Abstract
Human immunodeficiency virus type 1 (HIV-1) gene expression is controlled by a complex interplay between viral and host factors. We have previously shown that interferon-regulatory factor 1 (IRF-1) is stimulated early after HIV-1 infection and regulates promoter transcriptional activity even in the absence of the viral transactivator Tat. In this work we demonstrate that IRF-1 is also required for full NF-κB transcriptional activity. We provide evidence that IRF-1 and NF-κB form a functional complex at the long terminal repeat (LTR) κB sites, which is abolished by specific mutations in the two adjacent κB sites in the enhancer region. Silencing IRF-1 with small interfering RNA resulted in impaired NF-κB-mediated transcriptional activity and in repressed HIV-1 transcription early in de novo-infected T cells. These data indicate that in early phases of HIV-1 infection or during virus reactivation from latency, when the viral transactivator is absent or present at very low levels, IRF-1 is an additional component of the p50/p65 heterodimer binding the LTR enhancer, absolutely required for efficient HIV-1 replication.
Human immunodeficiency virus (HIV) replication is controlled mainly at the transcriptional level and depends upon a complex interaction between viral and cellular regulatory proteins acting on the viral promoter, the long terminal repeats (LTR). Among cellular factors, both NF-κB and interferon-regulatory factor 1 (IRF-1) have been shown to regulate LTR-driven transcription (12, 37).
The IRF family of transcription factors plays a key role in gene regulation by interferons, in viral infection, and in several immunological and growth-related cellular functions (18, 38). To date nine cellular members of this family have been identified based on a unique helix-turn-helix DNA binding motif located at the N terminus, which is responsible for binding to the interferon-stimulated responsive element. The less conserved C-terminal region acts as a regulatory domain and classifies IRFs into two groups: those that activate (IRF-3, IRF-7, and IRF-9/ISGF-3γ), and those that activate or repress (IRF-1, IRF-2, IRF-4/LSIRF/Pip, IRF-5, and IRF-8/ICSBP) gene transcription, depending on the target gene. IRFs, indeed, interact with each other and with other families of transcription factors, modifying both their sequence-specific binding activity and the formation of transcription initiation complexes (38).
Among IRFs, IRF-1 can associate with members of the NF-κB/Rel family, generating complexes that synergistically activate transcription. Accordingly, adjacent or overlapping binding sites for IRF-1 and NF-κB have been identified in the regulatory regions of many genes, including those for inducible nitric oxide synthase, interleukin-15 (IL-15), major histocompatibility complex class I, and vascular cell adhesion molecule I (4, 11, 30, 36).
The mammalian NF-κB/Rel proteins are a family of ubiquitous transcription factors that are activated in response to inflammatory stimuli and environmental stressors and involved in the innate and adaptive immune responses. The five family members, C-Rel, Rel-A (p65), Rel B, NF-κB1 (p50 and its precursor p105), and NF-κB2 (p52 and its precursor p100), exist only as homo- or heterodimers in resting cells, where they are associated with the IκB family of inhibitory proteins that act as chaperons to prevent NF-κB DNA binding (19). The sudden activation of NF-κB by a range of inducers determines the release and the degradation of IκBα after its phosphorylation, thereby allowing NF-κB to translocate into the nucleus and to activate transcription of target genes (19). NF-κB dimers bind a family of 9 to 11 DNA base pairs known as the κB binding site, and each target gene requires a specific combination of NF-κB proteins for activation (17).
The LTR enhancer region of HIV type 1 (HIV-1) subtype B contains two adjacent high-affinity binding site for NF-κB (28) that are critical for LTR promoter activity and important for optimal HIV-1 replication (2, 5, 10, 12, 29, 31). In activated T cells the predominant complex binding to the HIV-1 LTR enhancer is the heterodimer p50/p65 (2). We have previously shown that IRF-1 is stimulated early after HIV-1 infection and activates LTR transcription irrespective of the presence of Tat (6, 27, 37), suggesting that IRF-1, like NF-κB, has a key role in the early phases of viral replication and during reactivation from latency when the viral transactivator is absent or present at very low levels.
Accordingly, inflammatory mediators and cytokines, including IL-1, IL-6, and tumor necrosis factor alpha (TNF-α), secreted during immune responsiveness to infections that stimulate HIV-1 gene expression and replication, are also potent inducers of NF-κB and IRF-1 (15, 16, 22, 29).
Despite the evidence that NF-κB and IRF-1 are important regulators of the inducible expression of HIV-1, it remains unclear whether these two transcription factors function independently or cooperatively to regulate HIV-1 gene expression.
In the present study we investigated the interactions between NF-κB and IRF-1 in HIV-1 LTR transcription regulation and showed that IRF-1 binds to the enhancer κB sites in combination with NF-κB p50/p65 heterodimer and is required for full NF-κB transcriptional activity at the HIV-1 LTR enhancer. Silencing IRF-1 by small interfering RNA (siRNA) resulted, indeed, in impaired NF-κB-mediated transcriptional activation of the HIV-1 LTR early after infection of T cells. Taken together our data are consistent with a model of evolutionarily conserved physical interactions between NF-κB and IRF-1 in the regulation of cellular and viral genes and stress the critical role of IRF-1 in HIV-1 replication in response to viral infection and T-cell activation.
MATERIALS AND METHODS
Cell cultures, and treatments.
Jurkat and 1G5 T cells were grown in RPMI 1640, and HEK293 and HLM-1 cells were grown in Dulbecco modified Eagle medium (DMEM) (Bio-Whittaker, Cambrex Bio Science, Verviers, Belgium); all cell line media contained 10% fetal calf serum (FCS) and antibiotics. 1G5 Jurkat T cells with an integrated LTR and HLM-1 cells harboring an integrated Tat-defective provirus have been described previously (1, 35). Recombinant TNF-α (Pepro Tech EC Ltd., London) was used at 10 ng/ml.
Plasmids.
CMVBL and CMVBL IRF-1 (26) and CMVBL p50 and CMVBL p65 (25) have been described previously, HIV-1 LTR κB-enhancer-luc was obtained from BH10-LD1 (24) by PCR amplification inserting the KpnI and SmaI restriction sites and subcloned into the pGL3basic vector (Invitrogen Corp., San Diego CA) digested with KpnI and SmaI restriction enzymes. HIV-1 LTR κB enhancer-pmut-luc, HIV-1 LTR κB enhancer-dmut-luc, and HIV-1 LTR κB enhancer-pdmut-luc were obtained by site-directed mutagenesis using the oligonucleotides κB LTR-pmut (5′-GGACTTTCCGCTGGGGACTATCCAGGGAGGCGTGGCC-3′), κB LTR-dmut (5′-GCTTGCTACAAGGGACTATCCGCTGGGGACTTTCC-3′), and κB LTR-pdmut (5′-GCTACAAGGGACTATCCGCTGGGGACTATCCAGGGAGGCG-3′) and the QuikChange TM XL site-directed mutagenesis kit (Stratagene, La Jolla, CA) according to the manufacturer's protocol. pRLβactin was a gift of T. Matsuyama, Nagasaki University School of Medicine, Nagasaki, Japan.
Transient transfection and reporter gene assay.
Transient-transfection experiments were performed using FuGENE 6 transfection reagent (Roche Diagnostics Mannheim, Germany), PolyFect transfection reagent (Qiagen GmbH, Hilden, Germany), or the calcium phosphate transfection system (Life Technologies, Invitrogen Corporation, Carlsbad, CA) according to the manufacturer's protocol. Amounts of transfected DNA were normalized by using CMVBL empty vector. Reagents from Promega (Promega Corporation Madison, WI) were used to assay extracts for dual-luciferase activity in a Lumat LB9501 luminometer (E&G Berthold, Bad Wildbad, Germany).
