; the actual Tokyo/3/67 is a subtype H2N2 isolate. The sequence published under the accession number U38242 is an H1, not H2, sequence and, as described in the Los Alamos National Laboratory influenza virus database (www.flu.lanl.gov), it was likely derived from a laboratory H1 × N2 reassortant strain whose HA gene came from a WS/33-derived source (Fig. 1).
Hence, the claim (20, 21) that the “ice viruses” are related to Asian strains that circulated in birds or humans in the 1960s is incorrect. The two closest relatives of the “ice viruses” were not only human viruses but were, specifically, WS/33-derived strains. The erratum did not address how an apparently human virus from the 1930s came to be in two Siberian lakes, an observation that undermines the notion of migratory birds depositing influenza viruses into the lakes.
A rigorous phylogenetic reanalysis of the “ice viruses” provides an explanation (Fig. 1). First, the sequences described by Zhang et al. as Lake Park “ice viruses” and the one supposedly originating from water from Lake Edoma are positioned on the human H1N1 lineage. It is worth noting that distinct and strongly supported human, swine, and avian H1N1 lineages emerged when BMCMC or ML methods, and relatively realistic substitution models, were used (Fig. 1). In contrast, the neighbor-joining and maximum parsimony trees inferred by Zhang et al. (20) obscured the existence of these distinct lineages, making it more difficult to detect the problematic placement of putatively avian influenza virus strains on the human H1N1 lineage. For example, the Brevig-Mission/1/18 “Spanish flu” sequence was placed on the swine lineage in their tree, presumably due to the lack of an explicit, realistic model of nucleotide substitution.
Furthermore, the “ice viruses” are not merely on the human lineage; they form a monophyletic clade with published sequences from laboratory strains derived from the original Wilson Smith 1933 isolate. The unequivocal support for this clade (posterior probability, 1.0; bootstrap support, 100%) and the intermingling of “ice virus” and WS/33-derived sequences (Fig. 1) indicates that the “ice virus” (and the Lake Edoma virus) sequences are ultimately derived from the ancestor of the WS/33 sequences—the original Wilson Smith isolate itself. No other interpretation for the origin of these sequences is supported by these phylogenetic results.
Although the “ice virus” sequences are characterized by fairly long terminal branches, indicating considerable evolutionary change from the WS/33 ancestor, other WS/33-derived sequences exhibit equally long branches. For example, WSN/1933 (CY010788) and Wilson-Smith/1933 (DQ508905), as shown in Fig. 1, fall among Zhang et al.'s sequences and have also accumulated a considerable genetic distance from the WS/33 ancestor. Such long branches are expected for laboratory-adapted viruses that have experienced many rounds of replication during growth in cell culture or chicken eggs; the topological pattern, nevertheless, clearly indicates that they diverged from the WS/33 ancestor.
Finally, perhaps the clearest indication of the source of the “ice viruses” is the single clone of the positive control reference strain used by Zhang et al. (20): WS/33, clone p1.9 (Fig. 1). It is phylogenetically indistinguishable from the “ice viruses” in that it too falls in the WS/33 clade but also has a long terminal branch (Fig. 1).
In their Materials and Methods section, Zhang et al. (20) stated that they employed a nested PCR approach with multiple positive controls included for every set of RT-PCR experiments. Hence, every supposedly positive result from a Siberian ice or lake water sample came from a tube that had been manipulated in the presence of a concentrated WS/33 positive control and potentially vast amounts of first-round PCR product from the control. Evidently, approximately 4% of the time this led to contamination of test samples by the positive control. The alternative explanation—that two separate Siberian lakes actually contained laboratory-adapted, human H1N1 influenza A viruses derived from the same source as Zhang et al.'s positive control and that they were deposited there by migratory birds—is unworthy of serious consideration.
