Abstract
There are more than 100 known adenovirus (AdV) serotypes, including 50 human serotypes. Because AdV-induced disease is relatively species specific, vectors derived from nonhuman serotypes may have wider clinical potential based, in part, on the lack of ubiquitous memory immunity. Whereas a few of the human serotype capsids have been studied at the structural level, none of the nonhuman serotypes has been analyzed. The basis laid by the analysis of human AdV (hAdV) has allowed us to determine and compare the three-dimensional structure of the capsid of canine serotype 2 (CAV-2) to that of hAdV serotype 5 (hAdV-5). We show that CAV-2 capsid has a smoother structure than the human serotypes. Many of the external loops found in the hAdV-5 penton base and the hexon, against which the antibody response is directed, are shorter or absent in CAV-2. On the other hand, the CAV-2 fiber appears to be more complex, with two bends in the shaft. An interesting difference between the human and canine viruses is that the C-terminal part of protein IX is in a different position, making an antenna sticking out of the CAV-2 capsid. The comparison between the two viruses allows the identification of sites that should be easy to modify on the CAV-2 capsid for altering tissue tropism or other biological activities.
Adenoviridae are nonenveloped icosahedral viruses with 26- to 44-kb double-stranded DNA genomes (26). They infect a variety of vertebrates including mammals, fish, birds, and reptiles (12). The more than 50 human adenovirus (hAdV) serotypes are the most extensively studied because of their ability to induce respiratory, ocular, and enteric infections in immunocompetent individuals. hAdV infections are a significant cause of morbidity and mortality in newborns and immunocompromised individuals (28). AdVs are also used for gene therapy trials and anticancer treatments (6) although the widespread immunity against human serotypes severely compromises their use as vectors. Because AdV-induced morbidity is relatively species specific, vectors derived from nonhuman AdVs may be more clinically useful than those derived from human serotypes, based, in part, on the potential lack of ubiquitous memory immunity (cellular and humoral) (5, 11, 18, 30, 35, 36).
One of the best characterized nonhuman AdVs is the canine serotype 2 (cAdV-2, commonly referred to as CAV-2) (29). We previously showed that CAV-2 vectors could lead to preferential transduction of neurons in vivo (49), long-term (>1 yr) expression in the brain (50), and a lower cellular immune response (27, 49). The majority of donors in a random cohort harbored no or only low levels of anti-CAV-2 neutralizing antibodies (30, 37), and less than half of this cohort harbored anti-CAV-2 memory T cells (38). When the donors harbored an anti-CAV-2 cellular response, it was on the average 10-fold lower than the response to hAdV serotype 5 (hAdV-5). Finally, in contrast to hAdV-5-based vectors, CAV-2 vectors poorly transduce or mature human dendritic cells or activate the complement cascade (37, 40), both of which play pivotal roles in orchestrating and bridging innate, adaptive, and memory immune responses. These findings suggest that CAV-2 vectors may have specific clinical advantages, especially in terms of safety (39).
However, there exist certain enigmas concerning various aspects of CAV-2 biology. For example, the in vitro paradigm for AdV tropism is predominately based on studies using hAdV-2 or hAdV-5 in epithelial cells. Initially, the fiber knob binds to the coxsackievirus AdV receptor (CAR) (4, 52). Then, a conserved Arg-Gly-Asp (RGD) motif in the penton base binds to integrins, triggering endocytosis (24). Although CAV-2 uses CAR to infect cells (48), the capsid does not contain an identifiable integrin-interacting motif, yet it traffics in epithelia cells with similar kinetics (8). Furthermore, although both hAdV-5 and CAV-2 use CAR, CAV-2 preferentially transduces neurons when injected into the brain while hAdV-5 preferentially transduces astrocytes (1, 32). CAV-2 is also capable of an impressive level of axonal transport in neurons that can be up to 100-fold greater than that of hAdV-5 (49). In addition, in contrast to previous studies with hAdV-5 (16, 17), we have been unable to complex CAV-2 virions with divalent cations or lipoplexes (unpublished data).
To resolve these and other CAV-2 enigmas, we need to better understand the capsid structure. Based on the data from different hAdVs, the AdV capsid structure appears to be relatively well conserved. The capsid contains 7 out of the 12 polypeptides present in the virion. The major structural components are the trimeric hexons, 240 of which form the 20 facets of the icosahedron (51). At each of the 12 vertices are a pentameric penton base and an externally projecting trimeric fiber. The atomic structures of these three individual components are known for hAdV-2 or hAdV-5 (44, 54, 58).
The hexon trimer has a pseudohexagonal base with three towers extending upwards. The triangular top is rotated ∼10° counterclockwise with respect to the hexagonal base. The loops extending from the irregularly shaped towers (44) have not been solved by X-ray crystallography, although they have been visualized and modeled in recent cryoelectron microscopy (cryo-EM) structures (15, 45).
In contrast to the hexon, the penton base is wide at the top (58). It has an overall tulip shape, with each of its five petals having a right-handed twist around the fivefold axis. The top of the base is almost flat with the exception of two protuberances. One of these extensions, which are variable in size depending on the serotype, contains the above-mentioned integrin-interacting motif. The other extension is a hypervariable loop that also varies in size depending on the serotype (20).
The trimeric AdV fiber contains a highly conserved N-terminal sequence that binds to the base, a thin shaft of variable length, and a globular knob at the distal end. The length, the flexibility of the fiber shaft, and the sequence of the knob domain also vary between serotypes (9). Four of the 23 repeats of the hAdV-2 fiber shaft were crystallized, and the resolved structure demonstrated a novel triple β-spiral (54). Notably though, the entire fiber was not crystallized, probably due to its flexible structure. The number of repeats can vary from a minimum of five for hAdV-3 to at least 46 for bovine AdV-3 (BAV-3) (43).
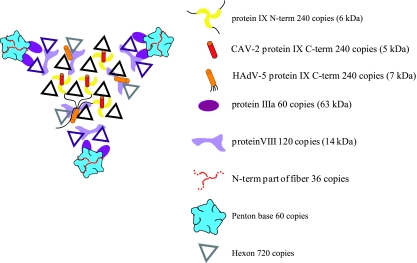
Apart from these major proteins, the capsid is stabilized by hexon-associated proteins (51). Using high-resolution cryo-EM three-dimensional (3D) reconstructions combined with secondary structure predictions, a new location of the minor structural proteins has been proposed. Protein IX forms a connected complex of four trimers in the middle of each facet (45). Protein IIIa is located inside the capsid underneath the penton base, connected to its N-terminal part (45). A total of 120 copies of protein VIII are located on the inner face of the capsid, and each makes the link between four hexon trimers around the three- and fivefold axes (15).
Here, we have determined the 3D structure of the CAV-2 capsid and compared it to the known X-ray and EM structures of hAdV-5 or hAdV-2 by fitting the different atomic resolution structures into the EM density. Globally, we found that CAV-2 capsid has a smoother structure than the human serotypes. Many of the external loops found in hAdV-2 and hAdV-5 (hAdV-2/5) are either shorter or absent in CAV-2. On the other hand, the CAV-2 fiber appears to be more complex, with two bends in the shaft. The knowledge of the structure will allow the identification of sites where functional loops may be added to CAV-2 to potentially modify tissue tropism.
MATERIALS AND METHODS
CAV-2 vectors and recombinant fiber.