Western blot analysis.
Aliquots of nuclear or whole-cell extracts prepared as previously described (36, 37) were separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to a nitrocellulose membrane (Amersham Pharmacia Biotech, United Kingdom). Western blot analysis was performed by probing blots with polyclonal antibodies against IRF-1, NF-κB p50, NF-κB p65, TFIIH p89, and actin (Santa Cruz Biotechnology Inc, Santa Cruz, CA) and then with anti-rabbit, -goat, or -mouse horseradish peroxidase-coupled secondary antibody (1:2,000 dilution) (Calbiochem, brand of EMD Biosciences, Inc, an affiliate of Merck KGaA, Darmstadt, Germany). The detection reaction was performed using the ECL Western blotting detection reagents (Amersham Pharmacia Biotech).
Coimmunoprecipitation and immunoblot analysis.
Nuclear extracts from Jurkat T cells treated with TNF-α (10 ng/μl) were incubated overnight with 1 μg polyclonal anti-IRF-1 antibody (Santa Cruz Biotechnology) and then incubated with Ultralink immobilized protein A/G-Sepharose (Pierce Biotechnology, Rockford, United Kingdom) for 2 h at room temperature. After extensive washing, immunoprecipitates were eluted by boiling the beads for 5 min in 1× SDS sample buffer. Eluted proteins were separated by SDS-PAGE and subjected to Western blotting with polyclonal antibodies against IRF-1, NF-κB p65, and TFIIH p89 (Santa Cruz Biotechnology).
EMSA.
Aliquots of whole or nuclear cell extracts were subjected to electrophoretic mobility shift assay (EMSA) as previously described (37) by using double-stranded 32P-labeled oligonucleotides corresponding to the sequences κB LTR WT (5′-ACAAAGGGACTTTCCGCTGGGGACTTTCCAGGGAG-3′), κB LTR pmut (5′-ACAAAGGGACTTTCCGCTGGGGACTATCCAGGGAG-3′), κB LTR dmut (5′-ACAAAGGGACTATCCGCTGGGGACTTTCCAGGGAG-3′), κB LTR pdmut (5′-ACAAAGGGACTATCCGCTGGGGACTATCCAGGGAG-3′), and immunoglobulin G-κB (5′ AGTTGAGGGGACTTTCCCAGGC 3′).
Supershift analysis was performed by incubating the reaction mixture with 2 μg of polyclonal antibodies against IRF-1, NF-κB p50, and NF-κB p65 (Santa Cruz Biotechnology).
HIV virus stock preparation, infection, viral DNA, and p24 detection.
Replication-competent T-cell-tropic virus was generated by calcium phosphate-mediated transient transfection of HEK293 cells with the pHXBc2 molecular clone. Virus-containing supernatant was filtered, frozen in aliquots at −70°C, and titrated on C8166 cells. Jurkat T cells were inoculated with HIV-1/HXBc2 at multiplicity of infection of 0.05 50% tissue culture infective dose (TCID50) per cell as previously described (37).
Virus production in HLM-1 cells was then monitored by measuring the levels of p24 in the culture supernatants with the p24/27 core antigen assay kit (Innogenetics NV, Gent, Belgium) as specified by the manufacturer.
To assess viral DNA amounts in infected cells, virus was produced in human peripheral blood mononuclear cells, harvested before the occurrence of cytopathic effects, and treated with DNase in order to minimize contamination of HIV stock with plasmidic or cellular DNA. Briefly, laboratory-adapted HXBc2 virus was grown in mitogen-activated peripheral blood mononuclear cells with 1 μg/ml phytohemagglutinin in RPMI 1420, 10% FCS, and IL-2 (10 U/ml) (Boehringer, Mannheim, Germany). Supernatants were filtered (0.2-μm filter; Schleicher and Schuell, Keene, NH) and stored at −70°C. The virus stock was at 5 × 105 TCID50/ml. Virus was treated with 50 U/ml RNase-free DNase (Boehringer Mannheim) in 10 mM MgCl2 at 37°C for 30 min.
Jurkat T cells (5 × 106) were inoculated with 0.05 TCID50 of HXBc2 virus/cell at 37°C for 5 and 10 h. Cells were extensively washed, and total cell DNA was extracted with a QIAmp DNA extraction minikit (Qiagen). The HIV-1 Gag full reverse transcript was quantified by real-time PCR with an ABI7700 instrument (Applied Biosystems, Foster City, CA). Primers used for HIV-1 Gag amplification (Applied Biosystems) were used at 100 μM. For each experiment, a standard curve of the amplicon being measured was run in duplicate, ranging from 10 to 103 copies, plus a no-template control. Data are expressed as the number of viral DNA copies/μg DNA.
SeV infection.
Jurkat T cells (3 × 106) expressing control siRNA or IRF-1 siRNA were infected with 500 hemagglutinating units of Sendai virus (SeV) and placed for 1 h in serum-free medium. Cells were than washed and placed in growth medium containing 10% fetal bovine serum. After 10 h, cells were lysed, RNA was extracted, and the nucleoprotein was quantified by real-time PCR as described below.
siRNA design and vector construction.
IRF-1 siRNA oligonucleotide target sequences were selected by using the siRNA design tool Xeragox (Qiagen Inc.). The selected target sequences were tested so as not to match any known human or viral gene (other than human IRF-1) sequences by using NCBI nucleotide BLAST at http://www.ncbi.nlm.nih.gov/BLAST/. Retroviral vectors for siRNA expression were generated using the BD knockout RNA interference (RNAi) system (RNAi ready pSIREN-RetroQ system; BD Biosciences, Clontech, Palo Alto, CA). Briefly, two complementary oligonucleotides specific for IRF-1siRNA, For (5′-GATCCGGGATGCCTGTTTGTTCCGGTTCAAGAGACCGGAACAAACAGGCATCCTTTTTTCTAGAG-3′) and Rev (5′-AATTCTCTAGAAAAAAGGATGCCTGTTTGTTCCGGTCTCTTGAACCGGAACAAACAGGCATCCCG-3′), were designed and used together with control siRNA and Luc siRNA (negative control and luciferase siRNA annealed oligonucleotides; BD Biosciences, Clontech), including a 5′-BamHI restriction site overhang on the top strand, the 19-base oligonucleotide sequence of the siRNA target site, a 9-nucleotide hairpin loop sequence, a 19-base antisense oligonucleotide sequence, a 6-nucleotide poly(T) RNA polymerase III terminator sequence, an XbaI unique restriction site immediately downstream of the terminator sequence for restriction digest analysis to confirm the presence of the cloned insert, and an EcoRI site overhang on the bottom strand according to the specific manufacturer's protocol. Each forward oligonucleotide was annealed with its complementary reverse counterpart and subcloned into the RNAi ready pSIREN-RetroQ vector digested with BamHI and EcoRI restriction enzymes. The resulting DNA constructs were tested for their ability to silence IRF-1 protein expression upon transient cotransfection with IRF-1 expression vector (CMVBL IRF-1) in HEK293 cells. IRF-1 protein expression was tested by Western blotting using whole-cell extracts from cotransfected HEK293 cells harvested 48 h after transfection.
Generation of stably expressing siRNA Jurkat T cells.