This instance of accidental contamination of ice samples with positive control sequences calls into question other results by the same group in which they purportedly identified ancient viruses preserved in ice (3). For example, Castello et al. (3) reported tobacco mosaic virus (TMV) amplification from ice cores up to 140,000 years old and many times that old according to unpublished results (20). These “ice viruses” were virtually identical to modern TMV sequences, a finding that is at odds with the considerable accumulation of nucleotide substitutions expected after 140,000 years of evolution in such a rapidly evolving virus. Moreover, the experimental design involved purified TMV positive controls and a nested PCR approach. Each “ancient” sequence was thus generated in the presence of an obvious source of PCR contamination (in addition to residual TMV RNA, which can potentially be present on the hands of anyone who has recently held a cigarette). The authors explained the presence of modern-looking RNA virus sequences in 140,000-year-old ice cores as being “due to ancient forms continually returning to the atmosphere and hydrosphere from glacial meltwater or from ablated glacial surfaces.” Contamination is a more parsimonious explanation.
These “ice virus” examples stand in contrast to the phylogenetic patterns observed with authentic “fossil” sequences, such as the 1918 Spanish flu strains (Fig. 1) and the human immunodeficiency virus type 1 sequence recovered from a frozen blood sample from 1959 (23). In these cases, the authenticity of the putatively ancient viral sequences is strongly supported by the fact that they had accumulated substantially fewer nucleotide substitutions than their modern relatives, as illustrated by their short root-to-tip branch lengths. This is exactly what would be expected if they were sampled many generations earlier than modern strains, whose subsequent evolutionary change is recorded in their longer branches.
Other anomalies.
Similar laboratory contamination artifacts may explain other cases of influenza viruses that appear perplexingly out of place on phylogenetic trees. For example, Bikour et al. (2) reported a 1930-like swine influenza A (H1N1) virus supposedly present during an outbreak of respiratory disease in swine in Quebec in 1990-1991. The HA sequence from that isolate, swine/St-Hyacinthe/148/90, was indeed more like a 1930 swine virus than what one might have expected in the 1990s (Fig. 1). In fact, it is virtually identical to swine/Iowa/15/30 and is nested among published sequences from this 1930 isolate. Like WS/33, swine/Iowa/15/30 is a common laboratory reference strain, and it was used as a positive control by Bikour et al. (2). In other words, this appears to be another case of laboratory contamination with a reference strain present in the same laboratory reporting the extraordinary result. The authors suggested that influenza viruses “can be maintained for long periods in swine, perhaps in geographically isolated pockets.” That idea fails to explain, mechanistically, how any strain of influenza virus replicating in swine could exhibit complete evolutionary stasis over 60 years. Accidental contamination of one culture with a reference strain, or simple PCR contamination, on the other hand, easily explains the pattern. As with the more recent “Korean pig flu” scare (8), this seemingly exceptional result appears to reflect carelessness with a positive control sample.
Similarly, the human-swine H1N1 hybrid virus reportedly circulating among humans and animals in Alma Ata in the 1980s (7) had an HA gene sequence closely related to a published swine/Iowa/15/30 sequence (Fig. 1). This suggests a laboratory error of one kind or another: perhaps this was another case of simple contamination within the lab reporting the sequence, and no such virus actually circulated. Alternatively, this might represent a genuine escape of an experimental swine/Iowa/15/30 HA-containing virus which then temporarily circulated in humans. (A similar unintentional release of an archival influenza virus isolate may have led to the reemergence of the H1N1 subtype in humans in the 1970s [15].) Either way, the presence of a 1930-like virus at a much later period bears the unmistakable stamp of human-influenced, not natural, processes.
Anchlan et al. (1) reported influenza virus isolates from humans in Mongolia, in 1988 and 1991, with close similarities to another common lab strain, PR/8/34, derived from the original Puerto Rico isolate from 1934. Again, the phylogenetic analysis indicates that the Mongolian isolates from the 1980s and 1990s are nested within the clade of sequences derived from the original PR/8/34 isolate (Fig. 1). In this case, there is some evidence in favor of PR/8/34-related viruses actually circulating in Mongolia in the 1980s and 1990s. First, a (perhaps not completely) UV light-inactivated reassortant vaccine (PR/8/34 × USSR/77), prepared in Leningrad in 1978, was apparently used in the Mongolian population around 1978 (1, 19). The Mongolian isolate from 1988 was found to be a reassortant between PR/8/34 and USSR/77, while the one from 1991 was PR/8/34-like in all genes (1). Moreover, Anchlan et al. found that 12% of sera from various parts of Mongolia apparently contained antibodies against PR/8/34 (1). However, the fact that the sequences recovered in 1988 and 1991 were virtually identical to published PR/8/34 sequences and had apparently not accumulated the approximately 10 years' worth of substitutions expected in the decade or so since the experimental vaccine had been administered suggests that laboratory contamination rather than an escaped vaccine strain is the more likely explanation for these results.
