Abstract
In this report we propose a model in which after the herpes simplex virus (HSV) capsid docks at the nuclear pore, the tegument protein attached to the capsid must be cleaved by a serine or a cysteine protease in order for the DNA to be released into the nucleus. In support of the model are the following results. (i) Exposure of cells at the time of or before infection to l-(tosylamido-2-phenyl) ethyl chloromethyl ketone (TPCK), a serine-cysteine protease inhibitor, prevents the release of viral DNA or expression of viral genes. TPCK does not block viral gene expression after entry of viral DNA into the nucleus. (ii) The tegument protein VP1-2, the product of the UL36 gene, is cleaved shortly after the entry of the HSV 1 (HSV-1) virion into the cell. (iii) The proteolytic cleavage of VP1-2 does not occur in cells that are infected with HSV-1 under conditions that prevent the release of the viral DNA into the nucleus. (iv) The proteolytic cleavage of VP1-2 occurs only after the capsid is attached to the nuclear pore. Thus, TPCK prevented the release of HSV-1 DNA into the nucleus when added to medium 1 hour after infection with tsB7 at 39.5°C followed by a shift down to the permissive temperature. The ts lesion maps in the UL36 gene. At the nonpermissive temperature, the capsids accumulate at the nuclear pore but the DNA is not released into the nucleus.
In this report we present supporting evidence for a model for the release of herpes simplex virus 1 (HSV-1) DNA from the capsid into the nucleus of an infected cell. Briefly, following the attachment of the capsid to the nuclear pore, VP1-2, the largest tegument protein, undergoes a proteolytic cleavage that cleaves a 55-kDa N-terminal fragment. This proteolytic cleavage is essential for the subsequent conformational changes in the capsid that allow the release of the HSV-1 DNA into the nucleus. Relevant to the presentation of the data supporting the model are the following.
HSV-1 initiates infection by attachment to the cell membrane via the interaction between viral glycoproteins gB and gC with heparan sulfate (39, 43). The initial attachment is followed by the fusion of the viral envelope with the plasma membrane, a process initiated by the interaction of gD with a specific receptor and mediated by gB, gH, and gL (11, 24, 35). The capsid with associated tegument proteins is released into the cytosol and transported via dynein and microtubule-directed transport toward the nucleus (40). During the transport some tegument proteins, most notably VP16 (2) and virion host shutoff protein (23), the products of UL48 and UL41, respectively, are released, whereas some, such as VP1-2, the product of the UL36 gene, remain associated with the capsid (16, 44). Once the capsid arrives at the nuclear pore, viral DNA is rapidly released into the nucleus to enable the expression of its genes. Information regarding the process of attachment to the nuclear pore and the release of viral DNA into the nucleus is scarce. Attachment of capsid to the nuclear pores and translocation of viral DNA into the nucleus require importin-β but not importin-α. Ran GTPase is also essential for this process (33). HSV-1 tegument protein VP1-2 has been suggested to play a role in this process, based on studies of the temperature-sensitive mutant tsB7, which carries a temperature-sensitive mutation in the UL36 gene (1). When cells are infected with the tsB7 mutant at a nonpermissive temperature, capsids are accumulated at the nuclear pores but DNA fails to be released. DNA is released from tsB7 capsid only upon a shift to the permissive temperature (1).
The UL36 gene is conserved among alpha-, beta-, and gammaherpesvirus subfamilies (36). VP1-2 plays an essential role in the formation of mature virions, as the deletion of the UL36 gene leads to the accumulation of DNA-filled, unenveloped capsids in the cytoplasm of infected cells (6). VP1-2 is tightly associated with capsid (15, 41), and based on cryoelectron microscopy images it appears that it is localized at the penton structures (44). VP1-2 interacts with a 140-kDa viral protein that binds the a sequence of HSV-1 DNA (5) and is suggested to interact with VP5 (44).
VP1-2 is the largest protein encoded by the UL36 gene. It is expressed as a γ (late) gene (28), and based on its sequence, it is predicted to be 366 kDa in mass. However, its apparent molecular mass is about 270 kDa, based on electrophoretic mobility in denaturing gels (41). The reasons for the difference in the predicted and observed sizes are not clear. Kattenhorn et al. (22) have identified a 450- to 550-amino-acid (aa) N-terminal fragment of VP1-2 as a peptide that appears later during the HSV-1 infection and which was shown to exhibit deubiquitinating activity.
MATERIALS AND METHODS
Cells and viruses.
Vero, SK-N-SH, and Hep-2 cells were obtained from the American Type Culture Collection. The telomerase-transformed human embryonic lung fibroblasts (HEL cells) were obtained from T. Shenk (Princeton). Cells were maintained in Dulbecco's modified Eagle's minimal essential medium supplemented with 10% (vol/vol) newborn calf serum (Vero and Hep-2 cells) or 10% fetal bovine serum (SK-N-SH and HEL cells). HSV-1(F) is the prototype HSV-1 strain used in our laboratory (10). Isolation and characterization of HSV-1 (HFEM) tsB7 were described elsewhere (1).
Virion purification.
HSV-1(F) and tsB7 virions were purified as previously described (41). Briefly, cells grown in roller bottles to 100% confluence were infected and scraped into the medium when 100% cytopathic effect was reached. Cells were collected by centrifugation at 2,000 rpm for 5 min and resuspended in 1 mM sodium phosphate buffer, pH 7.4. Cell membranes were disrupted in a Dounce homogenizer with a tight pestle, the medium was supplemented with 0.2 M sucrose, and nuclei were pelleted by centrifugation at 3,000 rpm for 5 min. Viral particles from cell lysates and cell medium were collected by ultracentrifugation at 20,000 rpm for 2 h in an SW28 rotor. Viral particles were resuspended in 1 mM sodium phosphate buffer (pH 7.4), layered on a dextran-10 gradient (1.04 to 1.09 g/cm3), and centrifuged for 2 h at 20,000 rpm in an SW41 rotor. The light-scattering band at the middle of the gradient was isolated, diluted in 1 mM phosphate buffer (pH 7.4), and pelleted at 20,000 rpm for 2 h. The virion pellet was resuspended in 199V medium (Sigma-Aldrich, St. Louis, MO) and stored at 4°C.
GST protein purification and antibody generation.
Codons 109 to 333 of UL36 were PCR amplified using primers EN-UL36-325F (5′GAA TTC CCG CCG CGC ATG TGT TCG A3′) and EN-UL36-999B (5′ GAA TTC GGA TGT CTC GGA GGC CCT G 3′) from HSV-1(F) DNA. The fragment was ligated into the pGEX4T-1 EcoRI restriction site. The construct was then designated UL36(109-333aa)-pGEX4T-1. Codons 3040 to 3165 of UL36 were PCR amplified from HSV-1(F) DNA by using primers EC-UL36-9118F (5′ GAA TTC TCG CTG CTT CAG ACC CTG TAT G 3′) and E-CUL36-B (5′ GAA TTC AAT AAT CGA GCG CGT CTA GC 3′). The fragment was then ligated into the pGEX4T-1 EcoRI restriction site. The construct was then named UL36(3040-3065aa)-pGEX4T-1. Both UL36-glutathione S-transferase (GST) constructs were then transformed into BL21 cells, and UL36-GST fusion proteins were purified as described previously (12). Around 10 mg of UL36(3040-3165aa)-GST fusion protein was cut out of the gel and, along with 10 mg of the eluted UL36(109-333aa)-GST fusion protein, was sent to Josman, LLC, for rabbit polyclonal antibody generation.