The Gp2 293 pantropic packaging cell line (BD Biosciences, Clontech) grown in DMEM containing 10% FCS and antibiotics was transiently transfected with pSIREN-RetroQ retroviral vectors together with pVSVG expression vector (BD Biosciences, Clontech) using calcium phosphate transfection. DMEM was replaced with RPMI 1640 containing 10% FCS and antibiotics. Retroviral supernatants were collected at 48 h. Jurkat T cells were transduced with the retroviral supernatants supplemented with 4 μg/ml Polybrene (Sigma-Aldrich Chemie GmbH, Steinheim, Germany). Transduced cells were selected with 2 μg/ml puromycin (Sigma-Aldrich Chemie) 5 days after transduction.
Quantitative real-time reverse transcription-PCR (RT-PCR).
Total cellular RNA was isolated from 3 × 106cells using the RNA Easy total RNA extraction kit from Qiagen. RNA was treated with RNase-free DNase (Qiagen), and first-strand cDNA was synthesized with Superscript II (Invitrogen, Carlsbad, CA) according to the manufacturer's instruction. Real-time PCR was performed using an ABI 7000 sequence detection system (PE Applied-Biosystems, Warrington, United Kingdom) and SYBR green PCR master mix (Applied Biosystems). The optimization of the real-time PCR was performed according to the manufacturer's instructions but scaled down to 25 μl per reaction mixture. For quantification of HIV-1 early transcripts, a 123-bp fragment corresponding to double-spliced Tat/Rev RNA was amplified using the LA41/45 primers described previously (37). Primer sequences for the SeV nucleoprotein gene were as follows: For, 5′-TGCCCTGGAAGATGAGTTAG-3′; Rev, 5′-GCCTGTTGGTTTGTGGTAAG-3′ (40). The transcript levels were normalized against the expression levels of glyceraldehyde-3′-phosphate dehydrogenase (GAPDH) (For, 5′-GGGTGTGAACCATGAGAAG-3′; Rev, 5′-GCTAAGCAGTTGGTGGTGC-3′) selected with the online program Real-Time PCR Primer Design (Genscript Corp.).
ChIP.
Chromatin immunoprecipitation (ChIP) assays were performed as previously described (33). Briefly, 1G5 Jurkat T cells with an integrated LTR (1) were treated with TNF-α (10 ng/ml) for 15 min. Formaldehyde (final concentration, 1%) was then added to cross-link proteins and DNA. The cells were sonicated and lysates immunoprecipitated with normal rabbit serum, p65, or IRF-1 antibodies (Santa Cruz Biotechnology). Protein complexes were collected with protein A/G ultralink immobilized (Pierce, Rockford, IL). Samples were then eluted, and after reverse cross-linking, proteins were digested with protease K and RNA was removed with RNase A. DNA was purified with the QIAquick PCR purification kit (Qiagen) and resuspended in 30 μl of H2O, and 4 μl was used for PCR analysis. The sequences of specific primers used for amplification of the LTR NF-κB enhancer were as follows: For, 5′-CGAGAGCTGCATCCGGAGTA-3′; Rev, 5′-GAGGCTTAAGCAGTGGGTTC-3′. The PCR product (231 bp) was analyzed on a 1.0% agarose gel. The sequences of specific primers used for amplification of the human IκBa gene promoter were as follows: For, 5′-GGGTTTAGGCTTCTTTTTCCCCCTAGCAG-3′; Rev, 5′-TGGGGATTTCTCTGGGGCGGGGTCAGGCT-3′ (3). The PCR product (111 bp) was analyzed on a 2.0% agarose gel.
RESULTS
IRF-1 and NF-κB form an active complex on the HIV-1 LTR enhancer.
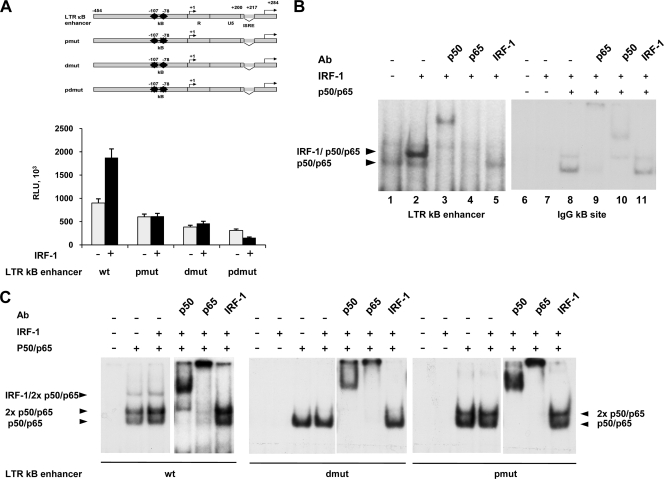
We have previously reported that IRF-1 activates LTR transcription even in the absence of the viral transactivator Tat and that an IRF consensus site mediated this effect. However, deletions of this site indicated that the IRF-1 effect was also dependent on a functional NF-κB binding site (37). To gain further insights into the mechanism underlying the enhancer-mediated effect of IRF-1, HIV-LTR κB enhancer reporter constructs bearing the two tandemly repeated κB binding sites corresponding to the LTR clade B virus enhancer region (LTR κB enhancer) (28), wild type (wt) or mutated in the proximal (pmut), distal (dmut), or both (pdmut) κB sites as indicated in Table 1 and depicted in Fig. 1A, were transiently transfected in HEK293 cells along with an IRF-1 expressing vector. IRF-1 overexpression doubled basal activity of the wt LTR enhancer, but this increase was almost abolished when both the proximal and the distal NF-κB sites were mutated. Mutations in either the proximal or the distal κB site also greatly impaired the basal transcriptional activity. Since no consensus sequence for IRF-1 is evident on the LTR enhancer, to assess whether the IRF-1 effect was mediated by cooperation with NF-κB, we carried out gel retardation assays using double-stranded DNA probes corresponding to the LTR κB enhancer and total cell extracts from HEK293 cells expressing IRF-1. As previously reported, IRF-1 overexpression also induces NF-κB nuclear translocation and activation (20). As shown in Fig. 1B, a low basal p50/p65 expression was evident in control cells (lane 1), and overexpression of IRF-1 (lane 2) led to the appearance of a major complex containing IRF-1 and the heterodimer p50/p65 as assessed by supershift analysis with specific antibodies (lanes 3 to 5). Interestingly, when a probe corresponding to the canonical κB site from the immunoglobulin G light chain was used in EMSA with the same cell extracts, overexpression of IRF-1 did not induce the formation of any complex (Fig. 1B, lane 7). Similarly, the overexpression of p50/p65 subunits together with IRF-1 resulted in the formation of the p50/p65 complex but not of the composite IRF-1/p50/p65 complex as assessed by supershift analysis (lanes 9 to 11). These results indicate that the IRF-1/NF-κB complex specifically formed only on the κB site present on the HIV-1 LTR enhancer, whereas canonical κB sites bind only the p50/p65 complex (lanes 8 to 11).
TABLE 1.
Sequences of wt and mutant NF-κB DNA binding sites on the HIV-1 LTR
| LTR κB enhancer | Sequence (5′→3′) |
|---|---|
| wt | ACAAGGGACTTTCCGCTGGGGACTTTCCAGGGAG |
| pdmut | ACAAGGGACTATCCGCTGGGGACTATCCAGGGAG |
| pmut | ACAAGGGACTTTCCGCTGGGGACTATCCAGGGAG |
| dmut | ACAAGGGACTATCCGCTGGGGACTTTCCAGGGAG |
a Underlining and bold face indicate the core sequence of NF-κB binding sites and the nucleotide substitutions in the mutants, respectively.
FIG. 1.
