Abstract
Electron cryomicrographs of intact parainfluenza virus 5 (PIV5) virions revealed two different surface structures, namely, a continuous layer and distinct individual spikes. The structure of these spikes reconstructed from intact virions was compared with known F ectodomain structures and was found to be different from the prefusion PIV5 F0 structure but, surprisingly, very similar to the human PIV3 F postfusion structure. Hence, we conclude that the individual F1+F2 spikes in intact PIV5 virions also correspond to the postfusion state. Since the observed fusion activity of PIV5 virions has to be associated with prefusion F1+F2 proteins, they have necessarily to be localized in the continuous surface structure. The data therefore strongly suggest that the prefusion state of the F1+F2 protein requires stabilization, most probably by the association with hemagglutinin-neuraminidase. The conversion of F1+F2 proteins from the prefusion toward the postfusion state while embedded in the virus membrane is topologically difficult to comprehend on the basis of established models and demands reconsideration of our current understanding.
Infection of eukaryotic cells by enveloped viruses requires the release of the viral genome into the host cell. To this end, specific viral glycoproteins mediate binding to host cells and/or fusion of the viral envelope with the respective target membrane. For paramyxoviruses, e.g., Sendai virus, parainfluenza virus 5 (PIV5; formerly known as simian virus 5), Newcastle disease virus, and human respiratory syncytial virus, the following two proteins of the envelope are responsible for this early phase of virus infection: (i) a protein for binding of viruses to the host cell, such as the hemagglutinin-neuraminidase (HN) protein (13, 31); and (ii) the homotrimeric fusion protein (F). For priming of the F0 precursor for fusion, its cleavage into two covalently linked subunits, F1 and F2, is necessary. The F1+F2 protein, which is supposed to be in a metastable conformation, mediates the merger between the viral envelope and the host cell membrane by undergoing a conformational change. Typically, the F protein mediates membrane fusion only in the presence of the homotypic HN protein. The precise role of HN with respect to the conformational changes of F1+F2 is not known, and different models are discussed. Initially, it was suggested that binding of HN to cell receptors eventually leads to an interaction of HN and F, triggering the conformational change of F that mediates fusion. A more recent model proposes that HN-F complexes formed during virus maturation stabilize the metastable prefusion conformation of F1+F2 (21), while interaction of HN with cellular receptors destabilizes the complexes and the release of metastable F1+F2 causes refolding to the fusion conformation and, subsequently, to the stable postfusion conformation.
Sequences of the F protein which are thought to be essential for the fusion process, such as the hydrophobic N terminus, the fusion peptide, and the two 4-3 heptad repeat segments (HRC and HRN) of the membrane-anchored F1 subunit, are highly conserved among Paramyxoviridae (18). The HRC segment, located proximal to the transmembrane domain, is separated by a long stretch from the HRN segment, located adjacent to the fusion peptide. The current model of F1+F2-mediated fusion describes conformational changes of the F1+F2 trimer, at which the fusion peptide is supposed to insert into the target membrane (11, 23, 32, 33), while the high affinity between the two heptad repeat segments causes the contiguous placement of the HRC-containing helices in an antiparallel orientation, resulting in the formation of a thermodynamically stable six-helix bundle (1, 39). In consequence, the transmembrane domain and the fusion peptide are transposed in close association, enabling the convergence of the viral envelope and the target membrane (1, 22, 27, 29, 39). Indeed, soluble derivatives of HRN and HRC inhibit fusion, presumably by binding to the HRC and HRN domains of the F protein, respectively (12, 17, 19, 28, 29). The six-helix bundle is considered a structural element of the postfusion state of F1+F2 and turned out to be remarkably stable (39).
To unravel the three-dimensional (3D) structure of the F protein, studies using either X-ray crystallography or electron cryomicroscopy-based 3D reconstruction techniques have been undertaken in recent years (6, 20, 37, 38). These studies revealed the principal organization of the ectodomain as containing a head, a neck, and a distal stalk domain. However, looking closer at these structures, the stalk of the F ectodomain of human PIV3 (hPIV3) is surprisingly formed by a six-helix bundle motif (37) thought to correspond to the postfusion state, as the F protein was employed in its uncleaved form. Although this result may suggest that cleavage is not required for the formation of the six-helix bundle, it was surmised that the six-helix bundle was formed due to the absence of the transmembrane and/or cytoplasmic domain and that these domains would be essential for the stability of the prefusion state. Support for this assumption was given by recently published studies on the uncleaved F0 ectodomain of PIV5 (38). To preserve the soluble F0 protein in the prefusion state, an isoleucine zipper domain (GCNt) in the heptad repeat phase was appended to the HRC region, mimicking the putative function of the transmembrane domain. Indeed, the GCNt and HRC sequences formed a trimeric coiled-coil stalk, while the HRN sequence was expectedly absent from this region and was instead localized in the head region. These studies underscore the potential role of the transmembrane domain in the 3D structure of the F ectodomain.