Antibodies.
Mouse anti-ICP4 monoclonal antibody H640 was obtained from the Goodwin Institute for Cancer Research Inc. (Plantation, FL). Rabbit anti-US3 polyclonal antibody was described previously (29). Mouse anti-actin monoclonal antibody was purchased from Sigma-Aldrich (St. Louis, MO).
Protease inhibitors.
l-(Tosylamido-2-phenyl) ethyl chloromethyl ketone (TPCK) (Sigma-Aldrich, St. Louis, MO) was dissolved in dimethyl sulfoxide (Sigma-Aldrich, St. Louis, MO) to prepare a 150 mM stock solution. The stock solution was further diluted in 199V medium to prepare working solutions of the desired concentrations. For all infections with HSV-1 only (i.e., in the absence of TPCK), dimethyl sulfoxide was added to the medium to a final concentration equivalent to that present in TPCK-treated cells.
Generation of recombinant baculoviruses.
Recombinant baculoviruses carrying the HSV-1 UL6 or UL38 gene fused at the C terminus to green fluorescent protein (GFP) were generated using the PharMingen baculovirus expression system as previously described (18, 29). An MTS-1 vector (38) carrying the GFP gene (MTS-1/GFP vector) was generated by insertion of a DNA fragment containing the GFP gene between the PstI and BglII restriction sites of the MTS-1 vector. DNA fragments containing the UL6 or UL38 gene were amplified by PCR using HSV-1 genomic DNA as a template and cloned into MTS-1/GFP in frame with GFP. Plasmids were transfected together with the BaculoGold baculovirus DNA (PharMingen) into Sf9 insect cells according to the manufacturer's instructions. Five days after the transfection, supernatant containing the recombinant baculovirus was collected by centrifugation at 1,500 rpm for 10 min. Baculovirus was subsequently amplified in Sf9 cells grown in a 150-cm2 tissue culture flask.
Immunoblots.
For immunoblots, cells were harvested, washed with cold phosphate-buffered saline (PBS) supplemented with 1 mM EDTA and Complete mini protease inhibitor cocktail (Roche, Indianapolis, IN), and resuspended in lysis buffer (20 mM Tris [pH 8.8], 1 mM EDTA, 1% [vol/vol] NP-40, 0.4 M NaCl, 2 mM dithiothreitol, 0.1 mM Na3VO4, 10 mM NaF, Complete mini protease inhibitor cocktail). Samples were briefly sonicated, and protein concentrations were determined with the Bio-Rad protein assay (Bio-Rad Laboratories, Hercules, CA). Approximately 100 μg of each sample was electrophoretically separated by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis and electrically transferred onto nitrocellulose membrane. Membranes were blocked with 5% nonfat dry milk in PBS supplemented with 0.05% (vol/vol) Tween 20 (PBT) and reacted with primary antibody diluted in PBT overnight at 4°C. Anti-ICP4 and anti-Us3 antibodies were used at a 1:1,000 dilution, anti-actin antibody was diluted according to the manufacturer's instructions (1:500), and anti-VP1-2 N antibody was diluted 1:20,000. After several rinses with PBT, the membranes were exposed to goat anti-mouse or anti-rabbit alkaline phosphatase (AP)-conjugated secondary antibody diluted in PBT according to the manufacturer's instructions for 1 h at room temperature. For development of AP-conjugated secondary antibody, immunoblots were incubated with AP buffer (100 mM Tris [pH 9.5], 100 mM NaCl, 5 mM MgCl2) containing 5-bromo-4-chloro-3-indolyl phosphate and nitroblue tetrazolium.
Nuclease assays.
Cells were washed with PBS and with low-pH citrate buffer (40 mM sodium citrate, 10 mM KCl, 135 mM NaCl, pH 3.0) to inactivate unpenetrated virions and then again with PBS. Cells were then incubated with 2 mg of proteinase K (Invitrogen, Carlsbad, CA)/ml at 37°C for 1 h to remove unpenetrated, attached virions (32). Nuclease assay was performed as previously described (42), with some modifications. Cells were washed in PBS three times, collected by centrifugation (2,000 rpm, 5 min), resuspended in Tris-buffered saline (137 mM NaCl, 5 mM KCl, 0.7 mM Na2HPO4, 5.5 mM glucose, 25 mM Tris-HCl, pH 7.4), and divided into two equal samples which were used to prepare total cellular and DNase-resistant (encapsidated) DNA isolates. The cells from both samples were pelleted and resuspended in 184 μl of reticulocyte standard buffer (10 mM Tris-HCl [pH 7.5], 10 mM KCl, 1.5 mM MgCl2) containing 0.5% (vol/vol) NP-40. Water (for total DNA) or 200 μg DNase I/ml (90% protein, ≥2,000 U/mg protein; Sigma-Aldrich, St. Luis, MO) (for encapsidated DNA) was added and samples incubated for 30 min at 37°C. An equal volume of 2× CLB (20 mM Tris-HCl [pH 7.5], 2 mM EDTA, 1.2% SDS, 1 mg of pronase [Sigma-Aldrich, St. Louis, MO]/ml) was added and samples incubated for additional 1 h at 37°C. DNA was isolated by phenol-chloroform extraction and dissolved in water. Viral DNA was detected by PCR, using primers specific for HSV-1 DNA (forward, ATGCTGGCGGACGGCTTTGAAACTGACATCGCG; reverse, TGGGATAGCGTATAACGGGGGCCATG). Cycling conditions were the following: 2 min at 94°C; followed by 27 cycles of 30 s at 94°C, 30 s at 62°C, and 2 min at 68°C; and a final 10-min extension at 68°C. PCR products were separated on a 1% agarose gel containing 1 μg/ml of ethidium bromide and visualized under UV light. Additionally, HSV-1 DNA was detected by Southern blotting, for which DNA was digested with BamHI, separated on a 0.7% agarose gel, transferred to a positively charged nylon membrane (Ambion, Austin, TX), and hybridized with radiolabeled BamHI/StuI fragment of plasmid pRB115.
Immunofluorescence.
U2OS cells were grown on four-well slides (Erie Scientific, Portsmouth, NH) and transduced with baculoviruses carrying UL6-GFP or UL38-GFP constructs. Sixteen hours later, cells were rinsed with PBS, fixed in methanol at −80°C for 2 h and air dried. The samples were mounted in Vectashield mounting medium for fluorescence (Vector Laboratories, Inc., Burlingame, CA), and cellular localization of GFP constructs was determined by fluorescence microscopy using a Zeiss (Thornwood, NY) confocal microscope.
RESULTS
TPCK prevents the expression of ICP4 protein.