CAVGFP and CAVpIX-GFP were prepared as previously described (30, 31). Briefly, CAVGFP is a CAV-2 vector with a deletion of the E1 region and with a green fluorescent protein (GFP) expression cassette. The capsid contains no modifications. CAVpIX-GFP is a vector containing GFP fused to the C-terminal end of protein IX. All vectors were purified by double banding on CsCl gradients, and CsCl was removed using PD-10 columns (Pharmacia). The vectors were stored in phosphate-buffered saline containing 10% glycerol. Stocks were 2 × 1012 physical particles/ml with at least 1 infectious particle/5 physical particles. The approximately 1.7-kb CAV-2 fiber coding region was cloned in pBlueBac4.5/V5-His-TOPO (Invitrogen). Recombinant baculovirus was generated, isolated, amplified, and purified following the manufacturer's protocol. The His-tagged fiber was purified using standard protocols.
Negative staining.
Four microliters of the recombinant CAV-2 fiber sample (∼0.1 mg/ml) was loaded in the mica-carbon interface as described previously (46). The sample was stained using 2% sodium silico tungstate, pH 7.5, and air dried. Images were taken under low-dose conditions in an EX1200-II JEOL electron microscope working at 100 kV and with a nominal magnification of 40,000. The images were scanned on a Z/I Imaging scanner (Photoscan TD) with a pixel size of 14 μm (3.5 Å per pixel at the sample level). A total of 800 fibers were selected using X3d (10), cut into 200- by 200-pixel squares, transferred into the SPIDER program (19), and subjected to classification and averaging.
Cryo-EM.
For cryo-EM, the glycerol was removed, and the sample was concentrated to ∼1 mg/ml. Four microliters of sample was loaded onto a Quantifoil R2/1 holey grid (Quantifoil Micro Tools GmbH, Germany), blotted for 1 to 2 s to remove the excess liquid, and then rapidly plunged into liquid ethane cooled using liquid nitrogen. The frozen grid was transferred into an LaB6 CM200 Philips electron microscope using a GATAN 626 cryoholder. The images were taken under low-dose conditions at 200 kV (less than 10 e−/Å2) and with a nominal magnification of 27,500. Each area was imaged twice using two different defocus values, starting with the image closest to focus. The negatives were developed in full-strength D19 developer for 12 min.
Image analysis.
Negatives were screened for astigmatism and drift by optical diffraction, and only those showing information up to 12 Å were digitized using the Photoscan TD with a pixel size of 7 μm (2.54 Å per pixel at the specimen level as calibrated using tobacco mosaic virus). The particles were selected interactively using X3d (10) and boxed into 447- by 447-pixel squares with a circular mask applied of radius 219 pixels plus four fade pixels. The images were corrected for the contrast transfer function effect, as described previously, using the program CTFMIX (10). A total of 5,350 particles from 21 negatives were used in the image analysis process. Determination of particle origin and orientation were performed with the model-based polar Fourier transform (PFT) programs using the previously determined 3D structure of hAdV-5 (3, 15). After PFT refinement was completed, a supplementary cycle was carried out using a new version of PFT, PFT2 (D. M. Belnap, J. B. Heymann and J. F. Conway, unpublished results). The final reconstruction was obtained using the PFT2 and EM3DR2 programs with a total of 2,780 particles (about half of the total). The resolution of the final map was estimated to 12 Å by Fourier shell correlation (53) calculated between independent half-data set maps and by applying a correlation limit of 0.3 (not shown). The same process was essentially used for CAVpIX-GFP, but we used Quantifoil grids covered by a thin layer of continuous carbon to increase the visible virus concentration. Only 800 particles were used in this reconstruction, which limited the resolution to 25 Å. Map visualization was performed using the programs WEB (19) and PYMOL (14), and the contour level was chosen to include an average protein density of 0.84 Da/Å3. The EM maps were uploaded into the EM database associated with the Macromolecular Structure Database (see below).
Fitting of the atomic structures of hexon and penton base into the EM density.
The atomic structure of the hAdV-5 hexon and that of the hAdV-2 penton base were placed manually into the corresponding CAV-2 EM densities using the program PYMOL (14). These manual fits were then optimized using SITUS (55).
Molecular modeling of the 3D structure of CAV-2 fiber.
To model the 3D structure of the CAV-2 fiber, we used the crystal structure of the homotrimeric C-terminal CAV-2 fiber head (residues 361 to 542) (47) and two known crystal structures of homologous proteins: reovirus attachment protein σ1 (7) and the C-terminal part of the hAdV-2 fiber containing four repeats of the shaft domain followed by the head domain (54). The hAdV-2 structure allowed us to model the last four repetitive elements of the CAV-2 fiber shaft and the region where this shaft is linked to the head. The N-terminal repetitive structure of the CAV-2 fiber shaft was also modeled by using the β-spiral fragment of the known hAdV-2 structure as a template and in accordance with the repeat alignment shown on Fig. 3. The two longer repeats (see Fig. 3C, repeats 4 and 10) of hAdV-2 having 7- and 6-residue insertions in the middle of the repeats were modeled by using the crystal structure of reovirus attachment protein σ1 (7). The σ1 protein has one such long repeat containing a similar 5-residue insertion. The model was built by using the Insight II program (13).
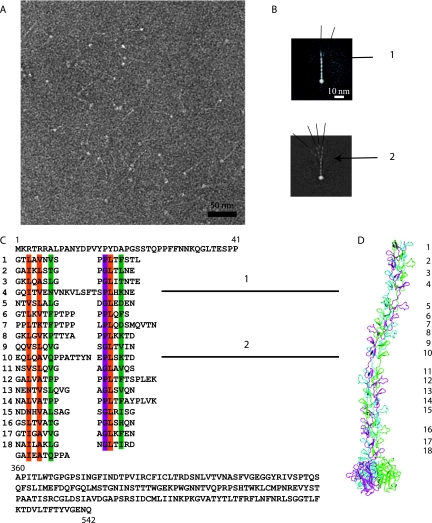
FIG. 3.
The CAV-2 fiber. (A) Electron micrograph of negatively stained CAV-2 fibers. The thin shafts and their globular knobs are clearly visible. Scale bar, 50 nm. (B) Averaged image of fibers selected from micrographs like that shown in panel A. The fiber is generally straight but can exhibit two kinks as indicated by the numbers 1 and 2. The different lines show the angle that the N-terminal part makes with the rest of the shaft. (C) Sequence of the CAV-2 fiber shaft showing the repeat motifs that correspond to the location of the bends. The top part of the sequence is the N-terminal tail of the fiber, and the lower part is the C-terminal knob. The repeat residues involved in the hydrophobic core are in orange, the ones forming the peripheral hydrophobic patches are in green, and the conserved glycine or proline residues are highlighted in purple according to van Raaij et al. (54). (D) Hypothetical model of the CAV-2 fiber showing the predicted bends at repeats 4 and 10. The repeats are indicated with the same numbers as in the sequence shown in panel C.
Comparison with hAdV-5.
All comparisons between the CAV-2 and the hAdV-5 EM structures were done using the hAdV-5 3D reconstruction previously determined in the laboratory and freely available in the EM database associated with the Macromolecular Structure Database (http://www.ebi.ac.uk/msd-srv/emsearch/index.html) under accession number 1111.
Sequence alignments and structure prediction.
All sequence alignments were carried out using CLUSTAL (http://www.ebi.ac.uk/clustalw/). Secondary structure predictions were made using PREDATOR (http://bioweb.pasteur.fr/seqanal/interfaces/predator-simple.html) and coil-coiled predictions were made using http://www.russell.embl-heidelberg.de/cgi-bin/coils-svr.pl.
Protein structure accession numbers.
EM density maps have been deposited in the Macromolecular Structure Database at the European Bioinformatics Institute under accession numbers 1462 and 1463 for CAV-2 and CAVpIX-GFP, respectively.