IRF-1 and NF-κB form an active complex on the HIV-1 enhancer region. (A) HEK293 cells were transiently cotransfected with the indicated reporter constructs (2.5 ng) corresponding to the wt or mutated LTR κB enhancer and an expression vector for IRF-1 (10 ng). Twenty-four hours after transfection, luciferase activity was measured as described in Materials and Methods. Means ± standard deviations from three separate experiments after normalization with Renilla activity are shown. RLU, relative light units. (B) Whole-cell extracts (15 μg) from IRF-1-transfected HEK293 cells were subjected to EMSA using the wt κB LTR probe or a canonical κB site. Supershift analysis was performed using specific anti-IRF-1, anti-p65, and anti-p50 antibodies. (C) Whole-cell extracts (15 μg) from IRF-1- and p50/p65-transfected HEK293 cells were subjected to EMSA using the wt κB LTR probe or probes mutated as shown in Table 1. Supershift analysis was performed as for panel B.
Interestingly, despite the absence of IRF-1 in HEK293 cells, when p50 and p65 κB subunits were overexpressed, an IRF-1/p50/p65 complex with the wt κB enhancer was formed even in the absence of IRF-1 overexpression (Fig. 1C, second lane). This is in accordance with the stimulatory activity of NF-κB on the IRF-1 gene promoter (32). Coexpression of IRF-1 together with p50/p65 further stimulated the formation of the complex (Fig. 1C, third lane).
To assess the specific contributions of the two NF-κB binding sites in the formation of the IRF-1/NF-κB complex, EMSA was performed with cell extracts from HEK293 cells overexpressing both IRF-1 and p50/p65 and oligonucleotides bearing mutations in the enhancer region, as depicted in Table 1. As shown in Fig. 1C, mutations in the distal NF-κB site (dmut) (middle panel) allowed the formation of a single complex containing the monomeric p50/p65 heterocomplex, whereas mutations in the proximal NF-κB site (pmut) (right panel) allowed the formation of both a mono- and a dimeric heterocomplex p50/p65, as assessed by supershift analysis with specific antibodies. Interestingly, mutations in either the proximal or the distal NF-κB site abolished the formation of the slowest-migrating complex containing IRF-1, suggesting that the integrity of the two NF-κB binding sites in the enhancer region of the HIV-1 LTR is required for the formation of the IRF-1/NF-κB complex. Mutations in both κB sites abolished the formation of all the specific complexes (not shown).
TNF-α treatment induces NF-κB association with IRF-1 on the LTR κB sites.
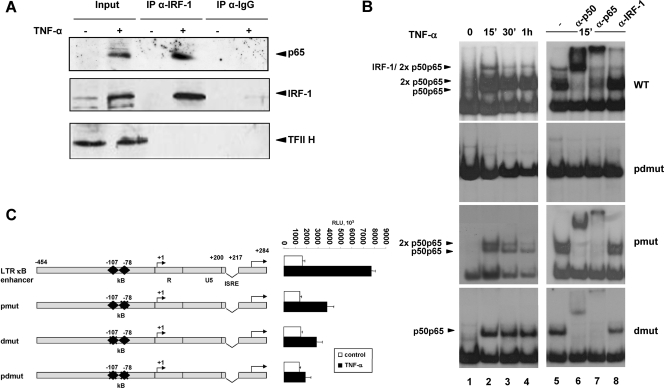
Previous studies have shown that HIV-1 is unable to productively infect resting T cells and that latently infected T cells can reactivate virus only upon treatment with mitogens and cytokines (14, 21, 41), including TNF-α, one of the most powerful HIV-1 inducers, whose major effect is NF-κB activation. To assess whether the IRF-1/NF-κB complex plays a role also in the context of cytokine-mediated virus reactivation, Jurkat T cells, which have basal IRF-1 levels, were treated with TNF-α in order to activate NF-κB. To determine in vivo interactions between IRF-1 and NF-κB, coimmunoprecipitation experiments were performed. As shown in Fig. 2A, the p65-NF-κB subunit was readily detected in the anti-IRF-1 immunocomplexes obtained from cells treated with TNF-α for 4 h, indicating that IRF-1 and p65 associate intracellularly. EMSA performed using the LTR κB probe indicated that no specific complexes were present in extracts from untreated cells, but starting from 15 min of TNF-α treatment and progressively disappearing, three major complexes were evident (Fig. 2B, upper panels, lanes 1 to 4). Supershift analysis with specific antibodies directed against p50, p65, and IRF-1 demonstrated that the slowest-migrating complex contained both IRF-1 and the heterodimer p50/p65, whereas the faster-migrating complexes were both formed by the p50/p65 heterodimers (Fig. 2B, upper panels, lanes 5 to 8).
FIG. 2.
TNF-α-activated NF-κB associates with IRF-1 on the HIV-1 LTR enhancer, and the complex is required for full LTR enhancer transcriptional stimulation. (A) Jurkat T cells left untreated or treated with TNF-α (10 ng/ml) for 4 h. Nuclear cell extracts were immunoprecipitated (IP) with anti-IRF-1 specific antibodies, and the complexes were separated by 10% SDS-PAGE and subsequently probed with anti-IRF-1 and anti-p65 specific antibodies. Western blotting with anti-TFII H in the input is shown as a control of sample loading. IgG, immunoglobulin G. (B) Jurkat T cells left untreated or treated for the indicated time with TNF-α (10 ng/ml) and nuclear cell extracts (15 μg) incubated with oligonucleotides corresponding to HIV-1 wt κB binding sites or sites mutated as indicated in Table 1 were analyzed by EMSA (lanes 1 to 4). Supershift assays with specific antibodies against p50, p65, and IRF-1 were performed on cell extracts from cells treated with TNF-α for 15 min (lanes 5 to 8). (C) Jurkat T cells were transfected with the indicated reporter constructs (1 μg) corresponding to wt κB enhancer or enhancers mutated as for panel B, and luciferase activity was measured as described in Materials and Methods before or after stimulation with TNF-α for 9 h. Means ± standard deviations from three separate experiments after normalization with Renilla activity are shown. RLU, relative light units.
To assess the specific contributions of the two NF-κB binding sites in the formation of the IRF-1/NF-κB complex, EMSA was performed with the same cell extracts and oligonucleotides mutated in the proximal κB site (pmut), the distal κB site (dmut), or both (pdmut) (Table 1). As shown in Fig. 2B, mutations in both κB sites abolished the formation of all specific complexes; mutations in the distal NF-κB site allowed the formation of a single complex containing the monomeric p50/p65 heterocomplex, whereas mutations in the proximal NF-κB site still allowed the formation of both the mono- and dimeric heterocomplex p50/p65, as assessed by supershift analysis (lanes 5 to 8) and as already shown in Fig. 1C with p50 and p65 overexpression. Mutations in either the proximal or the distal NF-κB site abolished the formation of the slowest-migrating complex containing IRF-1.
The IRF-1/NF-κB complex is necessary for full LTR-κB site-mediated transcriptional activity.
The correlation between the formation of the IRF-1/NF-κB complex and the κB enhancer transcriptional activity was assessed in Jurkat T cells transiently transfected with reporter constructs bearing mutations indicated in Table 1 and treated with TNF-α. As shown in Fig. 2C, the threefold-TNF-α-induced stimulation of wt LTR κB enhancer basal activity was abolished when both the proximal and the distal NF-κB sites were mutated. Mutations in either the proximal or the distal κB site significantly reduced (50% ± 10%) the TNF-α-induced stimulation.
These results indicate that the integrity of both NF-κB sites in the enhancer region is necessary for IRF1/p50/p65 complex formation and for full stimulation of LTR transcriptional activity by proinflammatory cytokines.
IRF-1 binds the κB enhancer in vivo and activates an integrated provirus synergistically with NF-κB.
