Abstract
The transmission of highly pathogenic avian influenza H5N1 virus to Southeast Asian countries triggered the first major outbreak and transmission wave in late 2003, accelerating the pandemic threat to the world. Due to the lack of influenza surveillance prior to these outbreaks, the genetic diversity and the transmission pathways of H5N1 viruses from this period remain undefined. To determine the possible source of the wave 1 H5N1 viruses, we recently conducted further sequencing and analysis of samples collected in live-poultry markets from Guangdong, Hunan, and Yunnan in southern China from 2001 to 2004. Phylogenetic analysis of the hemagglutinin and neuraminidase genes of 73 H5N1 isolates from this period revealed a greater genetic diversity in southern China than previously reported. Moreover, results show that eight viruses isolated from Yunnan in 2002 and 2003 were most closely related to the clade 1 virus sublineage from Vietnam, Thailand, and Malaysia, while two viruses from Hunan in 2002 and 2003 were most closely related to viruses from Indonesia (clade 2.1). Further phylogenetic analyses of the six internal genes showed that all 10 of those viruses maintained similar phylogenetic relationships as the surface genes. The 10 progenitor viruses were genotype Z and shared high similarity (≥99%) with their corresponding descendant viruses in most gene segments. These results suggest a direct transmission link for H5N1 viruses between Yunnan and Vietnam and also between Hunan and Indonesia during 2002 and 2003. Poultry trade may be responsible for virus introduction to Vietnam, while the transmission route from Hunan to Indonesia remains unclear.
It has been 10 years since the first avian-to-human transmission of influenza virus was confirmed in Hong Kong (30). In the past decade, the “bird flu” caused by highly pathogenic avian influenza (HPAI) H5N1 virus has developed from an endemic disease in China to a panzootic disease affecting 60 countries across Asia, Europe, and Africa (11). The precursor virus of all of these H5N1 viruses, A/goose/Guangdong/1/1996 (Gs/GD), initially became established in domestic geese in southern China from 1996 to 1999 (36, 39). Since the first detection of Gs/GD-like viruses in ducks in 2000, H5N1 viruses underwent extensive genetic reassortment, wherein many different reassortants, or genotypes, were generated (12, 13, 22). Many of these genotypes were first recognized during the second and third major HPAI H5N1 outbreaks in Hong Kong live-poultry markets from 2001 to 2002 (10, 13). During the same period, some of these reassortants were also detected from poultry or poultry products in Korea and Japan (21, 23).
One of these novel reassortants (genotype Z) became dominant in southern China since early 2002 (5, 22) and was subsequently transmitted to neighboring countries in East and Southeast Asia in 2003 and 2004 (22). These H5N1 genotype Z viruses caused extensive outbreaks in poultry and were repeatedly introduced into humans, eventually becoming endemic in poultry in Vietnam and Indonesia (27). The persistent introduction of H5N1 virus into humans raises the possibility that it emerges as a human pandemic strain, either as a purely avian virus adapting to human-to-human transmission or through reassortment with current human influenza virus strains (32, 35). Although surveillance in market poultry was strengthened after outbreaks in these regions, the source and virus dissemination pathway in Southeast Asia remains uncertain due to the lack of convincing information prior to this spread.
The second major transmission wave of those HPAI H5N1 viruses occurred in 2005 after a major outbreak in migratory waterfowl at Qinghai Lake in northern China (2, 9). Systematic influenza surveillance in southern China provided sufficient information to understand the evolutionary pathway of these Qinghai-like viruses (clade 2.2) and to predict the development of this lineage (4, 5, 22). Similarly, the evolutionary pathway of the currently dominant H5N1 variant (clade 2.3.4 or Fujian-like) was also detected as early as March 2005 (26). These incidents highlight the importance of long-term surveillance to understand the development of H5N1 influenza viruses.
The lack of sufficient information prior to the outbreak of 2003 has led to controversy regarding the wave 1 transmission pathway in Southeast Asia. Previously, analysis based on the available sequence data concluded that all of the H5N1 viruses detected in Southeast Asia had originated from either Guangdong or Hong Kong (22, 27, 34). To clarify this question and further understand the genesis and evolutionary pathway of H5N1 viruses, we characterized 73 H5N1 influenza viruses isolated from our surveillance program in southern China from January 2001 to February 2004. During that period H5N1 viruses evolved rapidly and were genetically diversified; however, antigenic and genetic analyses clearly show that transmission of H5N1 viruses between Yunnan and Vietnam and also between Hunan and Indonesia in 2002 and 2003 initiated the wave 1 outbreaks. The phylogenetic relationships between the viruses from southern China and Southeast Asia, along with their geographic distribution, suggest that poultry trade may be responsible for virus introduction into Vietnam, while the transmission route from Hunan to Indonesia could be either via migratory birds or poultry movement. The present study also revealed that extensive genetic reassortment not only resulted in increased genetic diversity of those HPAI H5N1 viruses but also caused subsequent outbreaks that directly led to the dissemination of these viruses both within China and to neighboring countries.
MATERIALS AND METHODS
Surveillance and viruses studied.
Influenza surveillance in live-poultry markets has been conducted in Shantou, Guangdong Province, since July 2000. In October 2002, our surveillance was extended to the Hunan and Yunnan provinces. Tracheal, cloacal, or fecal swabs were sampled in three major types of poultry (chicken, duck, and goose) and also in different types of minor poultry in those markets. Sampling and virus isolation were carried out at 10-day intervals. Virus isolation and identification have been described in our previous reports (15).
Antigenic analysis.
Virus isolates were subtyped by standard hemagglutination inhibition (HI) tests using a panel of the World Health Organization reference antisera, while the antigenic characteristics of the H5N1 influenza viruses were compared by an HI assay with five ferret polyclonal antisera and five monoclonal antibodies (MAbs) as previously described (15). The MAbs 3C8, 1F1B8, 10DD2, and 14D4 to the hemagglutinin (HA) of Ck/HK/YU22/02 were produced in our laboratories, and MAb CP176/26 to the HA of Ck/Pennsylvania/1/83 and the five ferret antisera were produced by the Department of Infectious Diseases at St. Jude Children's Research Hospital, Memphis, TN. The HI assay started at 1:40 dilutions for ferret antisera and at 1:100 dilutions for all MAbs.