RESULTS AND DISCUSSION
EM model of CAV-2.
We determined the 3D structure of CAV-2 capsid by cryo-EM at 12-Å resolution. The dimension and the thickness of the CAV-2 capsid shell are similar to those of hAdV-5 (Fig. 1). The icosahedral shape is well defined and exhibits the pseudo T=25 triangulation number imposed by the trimeric hexon protein (Fig. 1A and B). There are 12 triangular hexons per facet and one penton on each vertex. The hexons are packed in an orientation similar to that of hAdV-5, including the slight tilt at each of the 20 edges of the virion (30° versus 10° for the other hexons belonging to the same facets). The penton base also has roughly the same shape in both capsids. The CAV-2 fiber is flexible, which explains why only part of it was visible in the 3D reconstruction (Fig. 1B, star). Finally, densities are present in the middle of the facet between the hexons. This density, attributed to protein IX, exhibits a triskelion-like shape, which is similar to that of hAdV-5 (Fig. 1; see also Fig. 5).
FIG. 1.
3D reconstruction of CAV-2 at 12-Å resolution and comparison with the previously determined structure of hAdV-5 (MSD accession number 1111) at 10-Å resolution. (A) Isosurface representation of CAV-2 as viewed down the threefold axis. The pentons are shown in petrol blue, and the rest of the capsid is shown in yellow. The icosahedral axes are indicated by an ellipse (twofold), a triangle (threefold), and a pentagon (fivefold). A continuous line connecting the three pentons pointing in your direction delimits a facet. (B) A central slice through the CAV-2 EM reconstruction showing that the capsid density is well defined. One of the fibers sticking out radially from the capsid is indicated by a star. A circle surrounds the nonattributed extra density present under the penton base. The arrow indicates protein IX density. Scale bar, 10 nm. (C) Isosurface representation of hAdV-5 view down the threefold axis. The pentons are shown in blue, and the rest of the capsid is shown in gray. (D) A central slice through the hAdV-5 EM reconstruction showing that the capsid density is well defined. The fibers sticking out radially from the capsid are indicated by a star. The RGD loop is indicated by an arrow. Scale bar, 10 nm.
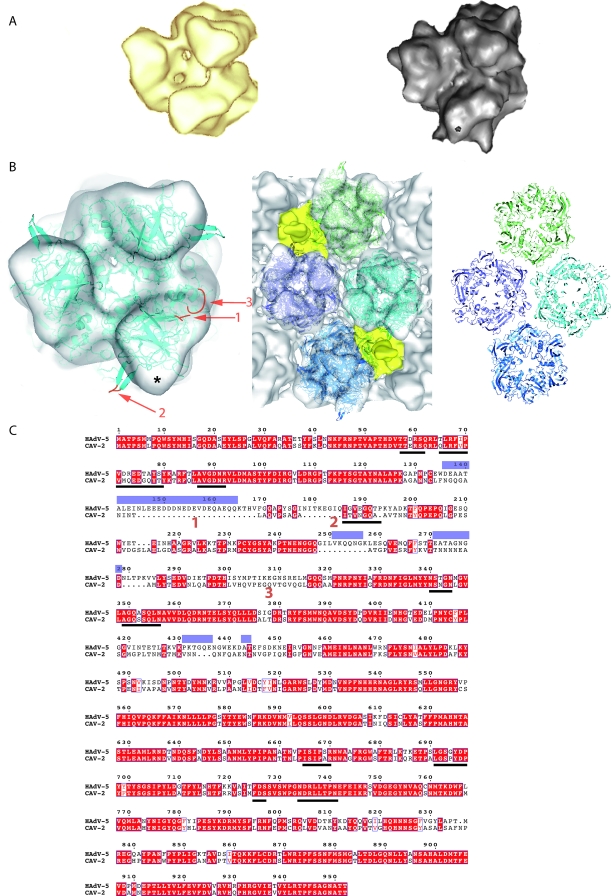
FIG. 5.
The minor protein IX. (A) Detailed view of one facet of CAV-2 seen down the threefold axis of the virion. The pentons are located on the vertices of the triangle and are shown in petrol blue, and the hexons are shown in pale yellow as in Fig. 1. Additional densities present as four groups of trimeric triskelions assigned to be protein IX are yellow (N-terminal part) and red (C-terminal part). (B) The top part represents the center of the facet described as in panel A but slightly tilted to see the extension of the C terminal part of protein IX form its N terminal part. The lower part is a slice through the capsid along the line shown in panel A. The color codes are the same as in panel A. (C) Detailed view of one facet of hAdV-5 seen down the threefold axis of the virion. The pentons are located on the vertices of the triangle and are in blue, and the hexons are gray. Protein IX is shown in yellow, and the cylindrical density that we initially assigned to protein IIIa (15) but that was recently reassigned to the C-terminal part of protein IX (45) is in red. (D) Detailed view of the threefold axis of the CAVpIX-GFP reconstruction. The penton and the hexons are as described in panel A. Protein IX is in yellow (N-terminal part) and red (C-terminal part), and the extra density assigned to GFP is in green. (E) Sequence alignment of hAdV-5 and CAV-2 protein IX. Conserved residues are in white text and highlighted in red; partially conserved residues are in red text. The arrows and open bars represent predicted β-strands and α-helices, respectively, as given by the secondary structure prediction software (http://bioweb.pasteur.fr/seqanal/interfaces/predator-simple.html). The predicted coil-coiled is boxed into a rectangle.
The CAV-2 penton base is smooth.
Sequence alignment between the hAdV-5 and CAV-2 penton bases shows that 310 of the 571 amino acids are identical and that 14 amino acids are similar between the two species. However, the CAV-2 penton base sequence is ∼20% shorter than the hAdV-5 sequence (477 versus 571) (Fig. 2C). Nonetheless, the overall shape of the two pentagons is similar (Fig. 2A). A slice through the density (Fig. 1B) shows a cone-like geometry that fits perfectly into the vertices surrounded by the hexons.
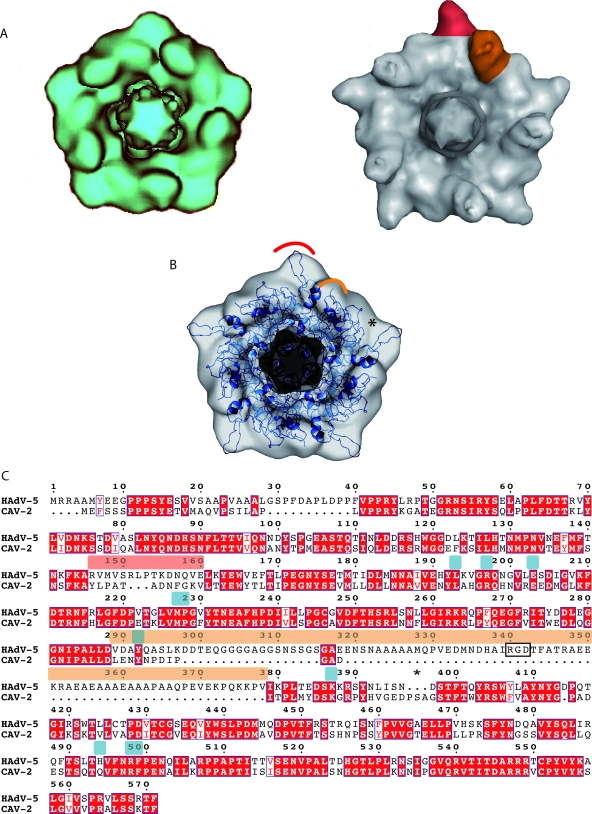
FIG. 2.