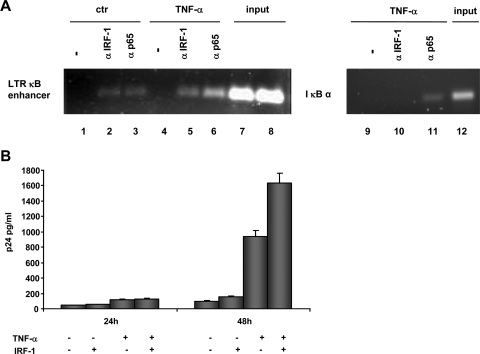
To demonstrate the in vivo binding of the IRF-1 on the HIV-1 LTR, ChIP assays were employed using 1G5 cells, a Jurkat cell line containing an integrated LTR (1). Sheared formaldehyde-cross-linked chromatin extracts prepared from cells untreated or treated with TNF-α in order to activate NF-κB were immunoprecipitated with antibodies specific for IRF-1 and p65. The abundance of LTR DNA in immunoprecipitates was assessed after amplification by PCR with primers specific for the HIV-1 LTR. Ethidium bromide visualization of PCR products showed that p65 and IRF-1 binding to HIV-1 LTR DNA was barely detectable in untreated cells (Fig. 3A, left panel). However, both p65 and IRF-1 binding significantly increased 15 min after stimulation with TNF-α. Levels of input LTR DNA were similar, and nonspecific DNA sequences, as well as no signal with unrelated antibodies, supported specificity in the ChIP procedure. To confirm the specificity of IRF-1 and NF-κB association only on the κB site of the HIV-1 LTR promoter, we amplified in immunoprecipitates the κB site present on the IκBα gene promoter (3). As shown in Fig. 3A (right panel), after 15 min of TNF-α stimulation, p65 but not IRF-1 was recruited on the IκBα promoter.
FIG. 3.
IRF-1 binds the κB enhancer in vivo and synergistically with NF-κB activates an integrated provirus. (A) 1G5 cells untreated or treated with TNF-α (10 ng/ml) for 15 min were subjected to ChIP as described in Materials and Methods, using either normal rabbit serum or anti-IRF-1 and anti-p65 antibodies. Samples were amplified by PCR, using primers specific for LTR NF-κB enhancer (lanes 1 to 8) and for the κB1 site on the IκBα gene promoter (lanes 9 to 12). (B) HLM-1 cells were transfected with an IRF-1 expression vector and treated with TNF-α (10 ng/ml). After 24 and 48 h, p24 antigen accumulation was determined in the cell supernatants as indicated in Materials and Methods. Error bars indicate standard deviations.
Finally, to better evaluate the combined effect of IRF-1 and NF-κB on the transcriptional activity of the HIV-1 LTR promoter integrated in the context of a chromatin structure, we took advantage of a modified HeLa cell line (HLM-1) harboring an integrated provirus (35). These cells, which display very low basal IRF-1 expression (not shown), were transfected with an IRF-1 expression vector and treated with TNF-α in order to activate NF-κB. Virus production was monitored, at different time points, by measuring the levels of p24 in the culture supernatants. As shown in Fig. 3B, upon TNF-α treatment, starting from 24 h onward, IRF-1 overexpression resulted in stimulation of p24 accumulation compared with control cells, while a synergistic effect was observed after TNF-α treatment, confirming the relevance of the IRF-1/NF-κB interactions on the HIV-1 LTR promoter from an integrated provirus.
Inhibition of IRF-1 expression impairs the formation of IRF-1/p50/p65 complex and HIV-1 LTR transcriptional activity.
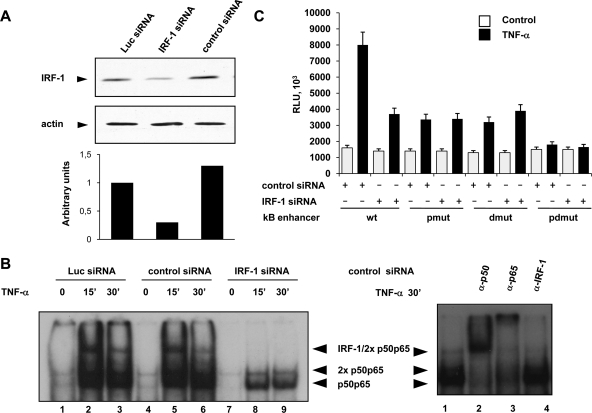
To further assess the importance of the IRF-1/p50/p65 complex in LTR transactivation, Jurkat T cells stably expressing a specific IRF-1-targeting siRNA were generated. The efficacy of siRNA on IRF-1 expression was assessed by Western blotting (Fig. 4A). IRF-1 was significantly reduced (80%) in cells expressing IRF-1-specific siRNA (lane 2) compared with cells expressing luciferase (lane 1) or control siRNA (lane3). EMSA performed with cell extracts from siRNA-expressing cells treated with TNF-α indicated that IRF-1 knockdown completely abolished the formation of the IRF-1/p50/p65 complex binding the HIV-1 LTR enhancer (Fig. 4B, lanes 8 and 9). The complex was instead evident in cells expressing luciferase siRNA (lanes 2 and 3) or control siRNA (lanes 5 and 6), as assessed by supershift analysis with specific antibodies (right panel, lanes 2 to 4).
FIG. 4.
Knockdown of IRF-1 by siRNA abolishes the formation of the IRF-1/p50/p65 complex and impairs HIV-1 LTR transcriptional activity. (A) IRF-1 expression was determined in whole extracts from Jurkat T cells stably expressing either IRF-1-targeting or control siRNAs (control siRNA or Luc siRNA) by Western blotting using specific antibody against IRF-1. Western blotting using an antiactin antibody was used as control of sample loading. The intensity of the specific bands was measured by densitometry and reported as percentage of IRF-1 expression in IRF-1 siRNA-expressing cells relative to control cells after normalization with actin levels. (B) Left panel, nuclear cell extracts prepared from Jurkat T cells, stably expressing Luc siRNA (lanes 1 to 3), control siRNA (lanes 4 to 6), or IRF-1 siRNA (lanes 7 to 9) and stimulated with TNF-α for the indicated time, were subjected to EMSA using an LTR κB probe. Right panel, nuclear cell extracts from control siRNA-expressing cells stimulated for 30 min with TNF-α were further analyzed by supershift with specific antibodies against p65, p50, and IRF-1. (C) Jurkat T cells stably expressing either IRF-1 or control siRNA were transiently transfected with the wt LTR κB enhancer reporter construct (1 μg) or with constructs mutated as in Fig. 1A and stimulated with TNF-α for 9 h. Twenty-four hours after transfection, luciferase activity was measured as described in Materials and Methods. Means ± standard deviations from three separate experiments were calculated after normalization with Renilla activity. RLU, relative light units.
Next, to determine whether IRF-1 down-modulation affected TNF-α-induced transcription, luciferase assays were performed with Jurkat T cells expressing IRF-1-targeting or control siRNA, treated with TNF-α, and transfected with the wt LTR κB reporter construct or with constructs mutated as in Fig. 2C. The results (Fig. 4C) indicated that inhibition of IRF-1 in IRF-1 siRNA expressing cells caused a significant decrease (40% ± 8%) in the wt LTR enhancer transcriptional activity in response to TNF-α. Mutations either in the proximal or the distal κB site caused a 50% ± 10% decrease in TNF-α-induced transcriptional activity in control cells that was not affected by the inhibition of IRF-1 in IRF-1 siRNA-expressing cells.
Together, these results indicate that IRF-1 is not dispensable for full NF-κB-directed LTR transactivation.
HIV-1 gene transcription is inhibited by IRF-1 siRNA expression.