Being aware of an important role of the transmembrane domain, we recently studied the 3D structure of the ectodomain of the full-length Sendai F1+F2 protein reconstituted into liposomes by electron cryomicroscopy (20). The appearance of the 3D structure of the Sendai F1+F2 ectodomain was not compatible with that of CGNt-PIV5 F0 assigned to the prefusion state (38) but was very similar to the crystal structure of the hPIV3 F0 ectodomain (37), which is supposed to correspond to the postfusion state due to the presence of the six-helix bundle. Yin et al. (37) argued that in our previous study the solubilization and reconstitution of the Sendai F1+F2 protein may have affected the conformation of the ectodomain. In order to address this concern, we aimed to study the 3D structure of the ectodomain of the full-length F proteins in intact Sendai and PIV5 virions in the cleaved F1+F2 state, using electron cryomicroscopy and 3D reconstruction techniques with single particles. In principle, we expected that the presence of HN might hamper the determination of the F1+F2 protein structure. This was indeed the case for Sendai virus. However, although it was also true for the majority of PIV5 virions, we found a sufficient number of well-resolved spike-like proteins protruding from the viral membrane to be suitable for single-particle analysis and 3D reconstruction. Since we selected the virus strain PIV5 W3A, for which it has been shown that the cleaved F protein also mediates cell-cell fusion in the absence of HN (16), we expected the conformation of the spikes to correspond to the prefusion state. Very surprisingly, elaborate structural analysis based on multivariate statistical analysis of >5,700 single spikes presented an F protein 3D structure equivalent to the postfusion state. This result is supported by the observation that heat treatment of the virus sample strongly enhanced the number of individual spikes but did not affect the morphology of the ectodomain. In addition, Sendai virus, which shows an undifferentiated surface corona in the fusion active state, produced individual spike proteins with postfusion state morphology upon heat treatment at 65°C, a temperature at which the fusion activity is known to be abolished. Hence, in context with earlier studies, we propose that the prefusion state cannot be preserved for those F1+F2 protein molecules which are not in complex with HN, even as integral constituents of intact nonfused virions.
MATERIALS AND METHODS
Cell culture and virus preparation.
Sendai virus (strain Z) was grown for 48 h in 10-day-old embryonated hen eggs as described previously (20). PIV5 (strain W3A) was kindly supplied by H. Klenk, Institute of Virology, Philipp-University Marburg. Monolayer cultures of MDBK cells (BioWhittaker, Europe, Verviers, Belgium) were grown to confluence in Dulbecco's modified Eagle medium (DMEM; Gibco Invitrogen Corp., Karlsruhe, Germany) supplemented with 5% (vol/vol) fetal calf serum, 2 mmol/liter glutamine, and antibiotics (100 IU of penicillin and 100 mg/liter of streptomycin) (Biochrome KG, Berlin, Germany) in tissue petri dishes (internal diameter, 15 cm; Falcon, Biosciences Labware). Cells were overlaid with 2 ml of PIV5-containing DMEM suspension (0.1 to 0.3 PFU/cell). After 1 hour of incubation at room temperature, 18 ml of DMEM with 2% (vol/vol) fetal calf serum was added, and cells were incubated at 37°C for 3 days. Cells were always kept at 5% CO2. Subsequently, after centrifugation at 1,000 × g and 4°C for 30 min, the supernatant was centrifuged at 100,000 × g and 4°C for 1.5 h. The virus-containing sediment was resuspended in a small volume of phosphate-buffered saline (PBS; pH 7.4) and centrifuged on a discontinuous saccharose gradient (60%, 40%, and 20% in 5 mM HEPES, 150 mM NaCl, 0.1 mM EDTA, pH 7.4) at 4°C for 16 h. The virus layer was resuspended in 3 volumes of PBS and centrifuged at 100,000 × g and 4°C for 1 hour. Viruses were stored in PBS at −78°C.
Fusion assay.
Virions were labeled with the fluorophore octadecylrhodamine B chloride R18 (Molecular Probes, Eugene, OR) as described previously (14, 20). Twenty microliters of R18-labeled PIV5 (1 mg virus protein/ml) was mixed with 50 μl of human erythrocyte ghosts (5 mg protein/ml) and incubated for 30 min at 4°C. Subsequently, the suspension was added to 1.93 ml of prewarmed PBS (pH 7.4, 37°C), and fusion was monitored and quantified by fluorescence dequenching of R18 (3), using an Aminco Bowman 2 fluorescence spectrometer with 560- and 590-nm excitation and emission wavelengths, respectively. At the end of measurements, 0.5% Triton X-100 was added to obtain maximal dequenching.
Electron microscopy.
Droplets of the sample (5 μl) were applied to hydrophilized carbon-covered microscopic copper grids (400-mesh), and supernatant fluid was removed with a filter paper until an ultrathin layer of the sample was obtained. Again, a droplet of contrasting material (1% phosphotungstic acid at pH 7.4) was added and blotted. A heavy-metal-stained layer of the protein assemblies was thus prepared and was subsequently plunged into liquid ethane in order to preserve the protein structure in its hydrated state.
A Gatan-626 specimen holder/cryotransfer system was used in a Tecnai F20 transmission electron microscope (FEI Company, OR) equipped with a field emission gun keeping the sample temperature at −180°C. Imaging was performed under low-dose conditions, using a primary magnification of ×51,064 at an accelerating voltage of 160 kV. The defocus value was chosen to be 1.8 μm.
Reconstruction of 3D structure.