The CAV-2 penton base. (A) Comparison between the computationally isolated penton from the CAV-2 capsid in blue (left) and the hAdV-5 capsid in gray (right). One of the five hypervariable regions is highlighted in red, and one protuberance harboring the RGD sequence is shown in orange in the hAdV-5 penton. (B) Fitting of the hAdV-2 penton base X-ray structure in blue (PDB accession number 1X9T) into the 12-Å CAV-2 cryo-EM envelope. The red and orange parts correspond to the regions indicated in panel A. The star indicates a region where the X-ray structure did not fill the EM density. (C) Sequence alignment of the hAdV-5 and CAV-2 penton bases visualized with ESPript (23). The RGD and hypervariable loops are indicated by bars above the sequence in the same colors as used in panels A and B. The RGD sequence is boxed with a black rectangle. Conserved residues are in white text and highlighted in red; partially conserved residues are in red text. The star highlights three extra amino acids present in CAV-2 compared to hAdV-5 that could fill up the density indicated by the star in panel B. The amino acids involved in fiber binding for hAdV-5 are indicated by blue bars above the sequence.
The CAV-2 penton base isosurface representation is smoother than that of hAdV-2/5 (Fig. 2A). By comparing the EM structures (Fig. 2A), we identified two protuberances that are smaller in the CAV-2 base (Fig. 2A, right panel, red and orange areas). The atomic structure of the hAdV-2 penton base (99% identical to that of hAdV-5) was fitted into the CAV-2 penton base EM density in order to identify the residues that are responsible for the smooth appearance of the CAV-2 capsid (Fig. 2B). The first difference is located in the hypervariable loop (Fig. 2A, red area, and C, sequence indicated by a red bar), which is 3 amino acids shorter in the CAV-2 base. On average, the amino acids composing this loop are also smaller in size (PATADNFG) (Fig. 2C). The second difference (Fig. 2, orange area) lies on the protuberances located on the top of the base that contains the RGD sequence in most hAdV (residues 289 to 379 for hAdV-5) (Fig. 2C). Due to its flexibility, the entire RGD loop structure is not visible either in the atomic structure of the hAdV-2 penton base or in the hAdV-5 cryo-EM structure. This protuberance is also present in the CAV-2 base but is much less pronounced because it is only 12 amino acids long. The CAV-2 loop does not contain an RGD sequence. Despite the small size of the protuberance, it is clearly visible in the cryo-EM map. This reinforces the idea that the N-terminal part of the fiber interacts with the base of the RGD loop (20) and contributes to the size of the protuberance.
Next to the orange loop in the CAV-2 penton base there are three extra amino acids after the RGD loop, indicated by a star in the fit shown in Fig. 2B and in the sequence in panel C. This corresponds to an empty region in terms of both the fit and sequence (Fig. 2B and C, stars). This could easily explain the smoother pentagonal shape of the CAV-2 base (slightly larger on the middle of the edge and smaller on the pentagon vertex).
According to our sequence alignment (Fig. 2C), the N-terminal part of the CAV-2 penton base is also 15 amino acids shorter than that of hAdV-5. The currently unresolved N-terminal part of the penton base (amino acids 1 to 49) contains two highly conserved PPXY (where X is any residue) motifs implicated in interactions with host cell WW domain-containing ubiquitin ligases (21, 22). In the mature virion, this region is probably located under the penton base, where it interacts with other viral capsid proteins (20, 45). The PPXY sequences are also present in CAV-2, suggesting that these sequences are critical for some phase in the AdV life cycle (22). The 15-amino-acid difference is too small to be interpreted in the EM map because it is in contact with other proteins on the inside of the virus capsid.
On the CAV-2 penton base, 9 out of the 10 residues involved in fiber tail-penton base interaction are conserved compared to hAdV-5 (Fig. 2C, indicated by blue bars). The only difference is that histidine 494 of hAdV-5 is a glutamine in CAV-2. The fiber sequence that interacts with the base is also conserved: amino acids 13 to 19 follow the conserved sequence seen in hAdV (58). The binding of the fiber to the penton base should therefore be very similar to that described for hAdV-2 (58).
The CAV-2 fiber contains two bends.
The CAV-2 fiber is 542 amino acids long: the first 41 amino acids make up the tail, the central 320 amino acids comprise the shaft, and the last 182 amino acids form the knob (Fig. 3C). In hAdV-5 each of the ∼15-amino-acid repeats is 13 Å long (42) and induces a rotation of ∼50° along the fiber axis (54). Based on sequence analysis, Rasmussen et al. (41) predicted that the CAV-2 shaft had 18 repeats with 18 amino acids per repeat. Our sequence analysis suggests that there are 18.5 repeats (Fig. 3C). The number of amino acids/repeat varies: seven repeats have the quintessential 15-amino-acid motif, the two repeats with 16 amino acids have the “extra” amino acid on the outside of the repeat, the three repeats with 17 amino acids have the extra amino acids inside the repeat, one repeat has 18 amino acids (extra internal), and two repeats have 19 amino acids (extra external). Two of the three longest repeats (21 to 22 amino acids) contain six or seven extra internal amino acids. All hAdV fiber sequences contain an irregular, long repeat near the N-terminal attachment site on the penton base, which has been linked to a possible bend at this position (42). However, this irregular repeat tends to be the third repeat and not the fourth, as is the case for the CAV-2 fiber (9). The much longer fibers of BAV-3 and the avian chicken embryo lethal orphan virus have additional bends, also related to irregular repeats (25, 43). Wu et al. (57) showed that such a 6- to 7-amino-acid internal insert can introduce a bend in the hAdV-5 shaft that may allow greater access of the penton base's integrin-interacting motifs to the cell surface (56).
As mentioned previously, the CAV-2 fiber was too flexible to be reconstructed by cryo-EM on the entire virion. To complete our structural analysis, we imaged recombinant CAV-2 fibers by negative staining (Fig. 3A). After image averaging (the fiber head and the apical part of the different fibers are superposed but their N-terminal parts show the different positions they can adopt), we also found at least two bends in the shaft (Fig. 3B). The location of the hinges correlates well with the 21- and 22-amino acid-long repeats with six to seven internal amino acids (Fig. 3B, repeats 4 and 10).
To test the agreement of the EM data with the (i) suggested repeat alignment, (ii) dimensions of the predicted atomic structure, and (iii) locations and properties of the kink regions, we modeled the 3D structure of the fiber (Fig. 3D). Recently, the crystal structure of the homotrimeric C-terminal CAV-2 fiber head (residues 361 to 542) was determined (47). We also took advantage of two other known crystal structures of homologous proteins corresponding to different parts of the fiber to model its overall structure. The first structure that we used as a template was the C-terminal part of the hAdV-2 fiber containing four repeats of the shaft domain followed by the receptor-binding head domain (54). This structure allowed us to model the last four repetitive elements of the CAV-2 fiber shaft and the region where this shaft is linked to the head domains. Most of the CAV-2 repeats are similar to those in hAdV-2, and, therefore, the crystal structure of the hAdV-2 repeats was used to model the remaining part of the CAV-2 fiber. The exceptions were repeats 4 and 10 (Fig. 3C) having 7- and 6-residue insertions between two conserved β-strand motifs. These repeats were modeled by using the crystal structure of the reovirus attachment protein σ1 (7). In the structure of σ1, one repeat contains a similar 5-residue insertion. This insertion interrupts the progression of the regular β-spiral and introduces a bend into the σ1 trimer. The structure of the insertion is loose, with slightly different main chain conformations in each monomer extended along the fiber axis. This structure was used to model the two longer repeats of CAV-2.