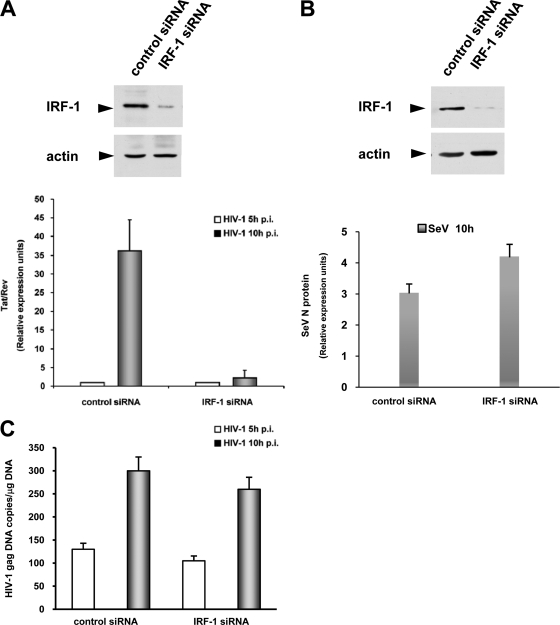
To assess the relevance of the above-reported findings in the context of HIV-1 infection, Jurkat T cells stably expressing IRF-1-targeting or control siRNA were infected with the HXBc2 strain. The inhibition of IRF-1 expression in IRF-1 siRNA was assessed by Western blot analysis (Fig. 5A, upper panel). Viral RNA amounts were measured at 5 and 10 h postinfection (p.i.) by real-time RT-PCR using primers designed to amplify HIV-1 primary transcripts (37). No significant accumulation of HIV-1 transcripts was observed at 5 h p.i. either in control or in IRF-1-targeting siRNA-expressing Jurkat T cells, and this value was used as the basis of comparative results. In contrast, at 10 h p.i., viral RNA was clearly detectable in control siRNA-expressing cells but not in cells in which IRF-1 expression was inhibited (Fig. 5A). In order to confirm that the effect of stable IRF-1 knockdown on the generation of HIV-1 RNA is specific, Jurkat cells stably expressing IRF-1-targeting or control siRNA were infected with SeV, and at 10 h p.i., expression of the viral nucleoprotein was assessed by real-time PCR. As shown in Fig. 5B, a slight increase in SeV N protein transcription was observed in IRF-1 knockdown compared with control siRNA-expressing cells, demonstrating that the dramatic decrease in HIV-1 transcription observed in IRF-1 knockdown cells is not reproduced in a different virus setting. Finally, to exclude the possibility that the observed HIV-1 transcription inhibition could be due to different amounts of retrotranscribed viral DNA in the two cell lines, total DNA was extracted from IRF-1-targeting and control siRNA-expressing cells infected with HIV-1, and HIV-1 gag DNA copies were measured. As shown in Fig. 5C, comparable levels of HIV-1 gag DNA were present in the two cell lines, irrespective of the presence of IRF-1.
FIG. 5.
(A) HIV-1 transcription is inhibited by IRF-1 siRNA expression. Total RNA was purified, at different time points, from Jurkat T cells stably expressing either IRF-1 or control siRNA, infected with the HIV-1 pHXBc2 molecular clone. Primary HIV-1 transcripts were monitored by real-time RT-PCR using specific primers for early transcripts (37) Levels from uninfected cells at 5 h were set as the basis of comparative results and for GAPDH, whose sequence is indicated in Materials and Methods. (B) RT-PCR analysis was performed for SeV N protein on RNA extracts from 10-h SeV-infected control and IRF-1 siRNA-expressing Jurkat cells. Data are normalized by the level of GAPDH mRNA expression in each sample and shown as relative expression units. Western blot analysis of IRF-1 levels was performed on whole-cell extracts from control and IRF-1 siRNA-expressing Jurkat cells infected with HIV-1 (A, upper panel) or SeV (B, upper panel); actin levels are also shown as loading control. (C) Total DNA from Jurkat T cells stably expressing either IRF-1 or control siRNA and infected with HIV-1 at the indicated time points was purified, and the HIV-1 gag DNA full reverse transcript was quantified by real-time PCR as indicated in Materials and Methods. Error bars indicate standard deviations.
DISCUSSION
The NF-κB and IRF pathways are prime targets for viral evasion, and most viruses have evolved strategies to affect this early immune response of the host (16). In particular, HIV-1 has diverted immune regulatory functions of these transcription factors, incorporating and using their DNA binding sites in the LTR viral promoter (16, 37, 39). The HIV-1 enhancer is the most widely studied element in the LTR and consists of two conserved binding sites for the NF-κB/Rel B family of transcription factors. The role of NF-κB in controlling transcription from the LTR in both monocytes and T cells in conjunction with NF-AT has been well defined (for a review, see reference 34).
In the present study, using EMSA, reporter assays, and IRF-1-targeting siRNA, we have demonstrated that physical interactions between IRF-1 and NF-κB occur on the HIV-1 enhancer and that the formation of the IRF-1/NF-κB complex is required for full LTR transcription in T cells.
Initiation of transcription on the HIV-1 LTR appears to follow the normal eukaryotic transcriptional pathway, with recruitment of RNA polymerase II holoenzyme through its interaction with components of the basal transcription apparatus. This pathway has been clearly defined in the context of Tat transactivation (8); however, this may also occur in the early phases of infection, when the viral transactivator is still absent or in small amounts and cellular transcription factors, including NF-κB and IRF-1, are activated and able to stimulate LTR transcription per se (6, 37). Here we also demonstrated that IRF-1 binds in vivo the HIV-1 LTR and stimulates its transcriptional activity synergistically with NF-κB in the context of a chromatin structure. The functional relevance of the IRF-1/NF-κB complex in HIV-1 LTR transcriptional activation was further stressed by the observation that although the level of the IRF-1/NF-κB complex was significantly lower than that of the p50/p65 heterocomplex (Fig. 2B, upper panel), it accounted for approximately 50% of LTR activity (Fig. 2C).
Association of IRF-1 and NF-κB and synergistic activation of several cellular genes, including those for inducible nitric oxide synthase, IL-15, major histocompatibility complex class I, and vascular cell adhesion molecule I, has been already reported (4, 11, 30, 36). However, in those gene promoters, overlapping or adjacent binding sites for the two transcription factors are present. In contrast, on the HIV-1 enhancer no consensus sequence for IRF-1, close to the NF-κB sites, is evident. Thus, it would be interesting to assess whether other viral promoters, stimulated by NF-κB, are fully activated by IRF-1/NF-κB complexes as we reported here for the HIV-1 LTR enhancer.
In the present study we have also shown that mutations in both the distal and proximal κB site in the enhancer region prevented the formation of the IRF-1/NF-κB complex and resulted in decreased LTR transcriptional activity in T cells. These findings may provide a mechanistic insight to the observation that the molecular organization of LTR in different naturally occurring HIV-1 subtypes appears to mirror variations in the HIV-1 enhancer that influence viral expression and replication. In this respect, HIV-1 LTR clade E, the predominant subtype in Asia, which contains a single NF-κB site in the enhancer, has reduced TNF-α and T-cell activation responsiveness (23, 28). This is in accordance with our observation that both NF-κB sites are required for the formation of the IRF-1/NF-κB complex and for full LTR transcriptional activity. Therefore, it would be interesting to assess whether IRF-1/NF-κB interactions also occur on the HIV-1 LTR enhancer from clade C, which is predominant in Africa and which contains three NF-κB binding sites and has higher transcriptional activity than other subtypes.