Laser-optically checked micrographs were digitized using a Heidelberger Primescan drum scanner (Heidelberger Druckmaschinen AG, Heidelberg, Germany), with a nominal pixel resolution of 0.78 Å for the digitized images.
All image processing was performed using the Imagic-5 software package (Image Science GmbH, Berlin, Germany). Molecules were interactively selected and extracted from the digitized micrographs as 400- by 400-pixel fields. All fields were then contrast transfer function corrected following the protocol of Sander et al. (30). For computational efficiency, the resulting phase-corrected single images were interpolated to a 1.56-Å pixel size for all subsequent steps. As a first reference, a Gaussian-filtered rectangle was used for a prealignment of the spikes. For further alignment steps, the prealigned proteins were roughly contoured to mask neighboring proteins as well as the lower part of the stalk adherent to the viral membrane. In this way, it could be ensured that only the protein head region was considered for the alignment. Multivariate statistical analysis and hierarchical classification of aligned images were performed as described earlier (20). The 3D reconstruction (EMD-1421) was calculated based on the “angular reconstitution” approach (34).
Fourier shell correlation (35) of two different 3D reconstructions, each of which included half of the final class averages, was done to assess the resolution. For the 3D structure, the resolution obtained in the final 3D reconstruction was determined to be ∼15 Å, applying the 3σ threshold criterion (∼23 Å at a 0.5 criterion) and taking C3 point-group symmetry into account (24).
RESULTS
Fusion activity of PIV5 and Sendai virus.
It has already been shown that the F protein of PIV5 W3A is cleaved when PIV5 is grown in tissue culture (MBDK cells) (25, 26). Likewise, the F protein of egg-grown Sendai virus is known to be cleaved. Cleavage of F proteins has been confirmed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (data not shown).
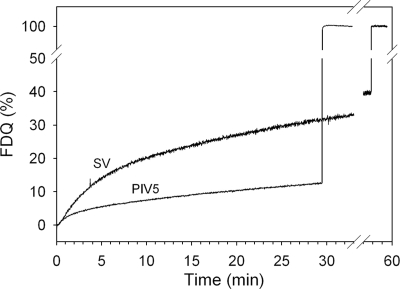
To assess the fusion activity of virions, viruses labeled with the lipid-like fluorescent probe R18 at a self-quenching concentration were incubated with human erythrocyte ghosts as a target at 37°C. As shown in Fig. 1, significant fusion was revealed from a relief of self-quenching due to redistribution of R18 to ghost membranes.
FIG. 1.
Fusion kinetics of intact PIV5 and Sendai virus (SV) with human erythrocyte ghosts at 37°C and pH 7.4. R18-labeled PIV5 was bound to ghosts for 30 min at 4°C. Subsequently, fusion was initiated (0 min) by transfer of virosome cell complexes to prewarmed buffer (37°C). Fusion is measured by the release of self-quenching of R18 caused by redistribution of the fluorescent lipid analogue to the ghost plasma membrane. Fluorescence dequenching (FDQ) was normalized to the fluorescence intensity obtained after the addition of 0.5% Triton X-100 at the end of the kinetics (infinite dilution of the analogue).
Electron microscopy and image analysis.
Samples were embedded in a matrix with a somewhat higher contrast than that of a conventional vitreous ice preparation (8). Despite the relatively high acceleration voltage of the microscope, the signal-to-noise ratio was improved and the beam sensitivity was reduced (8). This specimen preparation procedure was essentially the same as that described earlier for the 3D structure determination of influenza virus hemagglutinin (HA) (4). With this preparation technique, it has been shown by comparison of both the X-ray crystal structure and the electron microscopic reconstruction of HA that there is no significant structural difference within the limits of the achieved resolution of 10 Å. In order to meet objections against the use of a heavy metal stain, which might have caused alterations in the protein structure, we complemented our studies by “classic” electron cryomicroscopy measurements (no staining) and found no differences in the appearance of the viral protein layer. A similar in situ approach was applied very recently for determination of the structure of the S fusion protein of severe acute respiratory syndrome coronavirus (2).
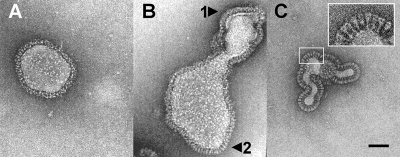
Typical electron micrographs of pleomorphic PIV5 virions are shown in Fig. 2. As shown, the surface structure of the virus envelopes is heterogeneous. Two different appearances of the surface were observed, with (i) a continuous layer (Fig. 2A and B, arrow 1), as observed exclusively for Sendai virus; or (ii) a continuous layer with additional individual spikes (Fig. 2B, arrow 2) or exclusively distinct spikes (Fig. 2C). These spikes appear morphologically very similar to mace-like analogues we recently observed for full-length cleaved Sendai F protein reconstituted in virosomes (20). The described structural phenomena were observable for fresh virus preparations investigated immediately after isolation and even for preparations examined after prolonged storage (up to 4 years at −78°C).
FIG. 2.
Electron micrographs of a cryo-negative stain preparation of native PIV5 virions (strain W3A). Intact fusion-active virions were embedded in a matrix of vitreous ice in the presence of 1% (wt/vol) phosphotungstic acid. The surface structure of the envelopes is heterogeneous: either a continuous layer with no identifiable ultrastructure (A and B, arrow 1) or distinct individual spikes (B, arrow 2, and C) are detectable. Bar, 500 Å.