Anchlan et al. (1) stated that, “mutational and evolutionary rates of the Mongolian strains seem to be significantly lower when compared to the rates of human influenza A strains isolated in other parts of the world… . Thus, viruses from remote areas might keep the potential to reappear in the human population after several years to cause a pandemic.” Assuming that these isolates were derivatives of an incompletely inactivated vaccine (rather than simple contamination), their apparently low evolutionary rate (their similarity to 1930s era strains) would instead be a straightforward result of recent human exposure to a laboratory strain isolated in 1934. As with each of the above cases, there is no need to invoke evolutionary stasis or natural abiotic reservoirs. Rather, this case and the others involve laboratory contamination or escape of viruses that had been in cold storage in freezers for several decades and were thus out of phase with viral lineages that had accumulated changes without interruption over those decades.
To our knowledge—aside from the “Korean pig flu” case, which seems to represent another example of laboratory contamination by a WS/33-derived reference strain (8)—there are no further examples of human or swine H1N1 that appear to involve primitive viruses circulating at the “wrong” time. Hence, every anomaly in the human and swine H1N1 lineages is apparently explained by human error of one kind or another, whether it be a labeling error, contamination of cell culture or RT-PCR, or unintentional escape of a laboratory strain from an earlier era.
Avian influenza virus from 1917?
Regarding the modern-looking avian virus sequence from 1917 (10), despite the unquestionable success those authors have had recovering ancient human influenza viruses, it is practically impossible to avoid the conclusion that this sequence represents an artifact of contamination by an avian influenza virus isolate from the late 1990s. In this case, instead of an unexpectedly short branch suggestive of a primitive virus circulating in the present, the virus ostensibly representative of 1917 exhibited an unexpectedly long branch length, suggestive of a genetically modern virus circulating in the past. Fanning et al. (10) concluded that there has been little or no evolutionary change in avian influenza virus genes over nearly a century. This idea is more radical than is sometimes appreciated, because even if there has been extremely strong negative selection on avian influenza virus proteins, an RNA virus with such a high mutation rate and replication rate would still be expected to accumulate many synonymous substitutions over such a long interval. This is not merely a theoretical assertion; it is now clear that avian influenza virus, like the human and swine varieties, evolves to a molecular clock (4).
Crucially, the “1917” strain is identical in the available HA1 fragment to four strains isolated from wild birds in Ohio in 1999 (Fig. 1), which were not available for analysis when the “1917” strain was published. Moreover, its partial HA2 sequence is 99.7% identical to the Ohio 1999 strains, and its partial NP gene is 100% identical (data not shown). Inspection of the branch lengths of the avian H1N1 lineage in Fig. 1 reveals the unmistakable pattern of a molecular clock, with increasing genetic distance with year of sampling. This means that the long and modern-looking branch leading to the supposedly 1917 virus cannot be explained, as Fanning et al. (10) propose, by evolutionary stasis in avian flu virus from 1917 until the present. The Ohio 1999 strains are clearly descendants of an ancestor from the mid-1990s, and the whole avian lineage appears to emanate from an ancestor that existed around the 1950s. If the “1917” virus really existed, then the avian H1N1 lineage must have reevolved the 1917-like form over the last half-century or so.
Extensive convergent evolution is highly improbable. Given the strong purifying selection in this lineage, it is virtually impossible: almost all the nucleotide substitutions that have occurred in the avian clade depicted in Fig. 1 are silent at the amino acid level and could have had little or no exposure to the forces of natural selection. The idea that the strains that circulated among birds in Ohio in 1999 had retraced the evolutionary pathway to match the “1917” strain across a multitude of silent substitutions is untenable. Rather, the RT-PCR analysis of the 1917 strain was evidently compromised by an isolate closely related to the Ohio 1999 ones. It follows that the “1917” sequence tells us nothing about the origins or emergence of the 1918 Spanish flu pandemic or the potential for future such pandemics.