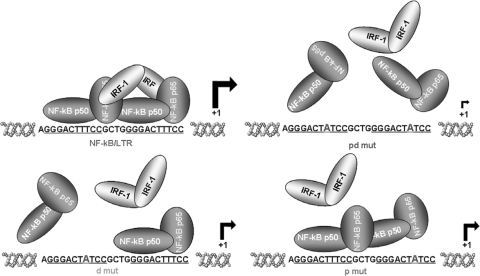
Our data have also unraveled a hierarchy of the two κB binding sites in terms of NF-κB binding ability. In our model (Fig. 6), mutations in the proximal κB site still allow the formation of the double p50/p65 NF-κB heterodimer, while when the distal κB site was mutated, only a single p50/p65 heterodimer bound. Even though the structural basis of these differences has not been investigated, either the distal or the proximal mutation does not allow the formation of the IRF-1/NF-κB complex, and the integrity of both sites is required for IRF-1/NF-κB complex formation. At the functional level, both mutations result in a similar decrease in transcriptional activity, indicating that the formation of the IRF-1/NF-κB complex is essential for full stimulation of the HIV-1 LTR enhancer. This conclusion is further stressed by the data obtained with cells knocked down for IRF-1. In these cells, upon stimulation with TNF-α, a substantial reduction in LTR transcriptional activity was observed, and even more impressively, in de novo HIV-1 infected cells, HIV-1 transcription was completely abolished early in infection when virus replication is essentially dependent on cellular factors. Accordingly, the IRF-1/NF-κB complex was also observed in de novo HIV-1-infected Jurkat T cells (data not shown). Since NF-κB is suddenly activated by the interaction between the gp120 envelope protein and the cellular CD4 receptor (7, 9, 13) and, similarly, IRF-1 expression is induced very soon after HIV infection (37), our data argue that, early in infection, IRF-1 is pivotal in HIV-1 replication by binding, in combination with NF-κB, to the κB sites.
FIG. 6.
Schematic representation of IRF-1/NF-κB interactions on the HIV-1 LTR enhancer region. The enhancer region present in the HIV-1 LTR promoter, containing two adjacent high-affinity binding sites for NF-κB that are critical for LTR promoter activity and important for optimal HIV-1 replication, is shown. A complex between IRF-1 and NF-κB is formed upon HIV-1 infection and TNF-α treatment. Mutations in either the distal or proximal κB site prevented the formation of the IRF-1/NF-κB complex and resulted in decreased LTR transcriptional activity. A hierarchy of the two κB binding sites in terms of NF-κB binding ability is also indicated: mutations in the proximal κB site still allowed the formation of the double p50/p65 NF-κB heterodimer, whereas when the distal κB site was mutated, only a single p50/p65 heterodimer bound.
The ability of IRF-1 to bind the κB sites on the HIV-1 LTR enhancer as a partner of NF-κB and the fact that the IRF-1/NF-κB complex is required for full activation of the HIV-1 LTR enhancer represent a new finding that further emphasizes the role of IRF-1 in HIV-1 replication and points to this transcription factor as a primary cellular target for therapeutic intervention aimed at reducing HIV-1 replication or eliminating viral reservoirs.
Acknowledgments
This work was supported in part by grants from the Italian AIDS Project, the Italian Ministry of Health, and the ISS-NIH Scientific Cooperation agreement to Angela Battistini and Barbara Ensoli.
We thank Roberto Gilardi for artwork and Sabrina Tocchio for editorial assistance.
Footnotes
Published ahead of print on 23 January 2008.
REFERENCES
- 1.Aguilar-Cordova, E., J. Chinen, L. Donehower, D. E. Lewis, and J. W. Belmont. 1994. A sensitive reporter cell line for HIV-1 tat activity, HIV-1 inhibitors, and T cell activation effects. AIDS Res. Hum Retroviruses 10295-301. [DOI] [PubMed] [Google Scholar]
- 2.Alcami, J., T. Lain de Lera, L. Folgueira, M. A. Pedraza, J. M. Jacque, F. Bachelerie, A. R. Noriega, R. T. Hay, D. Harrich, R. B. Gaynor, et al. 1995. Absolute dependence on kappa B responsive elements for initiation and Tat-mediated amplification of HIV transcription in blood CD4 T lymphocytes. EMBO J. 141552-1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Algarte, M., H. Kwon, P. Genin, and J. Hiscott. 1999. Identification by in vivo genomic footprinting of a transcriptional switch containing NF-κB and Sp1 that regulates the IκBalpha promoter. Mol. Cell. Biol. 196140-6153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Azimi, N., K. M. Shiramizu, Y. Tagaya, J. Mariner, and T. A. Waldmann. 2000. Viral activation of interleukin-15 (IL-15): characterization of a virus-inducible element in the IL-15 promoter region. J. Virol. 747338-7348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bachelerie, F., J. Alcami, F. Arenzana-Seisdedos, and J. L. Virelizier. 1991. HIV enhancer activity perpetuated by NF-kappa B induction on infection of monocytes. Nature 350709-712. [DOI] [PubMed] [Google Scholar]
- 6.Battistini, A., G. Marsili, M. Sgarbanti, B. Ensoli, and J. Hiscott. 2002. IRF regulation of HIV-1 long terminal repeat activity. J. Interferon Cytokine Res. 2227-37. [DOI] [PubMed] [Google Scholar]
- 7.Bossis, G., S. Salinas, C. Cartier, C. Devaux, and L. Briant. 2002. NF-kappaB activation upon interaction of HIV-1 envelope glycoproteins with cell surface CD4 involves IkappaB kinases. FEBS Lett. 516257-264. [DOI] [PubMed] [Google Scholar]
- 8.Brady, J., and F. Kashanchi. 2005. Tat gets the “green” light on transcription initiation. Retrovirology 269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Briand, G., B. Barbeau, and M. Tremblay. 1997. Binding of HIV-1 to its receptor induces tyrosine phosphorylation of several CD4-associated proteins, including the phosphatidylinositol 3-kinase. Virology 228171-179. [DOI] [PubMed] [Google Scholar]
- 10.Chen, B. K., M. B. Feinberg, and D. Baltimore. 1997. The κB sites in the human immunodeficiency virus type 1 long terminal repeat enhance virus replication yet are not absolutely required for viral growth. J. Virol. 715495-5504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Drew, P. D., G. Franzoso, K. G. Becker, V. Bours, L. M. Carlson, U. Siebenlist, and K. Ozato. 1995. NF kappa B and interferon regulatory factor 1 physically interact and synergistically induce major histocompatibility class I gene expression. J. Interferon Cytokine Res. 151037-1045. [DOI] [PubMed] [Google Scholar]
- 12.Duh, E. J., W. J. Maury, T. M. Folks, A. S. Fauci, and A. B. Rabson. 1989. Tumor necrosis factor alpha activates human immunodeficiency virus type 1 through induction of nuclear factor binding to the NF-kappa B sites in the long terminal repeat. Proc. Natl. Acad. Sci. USA 865974-5978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Flory, E., C. K. Weber, P. Chen, A. Hoffmeyer, C. Jassoy, and U. R. Rapp. 1998. Plasma membrane-targeted Raf kinase activates NF-κB and human immunodeficiency virus type 1 replication in T lymphocytes. J. Virol. 722788-2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Folks, T., J. Kelly, S. Benn, A. Kinter, J. Justement, J. Gold, R. Redfield, K. W. Sell, and A. S. Fauci. 1986. Susceptibility of normal human lymphocytes to infection with HTLV-III/LAV. J. Immunol. 1364049-4053. [PubMed] [Google Scholar]