To visualize similarity between the antigenic reaction patterns of different viruses, numerical analysis of HI titers was conducted by using PRIMER version 5.2.9 (PRIMER-E, Plymouth, United Kingdom). Titers in the HI assay below the detection level and greater than 12,800 for MAbs were converted to 0 and 25,600 in this analysis, respectively. The data were standardized and square-root transformed, and the Bray-Curtis coefficient (3) was used to construct a similarity matrix. Hierarchical agglomerative clustering with group average linking (28) was conducted, and a dendrogram was produced. Nonmetric multidimensional scaling (19) was also used to produce two- and three-dimensional ordinations over 100 iterations. The two-dimensional configuration with lowest overall stress is presented.
Phylogenetic and molecular analysis.
To identify the possible source and understand the evolutionary pathway of the established HPAI H5N1 influenza virus in this region, 73 representative H5N1 strains isolated from our surveillance program from January 2001 to February 2004, together with all publicly available sequence data, were phylogenetically analyzed. Virus isolates were selected to represent each of the sampling sites at different positive sampling occasions (Table 1).
TABLE 1.
H5N1 viruses isolated from poultry in southern Chinaa
| Sampling yr | Positive sampling site(s) | Positive sampling mo | Representative virus | No. of virus isolates sequenced/no. of H5N1 isolates
|
||
|---|---|---|---|---|---|---|
| ST | HN | YN | ||||
| 2001 | ST | Jan | Dk/ST/195/01 | 1/1 | ||
| Feb | Dk/ST/1101/01 | 2/4 | ||||
| Mar | Dk/ST/1437/01 | 1/1 | ||||
| May | Dk/ST/1930/01 | 1/1 | ||||
| Jun | Ck/ST/2535/01 | 1/1 | ||||
| Oct | Dk/ST/4407/01 | 2/15 | ||||
| Nov | Dk/ST/4912/01 | 3/7 | ||||
| Dec | Ck/ST/5738/01 | 4/17 | ||||
| 2002 | ST | Jan | Gs/ST/157/02 | 5/18 | ||
| Feb | Dk/ST/700/02 | 2/2 | ||||
| Mar | Pa/ST/1075/02 | 1/1 | ||||
| Aug | Qa/ST/3054/02 | 2/5 | ||||
| ST, HN | Oct | Qa/ST/3846/02 | 2/6 | 1/1 | ||
| Ck/HN/23/02 | ||||||
| ST, HN, YN | Nov | WDk/ST/4124/02 | 3/4 | 1/16 | 1/8 | |
| Dk/HN/795/02 | ||||||
| Dk/YN/862/02 | ||||||
| ST, HN, YN | Dec | Ph/ST/4567/02 | 1/1 | 1/20 | 1/7 | |
| Dk/HN/1340/02 | ||||||
| Dk/YN/971/02 | ||||||
| 2003 | ST, HN, YN | Jan | Ph/ST/40/03 | 2/4 | 3/74 | 2/20 |
| Dk/HN/300/03 | ||||||
| Dk/YN/119/03 | ||||||
| ST, HN, YN | Feb | SCk/ST/540/03 | 2/3 | 1/13 | 4/39 | |
| Dk/HN/782/03 | ||||||
| Ck/YN/1252/03 | ||||||
| YN | Mar | Dk/YN/215/03 | 1/16 | |||
| YN | Apr | Dk/YN/1628/03 | 1/2 | |||
| YN | Jul | Dk/YN/4072/03 | 1/1 | |||
| ST, YN | Aug | Pa/ST/2886/03 | 1/1 | 1/2 | ||
| Dk/YN/4645/03 | ||||||
| YN | Sept | Dk/YN/5090/03 | 2/5 | |||
| ST, YN | Oct | Ph/ST/3535/03 | 1/1 | 3/34 | ||
| Ck/YN/5686/03 | ||||||
| ST, HN, YN | Nov | Ck/ST/3744/03 | 2/4 | 2/21 | 2/5 | |
| Dk/HN/5410/03 | ||||||
| Ck/YN/6255/03 | ||||||
| ST, YN | Dec | Cu/ST/4690/03 | 3/3 | 4/29 | ||
| Dk/YN/6603/03 | ||||||
| 2004 | ST, HN, YN | Jan | SCk/ST/475/04 | 2/3 | 2/122 | 5/65 |
| Dk/HN/101/04 | ||||||
| Ck/YN/207/04 | ||||||
| ST, HN, YN | Feb | Pa/ST/1207/04 | 1/4 | 2/13 | 2/9 | |
| Dk/HN/533/04 | ||||||
| Dk/YN/522/04 | ||||||
| Totalb | 45/107 | 13/280 | 30/242 | |||
Abbreviations: Ck, chicken; Cu, chukkar; Dk, duck; Gs, goose; HN, Hunan; Pa, partridge; Ph, pheasant; SCk, silky chicken; Qa, quail; ST, Shantou; WDk, wild duck; YN, Yunnan.
The total for the ST, HN, and YN columns combined is 88/629 (13.99%).
Viral RNA extraction, cDNA synthesis, PCR and sequencing were carried out as described previously (38). All sequences were assembled and edited with Lasergene 6.0 (DNASTAR, Madison, WI). BioEdit 7 was used for alignment and residue analysis (17). The program MrModeltest 2.2 was used to determine the appropriate DNA substitution model and rate heterogeneity (24). The generated model was used in all subsequent analyses. Neighbor-joining trees were constructed by using PAUP* 4.0 (31), and Bayesian analysis was conducted with MrBayes 3.1 (18) using two replicates of 1 million generations with six chains, sampling every 100 generations. Estimates of the phylogenies were calculated by performing 1,000 neighbor-joining bootstrap replicates, and Bayesian posterior probabilities were calculated from the consensus of 18,000 trees after exclusion of the first 2,000 trees as burnin. All eight genes were sequenced for each virus isolate.
Sequence similarity.
To confirm the interrelationship of viruses isolated in southern China in 2002 and 2003 and viruses that were isolated in Southeast Asia during 2003, the similarity of each gene segment was determined using MEGA 4.0 (20). To calculate their similarity (p distances), full-length amino acid alignments of each gene segments were used.
Nucleotide sequence accession numbers.
The nucleotide sequences obtained in the present study are available from GenBank under accession numbers CY028924 to CY029507.
RESULTS
Epidemiology.
No H5N1 influenza virus was detected among 2,336 samples in Shantou, Guangdong during the first year of surveillance (July to December 2000). However, HPAI H5N1 viruses were isolated from both aquatic and terrestrial poultry in 8 months of 2001 and 7 months of 2002 (Table 1). H5N1 influenza viruses were detected from the first month (October 2002) of sampling in Hunan and the second month in Yunnan (November 2002). In 2003, H5N1 influenza viruses could be isolated in all months except for May and June, and the number of viruses isolated increased during the winter months (Table 1). These findings suggest that H5N1 influenza viruses were broadly prevalent in southern China as early as 2002.