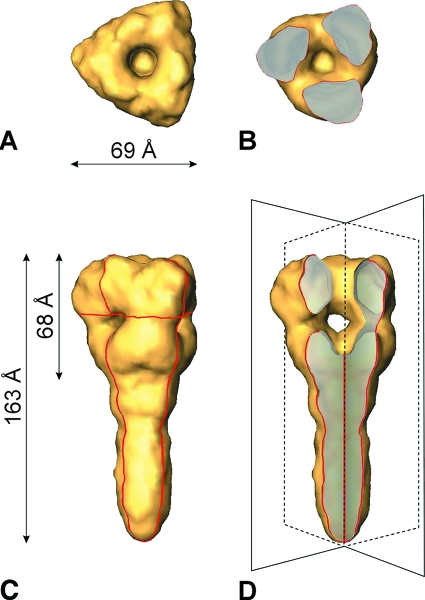
It is impossible to differentiate individual structural features on the dense corona-like surface layer, let alone to distinguish between F and HN proteins. In contrast, the less densely populated surface areas reveal distinct spikes. These virus-associated spikes were interactively selected, resulting in a data set of about 5,700 single particles. The 3D structure of PIV5 F1+F2 was reconstructed at a resolution of 15 Å (3σ criteria). In Fig. 3, a surface presentation of the obtained F1+F2 structure is shown. The length of the complete ectodomain is 163 Å. The distal head is 68 Å in length and 69 Å in width. The head is characterized by an axial channel and three radial channels. Both channels merge into a central domain of low density (central cavity).
FIG. 3.
Surface presentation of the 3D reconstruction of the ectodomain of the PIV5 F protein, determined from cryo-negative stain electron micrographs at a resolution of ∼15 Å. (A) Top view. (C) Side view. The viral membrane (not shown) is located at the lower end of the stalk region. (B) Top view of a horizontal section (the localization of the cross section is indicated by the horizontal red line in panel C). (D) Axial cutaway view (45°) of panel C to reveal the axial channel and the three radial channels merging into the central cavity. The imaginary cutting planes are drawn in fine black lines (compare to the red contours in panel C).
Surface structure of heat-inactivated virions.
Treatment of PIV5 at 65°C for 10 min is known to abolish the fusion activity of the F protein (9). Electron micrographs of heat-treated PIV5 virions revealed an enhanced number of individual spikes compared with untreated virus. Heat treatment of Sendai virus produced an almost quantitative yield of distinct individual spikes with a “postfusion” morphology comparable to that of untreated or heat-treated PIV5 virions (data not shown).
DISCUSSION
In recent years, several efforts have been made to determine the 3D structure of the F fusion protein of paramyxoviruses by X-ray crystallography. Investigations were based essentially on the use of the F0 protein, lacking both the transmembrane and the intraviral domain (6, 37). A more recent study used F0 (GCNt F0 protein of PIV5) elongated by an engineered coiled-coil domain replacing the transmembrane domain (38). The studies provided substantial progress in our understanding of both the prefusion and postfusion states of the F protein. However, it remained unclear to what extent the in vitro results would be transferable to the in situ situation. Therefore, it was challenging to directly study the F protein as an integral constituent of the viral membrane to (i) ensure a potential function of the transmembrane domain (38), (ii) determine the orientation of the fusion protein with respect to the viral membrane, and (iii) avoid any interfering influence by, e.g., detergent treatment or enzymatic cleavage.
Although we succeeded previously in determining the 3D structures of intact influenza virus HA (4) and of the intact Sendai virus F1+F2 protein (20) from rosette-like assemblies by using the single-particle approach, methodical transfer using native viruses failed due to the superposition of the very dense arrangement of spike proteins in the viral membrane. Likewise, in this study, we found densely packed corona-like surface structures for Sendai and PIV5 virions which did not allow for the resolution of individual structural entities. Since gel electrophoresis confirmed the presence of HN and F proteins, these coronas most probably correspond to densely populated domains of both proteins, which might include postulated (21) HN-F complexes. However, in the case of PIV5 (but not Sendai virus), we also found far less densely populated virus surface regions with prominent, structurally well-defined spikes suitable for single-particle analysis (Fig. 2). Although the pleomorphology of PIV5 virions complicates the selection of randomly oriented spikes, laborious data collection provided a sufficient number (5,700) of molecules for a 3D reconstruction. On this basis, we succeeded in determining the in situ 3D structure of the ectodomain of the full-length cleaved F1+F2 protein of native PIV5 virions not in complex with HN at a resolution level of 15 Å.
Comparison of F1+F2 proteins of PIV5 and Sendai virus by electron cryomicroscopy.
We previously reported the structure of the ectodomain of Sendai virus F1+F2 from electron cryomicrographs of reconstituted virions (lacking HN) at a resolution of ∼16 Å (20). The shape and size of the ectodomain of the full-length Sendai virus F1+F2 protein are very similar to those of the maze-like structured PIV5 F1+F2 protein presented here. Features such as the three wing-like protrusions, the axial and radial channels, and the low-density cavity are typical features for the distal part of both structures. Likewise, the stalks of both proteins are similar in size. These similarities suggest that the detergent solubilization and reconstitution of the Sendai virus F1+F2 protein did not affect the conformation within the frame of the resolution achieved.