If our model is correct, the CAV-2 trimeric fiber should be about 330 Å long. The β-spiral is 280 Å long with two flexible bends and a C-terminal globular head domain contributing 50 Å (Fig. 3D). This model, with respect to the number and location of the bends and the length, is in a good agreement with our EM data.
The CAV-2 hexon.
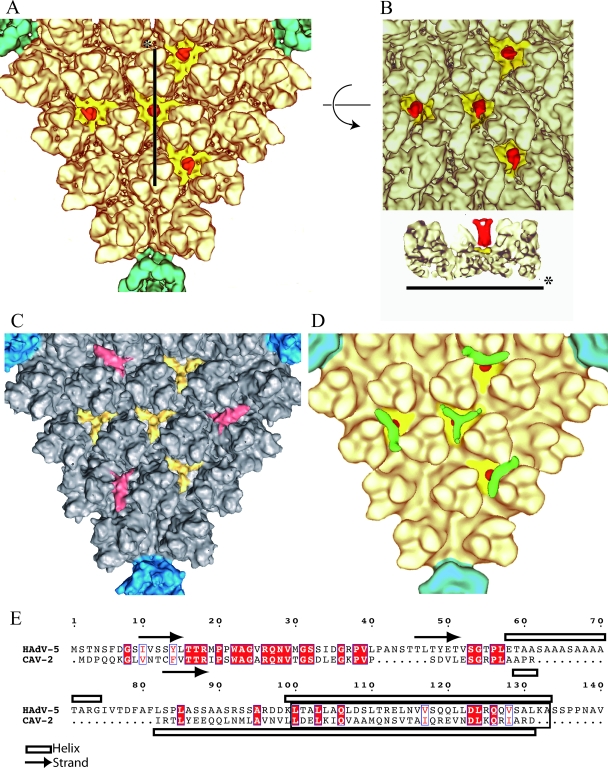
The general dimensions and twist of the homotrimeric CAV-2 hexon are very similar to those of hAdV-5 (Fig. 4A). Globally, the CAV-2 hexon sequence is shorter than that of hAdV-5 (Fig. 4C) (908 versus 952 amino acids) with all the differences located in the N-terminal half. The fit into the EM density of the CAV-2 hexon was carried out using the hAdV-5 X-ray structure (Protein Data Bank [PDB] 1P30) (Fig. 4B). Because the loops that are absent in the hAdV-5 X-ray structure (Fig. 4C, blue bars above the sequence) often correspond to a deletion in the CAV-2 sequence, the fit was good, and only one loop stays outside the density (Fig. 4B, left). This loop (amino acids 194 to 202 in hAdV-5 versus 153 to 161 in CAV-2) was visible in the atomic structure of the hAdV-5 hexon but not in that of the hAdV-2 hexon. The sequence just before this loop is absent in the CAV-2 hexon (Fig. 4B and C, region 2), which could explain the absence of density in the EM structure. The structural rearrangement caused by this difference could fill the empty part in the CAV-2 EM hexon structure (Fig. 4B, asterisk). Further, there is a 24-amino-acid gap after amino acid 144 (Fig. 4B and C, region 1). This region is located on the top of the hAdV-5 hexon and was not ordered in the X-ray structure. This may explain the smoother shape of the CAV-2 hexon compared to the EM density of the hAdV-5 hexon (Fig. 4A, right). The last structural difference, also located on the top of the hexon, is likely caused by the significant sequence divergence around amino acid 310 (Fig. 4B and C, region 3 in the hAdV-5 sequence). Interestingly, the protruding structures in the hAdV hexons, which contain most of the epitopes recognized by anti-hexon antibodies (44), are absent in CAV-2.
FIG. 4.
The CAV-2 hexon capsomer. (A) Comparison of computer-isolated hexons from CAV-2 (yellow) and hAdV-5 (gray) imaged in isodensity surface view. (B) Fitting of hAdV-5 hexon X-ray structure (PDB 1P30) into the CAV-2 cryo-EM envelope. Either one hexon alone (left) or the four hexons surrounding a twofold axis (middle) have been fitted using SITUS and imaged at the same time in the EM density. On the right, only the bottom wall of the pseudo-atomic model of the four hexons is shown. In the hexon alone, three regions are indicated on the X-ray structure; they correspond to large differences in the sequence of CAV-2 compared to that of hAdV-5 (see below). The red loops and regions 1 to 3 in the single hexon on the left are discussed in the text and correspond to sequence elements 1 to 3 indicated in panel C. The star indicates a region in the EM density that is not filled by the density derived from the crystal structure, as discussed in the text. (C) Sequence alignment between the CAV-2 and hAdV-5 hexon bases. Conserved residues are in white text and highlighted in red; partially conserved residues are in red text. The loops not resolved in the X-ray structure are indicated by blue bars above the sequences. The amino acid sequences involved in the contacts between hexons as described by Fabry et al. (15) are underlined with a bold line. They are conserved between CAV-2 and hAdV-5. Regions 1, 2, and 3 from panel B are indicated.
As mentioned above, we have been unable to complex CAV-2 vectors with divalent cations or lipoplexes, suggesting a more neutral charge of the external CAV-2 capsid. Because the capsid charge is dominated by the hexon charges, the data presented here support this prediction. For example, region 1 in Fig. 4C is composed mainly of negatively charged residues in hAdV-5, and, more generally, all the hAdV-5 loops that are missing in the CAV-2 capsid are rich in charged residues.
We also performed a multiple fitting on the capsid using four hexons surrounding the twofold axis of the virus. The regions involved in the interhexon contacts are the same as those previously determined for hAdV-5 (15) (Fig. 4C, bold lines). Furthermore, in most cases, the amino acids are conserved between the two species. The four hexons present around the twofold axis in CAV-2 interact using the same five loops that are used in hAdV-5: loops 65 to 70, 292 to 296, 614 to 619, 674 to 676, and 683 to 690 in CAV-2 corresponding to loops 65 to 70, 340 to 344, 665 to 670, 725 to 727, and 734 to 741 in hAdV-5 (Fig. 4B and C, bold lines). Only the interaction region 2 (Fig. 4B) is absent in CAV-2.
Minor proteins. (i) Protein IX.
A major difference in the 3D structure of hAdV and CAV-2 is located on the top of the triskelion structure attributed to the N-terminal domain of protein IX, where a rod-like density is present in CAV-2. This density is absent in the hAdV-5 reconstruction (Fig. 1B, arrow, and C and 5C). The sequence of protein IX is 25% shorter in CAV-2 than in hAdV-5 (105 versus 140 amino acids) (Fig. 5E). By comparing the 3D reconstructions of CAV-2 and hAdV, we should be able to locate protein IX within the capsid.
In 2005, Fabry et al. (15) published the 3D structure of the wild-type capsid of hAdV-5 as well as the 3D structure of a mutant virus with a deletion of protein IX. In the mutant structure the yellow and red densities shown in Fig. 5B were not present. At the time of that study the position represented by the red density was thought to be that of protein IIIa; the authors thus assumed that the lack of the red densities in the protein IX-deleted virus was due to a diminished stability of the capsid, i.e., the absence of protein IX could have resulted in the loss of protein IIIa (15). Recently, Saban et al. (45), using a 6-Å resolution map in which the α-helices in the capsid were visible plus secondary structure predictions, reassigned the red density to a four-helical bundle built from the C-terminal parts of four copies of protein IX. The triskelion structure was considered to be made up from the N-terminal parts of three copies of protein IX. The density for protein IIIa was reassigned to a highly helical density present underneath the penton base (45). The reassignment of the position of the C-terminal domain of protein IX was confirmed by Marsh et al., who showed that a protein IX mutant virus with GFP added at the C terminus had an additional density above the reassigned position for the C terminus (33), i.e., the position of the red domains shown in Fig. 5C.