Antigenic analysis.
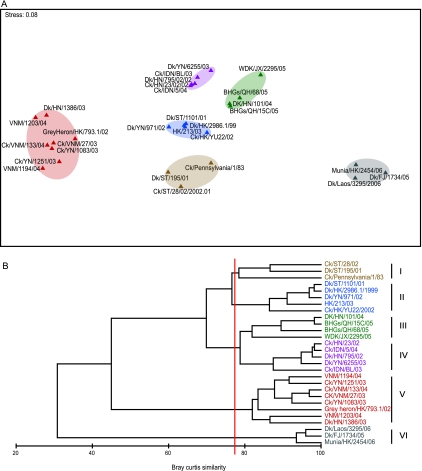
Antigenic analysis of representative strains with MAbs and ferret antisera against reference H5 viruses demonstrated a diversity of reaction patterns (Table 2). In all, six antigenic patterns or groups were recognized for the viruses characterized in the present study (Table 2 and Fig. 1). The antigenic reaction pattern of two viruses isolated from Shantou in 2001 and 2002 (Dk/ST/195/01 and Ck/ST/28/02) were similar to Ck/Pennsylvania/1/83 (antigenic group I), as noted in our previous study (8) that indicates establishment from early aquatic H5 subtype virus. Two viruses (Dk/ST/1101/01 and Dk/YN/971/02) were similar to those viruses isolated from Hong Kong from 2002 and 2003 (antigenic group II). Interestingly, two representative viruses isolated from Hunan (Ck/HN/23/02 and Dk/HN/795/02) had reaction patterns similar to those of early Indonesian viruses (e.g., Ck/IDN/BL/03) (antigenic group IV), while two representative viruses from Yunnan (Ck/YN/1083/03 and Ck/YN/1251/03) had reaction patterns similar to those of clade 1 viruses isolated from Vietnam (e.g., Ck/VNM/27/03) (antigenic group V). The close relationships between the viruses from southern China and Southeast Asia were confirmed in the phylogenetic analyses (see below).
TABLE 2.
HI titers from antigenic analyses of influenza A H5N1 virusesa
| Virus | Antigenic groupsb | Titer of MAb to:
|
Titer of ferret antiserum to:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Ck/Penn/1/83 (CP176/26) | Ck/HK/YU22/02
|
||||||||||
| 3C8 | 1F1B8 | 10DD2 | 14D4 | HK/213/03 | Dk/HN/101/04 | BHG/QH/68/05 | Ck/IDN/5/04 | VNM/1203/04 | |||
| Ck/Penn/1/83 | I | >12,800 | >12,800 | >12,800 | 800 | >12,800 | 80 | 40 | 640 | 320 | 160 |
| Ck/HK/YU22/02 | II | 200 | 6,400 | >12,800 | >12,800 | >12,800 | 40 | 80 | 320 | 640 | 80 |
| HK/213/03 | II | 400 | >12,800 | >12,800 | >12,800 | >12,800 | 2,560 | 640 | 2,560 | 2,560 | 2,560 |
| Dk/HN/101/04 | III | >12,800 | >12,800 | >12,800 | >12,800 | < | 160 | 160 | 1,280 | 1,280 | 160 |
| BHG/QH/68/05 | III | >12,800 | >12,800 | 6,400 | >12,800 | < | 80 | 80 | 640 | 640 | 40 |
| Ck/IDN/5/04 | IV | 400 | >12,800 | >12,800 | >12,800 | < | 80 | 160 | 1,280 | 1,280 | 160 |
| VNM/1203/04 | V | < | >12,800 | < | < | 800 | 40 | < | 80 | 160 | 320 |
| Dk/ST/195/01 | I | 800 | >12,800 | >12,800 | < | >12,800 | 160 | 80 | 640 | 80 | 160 |
| Ck/ST/28/02 | 800 | 6,400 | >12,800 | < | >12,800 | 160 | 160 | 640 | 320 | 640 | |
| Dk/HK/2986.1/99 | II | 1,600 | >12,800 | >12,800 | >12,800 | >12,800 | 320 | 320 | 1,280 | 640 | 640 |
| Dk/ST/1101/01 | 800 | >12,800 | >12,800 | >12,800 | >12,800 | 160 | 320 | 1,280 | 640 | 320 | |
| Dk/YN/971/02 | 200 | >12,800 | >12,800 | >12,800 | >12,800 | 160 | 160 | 640 | 640 | 320 | |
| MDk/JX/2295/05 | III | 6,400 | 3,200 | 6,400 | >12,800 | < | 40 | 40 | 320 | 320 | 40 |
| BHG/QH/15C/05 | >12,800 | >12,800 | >12,800 | >12,800 | 100 | 160 | 160 | 1,280 | 640 | 80 | |
| Ck/HN/23/02 | IV | 400 | >12,800 | >12,800 | >12,800 | < | 320 | 320 | 1,280 | 1,280 | 320 |
| Dk/HN/795/02 | 400 | >12,800 | >12,800 | >12,800 | < | 160 | 320 | 2,560 | 1,280 | 320 | |
| Ck/IDN/BL/03 | 400 | 6,400 | >12,800 | >12,800 | < | 80 | 160 | 640 | 640 | 160 | |
| Dk/YN/6255/03 | 400 | >12,800 | >12,800 | >12,800 | < | < | 80 | 640 | 640 | 40 | |
| GH/HK/793.1/02 | V | < | >12,800 | 800 | < | 1,600 | 320 | 160 | 1,280 | 1,280 | 640 |
| Dk/HN/1386/03 | < | >12,800 | < | < | 200 | 160 | 80 | 640 | 640 | 640 | |
| Ck/YN/1251/03 | < | 3,200 | < | < | 800 | 40 | < | 40 | 160 | 320 | |
| Ck/YN/1083/03 | < | 6,400 | < | < | 800 | 160 | 80 | 320 | 160 | 320 | |
| Ck/VNM/27/03 | < | 6,400 | < | < | 800 | 40 | 40 | 80 | 160 | 320 | |
| Ck/VNM/133/04 | < | 6,400 | < | < | 800 | 40 | < | 40 | 160 | 320 | |
| VNM/1194/04 | < | 1,600 | < | < | 400 | < | < | < | 80 | 80 | |
| Dk/FJ/1734/05 | VI | >12,800 | < | 1,600 | < | 100 | < | < | 40 | 640 | < |
| Dk/Laos/3295/06 | >12,800 | < | 1,600 | < | 200 | < | 40 | 80 | 640 | 40 | |
| Munia/HK/2454/06 | >12,800 | < | 3,200 | < | 100 | < | 40 | 160 | 640 | 40 | |
Abbreviations: BHG, bar-headed goose; Ck, chicken; Dk, duck; FJ, Fujian; GH, gray heron; HK, Hong Kong; HN, Hunan; IDN, Indonesia; JX, Jiangxi; MDk, migratory duck; Penn, Pennsylvania; QH, Qinghai; ST, Shantou; VNM, Vietnam; YN, Yunnan. <, HI titers are lower than 40 for antisera and lower than 100 for MAbs.