Conclusion.
Cooper and Poinar (5) proposed nine criteria for authenticating putative ancient DNA/RNA findings (extensive use of negative controls, cloning, independent replication, etc.). More recently, Gilbert et al. (11) argued in favor of a “cognitive” approach, suggesting that researchers validate the authenticity of their results on a case-by-case basis rather than using a checklist. With regard to viral work in general and in light of the above cases specifically, researchers wishing to provide convincing authentication might want to give special consideration to the following recommendations. (i) Researchers should physically separate pre- and post-PCR activities (5, 11); specifically, avoid nested PCR when attempting to authenticate the existence of viral nucleic acids from challenging sources. (ii) Researchers should ensure that putative sequences from ancient/degraded sources could not have arisen from virus cultures becoming contaminated by another strain from the same laboratory (17). (iii) Researchers should assess the quality and reliability of viral nucleic acids directly and by assaying host DNA/RNA (5, 11, 12). (iv) Researchers should use positive controls judiciously or not at all. If used, they should be genetically distinguishable from putative ancient/degraded sequences with 100% reliability. (v) Researchers should ensure that exceptional results are reproducible. (vi) Researchers should perform a thorough phylogenetic analysis, making use of state-of-the-art methods that employ explicit, data-supported nucleotide substitution models and a rigorous statistical framework. Report estimates of confidence (e.g., bootstrap support for ML, posterior probabilities for BMCMC) for key nodes on the tree(s) inferred. It is perhaps worth emphasizing that viral data sets sampled over a wide time interval can often provide a uniquely powerful test of authenticity; branches that are shorter or longer than expected for a sequence from a particular date can reveal possible errors (6, 22).
The sort of laboratory artifacts detected here have the potential to divert attention away from actual influenza virus reservoirs and the biological processes governing the emergence and spread of this pathogen. It is therefore important that every effort be made to avoid them in the future.
Acknowledgments
This work was supported by the David and Lucile Packard Foundation and NIAID/NIH (R21 AI065371).
Adam Bjork, Tom Gilbert, and two anonymous reviewers provided helpful comments.
Footnotes
Published ahead of print on 30 January 2008.
REFERENCES
- 1.Anchlan, D., S. Ludwig, P. Nymadawa, J. Mendsaikhan, and C. Scholtissek. 1996. Previous H1N1 influenza A viruses circulating in the Mongolian population. Arch. Virol. 1411553-1569. [DOI] [PubMed] [Google Scholar]
- 2.Bikour, M. H., E. H. Frost, S. Deslandes, B. Talbot, and Y. Elazhary. 1995. Persistence of a 1930 swine influenza A (H1N1) virus in Quebec. J. Gen. Virol. 762539-2547. [DOI] [PubMed] [Google Scholar]
- 3.Castello, J. D., S. O. Rogers, W. T. Starmer, C. M. Catranis, L. Ma, G. D. Bachand, Y. Zhao, and J. E. Smith. 1999. Detection of tomato mosaic tobamovirus RNA in ancient glacial ice. Polar Biol. 22207-212. [Google Scholar]
- 4.Chen, R., and E. C. Holmes. 2006. Avian influenza virus exhibits rapid evolutionary dynamics. Mol. Biol. Evol. 232336-2341. [DOI] [PubMed] [Google Scholar]
- 5.Cooper, A., and H. N. Poinar. 2000. Ancient DNA: do it right or not at all. Science 2891139. [DOI] [PubMed] [Google Scholar]