In the CAV-2 capsid (Fig. 5A and B), the triskelion N-terminal part of protein IX is present at the same position in the capsid as in hAdV-5 (Fig. 5C). The notable difference between the two reconstructions is that in CAV-2 a cylindrical density is present on the top of the triskelion, whereas the red density observed in the hAdV-5 capsid (Fig. 5C) was absent. The red density on top of the triskelion in CAV-2 (Fig. 5A and B) has more or less the same dimensions as the red rods in hAdV-5. It is therefore likely that the cylindrical density on top of the triskelion in the CAV-2 structure is also the C-terminal domain of protein IX. This interpretation is reinforced by comparing the sequences of the two proteins. Protein IX is a two-domain protein consisting of an N-terminal part that contains some β-strands and a C-terminal, α-helical part, both parts being connected by a linker, as shown by sequence alignment and secondary structure prediction (Fig. 5E). Saban et al. (45) did not detect any helices in the triskelion structure, confirming the assignment to the N-terminal domain of protein IX. This part of protein IX interacts with the hexon protein and should therefore be conserved. The N termini of the CAV-2 and the hAdV-5 sequences are the most similar between the two proteins (40% identity for the 52 first amino acids). However, the linker sequence (amino acids 40 to 81 in hAdV-5) is almost absent in CAV-2 (amino acids 39 to 53), and therefore the two domains must be close together in CAV-2. In contrast, in hAdV-5 the C and N termini could be quite far apart from each other, as suggested by Saban et al. (45). Although the CAV-2 C-terminal part of protein IX is slightly smaller and different from that of hAdV-5, this region is still predicted to be helical and organized as a coil-coiled domain (Fig. 5E). Therefore, we suggest that three C-terminal domains of protein IX make up the rod on top of the triskelion structure in the CAV-2 capsid. To test this hypothesis we performed cryo-EM and calculated a 3D reconstruction of a CAV-2 virion harboring a C-terminal pIX-GFP fusion protein (31). While the quality of the reconstruction (25-Å resolution) is lower than that of the native CAV-2 capsid, it is good enough to recognize the triskelion, the cylindrical red density, and, at the top of that, a small extra density (Fig. 5D, green areas). This latter density is smaller than the size of GFP, probably because it is flexible. The flexible linker between the C-terminal part of protein IX and GFP (30, 31) would allow movement, and therefore only the average position is visible in the 3D reconstruction. We believe that our combined data comparing the reconstructions of CAV-2 and the capsid harboring the protein IX-GFP fusion confirm the localization of the C-terminal domain of CAV-2 protein IX. Our results also confirm the assignment of Saban et al. (45) of the red rods in the hAdV structure shown in Fig. 5C as the C-terminal domain of human virus protein IX.
(ii) The interior of the capsid. (a) Protein IIIa.
As mentioned above, based on a 6-Å EM reconstruction and secondary structure predictions (45), the hAdV protein IIIa was reassigned to a helix-rich density lying under the penton base. This density is also present on the inside of the CAV-2 capsid (not shown). It exhibits the same topology as the human protein IIIa: there is an arm bound to the bottom part of the penton base, and, more toward the inside of the capsid, a globular region which resembles an empty sphere is visible and of same size as in hAdV-5. The CAV-2 protein IIIa is slightly smaller than the hAdV-5 protein (567 amino acids versus 585), but except for the 15 first amino acids, the N-terminal part predicted to be α-helical is very conserved. The C-terminal part is notably different and smaller in CAV-2. However, at the present resolution we are unable to see such differences.
(b) Protein VIII.
A total of 120 copies of protein VIII have been counted on the inner part of the hAdV-5 capsid (15). The same number of proteins can be counted in CAV-2, and each interacts with four hexons (not shown). The density corresponding to protein VIII seems to be slightly smaller and less continuous in CAV-2 than in hAdV-5. The sequences of these proteins are very similar (more than 66% similarity or identity), and therefore the differences observed in the density are probably due to a lack of resolution.
Finally, there is also an extra density just under the CAV-2 penton base (Fig. 1B, circle) that is absent in the hAdV-5 structure, but at this stage it remains unassigned.
Conclusion.
This work shows the first 3D structure of a nonhuman AdV. There are well over 100 AdV serotypes currently identified, with less than half of them human. We predict that this number of nonhuman AdVs is only the tip of the iceberg. We are just beginning to understand and uncover the potential of many of these viruses, in particular, of the nonhuman serotypes. Our study highlights the structural differences and similarities between hAdV-2/5 and CAV-2 and definitively localizes the N- and C-terminal parts of protein IX in this CAV-2 capsid. The molecular organization of the AdV capsid as it is known today is shown schematically in Fig. 6. The CAV-2 penton base and hexon have fewer and/or smaller loops around the conserved bodies of these capsid proteins. In the hAdV-2/5 penton base these loops are involved in internalization via the integrin-interacting domain (34), possibly binding coagulation factors (2), and are targeted by the humoral immune response following infection (37). The biological significance of many of the structural differences is likely to be complex. Yet we can now begin to rationally understand and resolve some of the CAV-2 enigmas. The capsid structure will allow us to rescan the external CAV-2 capsid for integrin-interacting motifs that could possibly induce internalization. Because of its C-terminal domain extending outside of the capsid wall, the CAV-2 protein IX may have an additional role during capsid disassembly at the nuclear pore by binding the anterograde motor kinesin (U. Greber, unpublished data). The smoother shape of the CAV-2 hexons and penton bases due to the lack of highly antigenic loops present in human serotypes explains the lack of cross-reacting human neutralizing antibodies (37).
FIG. 6.
Schematic view of one facet of the CAV-2 capsid seen from the outside of the capsid. The figure shows a summary of all structural data on the protein localization in the AdV capsid. The hexons highlighted in black belong to the group of nine whereas the violet ones are the peripentonal hexons (26). The different proteins occupy the same positions in the capsid (hAdV and the CAV-2) except for the C-terminal part of protein IX that can either be on the top of the N terminus of protein IX in the CAV-2 capsid or be near the twofold axis in the hAdV capsid. For the hAdV protein IX, a thin black line indicates the position of the linker between the N- and the C-terminal parts of protein IX as suggested by Saban et al. (45).
Biologically active loops can be added onto the CAV-2 penton base and hexon at the sites where such loops are present in the human viruses. In particular, it will be interesting to add an RGD loop to the CAV-2 penton base at the same site where this is found in the human virus. Apart from adding loops to pentons and hexons, the specific position of the C-terminal domain of CAV-2 protein IX is an ideal site to position additional protein domains for specific interactions with host cells. Finally, we may also be able to better understand and assay CAV-2 tropism by manipulating the rigidity of the flexible shaft of the fiber.
Acknowledgments
We thank Pascal Fender and Harry Wodrich for discussions and Emmanuelle Neumann for help with the electron microscope. We thank Vasiliki Kalatzis for critical reading of the manuscript. We thank the other members of our laboratories for constructive comments, suggestions, and help during the course of this study. We thank the IFR 122 recombinant protein platform for the production of the CAV-2 fiber.
E.J.K. is an INSERM fellow. G.S. was supported in part by a Jeunes Chercheurs grant from the Agence Nationale pour la Recherche. D.T.C. was funded by NIH award 5R01CA111569. This work was supported, in part, by the Association Française contre les Myopathies (AFM), Vaincre les Maladies Lysosomales, and the Fondation pour le Recherche Médicales. O.B. was an AFM postdoctoral researcher, M.E.B. is a Ph.D. student financed by the Rhone Alpes region, and C.M.S.F. is a Ph.D. student financed through a MENRT fellowship from the French government.