- 15.Fujita, T., L. F. Reis, N. Watanabe, Y. Kimura, T. Taniguchi, and J. Vilcek. 1989. Induction of the transcription factor IRF-1 and interferon-beta mRNAs by cytokines and activators of second-messenger pathways. Proc. Natl. Acad. Sci. USA 869936-9940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hiscott, J., H. Kwon, and P. Genin. 2001. Hostile takeovers: viral appropriation of the NF-kappaB pathway. J. Clin. Investig. 107143-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoffmann, A., T. H. Leung, and D. Baltimore. 2003. Genetic analysis of NF-kappaB/Rel transcription factors defines functional specificities. EMBO J. 225530-5539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Honda, K., A. Takaoka, and T. Taniguchi. 2006. Type I interferon [corrected] gene induction by the interferon regulatory factor family of transcription factors. Immunity 25349-360. [DOI] [PubMed] [Google Scholar]
- 19.Karin, M., and Y. Ben-Neriah. 2000. Phosphorylation meets ubiquitination: the control of NF-[kappa]B activity. Annu. Rev. Immunol. 18621-663. [DOI] [PubMed] [Google Scholar]
- 20.Kirchhoff, S., D. Wilhelm, P. Angel, and H. Hauser. 1999. NFkappaB activation is required for interferon regulatory factor-1-mediated interferon beta induction. Eur. J. Biochem. 261546-554. [DOI] [PubMed] [Google Scholar]
- 21.Krasnow, S. W., L. Q. Zhang, K. Y. Leung, L. Osborn, S. Kunkel, and G. J. Nabel. 1991. Tumor necrosis factor-alpha, interleukin 1, and phorbol myristate acetate are independent activators of NF-kappa B which differentially activate T cells. Cytokine 3372-379. [DOI] [PubMed] [Google Scholar]
- 22.Kroger, A., M. Koster, K. Schroeder, H. Hauser, and P. P. Mueller. 2002. Activities of IRF-1. J. Interferon Cytokine Res. 225-14. [DOI] [PubMed] [Google Scholar]
- 23.Lemieux, A. M., M. E. Pare, B. Audet, E. Legault, S. Lefort, N. Boucher, S. Landry, T. van Opijnen, B. Berkhout, M. H. Naghavi, M. J. Tremblay, and B. Barbeau. 2004. T-cell activation leads to poor activation of the HIV-1 clade E long terminal repeat and weak association of nuclear factor-kappaB and NFAT with its enhancer region. J. Biol. Chem. 27952949-52960. [DOI] [PubMed] [Google Scholar]
- 24.Liang, C., X. Li, Y. Quan, M. Laughrea, L. Kleiman, J. Hiscott, and M. A. Wainberg. 1997. Sequence elements downstream of the human immunodeficiency virus type 1 long terminal repeat are required for efficient viral gene transcription. J. Mol. Biol. 272167-177. [DOI] [PubMed] [Google Scholar]
- 25.Lin, R., D. Gewert, and J. Hiscott. 1995. Differential transcriptional activation in vitro by NF-kappa B/Rel proteins. J. Biol. Chem. 2703123-3131. [DOI] [PubMed] [Google Scholar]
- 26.Lin, R., A. Mustafa, H. Nguyen, D. Gewert, and J. Hiscott. 1994. Mutational analysis of interferon (IFN) regulatory factors 1 and 2. Effects on the induction of IFN-beta gene expression. J. Biol. Chem. 26917542-17549. [PubMed] [Google Scholar]
- 27.Marsili, G., A. L. Remoli, M. Sgarbanti, and A. Battistini. 2004. Role of acetylases and deacetylase inhibitors in IRF-1-mediated HIV-1 long terminal repeat transcription. Ann. N. Y. Acad. Sci. 1030636-643. [DOI] [PubMed] [Google Scholar]
- 28.Montano, M. A., V. A. Novitsky, J. T. Blackard, N. L. Cho, D. A. Katzenstein, and M. Essex. 1997. Divergent transcriptional regulation among expanding human immunodeficiency virus type 1 subtypes. J. Virol. 718657-8665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nabel, G., and D. Baltimore. 1987. An inducible transcription factor activates expression of human immunodeficiency virus in T cells. Nature 326711-713. [DOI] [PubMed] [Google Scholar]
- 30.Neish, A. S., M. A. Read, D. Thanos, R. Pine, T. Maniatis, and T. Collins. 1995. Endothelial interferon regulatory factor 1 cooperates with NF-κB as a transcriptional activator of vascular cell adhesion molecule 1. Mol. Cell. Biol. 152558-2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Osborn, L., S. Kunkel, and G. J. Nabel. 1989. Tumor necrosis factor alpha and interleukin 1 stimulate the human immunodeficiency virus enhancer by activation of the nuclear factor kappa B. Proc. Natl. Acad. Sci. USA 862336-2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pine, R. 1997. Convergence of TNFalpha and IFNgamma signalling pathways through synergistic induction of IRF-1/ISGF-2 is mediated by a composite GAS/kappaB promoter element. Nucleic Acids Res. 254346-4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Remoli, A. L., G. Marsili, E. Perrotti, E. Gallerani, R. Ilari, F. Nappi, A. Cafaro, B. Ensoli, R. Gavioli, and A. Battistini. 2006. Intracellular HIV-1 Tat protein represses constitutive LMP2 transcription increasing proteasome activity by interfering with the binding of IRF-1 to STAT1. Biochem. J. 396371-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roulston, A., R. Lin, P. Beauparlant, M. A. Wainberg, and J. Hiscott. 1995. Regulation of human immunodeficiency virus type 1 and cytokine gene expression in myeloid cells by NF-kappa B/Rel transcription factors. Microbiol. Rev. 59481-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sadaie, M. R., E. Tschachler, K. Valerie, M. Rosenberg, B. K. Felber, G. N. Pavlakis, M. E. Klotman, and F. Wong-Staal. 1990. Activation of tat-defective human immunodeficiency virus by ultraviolet light. New Biol. 2479-486. [PubMed] [Google Scholar]
- 36.Saura, M., C. Zaragoza, C. Bao, A. McMillan, and C. J. Lowenstein. 1999. Interaction of interferon regulatory factor-1 and nuclear factor kappaB during activation of inducible nitric oxide synthase transcription. J. Mol. Biol. 289459-471. [DOI] [PubMed] [Google Scholar]
- 37.Sgarbanti, M., A. Borsetti, N. Moscufo, M. C. Bellocchi, B. Ridolfi, F. Nappi, G. Marsili, G. Marziali, E. M. Coccia, B. Ensoli, and A. Battistini. 2002. Modulation of human immunodeficiency virus 1 replication by interferon regulatory factors. J. Exp. Med. 1951359-1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Taniguchi, T., K. Ogasawara, A. Takaoka, and N. Tanaka. 2001. IRF family of transcription factors as regulators of host defense. Annu. Rev. Immunol. 19623-655. [DOI] [PubMed] [Google Scholar]
- 39.Van Lint, C., C. A. Amella, S. Emiliani, M. John, T. Jie, and E. Verdin. 1997. Transcription factor binding sites downstream of the human immunodeficiency virus type 1 transcription start site are important for virus infectivity. J. Virol. 716113-6127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yount, J. S., T. A. Kraus, C. M. Horvath, T. M. Moran, and C. B. Lopez. 2006. A novel role for viral-defective interfering particles in enhancing dendritic cell maturation. J. Immunol. 1774503-4513. [DOI] [PubMed] [Google Scholar]
- 41.Zack, J. A., S. J. Arrigo, S. R. Weitsman, A. S. Go, A. Haislip, and I. S. Chen. 1990. HIV-1 entry into quiescent primary lymphocytes: molecular analysis reveals a labile, latent viral structure. Cell 61213-222. [DOI] [PubMed] [Google Scholar]