Antigenic groups are based on Fig. 1.
FIG. 1.
Numerical analysis of HI titers (see Table 2) by nonmetric multidimensional ordination in two dimensions (A) and using hierarchical agglomerative clustering (B). Each antigenic group (circled) has been labeled on the cluster (see also Table 2).
Phylogenetic analysis of the surface genes.
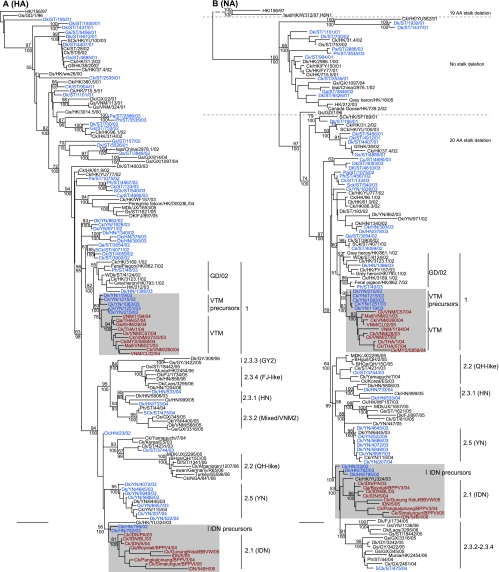
Phylogenetic analysis of the H5 HA gene showed that the Gs/GD-like viruses were highly diverse from 2001 to 2003 (Fig. 2A). One virus isolated from 2001 in Shantou, Guangdong (Dk/ST/195/01) fell at the base of the tree close to Gs/GD-like virus, while the remaining H5N1 viruses isolated in 2001 fell in multiple lineages that were most closely related to viruses isolated from poultry outbreaks in Hong Kong during that period. The viruses isolated in 2002 and 2003 from Guangdong, Hunan, and Yunnan showed increased genetic diversity, and even viruses isolated during the same period and from the same sampling site clustered into different sublineages, e.g., Ck/ST/3744/03 and Ph/ST/3535/03 (Fig. 2A).
FIG. 2.
Phylogenetic relationships of the HA (A) and NA (B) genes of representative influenza A viruses. The numbers above and below the branch nodes indicate neighbor-joining bootstrap values of ≥50% and Bayesian posterior probabilities of >95, respectively. Analyses were based on nucleotides 22 to 1,032 of the HA gene and nucleotides 1 to 1,090 of the NA gene. The HA and NA gene trees were rooted to duck/Hokkaido/51/96 and chicken/Scotland/59, respectively. Colors indicate viruses isolated from southern China (blue), Vietnam (red), and Indonesia (red). Scale bar, 0.01 nucleotide substitutions per site. Abbreviations: BHGs, bar-headed goose; BHgull, brown-headed gull; Ck, chicken; Cu, chukkar; Dk, duck; FJ, Fujian; Gf, Guinea fowl; Gs, goose; GD, Guangdong; GX, Guangxi; GY, Guiyang; HK, Hong Kong; HN, Hunan; IDN, Indonesia; JX, Jiangxi; Mall, mallard; MDk, migratory duck; MYS, Malaysia; NGA, Nigeria; Pa, partridge; Ph, pheasant; Qa, quail; QH, Qinghai; SCk, silky chicken; ST, Shantou; THA, Thailand; VNM, Vietnam; WDk, wild duck; YN, Yunnan.
Interestingly, eight viruses isolated from Yunnan in 2002 and 2003 (represented by Ck/YN/1215/02) fell as the progenitor to clade 1 viruses (Fig. 2A). Also, two viruses isolated from Hunan in 2002/2003 (represented by Dk/HN/795/02) were most closely related to clade 2.1 Indonesian viruses. Furthermore, two viruses isolated from Hunan and 15 viruses isolated from Yunnan during 2003 and 2004 formed monophyletic clades with previously described viruses from Hunan (clade 2.3.1) and Yunnan (clade 2.4), respectively (5), revealing a regionally dominant group of viruses in these provinces. These phylogenetic relationships show that during 2002 and 2003 the genetic diversity of H5N1 viruses in southern China increased dramatically and directly resulted in the subsequent establishment of these diverse sublineages in different regions.
Phylogenetic analysis of neuraminidase (NA) gene revealed two major lineages derived from the Gs/GD-like NA (Fig. 2B). The first lineage included 11 viruses detected in Shantou (Guangdong) in 2001 and 2002 and that did not have a deletion in the stalk region of the NA. It is noteworthy that the NA gene of Dk/ST/195/01 has the same 20-amino-acid (aa) deletion in its NA stalk region that is seen in all H5N1 genotype Z viruses. The majority of the viruses from the present study belonged to a second lineage in which the viruses had a 20-aa deletion in the NA stalk and were isolated in different regions since 2001 (Fig. 2B). The close phylogenetic relationship between viruses from Hunan (e.g., Dk/HN/795/02) and Indonesia and between viruses from Yunnan (e.g., Ck/YN/1215/02) and Vietnam were maintained for the NA gene (Fig. 2B).
Phylogenetic analyses of the internal genes.