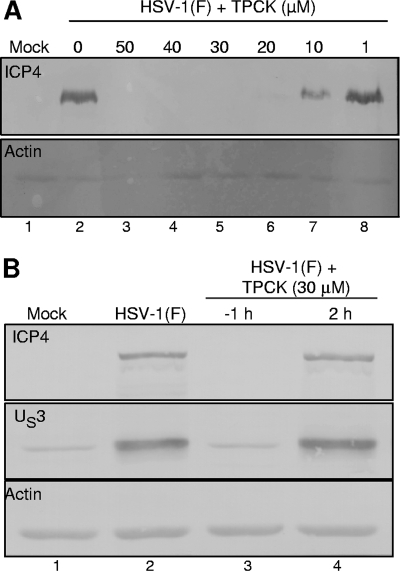
Experiments were done to test whether proteolytic cleavage is required for the release of HSV-1 genomic DNA into the nucleus of an infected cell. For that purpose we tested the effects of several protease inhibitors on the release of HSV-1 DNA. We chose protease inhibitors that were previously shown to interfere with the processes important for the HSV-1 replication cycle, more specifically, to inhibit the activity of viral protease encoded by the UL26 gene (25). Based on our initial studies (data not shown), we selected TPCK for our experiments. To test the effect of TPCK on the release of HSV-1 DNA into the nucleus, Vero cells were treated with TPCK at 10°C for 30 min and then exposed to 10 PFU of HSV-1(F) per cell. Cells were harvested and processed as described in Materials and Methods. As an indication of the release of viral DNA into the nucleus, we monitored the accumulation of ICP4. As shown in Fig. 1A, exposure of cells to TPCK before infection inhibited the accumulation of ICP4 in a TPCK dose-dependent manner. At a concentration of 20 μM TPCK, the accumulation of ICP4 was barely detectable. ICP4 was not detected in cells exposed to 30 μM TPCK. As a control, we tested phenylmethylsulfonyl fluoride, a trypsin inhibitor, and found that it did not interfere with the release of HSV-1 DNA from the capsid (data not shown).
FIG. 1.
TPCK prevents the expression of ICP4 and US3 only if added prior to the exposure of cells to HSV-1. (A) Vero cells were incubated for 30 min in the presence of TPCK at concentrations ranging from 0 to 50 μM and then exposed to 10 PFU of HSV-1(F) per cell at 10°C, followed by the shift to 37°C. After 3 h at 37°C, the expression levels of ICP4 were evaluated by Western blotting. (B) TPCK (30 μM) was added to Vero cells 1 h before (−1 h) or 2 h after the initiation of exposure to 10 PFU of HSV-1(F) per cell. The cells were harvested 8 h later, and the accumulation of ICP4 and US3 was determined by immunoblotting electrophoretically separated proteins in denaturing gels.
The expression of viral proteins is not inhibited by TPCK when it is added following the release of viral DNA into the nucleus.
The results in Fig. 1A indicate that exposure of cells prior to infection to 30 μM of TPCK prevents the accumulation of ICP4. In order to exclude the possibility that the lack of ICP4 expression in TPCK-treated cells infected with HSV-1 is a result of an inhibitory effect of TPCK on protein synthesis, cells were exposed to TPCK either 1 hour prior to or 2 hours after exposure of the cells to 10 PFU of HSV-1(F) per cell. The 2-hour time point was selected because it has been shown previously that this is sufficient time for HSV-1 DNA release and accumulation of empty capsids at the nuclear pores (40). The results in Fig. 1B show that exposure of cells to TPCK at 2 hours after infection had no effect on the accumulation of ICP4 or US3 (compare lanes 2 and 4). We conclude that the inhibition of accumulation of ICP4 or US3 in cells pretreated with TPCK prior to infection was due to inhibition of entry, transport to the nuclear pore, or release of viral DNA into the nucleus.
TPCK prevents the release of viral DNA from the capsid.
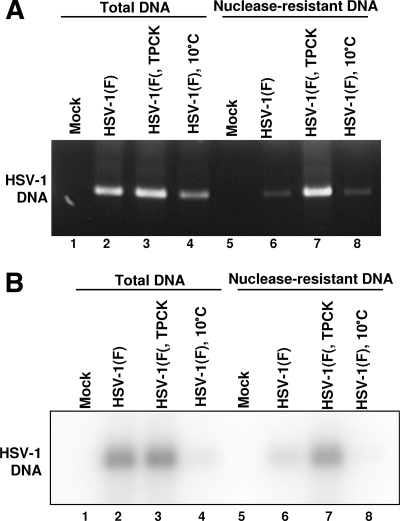
To test further the model that exposure of cells to TPCK prior to infection prevents the release of viral DNA into the nucleus, replicate cultures of Vero cells either treated with TPCK or left untreated were exposed to 10 PFU of HSV-1(F) per cell at 10°C. The cells were incubated for 2 h at 37°C and then harvested, lysed, subjected to DNase treatment, and probed for residual DNA as described in Materials and Methods. The results were as follows. In cells infected with HSV-1 but not treated with TPCK (Fig. 2, lane 6) most of the DNA was nuclease sensitive and therefore not in the capsid at the moment the cells were harvested. A small amount of DNA remained in the capsid, which is consistent with results published earlier (33). In lysates of cells treated with TPCK and then exposed to HSV-1, most of the viral DNA remained nuclease resistant (Fig. 2, lane 7), consistent with the hypothesis that the event blocked by TPCK occurs at or before the release of viral DNA from the capsids.
FIG. 2.
TPCK prevents the release of viral DNA from capsid and does not interfere with viral entry into the cell. Vero cells, untreated or treated with 30 μM TPCK for 30 min, were exposed to 10 PFU of HSV-1(F) per cell. After 2 h at 10°C, the temperature was changed to 37°C and incubation continued for additional 2 h. Cells were rinsed with citrate buffer, exposed to proteinase K for 1 h, and then lysed as described in Materials and Methods. The cell lysates were digested with DNase I followed by pronase and then extracted with phenol-chloroform. HSV-1 DNA was detected by PCR (A) and Southern blot analysis (B). As a control, cells were exposed to HSV-1 and kept at 10°C throughout the entire experiment.
Another issue addressed in this experiment was whether TPCK prevents the entry of viral particles into the cells, as that would also render viral DNA nuclease resistant. As a part of the nuclease assay, attached, unpenetrated virions were removed by proteinase K treatment prior to harvesting of the cells as described previously (32). Very little viral DNA was detected in control group kept at 10°C throughout the entire experiment, which allows attachment of viral particles to the cell membrane but does not allow viral entry, indicating that the removal of attached, unpenetrated virions was successful (Fig. 2, compare lanes 4 and 8). Therefore, viral DNA detected in infected cells treated with TPCK corresponded to viral DNA enclosed in the capsid that was protected from proteinase K digestion and hence inside the infected cell. This conclusion was revisited in additional experiments described below.
Nuclear import is not impaired in cells treated with TPCK.