Comparison of crystal structures and electron cryomicrograph structure of PIV5 F protein.
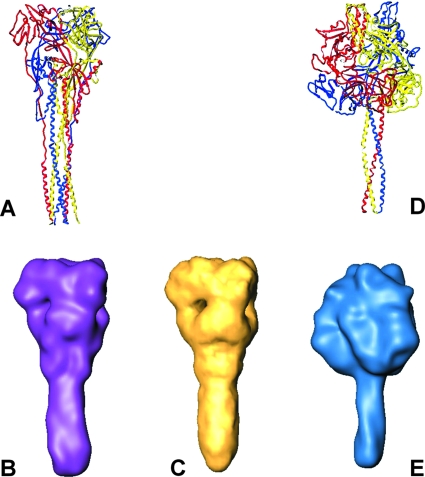
In Fig. 4, the crystal structures of the mushroom-like ectodomain of the uncleaved F0 protein of PIV5 (38) and of the maze-like hPIV3 F0 protein (37) are shown. While the hPIV3 F0 protein was expressed without the transmembrane and intraviral domains, the transmembrane domain of the PIV5 F0 protein was replaced by a coiled-coil domain (GCNt) (see the introduction). The latter engineering obviously had a significant impact on the overall structure of the protein. The crystal structure of the GCNt F0 protein of PIV5 revealed the threefold helical motif in the stalk which has been attributed to the prefusion state. It had been intended that the GCNt domain would mimic the role of the missing transmembrane domain of the PIV5 F0 ectodomain, as the latter domain has been proposed to be essential and sufficient to stabilize the prefusion conformation of the F0 protein (38). In the case of hPIV3 F0, the postfusion conformation was found, probably due to the removal of the transmembrane domain.
FIG. 4.
Comparison of X-ray and electron cryomicroscopy structures of paramyxovirus F proteins. (A and B) hPIV3 “postfusion” X-ray structure (Protein Data Bank entry 1ZTM), Gaussian filtered at 15-Å resolution (surface presentation is shown in purple). (C) PIV5 in situ structure at 15-Å resolution, reconstructed from electron cryomicroscopy data. (D and E) PIV5 “prefusion” X-ray structure (Protein Data Bank entry 2B9B), Gaussian filtered at 15-Å resolution (blue).
In order to enable a direct comparison of all structures, namely, those of hPIV3 F0, GCNt F0, and PIV5 F1+F2 protein, at the resolution level limited by the electron cryomicroscopy structure in the last case, we applied a Gaussian filter to the crystal data, adjusting to a comparable resolution level of 15 Å for all relevant protein structures.
A comparison of the isosurface of the PIV5 F1+F2 protein determined in our study with that of the GCNt F0 protein clearly shows significant differences in the shapes of the ectodomains and the sizes of the heads (Fig. 4). While the GCNt F0 protein is characterized by a prominent mushroom-like head and a comparatively slim stalk (diameter of 23 Å), the reconstructed head of the F1+F2 ectodomain adopts a maze-like shape with a broad stalk (diameter of nearly 30 Å). It is therefore reasonable to assume that the two structures of the PIV5 F protein differ significantly even at the comparatively low resolution level and thereby very likely reflect two different conformational states of the F protein.
Remarkably, while they are definitely distinct from the CGNt F0 protein structure, the morphologies of the PIV5 F1+F2 and hPIV3 F0 ectodomains are very similar (Fig. 4) (see Results). Interestingly, the diameter of the stalk of the reconstructed structure of PIV5 F1+F2 is only slightly different from that of Sendai virus F1+F2, which we reconstructed previously (20). Although the diameters of the six-helix bundles of PIV5 (1) and human respiratory syncytial virus (39) differed only marginally from our earlier reconstruction of Sendai virus F protein at 16-Å resolution (20), we could not unambiguously exclude at that time that the cleaved Sendai virus F protein would harbor a six-helix bundle. However, considering the significantly smaller diameter of the trimeric bundle of the GCNt F0 protein, we are now confident that the stalk of the reconstructed Sendai virus F protein (20) harbors a six-helix bundle.
Taken all together, the morphologies and dimensions of the head and stalk of the Sendai and PIV5 F1+F2 proteins coincide very well with the structure of hPIV3, which undoubtedly accommodates the postfusion six-helix bundle (see above). While it was assumed for a long time that the formation of the six-helix bundle occurs upstream of the fusion event to take viral and target membrane into close apposition (5, 36) it is now well accepted that the free energy release during the refolding into the six-helix bundle is used to induce fusion (1, 22). That means that the structural motif of the six-helix bundle is an indication of the postfusion state and the loss of fusion activity.
Hence, the structural analysis of our electron cryomicroscopy structure strongly implicates that the selected PIV5 F1+F2 protein adopts a postfusion rather than a prefusion state. Now, a similar conclusion can be drawn from our previously reconstructed structure of reconstituted Sendai F1+F2 protein (20). The assumption that F1+F2 of PIV5 is in a postfusion state is supported by the observation that heat treatment of the virus sample strongly enhanced the number of individual spikes but did not affect the morphology of the ectodomain. We rule out that the postfusion state is a specific consequence of the virus preparation procedure. Growth and isolation of virus were followed by a standard procedure which does not involve any step which may interfere with protein stability. If so, we would expect the same consequence for Sendai virus. However, the outer surface of Sendai virus appears structurally undifferentiated in the fusion active state. Interestingly, heat treatment of Sendai virus also produced individual spike proteins with postfusion state morphology. It is known that such a treatment of F1+F2 protein leads to inactivation of the fusion properties and thus promotes the formation of the postfusion state (7).