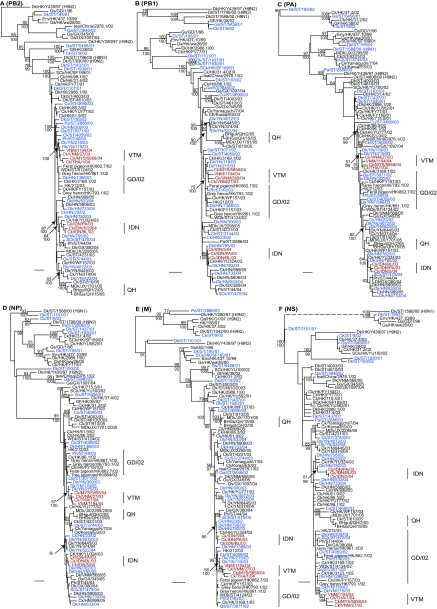
Phylogenetic relationships of the internal genes of representative viruses indicated that frequent reassortment between cocirculating viruses had resulted in a number of novel genotypes being detected in poultry from 2001 to 2003 (Fig. 3). The majority of the novel genotypes were detected in Shantou (Guangdong) live-poultry markets and clustered with other influenza subtype viruses (H9N1 and H9N2), confirming the frequent reassortment of cocirculating viruses, as detected in our previous studies of other subtypes (7, 12, 14, 15, 38). All internal genes of Dk/ST/195/01 consistently grouped with Gs/GD/1/96, providing the first record of a Gs/GD-like virus with the 20-aa NA stalk deletion (Fig. 2 and 3). The internal gene segments of those Hunan (clade 2.3.1) and Yunnan (clade 2.4) viruses maintained monophyletic groups, a finding consistent with their HA and NA phylogenies.
FIG. 3.
Phylogenetic relationships of the PB2 (A), PB1 (B), PA (C), NP (D), M (E), and NS (F) genes of representative influenza A viruses. The numbers above and below the branch nodes indicate neighbor-joining bootstrap values of ≥50% and Bayesian posterior probabilities of >95, respectively. Analyses were based on nucleotides 986 to 2288, 1 to 1480, 1404 to 2151, 1 to 990, 1 to 998, and 1 to 842, respectively. All gene trees were rooted to A/equine/Prague/1/56 except PB1 and PA, which were midpoint rooted. Colors indicate viruses isolated from southern China (blue), Vietnam (red), and Indonesia (red). Scale bar, 0.01 nucleotide substitutions per site. Abbreviations: BHGs, bar-headed goose; BHgull, brown-headed gull; Ck, chicken; Cu, chukkar; Dk, duck; FJ, Fujian; Gf, Guinea fowl; Gs, goose; GD, Guangdong; GX, Guangxi; GY, Guiyang; HK, Hong Kong; HN, Hunan; IDN, Indonesia; JX, Jiangxi; Mall, mallard; MDk, migratory duck; MYS, Malaysia; NGA, Nigeria; Pa, partridge; Ph, pheasant; Qa, quail; QH, Qinghai; SCk, silky chicken; ST, Shantou; THA, Thailand; VNM, Vietnam; WDk, wild duck; YN, Yunnan.
The internal genes of the 10 Hunan and Yunnan progenitor viruses also maintained their phylogenetic relationships to Vietnam and Indonesia isolates, thereby confirming the genesis and transmission pathway of those viruses (Fig. 3A to F). Phylogenetic analyses also revealed that these progenitor viruses belonged to H5N1 genotype Z.
Amino acid similarity.
In order to evaluate the genetic similarity of the Yunnan/Vietnam and Hunan/Indonesia viruses, we calculated the mean percent similarities of deduced amino acid sequence for each of the gene segments between these two groups of viruses (Table 3). The similarities of the HA and the NA genes were, respectively, 98.1 and 98.9% for Yunnan/Vietnam and 99.6 and 99.2% for Hunan/Indonesia. The mean percentage similarity for Yunnan/Vietnam was ≥99.5% for all internal genes, while for Hunan/Indonesia the mean percentage similarity also was ≥99.5% for all internal genes except for the NS gene (98.01%). The extremely high similarities between these two virus groups confirm that the viruses isolated from Yunnan and Hunan during 2002 are the likely direct progenitors of those clade 1 viruses in Vietnam and clade 2.2 viruses in Indonesia, respectively.
TABLE 3.
Amino acid similarity of viruses isolated in southern China and Southeast Asia
| Virus region | % Amino acid similaritya
|
|||||||
|---|---|---|---|---|---|---|---|---|
| PB2 | PB1 | PA | HA | NS | NA | M | NP | |
| Yunnan/Vietnam | 99.58 | 99.45 | 99.93 | 98.12 | 99.11 | 98.89 | 99.68 | 100 |
| Hunan/Indonesia | 99.93 | 100 | 99.44 | 99.64 | 98.01 | 99.16 | 100 | 99.89 |
The percent similarity was calculated based on p-distance measurements. For Yunnan/Vietnam, the similarity is based on eight virus isolates from Yunnan and three virus isolates from Vietnam. For Hunan/Indonesia, the similarity is based on two virus isolates from Hunan and three virus isolates from Indonesia.
Molecular characterization.
All of the 73 viruses characterized were highly pathogenic with variations of the multibasic cleavage site (PQRERRRKKR/G) in the HA molecule. The receptor-binding pocket of the HA1 retains amino acid residues 222-Gln and 224-Gly (H5 numbering used throughout) that have been shown to preferentially bind to α-2,3-NeuAcGal receptors (16, 29, 40). Other amino acid residues relevant to receptor-binding sites were identical to those of HK/156/97 and Gs/GD-like viruses in most isolates, but with some notable differences. Most of the progenitor Yunnan viruses characterized had an HA Ser129Leu substitution, which had been previously observed in clade 1 viruses (27), while the Hunan and Indonesian isolates maintained 129-Ser (Table 4) . In the NA amino acid sequences, all isolates characterized had 274-His, indicating sensitivity to oseltamivir (33). All eight clade 1 progenitor viruses from Yunnan had both the Leu26Ile and the Ser31Asn substitutions in the M2 protein (Table 4). These mutations may confer resistance to the amantadines, and both are present in all Vietnam clade 1 viruses characterized to date. No amantadine resistance mutations were observed in the two Hunan viruses that are progenitors to Indonesian isolates.
TABLE 4.
Genetic signature of viruses isolated from southern China and Southeast Asiaa
| Virus | HA RBSb
|
NAc
|
M2 ion channeld
|
PB2
|
NS1
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 129 | 212 | 217 | 274 | 279 | 26 | 27 | 30 | 31 | 34 | 627 | 92 | |
| Gs/GD | S | E | P | H | N | L | V | A | S | G | E | D |
| HK/97 | S | E | P | H | N | L | V | A | S | G | E | D |
| Dk/ST/195/01 | S | E | P | H | N | L | V | A | S | G | E | D |
| Dk/HN/1386/03 | L | K | S | H | N | L | V | A | S | G | E | D |
| Clade 1 (HK 02/03) | L | K | S | H | N | L | V | A | S | G | E | D |
| Dk/YN/1300/03 | L | K | S | H | N | I | V | A | N | G | E | D |
| Clade 1 (Vietnam) | L | R | S | H | N | I | V | A | N | G | E | D |
| Dk/HN/23/02 | S | K | S | H | N | L | V | A | S | G | E | D |
| Dk/HN/782/02 | S | K | S | H | N | L | V | A | S | G | E | D |
| Clade 2.1 (Indonesia) | S/L | K | S | H | N | L | V | A | S/N | G | E | D |
DISCUSSION
Our previous study suggested that HPAI H5N1 viruses had evolved into multiple regionally distinct sublineages in southern China and Southeast Asia (5). The circulation of different antigenic and genetic sublineages led the World Health Organization to prepare multiple pandemic vaccine candidates to deal with this situation (37). Currently, Vietnam (clade 1) and Indonesian (clade 2.1) H5N1 viruses maintain their antigenic and genetic traits and always cluster together in phylogenetic analysis (27). The viruses isolated from Europe and Africa have also not diversified greatly from the original Qinghai-like variant (2, 9). Although the differences among those regional sublineages complicate control efforts, they do provide the opportunity to trace the virus transmission pathways (5, 26).