The authors have no conflicting financial interests.
Footnotes
Published ahead of print on 23 January 2008.
REFERENCES
- 1.Akli, S., C. Caillaud, E. Vigne, L. D. Stratford-Perricaudet, L. Poenaru, M. Perricaudet, A. Kahn, and M. R. Peschanski. 1993. Transfer of a foreign gene into the brain using adenovirus vectors. Nat. Genet. 3224-228. [DOI] [PubMed] [Google Scholar]
- 2.Baker, A. H., J. H. McVey, S. N. Waddington, N. C. Di Paolo, and D. M. Shayakhmetov. 2007. The influence of blood on in vivo adenovirus bio-distribution and transduction. Mol. Ther. 151410-1416. [DOI] [PubMed] [Google Scholar]
- 3.Baker, T. S., and R. H. Cheng. 1996. A model-based approach for determining orientations of biological macromolecules imaged by cryoelectron microscopy. J. Struct. Biol. 116120-130. [DOI] [PubMed] [Google Scholar]
- 4.Bergelson, J. M., J. A. Cunningham, G. Droguett, E. A. Kurt-Jones, A. Krithivas, J. S. Hong, M. S. Horwitz, R. L. Crowell, and R. W. Finberg. 1997. Isolation of a common receptor for coxsackie B viruses and adenoviruses 2 and 5. Science 2751320-1323. [DOI] [PubMed] [Google Scholar]
- 5.Both, G. W. 2002. Xenogenic adenoviruses, p. 447-479. In D. Curiel and J. Douglas (ed.), Adenoviral vectors for gene therapy. Academic Press, San Diego, CA.
- 6.Campos, S. K., and M. A. Barry. 2007. Current advances and future challenges in adenoviral vector biology and targeting. Curr. Gene. Ther. 7189-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chappell, J. D., A. E. Prota, T. S. Dermody, and T. Stehle. 2002. Crystal structure of reovirus attachment protein signa1 reveals evolutionary relationship to adenovirus fibre. EMBO J. 211-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chillon, M., and E. Kremer. 2001. Trafficking and propagation of canine adenovirus vectors lacking a known integrin-interacting motif. Hum. Gene Ther. 121815-1823. [DOI] [PubMed] [Google Scholar]
- 9.Chroboczek, J., R. W. Ruigrok, and S. Cusack. 1995. Adenovirus fiber. Curr. Top. Microbiol. Immunol. 199163-200. [DOI] [PubMed] [Google Scholar]
- 10.Conway, J. F., and A. C. Steven. 1999. Methods for reconstructing density maps of “single” particles from cryoelectron micrographs to subnanometer resolution. J. Struct. Biol. 128106-118. [DOI] [PubMed] [Google Scholar]
- 11.D'Ambrosio, E., N. Del Grosso, A. Chicca, and M. Midulla. 1982. Neutralizing antibodies against 33 human adenoviruses in normal children in Rome. J. Hyg. 89155-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davison, A. J., M. Benko, and B. Harrach. 2003. Genetic content and evolution of adenoviruses. J. Gen. Virol. 842895-2908. [DOI] [PubMed] [Google Scholar]
- 13.Dayringer, H. E., A. Tramontano, S. R. Sprang, and R. J. Fletterick. 1986. Interactive program for visualization and modeling of protein, nucleic acids and small molecules. J. Mol. Graph. 482-87. [Google Scholar]
- 14.DeLano, M. C., and Y. Cao. 2002. High b-value diffusion imaging. Neuroimaging Clin. N. Am. 1221-34. [DOI] [PubMed] [Google Scholar]
- 15.Fabry, C. M., M. Rosa-Calatrava, J. F. Conway, C. Zubieta, S. Cusack, R. W. Ruigrok, and G. Schoehn. 2005. A quasi-atomic model of human adenovirus type 5 capsid. EMBO J. 241645-1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fasbender, A., J. H. Lee, R. W. Walters, T. O. Moninger, J. Zabner, and M. J. Welsh. 1998. Incorporation of adenovirus in calcium phosphate precipitates enhances gene transfer to airway epithelia in vitro and in vivo. J. Clin. Investig. 102184-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fasbender, A., J. Zabner, M. Chillon, T. O. Moninger, A. P. Puga, B. L. Davidson, and M. J. Welsh. 1997. Complexes of adenovirus with polycationic polymers and cationic lipids increase the efficiency of gene transfer in vitro and in vivo. J. Biol. Chem. 2726479-6489. [DOI] [PubMed] [Google Scholar]
- 18.Flomenberg, P., V. Piaskowski, R. L. Truitt, and J. T. Casper. 1995. Characterization of human proliferative T cell responses to adenovirus. J. Infect. Dis. 1711090-1096. [DOI] [PubMed] [Google Scholar]
- 19.Frank, J., M. Radermacher, P. Penczek, J. Zhu, Y. Li, M. Ladjadj, and A. Leith. 1996. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol. 116190-199. [DOI] [PubMed] [Google Scholar]
- 20.Fuschiotti, P., G. Schoehn, P. Fender, C. M. Fabry, E. A. Hewat, J. Chroboczek, R. W. Ruigrok, and J. F. Conway. 2006. Structure of the dodecahedral penton particle from human adenovirus type 3. J. Mol. Biol. 356510-520. [DOI] [PubMed] [Google Scholar]
- 21.Galinier, R., E. Gout, H. Lortat-Jacob, J. Wood, and J. Chroboczek. 2002. Adenovirus protein involved in virus internalization recruits ubiquitin-protein ligases. Biochemistry 4114299-14305. [DOI] [PubMed] [Google Scholar]
- 22.Garcel, A., E. Gout, J. Timmins, J. Chroboczek, and P. Fender. 2006. Protein transduction into human cells by adenovirus dodecahedron using WW domains as universal adaptors. J. Gene. Med. 8524-531. [DOI] [PubMed] [Google Scholar]
- 23.Gouet, P., X. Robert, and E. Courcelle. 2003. ESPript/ENDscript: Extracting and rendering sequence and 3D information from atomic structures of proteins. Nucleic Acids Res. 313320-3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Greber, U. F. 2002. Signalling in viral entry. Cell Mol. Life Sci. 59608-626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hess, M., A. Cuzange, R. W. Ruigrok, J. Chroboczek, and B. Jacrot. 1995. The avian adenovirus penton: two fibres and one base. J. Mol. Biol. 252379-385. [DOI] [PubMed] [Google Scholar]
- 26.Horwitz, M. 2001. Adenoviruses, p. 2149-2171. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 27.Keriel, A., C. Rene, C. Galer, J. Zabner, and E. J. Kremer. 2006. Canine adenovirus vectors for lung-directed gene transfer: efficacy, immune response, and duration of transgene expression using helper-dependent vectors. J. Virol. 801487-1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kojaoghlanian, T., P. Flomenberg, and M. S. Horwitz. 2003. The impact of adenovirus infection on the immunocompromised host. Rev. Med. Virol. 13155-171. [DOI] [PubMed] [Google Scholar]
- 29.Kremer, E. J. 2004. CAR chasing: canine adenovirus vectors-all bite and no bark? J. Gene. Med. 6(Suppl. 1)S139—S151. [DOI] [PubMed] [Google Scholar]
- 30.Kremer, E. J., S. Boutin, M. Chillon, and O. Danos. 2000. Canine adenovirus vectors: an alternative for adenovirus-mediated gene transfer. J. Virol. 74505-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Le, L. P., J. Li, V. V. Ternovoi, G. P. Siegal, and D. T. Curiel. 2005. Fluorescently tagged canine adenovirus via modification with protein IX-enhanced green fluorescent protein. J. Gen. Virol. 863201-3208. [DOI] [PubMed] [Google Scholar]