But why and how do the F1+F2 protein of PIV5 and the reconstituted F1+F2 protein of Sendai virus studied transform into the postfusion state? Yin et al. (38) proposed that the transmembrane and intraviral domains are essential for maintaining the prefusion conformation. However, our study using in situ full-length PIV5 F1+F2 implicates that these domains are obviously not sufficient for maintaining the prefusion state of the cleaved F protein. It is conceivable that the kinetic barrier of the transition from the prefusion to the postfusion state becomes comparatively low after cleavage. Cleavage of the F0 precursor might be the trigger to initiate the fusion and the eventual inactivation cascade even in the absence of the target membrane. Thus, the F1+F2 protein has to be stabilized to keep it in a prefusion state in order to avoid inactivation before infection of a cell. We surmise that the interaction with HN might be of importance for the stabilization of the prefusion state of the F1+F2 protein, as has recently been suggested by McGinnes and Morrison (21). The authors reported that upon attachment of the HN protein to cellular receptors the HN-(F1+F2) complexes dissociate and the F1+F2 protein undergoes conformational changes leading to fusion and eventually to formation of the postfusion conformation. Such a destabilizing effect due to HN-F protein complex disorganisation would be consistent with our results. Individual F1+F2 spikes used for single particle analysis and 3D reconstruction turned out not to be in complex with the HN protein (as they would be structurally distinguishable in individual complexes) and consequently adopted the postfusion state. While F1+F2 proteins in the postfusion state cannot be responsible for the observed fusion activity of PIV5, very likely fusion between PIV5 envelope and target membranes of e.g., erythrocyte ghosts has been mediated by cleaved F proteins in complex with HN. Although it has been reported for the PIV5 W3A strain used here that the F protein enables cell-cell fusion even in the absence of HN (15, 26), our data imply that HN plays at least an essential role for the stabilization of the prefusion state of the cleaved F1+F2 form in intact virions. This finding is also consistent with previous results of Russell et al. (29) showing that coexpression of HN and F of PIV5 W3A clearly promotes F-mediated cell-cell fusion.
Connolly et al. (7) have shown that after cleavage of the GCNt F0 protein the conformation of the uncleaved prefusion form is essentially preserved, which would not support the view that cleavage of the F0 protein in the absence of HN would automatically trigger the postfusion conformation. However, as noticed by the authors (7) the GCNt trimerisation domain may affect F activation properties. Thus, the GCNt domain may probably enhance the stability of the ectodomain if compared with the wild-type protein.
The transition from the pre to the postfusion state appears to be a thermodynamically favored inherent folding pathway of F1+F2. This is strongly supported by the observed heat driven changes which do not lead to an unordered state but instead to a structurally defined conformation (postfusion state). However, the refolding of virus membrane embedded F1+F2 into a postfusion state in the absence of a target membrane cannot be understood on the basis of current models as already pointed out previously by others (10, 27). According to these models an early step of the conformational change would be the association of the HRN peptides of a F1+F2 trimer. Together with the stalk harbored HRC peptides, the ectodomain forms an extended rod which would expose the fusion peptides away from the viral membrane. To adopt the postfusion state while still embedded in the viral membrane would require that both HRN and HRC trimers disassemble in order to form a six helix bundle, where HRN peptides eventually stand in the central antiparallel position with respect to the outer HRC helices. While there is clear evidence for a stable six-helix bundle, there is no evidence for such an assembly-disassembly process. In particular, it is hard to imagine what would be the driving forces in such a process. Further detailed studies are required to explore which alternative pathway may exist circumventing the assumption of an assembly-disassembly process which could also be of relevance for structural intermediates mediating fusion.
In summary, our study shows that the in situ 3D structure of the ectodomain of spike proteins embedded in intact envelope viruses such as the F protein from paramyxoviruses can be determined by electron microscopy. An essential conclusion to be drawn is that single F proteins not in complex with HN protein adopt the postfusion state pointing to the relevance of HN-F interaction for the stabilization of the prefusion conformation of the F protein. The characterization of the postfusion conformation of F1+F2 in intact virions may certainly not be the priority objective in order to understand the conformational changes driving membrane fusion. However, the presented results provide not only insights into the in situ stability of the prefusion state of the F1+F2 protein but also valuable clues on the refolding pathway toward the postfusion state. The finding that membrane anchoring is not sufficient to preserve the prefusion state of the F1+F2 protein has general implications for studies on the structure of F1+F2 in that the prefusion state of native full-length F1+F2 cannot be characterized on isolated proteins but always require the complex with HN. This does however not preclude that HN could be substituted by appropriate ligands.
Acknowledgments
We are indebted to Bärbel Hillebrecht for virus growing. R. Schmidt and M. Schatz (IMAGE Science GmbH, Berlin, Germany) are gratefully acknowledged for their continuous technical support.