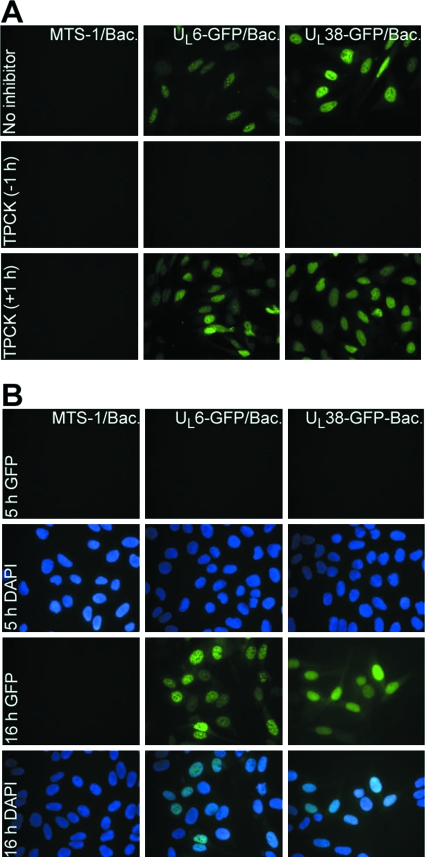
Since previous results showed that TPCK prevents the release of viral DNA into the nucleus and the release of viral DNA depends on the nuclear import machinery (33), experiments were carried out to show whether the effect of TPCK on the release of viral DNA is a consequence of a more general inhibitory effect toward nuclear import. For that purpose, U2OS cells were transduced with baculoviruses carrying the HSV-1 UL6 or UL38 gene fused C terminally to GFP (UL6-GFP and UL38-GFP, respectively), as described in Materials and Methods. UL6 and UL38 are proteins known to localize in the nucleus (3, 34). To evaluate the effect of TPCK on nuclear import of UL6-GFP and UL38-GFP, cells were treated with TPCK either 1 hour before or 1 hour after the exposure to baculoviruses. Cellular localization of UL6-GFP and UL38-GFP was determined 16 h later by fluorescence microscopy. The results in Fig. 3A show that in cells exposed to TPCK 1 h after the transduction with baculoviruses, UL6-GFP and UL38-GFP constructs were expressed and proteins localized in the nuclei of transduced, TPCK-treated cells, indicating that TPCK does not impair the nuclear import machinery. To eliminate the possibility that protein accumulated in the nucleus was expressed prior to the exposure to TPCK, a control experiment was performed in which U2OS cells were transduced with the described baculoviruses and the expression of constructs evaluated 5 or 16 h later. The results in Fig. 3B show that constructs were not expressed 5 h following the transduction, which is a time period shown to be more than sufficient for TPCK to inhibit the release of viral DNA into the nucleus. Interestingly, when cells were exposed to TPCK 1 h prior to transduction with baculoviruses, no GFP fluorescence could be detected.
FIG. 3.
TPCK does not affect nuclear import of proteins. (A) U2OS cells were transduced with baculoviruses carrying UL6-GFP or UL38-GFP constructs. TPCK was added either 1 h before (−1 h) or 1 h after (+1 h) the addition of baculoviruses, and cells were fixed 16 h later. (B) To exclude the possibility that UL6-GFP and UL38-GFP accumulate in the nucleus prior to the exposure of cells to TPCK, U2OS cells were transduced with baculoviruses carrying UL6-GFP or UL38-GFP constructs and fixed 5 or 16 h later. Cellular localization of UL6-GFP and UL38-GFP was determined by fluorescence microscopy with the aid of a Zeiss confocal microscope.
Virion VP1-2 is cleaved in infected cells.
The results presented so far demonstrate that a proteolytic cleavage is required for the release of HSV-1 DNA into the nucleus. It has been reported that in cells infected with the tsB7 HSV-1 mutant at the nonpermissive temperature, capsids are accumulated at the nuclear pores but viral DNA fails to be released (1). The temperature-sensitive mutation has been mapped to the UL36 gene, which encodes the VP1-2 protein. This result strongly suggests a role for VP1-2 in the release of viral DNA from the capsid. Several reports suggested that VP1-2 is proteolytically processed, and Kattenhorn et al. have demonstrated the presence of a 450- to 500-amino-acid N-terminal fragment of VP1-2 in cells infected with HSV-1 (22). In the light of these reports, several series of experiments were done to test the hypothesis that VP1-2 is subject to cleavage after viral entry into cells.
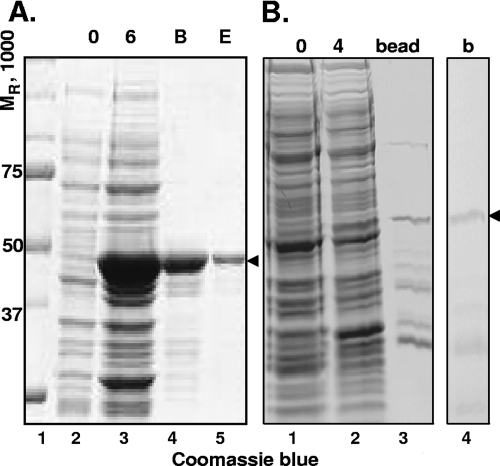
In the first series of experiments, we tested a variety of GST-VP1-2 fusion proteins for their properties. In the course of these studies we found that GST-UL36109-333 and GST-UL363040-3165 were soluble and suitable for purification, as shown in Fig. 4A, lane 5, and B, lane 4, respectively. Antibodies against the purified antigens were made in rabbits as described in Materials and Methods and designated VP1-2 N and VP1-2 C antibodies, respectively.
FIG. 4.
Purification of N- and C-terminal UL36 proteins for antibody generation. (A) BL21 cell culture expressing UL36(109-333aa)-GST protein was incubated for 6 h with 0.5 M IPTG (isopropyl-β-d-thiogalactopyranoside) at 30°C. Cells were lysed and incubated with glutathione-Sepharose. Protein was then eluted. Lane 2, uninduced cell culture; lane 3, cell culture induced with IPTG for 6 h; lane 4, UL36(109-333aa)-GST fusion protein bound to glutathione-Sepharose; lane 5, eluate of fusion protein from glutathione-Sepharose. Lanes 2 to 5, Coomassie blue-stained proteins. (B) BL21 cell culture expressing UL36(3040-3165aa)-GST protein was induced for 4 h with 0.5 M IPTG at 30°C. Lane 1, uninduced cell culture; lane 2, cell culture induced with IPTG for 4 h; lane 3, UL36(3040-3165aa)-GST protein bound to glutathione-Sepharose; lane 4, lane 3 immunoblotted with GST antibody. The protein of interest was cut out of the gel. Lanes 1 to 3, Coomassie blue-stained proteins; lane 4, AP detection of GST fusion proteins by immunoblotting with GST antibody. B, beads; E, eluent; b, blot.
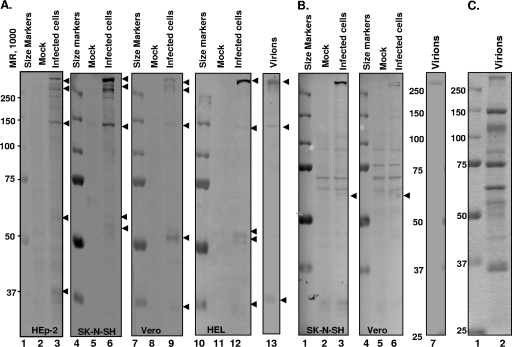
In the second series of experiments, we tested the reactivity of the VP1-2 N and VP1-2 C antibodies with purified virions. As illustrated in Fig. 5A, lane 13, and B, lane 7, both antibodies reacted with a protein with an electrophoretic mobility greater than 250 kDa and which comigrated with VP1-2 in electrophoretically separated lysates of purified virions stained with Coomassie blue (Fig. 5C lane 2).
FIG. 5.
Virion VP1-2 is cleaved upon entry into the cell. HEp-2, SK-N-SH, Vero, or HEL cells were exposed to 100 PFU of HSV-1 per cell at 10°C for 1 h. The temperature was shifted to 37°C and incubation continued for additional 2 h. Cell lysates or lysate of purified virion were probed with anti-VP1-2 N (A) or anti-VP1-2 C (B) antibody. Electrophoretically separated virion proteins were stained with Coomassie blue (C).