In the present study, our findings clearly demonstrate a direct precursor-descendant relationship between Yunnan and Vietnam (clade 1) and Hunan and Indonesia (clade 2.1) H5N1 viruses. All of the viruses from Hunan and Yunnan were isolated from November 2002 to April 2003, just prior to the onset of the first major transmission and outbreak wave in Southeast Asia that was first reported in November 2003 (22). These results suggest a transmission of H5N1 viruses from Yunnan to Vietnam and from Hunan to Indonesia.
The results of the antigenic analysis strongly correlated with the phylogenetic relationships. Molecular characterization at the amino acid level showed that the precursor-descendant viruses had almost identical amino acid sequences and identified consistent residue signatures between Yunnan-Vietnam and Hunan-Indonesia H5N1 viruses. For example, the dual Leu26Ile and Ser31Asn amantadine resistance mutations in the M2 protein of all Vietnam clade 1 viruses is also present in the Yunnan precursors (6). This provides evidence that these amantadine resistance mutations were present as early as 2002 in the viruses that were introduced into Vietnam to form clade 1, rather than resistance developing in Vietnam.
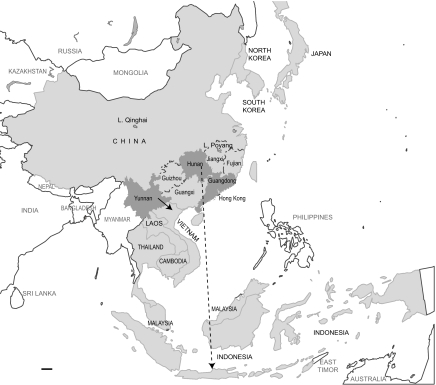
Since Yunnan shares a 600-km border with Vietnam (Fig. 4), it is reasonable to suggest that the virus was introduced into Vietnam through the trade of poultry at the border region. However, similar virus movement from Guangxi to Vietnam was observed in 2001 and 2005 and, more recently, with the Fujian-like variant (clade 2.3.1) (5, 26). Since we did not begin influenza surveillance in Guangxi until 2004, which also shares a long border with Vietnam (Fig. 4), we cannot exclude the possibility that viruses from Guangxi may also be clade 1 precursors.
FIG. 4.
Map of southern China showing the provinces under influenza surveillance and wave 1 transmission pathway of H5N1 viruses from southern China to other Southeast Asian regions.
The spread and transmission route from Hunan to Indonesia remains unclear. Our previous study suggested that the Indonesian H5N1 variant (clade 2.1) resulted from a single introduction in 2003, most likely to Java, where ca. 60% of poultry production in Indonesia is concentrated and where H5N1 outbreaks were first reported from (27). Java does not share any border with other countries, and it is approximately 3,600 km between Hunan and Java, which raises the possibility of virus transmission via migratory birds. However, since there is a lack of surveillance data from migratory birds in these regions in the period preceding the outbreaks, and there is also little information on poultry trade between China and Indonesia, it is unlikely that this transmission route can be fully determined.
The HPAI H5N1 viruses have been prevalent in poultry for 11 years in southern China (26, 39). At present, these viruses are endemic and panzootic in different types of poultry, especially in aquatic birds in many countries across Asia, Africa, and Europe (5, 9, 26). Historically, 24 HPAI H5 and H7 outbreaks have been recorded worldwide (1), but the prevalence of Gs/GD-like H5N1 variants is the first and only example of an HPAI virus that has persisted in poultry for an extended period and spread over such a huge geographical area. The Gs/GD-like H5N1 virus is also the only HPAI virus that has undergone such extensive reassortment to generate so many different variants (5, 13, 22). Interestingly, previous studies indicate that all of these reassortants have been generated in China (5, 13, 22). Since many H9N2 and H6N1 variants have also been simultaneously recognized from southern China, a reasonable explanation is that the cocirculation of different subtypes of influenza viruses has promoted reassortment events (7, 38). However, influenza viruses are broadly prevalent in their natural reservoirs, wild aquatic birds, in most of the H5N1-affected regions (1, 35). Therefore, why the current H5N1 viruses circulating in other affected regions have not undergone reassortment with “local” viruses remains a fascinating question.
The continuing endemicity of HPAI H5N1 virus in countries across three continents has given rise to a persistent and unprecedented pandemic threat. Our present study, together with previous reports, has repeatedly demonstrated that the only way to identify and understand the genesis and transmission pathways of this infectious agent is through systematic influenza surveillance (5, 8, 13, 15, 22, 26, 27). More importantly, systematic influenza surveillance can provide critical information in determining effective disease control measures. The successful intervention and control of HPAI H5N1 in Hong Kong in the last decade provides the best example of this.
Acknowledgments
This study was supported by the Li Ka Shing Foundation, the National Institutes of Health (NIAID contract HHSN266200700005C), and the Area of Excellence Scheme of the University Grants Committee (grant AoE/M-12/06) of the Hong Kong SAR Government. G.J.D.S. is supported by a career development award under NIAID contract HHSN266200700005C.
Footnotes
Published ahead of print on 23 January 2008.