We are grateful to the Deutsche Forschungsgemeinschaft for the Philips Tecnai F20 TEM and generous support to C.B. (BO 1000/4) and A.H. (HE 1928/4, SFB740/1-2007). B.B. was a recipient of the Heinrich-Böll-Stiftung. A supplementary benefit to A.H. and C.B. through SFB765 is gratefully acknowledged.
Footnotes
Published ahead of print on 23 January 2008.
REFERENCES
- 1.Baker, K. A., R. E. Dutch, R. A. Lamb, and T. S. Jardetzky. 1999. Structural basis for paramyxovirus-mediated membrane fusion. Mol. Cell 3309-319. [DOI] [PubMed] [Google Scholar]
- 2.Beniac, D. R., A. Andonov, E. Grudeski, and T. F. Booth. 2006. Architecture of the SARS coronavirus prefusion spike. Nat. Struct. Mol. Biol. 13751-752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blumenthal, R., A. Bali-Puri, A. Walter, D. Covell, and O. Eidelman. 1987. pH-dependent fusion of vesicular stomatitis virus with Vero cells. Measurement by dequenching of octadecyl rhodamine fluorescence. J. Biol. Chem. 26213614-13619. [PubMed] [Google Scholar]
- 4.Böttcher, C., K. Ludwig, A. Herrmann, M. van Heel, and H. Stark. 1999. Structure of influenza hemagglutinin at neutral and at fusogenic pH by electron cryo-microscopy. FEBS Lett. 463255-259. [DOI] [PubMed] [Google Scholar]
- 5.Chan, D. C., and P. S. Kim. 1998. HIV entry and its inhibition. Cell 93681-684. [DOI] [PubMed] [Google Scholar]
- 6.Chen, L., J. J. Gorman, J. McKimm-Breschkin, L. J. Lawrence, P. A. Tulloch, B. J. Smith, P. M. Colman, and M. C. Lawrence. 2001. The structure of the fusion glycoprotein of Newcastle disease virus suggests a novel paradigm for the molecular mechanism of membrane fusion. Structure 9255-266. [DOI] [PubMed] [Google Scholar]
- 7.Connolly, S. A., G. P. Leser, H.-S. Yin, T. S. Jardetzky, and R. A. Lamb. 2006. Refolding of a paramyxovirus F protein from prefusion to postfusion conformations observed by liposome binding and electron microscopy. Proc. Natl. Acad. Sci. USA 10317903-17908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Carlo, S., C. El-Bez, C. Alvarez-Rua, J. Borge, and J. Dubochet. 2002. Cryo-negative staining reduces electron-beam sensitivity of vitrified biological particles. J. Struct. Biol. 138216-226. [DOI] [PubMed] [Google Scholar]
- 9.Dutch, R. E., R. N. Hagglund, M. A. Nagel, R. G. Paterson, and R. A. Lamb. 2001. Paramyxovirus fusion (F) protein: a conformational change on cleavage activation. Virology 281138-150. [DOI] [PubMed] [Google Scholar]
- 10.Gaudin, Y., R. W. Ruigrok, and J. Brunner. 1995. Low-pH induced conformational changes in viral fusion proteins: implications for the fusion mechanism. J. Gen. Virol. 761541-1556. [DOI] [PubMed] [Google Scholar]
- 11.Gething, M. J., J. M. White, and M. D. Waterfield. 1978. Purification of the fusion protein of Sendai virus: analysis of the NH2-terminal sequence generated during precursor activation. Proc. Natl. Acad. Sci. USA 752737-2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ghosh, J. K., and Y. Shai. 1999. Direct evidence that the N-terminal heptad repeat of Sendai virus fusion protein participates in membrane fusion. J. Mol. Biol. 292531-546. [DOI] [PubMed] [Google Scholar]
- 13.Haywood, A. M. 1974. Characteristics of Sendai virus receptors in a model membrane. J. Mol. Biol. 83427-436. [DOI] [PubMed] [Google Scholar]
- 14.Hoekstra, D., T. de Boer, K. Klappe, and J. Wilschut. 1984. Fluorescence method for measuring the kinetics of fusion between biological membranes. Biochemistry 235675-5681. [DOI] [PubMed] [Google Scholar]
- 15.Horvath, C. M., R. G. Paterson, M. A. Shaughnessy, R. Wood, and R. A. Lamb. 1992. Biological activity of paramyxovirus fusion proteins: factors influencing formation of syncytia. J. Virol. 664564-4569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ito, M., M. Nisho, M. Kawano, S. Kusagawa, H. Komada, Y. Ito, and M. Tsurudome. 1997. Role of a single amino acid at the amino terminus of the simian virus F2 subunit in syncytium formation. J. Virol. 719855-9858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Joshi, S. B., E. E. Dutch, and R. A. Lamb. 1998. A core trimer of the paramyxovirus fusion protein: parallels to influenza virus hemagglutinin and HIV-1 gp41. Virology 24820-34. [DOI] [PubMed] [Google Scholar]
- 18.Lamb, R. A. 1993. Paramyxovirus fusion: a hypothesis for changes. Virology 1971-11. [DOI] [PubMed] [Google Scholar]
- 19.Lamb, R. A., and D. Kolakofsky. 2001. Paramyxoviridae: the viruses and their replication, p. 1205-1242. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology. Lippincott Williams & Wilkins, Philadelphia, PA.