The third series of experiments was done to determine whether VP1-2 was cleaved after infection. In this series of experiments, replicate cultures of HEp-2, Vero, SK-N-SH, or HEL cells were mock infected or exposed to 100 PFU of purified virus per cell. Two hours later, cells were harvested, solubilized, subjected to electrophoresis in a denaturing gel, and probed with either VP1-2 N or VP1-2 C antibody. The results (Fig. 5) were as follows.
(i) The VP-1-2 N antibody reacted with both full-length VP1-2 and protein in faster-migrating bands. In HEp-2, SK-N-SH, and Vero cells, we detected both full-length bands of the size predicted for intact VP1-2 and slightly faster-migrating bands significantly larger than 250 kDa. These faster-migrating bands were less obvious in the lysates of infected HEL cells.
(ii) The VP1-2 N antibody also reacted with proteins of approximately 150 kDa and 37 kDa in lysates of virions and in lysates of infected HEp-2, SK-N-SH, Vero, and HEL cells. We suspect that this antibody reacts in a nonspecific manner with virion proteins of approximately 150 and 37 kDa.
(iii) We noted that VP1-2 N antibody reacted with several bands migrating in the range of 50 to 60 kDa in electrophoretically separated lysates of infected cells.
(iv) In contrast to these results, we noted a very faint band with an electrophoretic mobility of approximately 60 kDa in electrophoretically separated lysates of cells probed with the VP1-2 C antibody.
The results presented in Fig. 5 were reproducible. The conclusion to be derived from these results is that VP1-2 is cleaved in multiple steps in a cell-dependent fashion. Since the polypeptide which served as an antigen for the production of the VP1-2 N antibody is close to the N terminus, the 50- to 60-kDa protein accumulating in infected cells most likely represents the amino-terminal domain of the protein. Even if the 55-kDa polypeptide is derived from the C terminus of the protein, the sum of the masses of the two polypeptides detected with our antibodies is much less than that of the intact VP1-2 protein, which suggests that the protein is cleaved into several polypeptides.
To verify that the 55-kDa protein reactive with the VP1-2 N antibody was indeed derived from VP1-2 protein, replicate roller bottle cultures of Vero cells were mock infected or exposed to 50 PFU of purified HSV-1(F) virions per cell for 4 h. The cells were harvested and lysed in 6 ml of lysis buffer containing 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.2% SDS, 5 mM dithiothreitol, 2 mM EDTA, 1% Triton X-100, 1% Sarkosyl, 10 mM NaF, 0.1 mM sodium vanadate, and fresh protease inhibitor cocktail (Complete; Roche). The total lysate was centrifuged at 10,000 rpm for 10 min. The supernatant fluid was preincubated with 30 μl preimmune rabbit antibody for 3 h at 4°C and incubated with 200 μl of protein A-agarose for an additional 2 h at 4°C. Lysates were clarified with centrifugation at 3,000 rpm for 10 min. The supernatant fluid was incubated with 50 μl of VP1-2 N antibody overnight at 4°C. The next day, 200 μl of protein A-agarose was added to the lysate and incubated for an additional 2 h. The immune complexes bound to agarose were rinsed five times with lysis buffer, eluted in SDS protein sample buffer, and separated on an 8% SDS-polyacrylamide gel in duplicate. The gels were either stained with Coomassie blue or transferred to nitrocellulose membranes, followed by immunoblotting with VP1-2N antibody. The Coomassie blue-stained protein of the correct size (50 to 55 kDa) was cut out of the gel and sent for mass spectrometry analysis to the Harvard mass spectrometry facility. The results were that the only HSV-1 proteins contained in the sample were derived from the amino-terminal domain of VP1-2.
The 55-kDa N-terminal cleavage product of VP1-2 does not accumulate in cells infected with HSV-1 under conditions that prevent the release of viral DNA from the capsid.
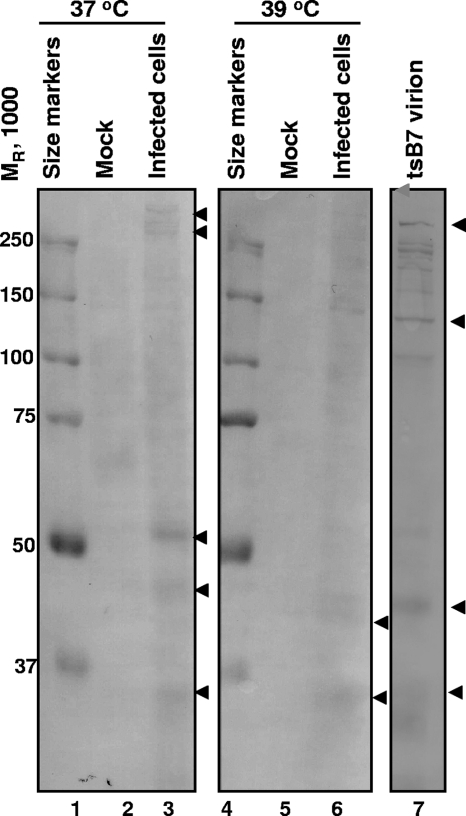
The objective of these experiments was to determine whether VP1-2 was cleaved under conditions in which viral DNA was blocked from being released from capsids. In the first series of experiments, replicate Vero cell cultures were exposed to 100 PFU of purified HSV-1(tsB7) at 10°C for 1 h. In this step the virions attached to the plasma membrane, but entry did not ensue. At the end of the adsorption interval, the medium was replaced and the cultures were rapidly transferred from 10°C to a water bath set at 39.5°C (nonpermissive temperature for tsB7) or 37°C (permissive temperature). The cells were harvested 1 h later, solubilized, subjected to electrophoresis in denaturing gels, and reacted with VP1-2 N antibody as described in Materials and Methods. The results shown in Fig. 6 indicate that VP1-2 N antibody reacted with a 55-kDa band that was present in lysates of cells maintained at 37°C (lane 3) but not in lysates of cells maintained at 39.5°C (lane 6). This band was not present in lysates of purified HSV-1(tsB7) virions (lane 7).
FIG. 6.
Virion VP1-2 is not cleaved in cells infected with tsB7 HSV-1 at the nonpermissive temperature. Vero cells were exposed to 100 PFU of tsB7 HSV-1 per cell at 10°C, and 1 hour later cells were placed in a 39°C or 37°C water bath for 1 h. Cells were harvested and lysates probed with anti-VP1-2 N antibody. For comparison, tsB7 virions were lysed, subjected to electrophoresis in a denaturing gel, and probed with the same antibody.
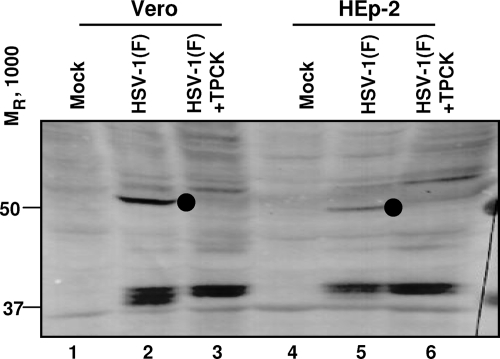
In the second series of experiments, we repeated the experiment shown in Fig. 2, except the lysates were electrophoretically separated in denaturing gels and probed with VP1-2 N antibody rather than digested with DNase. The results (Fig. 7) show that treatment of cells with TPCK, which has been shown earlier in this report to protect viral DNA from digestion with DNase, also blocked the cleavage of VP1-2 protein as shown by the absence of the 55-kDa protein.