- 32.Le Gal La Salle, G., J. J. Robert, S. Berrard, V. Ridoux, L. D. Stratford-Perricaudet, M. Perricaudet, and J. Mallet. 1993. An adenovirus vector for gene transfer into neurons and glia in the brain. Science 259988-990. [DOI] [PubMed] [Google Scholar]
- 33.Marsh, M. P., S. K. Campos, M. L. Baker, C. Y. Chen, W. Chiu, and M. A. Barry. 2006. Cryoelectron microscopy of protein IX-modified adenoviruses suggests a new position for the C terminus of protein IX. J. Virol. 8011881-11886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Medina-Kauwe, L. K. 2003. Endocytosis of adenovirus and adenovirus capsid proteins. Adv. Drug Deliv. Rev. 551485-1496. [DOI] [PubMed] [Google Scholar]
- 35.Olive, M., L. Eisenlohr, N. Flomenberg, S. Hsu, and P. Flomenberg. 2002. The adenovirus capsid protein hexon contains a highly conserved human CD4+ T-cell epitope. Hum. Gene. Ther. 131167-1178. [DOI] [PubMed] [Google Scholar]
- 36.Paillard, F. 1997. Advantages of nonhuman adenoviruses versus human adenoviruses. Hum. Gene. Ther. 82007-2009. [DOI] [PubMed] [Google Scholar]
- 37.Perreau, M., M. C. Guerin, C. Drouet, and E. J. Kremer. 2007. Interactions between human plasma components and a xenogenic adenovirus vector: reduced immunogenicity during gene transfer. Mol. Ther. 151998-2007. [DOI] [PubMed] [Google Scholar]
- 38.Perreau, M., and E. J. Kremer. 2005. Frequency, proliferation, and activation of human memory T cells induced by a nonhuman adenovirus. J. Virol. 7914595-14605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Perreau, M., and E. J. Kremer. 2006. The conundrum between immunological memory to adenovirus and their use as vectors in clinical gene therapy. Mol. Biotechnol. 34247-256. [DOI] [PubMed] [Google Scholar]
- 40.Perreau, M., F. Mennechet, N. Serratrice, J. N. Glasgow, D. T. Curiel, H. Wodrich, and E. J. Kremer. 2007. Contrasting effects of human, canine, and hybrid adenovirus vectors on the phenotypical and functional maturation of human dendritic cells: implications for clinical efficacy. J. Virol. 813272-3284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rasmussen, U. B., Y. Schlesinger, A. Pavirani, and M. Mehtali. 1995. Sequence analysis of the canine adenovirus 2 fiber-encoding gene. Gene 159279-280. [DOI] [PubMed] [Google Scholar]
- 42.Ruigrok, R. W., A. Barge, C. Albiges-Rizo, and S. Dayan. 1990. Structure of adenovirus fibre. II. Morphology of single fibres. J. Mol. Biol. 215589-596. [DOI] [PubMed] [Google Scholar]
- 43.Ruigrok, R. W. H., A. Barge, S. K. Mittal, and B. Jacrot. 1994. The fibre of bovine adenovirus is very long, but bent. J. Gen. Virol. 752069-2073. [DOI] [PubMed] [Google Scholar]
- 44.Rux, J. J., P. R. Kuser, and R. M. Burnett. 2003. Structural and phylogenetic analysis of adenovirus hexons by use of high-resolution X-ray crystallographic, molecular modeling, and sequence-based methods. J. Virol. 779553-9566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saban, S. D., M. Silvestry, G. R. Nemerow, and P. L. Stewart. 2006. Visualization of α-helices in a 6-angstrom resolution cryoelectron microscopy structure of adenovirus allows refinement of capsid protein assignments. J. Virol. 8012049-12059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schoehn, G., F. M. Vellieux, M. Asuncion Dura, V. Receveur-Brechot, C. M. Fabry, R. W. Ruigrok, C. Ebel, A. Roussel, and B. Franzetti. 2006. An archaeal peptidase assembles into two different quaternary structures: a tetrahedron and a giant octahedron. J. Biol. Chem. 28136327-36337. [DOI] [PubMed] [Google Scholar]
- 47.Seiradake, E., H. Lortat-Jacob, O. Billet, E. J. Kremer, and S. Cusack. 2006. Structural and mutational analysis of human Ad37 and canine adenovirus 2 fiber heads in complex with the D1 domain of coxsackie and adenovirus receptor. J. Biol. Chem. 28133704-33716. [DOI] [PubMed] [Google Scholar]
- 48.Soudais, C., S. Boutin, S. S. Hong, M. Chillon, O. Danos, J. M. Bergelson, P. Boulanger, and E. J. Kremer. 2000. Canine adenovirus type 2 attachment and internalization: coxsackievirus-adenovirus receptor, alternative receptors, and an RGD-independent pathway. J. Virol. 7410639-10649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Soudais, C., C. Laplace-Builhe, K. Kissa, and E. J. Kremer. 2001. Preferential transduction of neurons by canine adenovirus vectors and their efficient retrograde transport in vivo. FASEB J. 152283-2285. [DOI] [PubMed] [Google Scholar]
- 50.Soudais, C., N. Skander, and E. J. Kremer. 2004. Long-term in vivo transduction of neurons throughout the rat central nervous system using novel helper-dependent CAV-2 vectors. FASEB J. 18391-393. [DOI] [PubMed] [Google Scholar]
- 51.Stewart, P. L., R. M. Burnett, M. Cyrklaff, and S. D. Fuller. 1991. Image reconstruction reveals the complex molecular organization of adenovirus. Cell 67145-154. [DOI] [PubMed] [Google Scholar]
- 52.Tomko, R., R. Xu, and L. Philipson. 1997. HCAR and MCAR: the human and mouse cellular receptors for subgroup C adenoviruses and group B cocksakieviruses. Proc. Natl. Acad. Sci. USA 943352-3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Van Heel, M. 1987. Angular reconstitution: a posteriori assignment of projection directions for 3D reconstruction. Ultramicroscopy 21111-123. [DOI] [PubMed] [Google Scholar]
- 54.van Raaij, M. J., A. Mitraki, G. Lavigne, and S. Cusack. 1999. A triple beta-spiral in the adenovirus fibre shaft reveals a new structural motif for a fibrous protein. Nature 401935-938. [DOI] [PubMed] [Google Scholar]
- 55.Wriggers, W., and S. Birmanns. 2001. Using situs for flexible and rigid-body fitting of multiresolution single-molecule data. J. Struct. Biol. 133193-202. [DOI] [PubMed] [Google Scholar]
- 56.Wu, E., and G. R. Nemerow. 2004. Virus yoga: the role of flexibility in virus host cell recognition. Trends Microbiol. 12162-169. [DOI] [PubMed] [Google Scholar]
- 57.Wu, E., L. Pache, D. J. Von Seggern, T. M. Mullen, Y. Mikyas, P. L. Stewart, and G. R. Nemerow. 2003. Flexibility of the adenovirus fiber is required for efficient receptor interaction. J. Virol. 777225-7235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zubieta, C., G. Schoehn, J. Chroboczek, and S. Cusack. 2005. The structure of the human adenovirus 2 penton. Mol. Cell 17121-135. [DOI] [PubMed] [Google Scholar]