- 20.Ludwig, K., B. Baljinnyam, A. Herrmann, and C. Böttcher. 2003. The 3D structure of the fusion primed Sendai F-protein determined by electron cryomicroscopy. EMBO J. 223761-3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McGinnes, L. W., and T. G. Morrison. 2006. Inhibition of receptor binding stabilizes Newcastle disease virus HN and F protein-containing complexes. J. Virol. 802894-2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Melikyan, G. B., R. M. Markosyan, H. Hemmati, M. K. Delmedico, D. M. Lambert, and F. S. Cohen. 2000. Evidence that the transition of HIV-1 gp41 into a six-helix bundle, not the bundle configuration, induces membrane fusion. J. Cell Biol. 151413-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Novick, S. L., and D. Hoekstra. 1988. Membrane penetration of Sendai virus glycoproteins during the early stages of fusion with liposomes as determined by hydrophobic photoaffinity labeling. Proc. Natl. Acad. Sci. USA 857433-7437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Orlova, E. V., P. Dube, J. R. Harris, E. Beckmann, F. Zemlin, J. Markl, and M. van Heel. 1997. Structure of keyhole limpet hemocyanin type 1 (KLH1) at 15Å resolution by electron cryomicroscopy and angular reconstitution. J. Mol. Biol. 271417-437. [DOI] [PubMed] [Google Scholar]
- 25.Paterson, R. G., T. J. R. Harris, and R. A. Lamb. 1984. Fusion protein of the paramyxovirus simian virus 5: nucleotide sequence of mRNA predicts a highly hydrophobic glycoprotein. Proc. Natl. Acad. Sci. USA 816706-6710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paterson, R. G., S. W. Hiebert, and R. A. Lamb. 1985. Expression at the cell surface of biologically active fusion and hemagglutinin/neuraminidase proteins of the paramyxovirus simian virus 5 from cloned DNA. Proc. Natl. Acad. Sci. USA 827520-7524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peisajovich, S. G., O. Samuel, and Y. Shai. 2000. Paramyxovirus F1 protein has two fusion peptides: implications for the mechanism of membrane fusion. J. Mol. Biol. 2961353-1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rapaport, D., M. Ovadia, and Y. Shai. 1995. A synthetic peptide corresponding to a conserved heptad repeat domain is a potent inhibitor of Sendai virus-cell fusion: an emerging similarity with functional domains of other viruses. EMBO J. 145524-5531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Russell, C. J., T. S. Jardetzky, and R. A. Lamb. 2001. Membrane fusion machines of paramyxoviruses: capture of intermediates of fusion. EMBO J. 204024-4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sander, B., M. M. Golas, and H. Stark. 2003. Automatic CTF correction for single particles based upon multivariate statistical analysis of individual power spectra. J. Struct. Biol. 142392-401. [DOI] [PubMed] [Google Scholar]
- 31.Scheid, A., L. A. Caliguiri, R. W. Compans, and P. W. Choppin. 1972. Isolation of paramyxovirus glycoproteins. Association of hemagglutinating and neuraminidase activities with larger SV5 glycoprotein. Virology 50640-652. [DOI] [PubMed] [Google Scholar]
- 32.Scheid, A., and P. W. Choppin. 1974. Identification of the biological activities of paramyxovirus glycoproteins. Activation of cell fusion, hemolysis and infectivity by proteolytic cleavage of an inactive precursor protein of Sendai virus. Virology 50475-490. [DOI] [PubMed] [Google Scholar]
- 33.Scheid, A., and P. W. Choppin. 1977. Two disulfide-linked polypeptide chains constitute the active F protein of paramyxoviruses. Virology 8054-60. [DOI] [PubMed] [Google Scholar]
- 34.van Heel, M. 1987. Angular reconstitution: a posteriori assignment of projection directions for 3D reconstruction. Ultramicroscopy 21111-124. [DOI] [PubMed] [Google Scholar]
- 35.van Heel, M., and G. Harauz. 1986. Resolution criteria for three dimensional reconstructions. Optik 73119-122. [Google Scholar]
- 36.Weissenhorn, W., A. Dessen, S. C. Harrison, J. J. Skehel, and D. C. Wiley. 1997. Atomic structure of the ectodomain from HIV-1 gp41. Nature 387426-430. [DOI] [PubMed] [Google Scholar]
- 37.Yin, H.-S., R. G. Paterson, X. Wen, R. A. Lamb, and T. S. Jardetzky. 2005. Structure of the uncleaved ectodomain of the paramyxovirus (hPIV3) fusion protein. Proc. Natl. Acad. Sci. USA 1029288-9293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yin, H.-S., X. Wen, R. G. Paterson, R. A. Lamb, and T. S. Jardetzky. 2006. Structure of the parainfluenza virus 5 F protein in its metastable, prefusion conformation. Nature 43938-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao, X., M. Singh, V. N. Malashkevich, and P. S. Kim. 2000. Structural characterization of the human respiratory syncytial virus fusion protein core. Proc. Natl. Acad. Sci. USA 9714172-14177. [DOI] [PMC free article] [PubMed] [Google Scholar]