FIG. 7.
TPCK prevents the cleavage of VP1-2. Vero or HEp-2 cells, untreated or pretreated with 30 μM TPCK for 30 min, were exposed to 100 PFU of HSV-1(F) per cell at 10°C for 1 h. The temperature was shifted to 37°C and incubation continued for additional 2 h. Cell lysates were probed with anti-VP1-2 N.
Treatment of cells with TPCK during the maintenance of the cells at the nonpermissive temperature but after a significant time interval to enable transport of virus to the nuclear pore blocks the expression of viral genes after transfer of the cells to the permissive temperature.
The experiments described in this section were designed to determine at what point in the entry process the cleavage event blocked by TPCK takes place. The design of the experiments described in this section takes into account the following data presented here and in studies on HSV-1(tsB7) published years ago (1).
The studies described in the preceding sections showed that an event after viral entry involves proteolytic cleavage that is blocked by TPCK and that this cleavage is required for viral gene expression. We have also shown that addition of TPCK prior to infection does not affect virion entry into cells but does affect the availability of the DNA to digestion by DNase.
In earlier studies we have demonstrated that at 39.5°C the HSV-1(tsB7) mutant infects cells and that the capsids are rapidly transported to the nuclear pores but that viral gene expression does not ensue until after the cells are shifted to 34°C, the permissive temperature. The temperature-sensitive lesion of this mutant maps to the UL36 open reading frame. A persuasive argument that cleavage of UL36 protein is at least a candidate substrate of the critical cleavage event required for viral gene expression is based on the evidence that the protein is cleaved at the permissive temperature but not at the nonpermissive temperature.
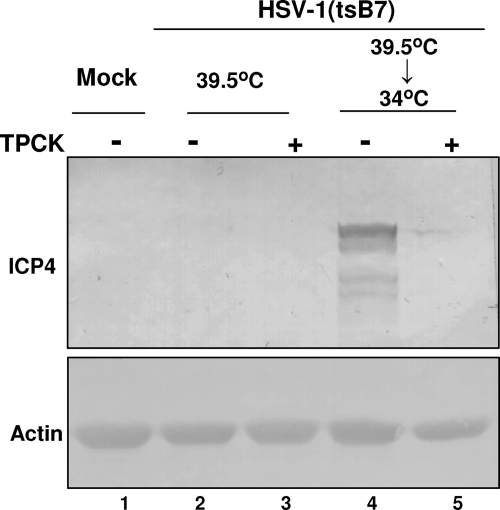
To test the hypothesis that the proteolytic cleavage event blocked by TPCK coincides with the temperature-sensitive event of HSV-1 tsB7, replicate cultures of Vero cells were exposed to 10 PFU of HSV-1(tsB7) mutant virus at 10°C, which allowed the attachment of virions to the cell membrane. After 1 h, the cultures were transferred to a 39.5°C water bath. One hour after the shift, one set of replicate cultures was exposed to 30 μM TPCK for 90 min. At that time subsets of treated and untreated cultures were shifted to 34°C and incubation continued for an additional 5 h. The cells were harvested, processed as described in Materials and Methods, and reacted with anti-ICP4 antibody. The results shown in Fig. 8 indicate that the untreated cultures shifted from 39.5 to 34°C expressed ICP4, whereas the treated cultures shifted to 34°C did not express ICP4. The results suggest that the critical proteolytic event required for viral gene expression takes place after transport of virions to the nuclear pore.
FIG. 8.
TPCK prevents the release of viral DNA into the nucleus even when added after arrival of capsids at the nuclear pore. Replicate cultures of Vero cells were exposed to10 PFU of HSV-1(tsB7) mutant virus per cell at 10°C. One hour later, the cultures were placed in 39.5°C water bath. After 1 h, some cultures were exposed to 30 μM TPCK for 1.5 h, and the temperature was shifted to 34°C or remained at 39.5C. After 5 h, cells were harvested and the expression of ICP4 in cell lysates determined by Western blotting.
DISCUSSION
The results presented in this report, together with results published earlier, support the following model for the release of HSV-1 DNA from capsids into the nucleus. Specifically, on docking at the nuclear pore, the capsid is oriented with a penton facing the lumen of the pore. VP1-2 is bound to the pentons. The attachment of the capsid to the nuclear pore triggers proteolytic cleavage of VP1-2, with the result that conformational changes in the capsid pentons trigger the release of viral DNA. The evidence that supports this model is as follows.
First, we showed that a proteolytic cleavage is essential for the release of viral DNA into the nucleus, as judged by the ability of protease inhibitor TPCK to prevent the release of viral DNA. This was demonstrated in two ways: (i) nuclease assay showed that in TPCK-treated cells infected with HSV-1, most of viral DNA remained nuclease resistant and therefore inside the capsid at the moment the cells were harvested, and (ii) TPCK prevented the expression of ICP4 in infected cells when added before the exposure of cells to HSV-1 but not when added 2 h after exposure, a time period sufficient for DNA entry and expression of α genes (20, 21). Several experiments were done to address the possibility that TPCK has an inhibitory effect on protein synthesis or nuclear import machinery (Fig. 1B and 3), and the results of these experiments demonstrated that this is not the case.
Second, we identified VP1-2 as a virion protein that has to be proteolytically cleaved in order for DNA to be released from the capsids. (i) We show that in addition to full-length VP1-2, anti-VP1-2 N antibody also reacts with several shorter polypeptides. Two of the polypeptides, approximately 37 and 150 kDa in molecular mass, were also present in the purified virion, and these bands are possibly a result of a nonspecific reaction of anti-VP1-2 N antibody with another viral protein, since other studies in which an anti-VP1-2 antibody was used did not show the presence of any additional forms of VP1-2 (27, 28). Another set of bands detected by VP1-2 N antibody had an apparent mass of approximately 55 kDa and are referred to as 55-kDa products below. This set of bands was not present in mock-infected cells or in lysates of virions but was present exclusively in infected cells, shortly upon viral entry, and was the result of proteolytic cleavage of virion VP1-2. The number of 55-kDa products was cell specific and was either one or two. This difference in the number of cleavage products could be explained by posttranslational modifications by differential proteolytic processing in these cell lines, since it has been shown previously that viral proteins can show different cleavage patterns in different cell lines (14, 19). (ii) We showed that if cells are infected under conditions that prevent the release of viral DNA from the capsid, N-terminal cleavage of VP1-2 does not occur (Fig. 6 and 7). The cleavage of VP1-2 does not take place in cells infected with the tsB7 mutant at the nonpermissive temperature or in cells infected with wild-type virus in the presence of TPCK, which was shown in this report to prevent the release of viral DNA.