REFERENCES
- 1.Alexander, D. J. 2007. An overview of the epidemiology of avian influenza. Vaccine 255637-5644. [DOI] [PubMed] [Google Scholar]
- 2.Bragstad, K., P. H. Jørgensen, K. Handberg, A. S. Hammer, S. Kabell, and A. Fomsgaard. 2007. First introduction of highly pathogenic H5N1 avian influenza A viruses in wild and domestic birds in Denmark, Northern Europe. Virol. J. 443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bray, R. J., and J. T. Curtis. 1957. An ordination of the upland forest communities of southern Wisconsin. Ecol. Monogr. 27325-349. [Google Scholar]
- 4.Chen, H., G. J. D. Smith, S. Y. Zhang, K. Qin, J. Wang, K. S. Li, R. G. Webster, J. S. M. Peiris, and Y. Guan. 2005. H5N1 virus outbreak in migratory waterfowl. Nature 436191-192. [DOI] [PubMed] [Google Scholar]
- 5.Chen, H., G. J. D. Smith, K. S. Li, J. Wang, X. H. Fan, J. M. Rayner, D. Vijaykrishna, J. X. Zhang, L. J. Zhang, C. T. Guo, C. L. Cheung, K. M. Xu, L. Duan, K. Huang, K. Qin, Y. H. Leung, W. L. Wu, H. R. Lu, Y. Chen, N. S. Xia, T. S. Naipospos, K. Y. Yuen, S. S. Hassan, S. Bahri, T. D. Nguyen, R. G. Webster, J. S. Peiris, and Y. Guan. 2006. Establishment of multiple sublineages of H5N1 influenza virus in Asia: implications for pandemic control. Proc. Natl. Acad. Sci. USA 1032845-2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheung, C. L., J. M. Rayner, G. J. D. Smith, P. Wang, T. S. P. Naipospos, J. Zhang, K. Y. Yuen, R. G. Webster, J. S. M. Peiris, Y. Guan, and H. Chen. 2006. Distribution of amantadine-resistant H5N1 avian influenza variants in Asia. J. Infect. Dis. 1931626-1629. [DOI] [PubMed] [Google Scholar]
- 7.Cheung, C. L., D. Vijaykrishna, G. J. D. Smith, X. H. Fan, J. X. Zhang, J. Bahl, L. Duan, K. Huang, H. Tai, J. Wang, L. L. M. Poon, J. S. M. Peiris, H. Chen, and Y. Guan. 2007. Establishment of influenza A virus (H6N1) in minor poultry in southern China. J. Virol. 8110402-10412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duan, L., L. Campitelli, X. H. Fan, Y. H. C. Leung, D. Vijaykrishna, J. X. Zhang, I. Donatelli, M. Delogu, K. S. Li, E. Foni, C. Chiapponi, W. L. Wu, H. Kai, R. G. Webster, K. F. Shortridge, J. S. M. Peiris, G. J. D. Smith, H. Chen, and Y. Guan. 2007. Characterization of low pathogenic H5 subtype influenza viruses from Eurasia: implications for the origin of highly pathogenic H5N1 viruses. J. Virol. 817529-7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ducatez, M. F., C. M. Olinger, A. A. Owoade, S. DeLandtsheer, W. Ammerlaan, H. G. M. Niesters, A. D. M. E. Osterhaus, R. A. M. Fouchier, and C. P. Muller. 2006. Introductions of H5N1 in Nigeria. Nature 44237. [DOI] [PubMed] [Google Scholar]
- 10.Ellis, T. M., R. B. Bousfield, L. A. Bissett, K. C. Dyrting, G. S. M. Luk, S. T. Tsim, K. Strum-Ramirez, R. G. Webster, Y. Guan, and J. S. M. Peiris. 2004. Investigation of outbreaks of highly pathogenic H5N1 avian influenza in waterfowl and wild birds in Hong Kong in late 2002. Avian Pathol. 33492-505. [DOI] [PubMed] [Google Scholar]
- 11.Food and Agricultural Organization of the United Nations. 2007. Avian Influenza Disease Emergency Bulletin, issue 46. http://www.fao.org/docs/eims/upload//228650/AIDENews_may07_no46.pdf.
- 12.Guan, Y., M. Peiris, K. F. Kong, K. C. Dyrting, T. M. Ellis, T. Sit, L. J. Zhang, and K. F. Shortridge. 2002. H5N1 influenza viruses isolated from geese in southeastern China: evidence for genetic reassortment and interspecies transmission to ducks. Virology 29216-23. [DOI] [PubMed] [Google Scholar]
- 13.Guan, Y., J. S. M. Peiris, A. S. Lipatov, T. M. Ellis, K. C. Dyrting, S. Krauss, L. J. Zhang, R. G. Webster, and K. F. Shortridge. 2002. Emergence of multiple genotypes of H5N1 avian influenza viruses in Hong Kong SAR. Proc. Natl. Acad. Sci. USA 998950-8955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guan, Y., K. F. Shortridge, S. Krauss, and R. G. Webster. 1999. Molecular characterization of H9N2 influenza viruses: were they the donors of the “internal” genes of H5N1 viruses in Hong Kong? Proc. Natl. Acad. Sci. USA 169363-9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guan, Y., K. F. Shortridge, S. Krauss, P. S. Chin, K. C. Dyrting, T. M. Ellis, R. G. Webster, and J. S. M. Peiris. 2000. H9N2 influenza viruses possessing H5N1-like internal genomes continue to circulate in poultry in southeastern China. J. Virol. 749372-9380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ha, Y., D. J. Stevens, J. J. Skehel, and D. C. Wiley. 2001. X-ray structures of H5 avian and H9 swine influenza virus hemagglutinins bound to avian and human receptor analogs. Proc. Natl. Acad. Sci. USA 9811181-11186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hall, T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 4195-98. [Google Scholar]
- 18.Huelsenbeck, J. P., and F. R. Ronquist. 2001. MrBayes: Bayesian inference of phylogenetic trees. Bioinformatics 17754-755. [DOI] [PubMed] [Google Scholar]
- 19.Kruskal, J. B. 1964. Nonmetric multidimensional scaling: a numerical method. Psychometrika 29115-129. [Google Scholar]
- 20.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: an integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5150-163. [DOI] [PubMed] [Google Scholar]
- 21.Lee, C. W., D. L. Suarez, T. M. Tumpey, H. W. Sung, Y. K. Kwon, Y. J. Lee, J. G. Choi, S. J. Joh, M. C. Kim, E. K. Lee, J. M. Park, X. Lu, J. M. Katz, E. Spackman, D. E. Swayne, and J. H. Kim. 2005. Characterization of highly pathogenic H5N1 avian influenza A viruses isolated from South Korea. J. Virol. 793692-3702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li, K. S., Y. Guan, J. Wang, G. J. D. Smith, K. M. Xu, L. Duan, A. P. Rahardjo, P. Puthavathana, C. Buranathai, T. D. Nguyen, A. T. S. Estoepangestie, A. Chaisingh, P. Auewarakul, H. T. Long, N. T. H. Hanh, R. J. Webby, L. L. M. Poon, H. Chen, K. F. Shortridge, K. Y. Yuen, R. G. Webster, and J. S. M. Pieris. 2004. Genesis of highly pathogenic and potentially pandemic H5N1 influenza virus in eastern Asia. Nature 430209-213. [DOI] [PubMed] [Google Scholar]
- 23.Mase, M., M. Eto, N. Tanimura, K. Imai, K. Tsukamoto, T. Horimoto, Y. Kawaoka, and S. Yamaguchi. 2005. Isolation of a genotypically unique H5N1 influenza virus from duck meat imported into Japan from China. Virology 339101-109. [DOI] [PubMed] [Google Scholar]
- 24.Nylander, J. A. A. 2004. MRMODELTEST 2. Evolutionary Biology Centre, Uppsala University, Uppsala, Sweden.