- 6.Corbitt, G., A. S. Bailey, and G. Williams. 1990. HIV infection in Manchester, 1959. Lancet 33651. [DOI] [PubMed] [Google Scholar]
- 7.Dem'ianenko, I. V., A. A. Shilov, Z. K. Chuvakova, O. V. Chaika, and E. I. Isaeva. 1988. Comparative characteristics of hemagglutinins of influenza viruses with antigenic structure Hsw1N1 isolated from man and animals. Vopr. Virosol. 33157-162. (In Russian.) [PubMed] [Google Scholar]
- 8.Enserink, M. 2005. Pig flu scare—case closed? Science 308339c. [DOI] [PubMed] [Google Scholar]
- 9.Evans, D. G. 1966. Wilson Smith: 1987-1965. Biographical memoirs of the Fellows of the Royal Society. 12478-487. [Google Scholar]
- 10.Fanning, T. G., R. D. Slemons, A. H. Reid, T. A. Janczewski, J. Dean, and J. K. Taubenberger. 2002. 1917 avian influenza virus sequences suggest that the 1918 pandemic virus did not acquire its hemagglutinin directly from birds. J. Virol. 767860-7862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gilbert. M. T. P., H.-J. Bandelt, M. Hofreiter, and I. Barnes. 2005. Assessing ancient DNA studies. Trends Ecol. Evol. 20541-544. [DOI] [PubMed] [Google Scholar]
- 12.Gilbert, M. T. P., T. Haselkorn, M. Bunce, J. J. Sanchez, S. B. Lucas, L. D. Jewell, E. Van Marck, and M. Worobey. 2007. The isolation of nucleic acids from fixed, paraffin-embedded tissues: which methods are useful when? PLoS ONE 2e537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huelsenbeck, J. P., and F. Ronquist. 2001. MrBayes: Bayesian inference of phylogeny (version 3.1). Bioinformatics 17754-755. [DOI] [PubMed] [Google Scholar]
- 14.Nakajima, K., U. Desselberger, and P. Palese. 1978. Recent human influenza A (H1N1) viruses are closely related genetically to strains isolated in 1950. Nature 274334-339. [DOI] [PubMed] [Google Scholar]
- 15.Palese, P. 2004. Influenza: old and new threats. Nat. Med. 10S82-S87. [DOI] [PubMed] [Google Scholar]
- 16.Swofford, D. L. 2003. PAUP*: phylogenetic analysis using parsimony (*and other methods), version 4.0b10. Sinauer Associates, Sunderland, MA.
- 17.Wain-Hobson, S., J.-P. Vartanian, M. Henry, N. Chenciner, R. Cheynier, S. Delassus, L. Pedrosa Martins, M. Sala, M.-T. Nugeyre, D. Guetard, D. Klatzman, J.-C. Gluckman, W. Rozenbaum, F. Barre-Sinoussi, and L. Montagnier. 1991. LAV revisited: origins of the early HIV-1 isolates from Institut Pasteur. Science 252961-965. [DOI] [PubMed] [Google Scholar]
- 18.Worobey, M., A. Rambaut, O. G. Pybus, and D. L. Robertson. 2002. Questioning the evidence for genetic recombination in the 1918 “Spanish flu” virus. Science 296211a. [DOI] [PubMed] [Google Scholar]
- 19.Yamnikova, S. S., J. Mandler, Z. H. Bekh-Ochir, P. Dachtzeren, S. Ludwig, D. K. Lvov, and C. Scholtissek. 1993. A reassortant H1N1 influenza virus caused fatal epizootics among camels in Mongolia. Virology 197558-563. [DOI] [PubMed] [Google Scholar]
- 20.Zhang, G., D. Shoham, D. Gilichinsky, S. Davydov, J. D. Castello, and S. O. Rogers. 2006. Evidence of influenza A virus RNA in Siberian lake ice. J. Virol. 8012229-12235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang, G., D. Shoham, D. Gilichinsky, S. Davydov, J. D. Castello, and S. O. Rogers. 2007. Evidence of influenza A virus RNA in Siberian lake ice (erratum). J. Virol. 812538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu, T., and D. D. Ho. 1995. Was HIV present in 1959? Nature 374503-504. [DOI] [PubMed] [Google Scholar]
- 23.Zhu, T., B. T. Korber, A. J. Nahmias, E. Hooper, P. M. Sharp, and D. D. Ho. 1998. An African HIV-1 sequence from 1959 and implications for the origin of the epidemic. Nature 391594-597. [DOI] [PubMed] [Google Scholar]