Third, several lines of evidence, previously published or presented in this report, indicate that proteolytic cleavage of VP1-2 occurs only after the capsid arrives and attaches to the nuclear pore. Batterson et al. (1) showed that DNA-filled tsB7 capsids are attached to the nuclear pore at the nonpermissive temperature. Our results in this report show that tsB7 virion VP1-2 is not cleaved at the nonpermissive temperature, indicating that attachment of the capsid to the nuclear pore does not require the cleavage of VP1-2 and that this cleavage occurs after the attachment of the capsid. Additionally, we show that when cells are infected with tsB7 virus at the nonpermissive temperature and TPCK is added only after capsids are attached to the nuclear pores, DNA is not released into the nucleus, as judged by the lack of ICP4 expression. This indicates that proteolytic cleavage essential for the release of HSV-1 DNA from the capsid occurs after the attachment of the capsid to the nuclear pore.
We suggest that proteolytic cleavage of VP1-2 induces conformational changes in capsid pentons that would trigger the release of viral DNA, based on several previously published observations. (i) It has been suggested that DNA is released from the capsid through a penton at capsid vertices, based on observations that purified capsids treated with trypsin (30) or guanidine·HCl (31) appear to loose their DNA through capsid vertices. (ii) HSV-1 capsids have been observed in electron micrographs to face nuclear pores with a vertex (33, 40). (iii) VP1-2 is very closely associated with capsids (15, 41); more specifically, VP1-2 was suggested to be associated with pentons of the virion capsid, occupying the space between the upper domains of two adjacent penton VP5 subunits (44). Such intimate association between VP1-2 and pentons makes it feasible that the conformation of pentons would be influenced by changes (i.e., proteolytic cleavage) in VP1-2.
A question resulting from these studies is the relation between the cleavage products of VP1-2 detected in the current studies and the 450- to 500-residue cleavage product, possessing deubiquitinating activity, of the N-terminal fragment of VP1-2 identified by Kattenhorn et al. (22). It is noteworthy that the 450- to 500-residue protein possessing deubiquitinating activity accumulated only late in infection. Although we cannot exclude the possibility that the cleavage of VP1-2 preceding viral release into cells generates a protein with potential deubiquitinating activity, the amounts of enzyme and timing of accumulation of the protein suggest that the enzyme is generated by the cleavage of newly translated protein rather than the VP1-2 brought into the cell in the course of infection.
We do not know, presently, which protease is responsible for the cleavage of VP1-2. Based on the specificity of TPCK, we can conclude that it is a serine or cysteine protease. Since VP1-2 is cleaved only after it arrives at the nuclear pore, it is likely that the protease is recruited to the capsid once it is attached to the nuclear pore. We cannot completely exclude the possibility that the protease responsible for the cleavage of VP1-2 is a viral protease. VP1-2 itself is a cysteine protease with deubiquitinating activity (22). It is possible that it has additional specificity, toward itself. Another protease encoded by HSV-1 genome is the product of the UL26 gene (26). It is a serine protease that cleaves itself and the product of the UL26.5 gene, and the cleavage sites specific for UL26 protease have been identified (7). Careful examination of the VP1-2 N-terminal sequence identifies three possible UL26 Pr cleavage sites: 474AVPAS478, 484VLAS487, and 507TVPAS511. Cleavage of VP1-2 at these sites would yield an N-terminal peptide with a molecular mass of 50 to 53 kDa. Selective cleavage at these three sites would yield multiple bands, as we have observed in our experiments. Although the protease has not been observed in mature, DNA-containing capsids, we cannot exclude the possibility that an undetectable amount of the protease is present in the mature capsids.
Proteolytic cleavage of viral proteins by cellular or viral proteases plays an important role in various stages of the viral life cycle. Later in the viral life cycle, proteolytic processing of viral proteins has been well documented as playing an important role in the assembly of newly formed viral particles. Examples of this are as follows: some families of RNA viruses, which require proteolytic cleavage of polypeptides encoded by viral genome in order to produce functional viral peptides (8), and herpesvirus, which requires the activity of viral protease encoded by the UL26 gene for the assembly and maturation of the capsid (13). A requirement for proteolytic processing is also important for early stages in viral infection: cleavage of spike protein of the severe acute respiratory syndrome virus by factor Xa is important for the entry of the severe acute respiratory syndrome virus (9), viral entry of Ebola virus requires endosomal proteolytic cleavage of its glycoprotein (4, 37), and proteolysis of adenoviral structural protein VI by the viral protease is required for the efficient capsid disassembly and transport of viral DNA into the nucleus (17). Our results describe an additional step in the HSV-1 life cycle that is regulated by a proteolytic cleavage.
Acknowledgments
These studies were aided by National Cancer Institute grant CA83939
Footnotes
Published ahead of print on 23 January 2008.
REFERENCES
- 1.Batterson, W., D. Furlong, and B. Roizman. 1983. Molecular genetics of herpes simplex virus. VIII. Further characterization of a temperature-sensitive mutant defective in release of viral DNA and in other stages of the viral reproductive cycle. J. Virol. 45397-407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Batterson, W., and B. Roizman. 1983. Characterization of the herpes simplex virion-associated factor responsible for the induction of alpha genes. J. Virol. 46371-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burch, A. D., and S. K. Weller. 2004. Nuclear sequestration of cellular chaperone and proteasomal machinery during herpes simplex virus type 1 infection. J. Virol. 787175-7185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chandran, K., N. J. Sullivan, U. Felbor, S. P. Whelan, and J. M. Cunningham. 2005. Endosomal proteolysis of the Ebola virus glycoprotein is necessary for infection. Science 3081643-1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chou, J., and B. Roizman. 1989. Characterization of DNA sequence-common and sequence-specific proteins binding to cis-acting sites for cleavage of the terminal a sequence of the herpes simplex virus 1 genome. J. Virol. 631059-1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Desai, P. J. 2000. A null mutation in the UL36 gene of herpes simplex virus type 1 results in accumulation of unenveloped DNA-filled capsids in the cytoplasm of infected cells. J. Virol. 7411608-11618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DiIanni, C. L., D. A. Drier, I. C. Deckman, P. J. McCann, 3rd, F. Liu, B. Roizman, R. J. Colonno, and M. G. Cordingley. 1993. Identification of the herpes simplex virus-1 protease cleavage sites by direct sequence analysis of autoproteolytic cleavage products. J. Biol. Chem. 2682048-2051. [PubMed] [Google Scholar]
- 8.Dougherty, W. G., and B. L. Semler. 1993. Expression of virus-encoded proteinases: functional and structural similarities with cellular enzymes. Microbiol. Rev. 57781-822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Du, L., R. Y. Kao, Y. Zhou, Y. He, G. Zhao, C. Wong, S. Jiang, K. Y. Yuen, D. Y. Jin, and B. J. Zheng. 2007. Cleavage of spike protein of SARS coronavirus by protease factor Xa is associated with viral infectivity. Biochem. Biophys. Res. Commun. 359174-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ejercito, P. M., E. D. Kieff, and B. Roizman. 1968. Characterization of herpes simplex virus strains differing in their effects on social behaviour of infected cells. J. Gen. Virol. 2357-364. [DOI] [PubMed] [Google Scholar]