- 25.Scholtissek, C., G. Quack, H. D. Klenk, and R. G. Webster. 1998. How to overcome resistance of influenza A viruses against adamantane derivatives. Antivir. Res. 3783-95. [DOI] [PubMed] [Google Scholar]
- 26.Smith, G. J. D., X. H. Fan, J. Wang, K. S. Li, K. Qin, J. X. Zhang, D. Vijaykrishna, C. L. Cheung, K. Huang, J. M. Rayner, J. S. M. Peiris, H. Chen, R. G. Webster, and Y. Guan. 2006. Emergence and predominance of an H5N1 influenza variant in China. Proc. Natl. Acad. Sci. USA 10316936-16941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith, G. J. D., T. S. P. Naipospos, T. D. Nguyen, M. D. de Jong, D. Vijaykrishna, T. B. Usman, S. S. Hassan, T. V. Nguyen, T. V. Dao, N. A. Bui, Y. H. C. Leung, C. L. Cheung, J. M. Rayner, J. X. Zhang, L. J. Zhang, L. L. M. Poon, K. S. Li, V. C. Nguyen, T. T. Hien, J. Farrar, R. G. Webster, H. Chen, J. S. M. Peiris, and Y. Guan. 2006. Evolution and adaptation of H5N1 influenza virus in avian and human hosts in Indonesia and Vietnam. Virology 350258-268. [DOI] [PubMed] [Google Scholar]
- 28.Sokal, R. R., and C. D. Michener. 1958. A statistical method for evaluating systematic relationships. University of Kansas Science Bulletin 381409-1438. [Google Scholar]
- 29.Stevens, J., O. Blixt, T. M. Tumpey, J. K. Taubenberger, J. C. Paulson, and I. A. Wilson. 2006. Structure and receptor specificity of the hemagglutinin from an H5N1 influenza virus. Science 312404-410. [DOI] [PubMed] [Google Scholar]
- 30.Subbarao, K., A. Klimov, J. Katz, H. Regnery, W. Lim, H. Hall, M. Perdue, D. Swayne, C. Bender, J. Huang, M. Hemphill, T. Rowe, M. Shaw, X. Xu, K. Fukuda, and N. Cox. 1998. Characterization of an avian influenza A (H5N1) virus isolated from a child with a fatal respiratory illness. Science 279393-396. [DOI] [PubMed] [Google Scholar]
- 31.Swofford, D. L. 2001. PAUP*: phylogenetic analysis using parsimony (and other methods) 4.0 beta. Sinauer Associates, Sunderland, MA.
- 32.Taubenberger, J. K., A. H. Reid, R. M. Lourens, R. Wang, G. Jin, and T. G. Fanning. 2005. Characterization of the 1918 influenza virus polymerase genes. Nature 437889-893. [DOI] [PubMed] [Google Scholar]
- 33.Treanor, J. J., F. G. Hayden, P. S. Vrooman, R. Barbarash, R. Bettis, D. Riff, S. Singh, N. Kinnersley, P. Ward, and R. G. Mills. 2000. Efficacy and safety of the oral neuraminidase inhibitor oseltamivir in treating acute influenza: a randomized controlled trial. JAMA 2831016-1024. [DOI] [PubMed] [Google Scholar]
- 34.Wallace, R. G., H. HoDac, R. H. Lathrop, and W. M. Fitch. 2007. A statistical phylogeography of influenza A H5N1. Proc. Natl. Acad. Sci. USA 1044473-4478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Webster, R. G., W. J. Bean, O. T. Gorman, T. M. Chambers, and Y. Kawaoka. 1992. Evolution and ecology of influenza A viruses. Microbiol. Rev. 56152-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Webster, R. G., Y. Guan, M. Peiris, D. Walker, S. Krauss, N. N. Zhou, E. A. Govorkova, T. M. Ellis, K. C. Dyrting, T. Sit, D. R. Perez, and K. F. Shortridge. 2002. Characterization of H5N1 influenza viruses that continue to circulate in geese in southeastern China. J. Virol. 76118-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.World Health Organization. 2007. Antigenic and genetic characteristics of H5N1 viruses and candidate H5N1 vaccine viruses developed for potential use as pre-pandemic vaccines. World Health Organization, Geneva, Switzerland. http://www.who.int/csr/disease/avian_influenza/guidelinestopics/en/index5.html.
- 38.Xu, K. M., G. J. D. Smith, J. Bahl, L. Duan, H. Tai, D. Vijaykrishna, J. Wang, J. X. Zhang, K. S. Li, X. H. Fan, R. G. Webster, H. Chen, J. S. M. Peiris, and Y. Guan. 2007. The genesis and evolution of H9N2 influenza viruses in poultry from southern China, 2000 to 2005. J. Virol. 8110389-10401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu, X., K. Subbarao, N. J. Cox, and Y. Guo. 1999. Genetic characterization of the pathogenic influenza A/Goose/Guangdong/1/96 (H5N1) virus: similarity of its hemagglutinin gene to those of H5N1 viruses from the 1997 outbreaks in Hong Kong. Virology 26115-19. [DOI] [PubMed] [Google Scholar]
- 40.Yamada, S., Y. Suzuki, T. Suzuki, M. Q. Le, C. A. Nidom, Y. S. Tagawa, Y. Muramoto, M. Ito, M. Kiso, T. Horimoto, K. Shinya, T. Sawada, M. Kiso, T. Usui, T. Murata, Y. Lin, A. Hay, L. F. Haire, D. J. Stevens, R. J. Russell, S. J. Gamblin, J. J. Skehel, and Y. Kawaoka. 2006. Hemagglutinin mutations responsible for the binding of H5N1 influenza A viruses to human type receptors. Nature 444378-382. [DOI] [PubMed] [Google Scholar]