- 11.Forrester, A., H. Farrell, G. Wilkinson, J. Kaye, N. Davis-Poynter, and T. Minson. 1992. Construction and properties of a mutant of herpes simplex virus type 1 with glycoprotein H coding sequences deleted. J. Virol. 66341-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frangioni, J. V., and B. G. Neel. 1993. Solubilization and purification of enzymatically active glutathione S-transferase (pGEX) fusion proteins. Anal. Biochem. 210179-187. [DOI] [PubMed] [Google Scholar]
- 13.Gao, M., L. Matusick-Kumar, W. Hurlburt, S. F. DiTusa, W. W. Newcomb, J. C. Brown, P. J. McCann III, I. Deckman, and R. J. Colonno. 1994. The protease of herpes simplex virus type 1 is essential for functional capsid formation and viral growth. J. Virol. 683702-3712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garmendia, A. E., Z. Lu, and E. R. Tulman. 1997. Discrete cleavage patterns of pseudorabies virus immediate early protein (IE180) seen in some cell lines upon extraction after cycloheximide reversal. J. Virol. Methods 64171-179. [DOI] [PubMed] [Google Scholar]
- 15.Gibson, W., and B. Roizman. 1972. Proteins specified by herpes simplex virus. 8. Characterization and composition of multiple capsid forms of subtypes 1 and 2. J. Virol. 101044-1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Granzow, H., B. G. Klupp, and T. C. Mettenleiter. 2005. Entry of pseudorabies virus: an immunogold-labeling study. J. Virol. 793200-3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Greber, U. F., P. Webster, J. Weber, and A. Helenius. 1996. The role of the adenovirus protease on virus entry into cells. EMBO J. 151766-1777. [PMC free article] [PubMed] [Google Scholar]
- 18.Hagglund, R., J. Munger, A. P. Poon, and B. Roizman. 2002. U(S)3 protein kinase of herpes simplex virus 1 blocks caspase 3 activation induced by the products of U(S)1.5 and U(L)13 genes and modulates expression of transduced U(S)1.5 open reading frame in a cell type-specific manner. J. Virol. 76743-754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heidner, H. W., T. A. Knott, and R. E. Johnston. 1996. Differential processing of Sindbis virus glycoprotein PE2 in cultured vertebrate and arthropod cells. J. Virol. 702069-2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Honess, R. W., and B. Roizman. 1974. Regulation of herpesvirus macromolecular synthesis. I. Cascade regulation of the synthesis of three groups of viral proteins. J. Virol. 148-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Honess, R. W., and B. Roizman. 1975. Regulation of herpesvirus macromolecular synthesis: sequential transition of polypeptide synthesis requires functional viral polypeptides. Proc. Natl. Acad. Sci. USA 721276-1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kattenhorn, L. M., G. A. Korbel, B. M. Kessler, E. Spooner, and H. L. Ploegh. 2005. A deubiquitinating enzyme encoded by HSV-1 belongs to a family of cysteine proteases that is conserved across the family Herpesviridae. Mol. Cell 19547-557. [DOI] [PubMed] [Google Scholar]
- 23.Kwong, A. D., J. A. Kruper, and N. Frenkel. 1988. Herpes simplex virus virion host shutoff function. J. Virol. 62912-921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ligas, M. W., and D. C. Johnson. 1988. A herpes simplex virus mutant in which glycoprotein D sequences are replaced by beta-galactosidase sequences binds to but is unable to penetrate into cells. J. Virol. 621486-1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu, F., and B. Roizman. 1992. Differentiation of multiple domains in the herpes simplex virus 1 protease encoded by the UL26 gene. Proc. Natl. Acad. Sci. USA 892076-2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu, F. Y., and B. Roizman. 1991. The herpes simplex virus 1 gene encoding a protease also contains within its coding domain the gene encoding the more abundant substrate. J. Virol. 655149-5156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McNabb, D. S., and R. J. Courtney. 1992. Analysis of the UL36 open reading frame encoding the large tegument protein (ICP1/2) of herpes simplex virus type 1. J. Virol. 667581-7584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McNabb, D. S., and R. J. Courtney. 1992. Characterization of the large tegument protein (ICP1/2) of herpes simplex virus type 1. Virology 190221-232. [DOI] [PubMed] [Google Scholar]
- 29.Munger, J., A. V. Chee, and B. Roizman. 2001. The U(S)3 protein kinase blocks apoptosis induced by the d120 mutant of herpes simplex virus 1 at a premitochondrial stage. J. Virol. 755491-5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Newcomb, W. W., F. P. Booy, and J. C. Brown. 2007. Uncoating the herpes simplex virus genome. J. Mol. Biol. 370633-642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Newcomb, W. W., and J. C. Brown. 1994. Induced extrusion of DNA from the capsid of herpes simplex virus type 1. J. Virol. 68433-440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nicola, A. V., and S. E. Straus. 2004. Cellular and viral requirements for rapid endocytic entry of herpes simplex virus. J. Virol. 787508-7517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ojala, P. M., B. Sodeik, M. W. Ebersold, U. Kutay, and A. Helenius. 2000. Herpes simplex virus type 1 entry into host cells: reconstitution of capsid binding and uncoating at the nuclear pore complex in vitro. Mol. Cell. Biol. 204922-4931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rixon, F. J., C. Addison, A. McGregor, S. J. Macnab, P. Nicholson, V. G. Preston, and J. D. Tatman. 1996. Multiple interactions control the intracellular localization of the herpes simplex virus type 1 capsid proteins. J. Gen. Virol. 772251-2260. [DOI] [PubMed] [Google Scholar]
- 35.Sarmiento, M., M. Haffey, and P. G. Spear. 1979. Membrane proteins specified by herpes simplex viruses. III. Role of glycoprotein VP7(B2) in virion infectivity. J. Virol. 291149-1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schlieker, C., G. A. Korbel, L. M. Kattenhorn, and H. L. Ploegh. 2005. A deubiquitinating activity is conserved in the large tegument protein of the Herpesviridae. J. Virol. 7915582-15585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schornberg, K., S. Matsuyama, K. Kabsch, S. Delos, A. Bouton, and J. White. 2006. Role of endosomal cathepsins in entry mediated by the Ebola virus glycoprotein. J. Virol. 804174-4178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sciortino, M. T., B. Taddeo, A. P. Poon, A. Mastino, and B. Roizman. 2002. Of the three tegument proteins that package mRNA in herpes simplex virions, one (VP22) transports the mRNA to uninfected cells for expression prior to viral infection. Proc. Natl. Acad. Sci. USA 998318-8323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shieh, M. T., D. WuDunn, R. I. Montgomery, J. D. Esko, and P. G. Spear. 1992. Cell surface receptors for herpes simplex virus are heparan sulfate proteoglycans. J. Cell Biol. 1161273-1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sodeik, B., M. W. Ebersold, and A. Helenius. 1997. Microtubule-mediated transport of incoming herpes simplex virus 1 capsids to the nucleus. J. Cell Biol. 1361007-1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Spear, P. G., and B. Roizman. 1972. Proteins specified by herpes simplex virus. V. Purification and structural proteins of the herpesvirion. J. Virol. 9143-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stow, N. D. 2001. Packaging of genomic and amplicon DNA by the herpes simplex virus type 1 UL25-null mutant KUL25NS. J. Virol. 7510755-10765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.WuDunn, D., and P. G. Spear. 1989. Initial interaction of herpes simplex virus with cells is binding to heparan sulfate. J. Virol. 6352-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou, Z. H., D. H. Chen, J. Jakana, F. J. Rixon, and W. Chiu. 1999. Visualization of tegument-capsid interactions and DNA in intact herpes simplex virus type 1 virions. J. Virol. 733210-3218. [DOI] [PMC free article] [PubMed] [Google Scholar]